Abstract
Auxin is a simple molecule with a remarkable ability to control plant growth, differentiation, and morphogenesis. The mechanistic basis for this versatility appears to stem from the highly complex nature of the networks regulating auxin metabolism, transport and response. These heavily feedback-regulated and inter-dependent mechanisms are complicated in structure and complex in operation giving rise to a system with self-organizing properties capable of generating highly context-specific responses to auxin as a single, generic signal.
Feedback loops and interdependent response mechanisms make auxin a remarkably versatile hormone in plants. The context-specific responses observed are the product of complex “landscapes” of auxin concentration and auxin responsiveness.
The regulation of plant development is dominated by specific distributions of the plant hormone auxin. From the specification of apical–basal axis in the embryo to the control of anisotropic growth in response to gravity and light, this simple molecule is able to regulate both pattern and growth throughout the life cycle of the plant (Leyser 2005). The principal reason for this capacity for control is that auxin, unlike other known plant hormones, can be actively transported in a coordinated, directional manner such that gradients may be formed that, in various guises, direct numerous distinct developmental phenomena (Tanaka et al. 2006). Although the phenomenology of auxin movement and distribution in the growing plant has been largely established ( Bennett et al. 1996; Benkova et al. 2003; Friml et al. 2003; Blilou et al. 2005; Bandyopadhyay et al. 2007; Grieneisen et al. 2007; Petersson et al. 2009), the link between differences in auxin concentration and outputs in terms of changes in growth and development is not well understood. What is abundantly clear is that the context in which the auxin signal is received is of utmost importance (Kieffer et al. 2010). These contexts can be developmental or environmental. The former is exemplified by the fact that accumulation of auxin in the pericycle of the root prompts the formation of a lateral root whereas a similar accumulation at the flank of the shoot apex gives rise to a leaf, or at a later stage, a flower (Benkova et al. 2003; Reinhardt et al. 2003). A different kind of contextual change is imposed by environmental fluctuation. Here, in addition to changes in auxin distribution such as those observed in response to gravity or light, the auxin sensitivity of a given developmental setting can be altered to modulate development (Perez-Torres et al. 2008).
Auxin responsiveness can be classed broadly as genomic or nongenomic. In the first, auxin controls development by altering programs of gene expression, whereas in the second auxin acts by regulating cell biology and physiology directly without requiring transcriptional change (Tromas et al. 2010a,b). Although our understanding of the latter is currently fragmentary, the picture is much clearer for the control of gene expression by auxin. This is represented by a “minimal” system of transcriptional regulators, which is simple in overall architecture but highly complex and nonlinear in operation with the capacity to generate a range of transcriptional control that is both qualitative (i.e., the type of output) and quantitative (i.e., the rate of change in output) in nature.
Importantly, auxin response and auxin distribution are themselves tightly linked (Leyser 2005). Multiple feedback loops mean that these phenomena are highly interdependent, giving rise to a system with self-organizing properties, capable of much higher levels of developmental control. Thus as a developmental regulator, auxin defies simple classification. In some situations it appears to act merely as a permissive signal, triggering a latent response appropriate to that cell-type once a certain threshold concentration of auxin is reached. In others, its influence is more complex, almost instructive in nature. Understanding how these different auxin response contexts are specified and how spatial differences in responsiveness are related to the active and directed movement of auxin is one of the key questions in plant developmental biology. This article seeks to explore these questions by examining the capacity of the auxin response system to generate complex and context-specific information and by highlighting the features of this system that enable it to act as an integrator of so many different signals controlling plant development.
BASIC MECHANISMS OF RESPONSE
The context-specific responses to auxin observed throughout the plant can be thought of as the product of two “landscapes,” a landscape of auxin concentration and a landscape of auxin responsiveness. The former is the net result of auxin metabolism and transport throughout the plant (Woodward and Bartel 2005). These are themselves complex subjects and so only the immediately relevant features will be considered briefly here. The first of these is that the steady state of the global pool of active auxin in the plant is determined not only by the balance of IAA synthesis and degradation but also by a system of IAA conjugation in which IAA can be inactivated by its conjugation to sugars or amino acids and peptides. Several of these conjugation events are reversible meaning that this is a mechanism by which active auxin can be temporarily removed from the system (Woodward and Bartel 2005).
Although spatial differences in the activity of these homeostatic mechanisms contribute significantly to patterns of auxin accumulation it is active and directed cell-to-cell movement of auxin that is the primary determinant of the gradients and highly asymmetric localizations of auxin that drive so much of development (Benkova et al. 2003; Tanaka et al. 2006). Active intercellular auxin transport is mediated by a system of membrane proteins prominent among which are the AUX1/LAX family of amino acid permeases for auxin influx and the PIN and ABCB (or PGP) families of transmembrane proteins for auxin efflux from the cell ( Bennett et al. 1996; Tanaka et al. 2006; Bandyopadhyay et al. 2007). Crucially the subcellular distribution and abundance of PIN proteins can be dynamically and coordinately regulated so that across a field of cells the direction and flux of auxin can be tightly controlled (Benkova et al. 2003; Tanaka et al. 2006).
In addition to controlled distribution, another important component of auxin's versatility as a signaling molecule is that, as mentioned above, responses to auxin are complex and highly context-dependent. These cell- and tissue-specific responses (the landscape of auxin responsiveness) are determined principally by spatial variation in the auxin signaling machinery that leads specifically to changes in gene expression (Lokerse and Weijers 2009; Kieffer et al. 2010). Experiments in which plants are treated with a variety of auxins show that several hundred genes are either up- or down-regulated in response (Okushima et al. 2005). These genomic responses can be extremely rapid, with new transcripts being detected in as little as 6 min after treatment (Abel et al. 1995). The induction of these rapidly induced genes is insensitive to the protein synthesis inhibitor cycloheximide confirming that these are primary early auxin response genes (Abel et al. 1995). Further, the fact that cycloheximide alone is sufficient to induce these genes indicates that they are under the control of a short-lived repressor protein (Abel et al. 1995). Understanding the nature and regulation of these repressors has led to the identification of a network of auxin receptors and transcription factors sufficient to account for how auxin can turn genes on, and by extension, turn others off.
The Aux/IAA and ARF Transcription Factors
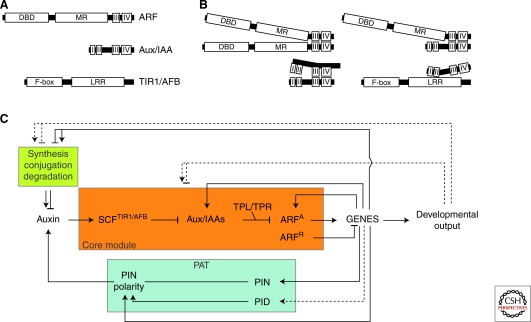
There are at least two families of transcription factors known to affect specifically transcriptional responses to auxin. These are the auxin response factors or ARFs, and the Aux/IAA corepressor proteins. Most Aux/IAA genes are themselves auxin inducible and encode highly unstable nuclear proteins with a characteristic four domain structure (Fig. 1A) (Abel et al. 1994; Abel et al. 1995). In contrast, only a handful of ARF genes are induced by auxin and encode relatively stable nuclear proteins with a three domain structure (Ulmasov et al. 1997a; Guilfoyle et al. 1998; Okushima et al. 2005; Salmon et al. 2008). These are both large families in Arabidopsis, with 29 Aux/IAAs and 23 ARFs (Liscum and Reed 2002).
Figure 1.
Schematic representation of the AFB-Aux/IAA-ARF system and feedback relationships with polar auxin transport (PAT) and auxin metabolism (A) ARF, Aux/IAA, and TIR1/AFB domain organization. (B) ARFs and Aux/IAAs can interact within their families through domain III and IV to form homo- and hetero-dimers (left) and interaction between families can also occur involving ARFs and Aux/IAAs (top right), and Aux/IAAs and TIR1/AFBs (bottom right). (C) The complexity of feedback regulation in auxin response. Lines ending in bars indicate inhibition, whereas lines ending in arrowheads indicate activation. Possible mechanisms of action are indicated by dashed lines.
ARFs can bind to so-called auxin response elements (AuxREs) in the promoters of auxin-regulated genes via a VP1-like B3 DNA-binding domain toward the amino terminus of the protein (Ulmasov et al. 1997a; Guilfoyle et al. 1998; Ulmasov et al. 1999b). These AuxREs are characterized by the occurrence of TGTCNC elements (frequently TGTCTC) that often involve coupling elements (Ulmasov et al. 1997b; Guilfoyle et al. 1998; Ulmasov et al. 1999b). Synthetic promoters containing palindromic or direct repeats of these six nucleotides are sufficient to confer auxin-inducibility to cis-regulated reporters (as in the DR5- class of auxin response reporters) (Ulmasov et al. 1997a; Ulmasov et al. 1997b). Once bound, current data suggest that ARFs can have one of two activities, either activation or repression of the target gene (Ulmasov et al. 1999a; Tiwari et al. 2003) (Fig. 1C). This appears to be dependent on the make-up of a variable region between the defined amino- and carboxy-terminal domains known as the middle region (MR) (Fig. 1A). MRs that are Q-rich promote transcription whereas MRs rich in P/S/T residues act as repressors (Ulmasov et al. 1999a; Tiwari et al. 2003). Of the 23 ARFs in Arabidopsis, only five (ARFs 5, 6, 7, 8, and 19) fall into the activating class whereas the remainder are repressors (Ulmasov et al. 1999a; Tiwari et al. 2003). It is important to note that this assignment of ARFs as activators or repressors is based almost entirely on a transient expression assay in Arabidopsis protoplasts and thus it is possible that ARFs may possess different functions in vivo and in different cellular contexts. For the purposes of discussion here, ARFs with a demonstrated capacity to act as transcriptional activators are referred to as ARFA and those which have been shown to be able to act as repressors, as ARFR to indicate the apparent inherent regulatory characteristics of the proteins.
At the carboxyl terminus of the ARF protein is a pair of protein-protein interaction domains known as domains III and IV. These domains allow a variety of dimers to be formed including homodimers and heterodimers within the ARF family and crucially, heterodimers with the second family of auxin-related transcription factors, the Aux/IAAs (Kim et al. 1997; Kepinski and Leyser 2002; Guilfoyle and Hagen 2007) (Fig. 1A,B).
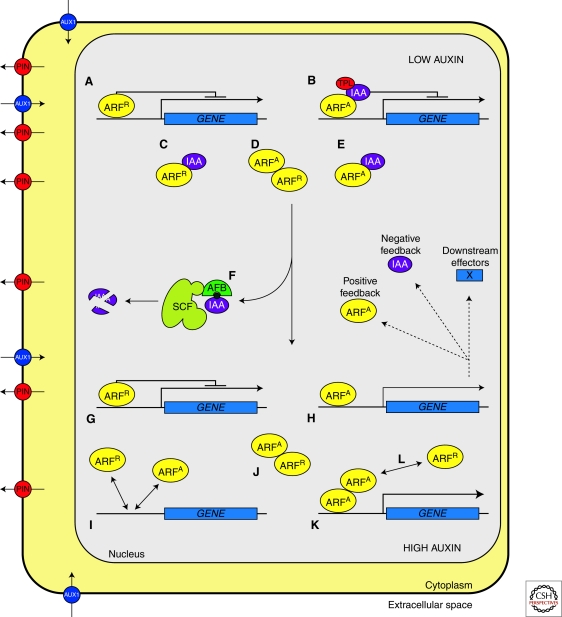
Aux/IAA proteins do not appear to bind DNA themselves but can affect the transcription of ARF-regulated genes by dimerising with ARFs (Tiwari et al. 2001; Guilfoyle and Hagen 2007). This is because Aux/IAAs possess a potent transcriptional repression activity conferred by domain I of the protein (Tiwari et al. 2004). Domain I contains an ERF-associated amphiphilic repression (EAR)-type motif which allows interaction with the corepressor protein TOPLESS (TPL) (Szemenyei et al. 2008) (Fig. 2B). TPL and TPL-related proteins (TPRs) are related to the Groucho/Tup1 family of co-repressors which act by recruiting histone deacetylases (HDACs) to the target locus thereby bringing about a repressed, transcriptionally inactive chromatin state (Szemenyei et al. 2008). TPL/TPR proteins are predicted to act in a similar way so that Aux/IAAs, by dimerising with AuxRE-bound ARFs, bring about repression by recruiting TPL and associated HDAC activities to that particular locus (Szemenyei et al. 2008). In theory, this is not the only way that Aux/IAAs can affect transcription of ARF-regulated genes. Because the regulatory events at the AuxRE depend so heavily on the balance of ARF-ARF and ARF-Aux/IAA dimers, the fact that Aux/IAAs appear to be able to interact readily with each other and many ARF family members means that there is significant scope for effects due to blocking and sequestration which may occur either at or away from the promoters of ARF-regulated genes (Fig. 2C,E).
Figure 2.
Schematic representation of AFB-Aux/IAA-ARF-mediated auxin response. When auxin levels are low, Aux/IAAs repress ARF-mediated transcriptional activation by dimerising with DNA-bound activating ARFs (ARFA) and recruiting the corepressor protein TPL (B), or theoretically by sequestering ARFA from their promoters (E). Although the occurrence and relevance in vivo of Aux/IAA-ARFR dimers is unknown, Aux/IAAs could affect ARFR activity by interfering with the formation of ARF R dimers through blocking or sequestration (C). When cellular auxin levels increase the affinity of the receptors TIR1/AFB for their substrate Aux/IAA increases, promoting the degradation of Aux/IAA via the proteasome (F). ARFA are then able to regulate positively transcription (H); it has been shown that some ARFs bind more stably to promoters as dimers (K) and this could increase their transcriptional activity. ARFR could repress transcription by directly binding to the promoter of target genes (A,G) and importantly, by competing with ARFA for common target AuxRE binding sites (I), or by interacting with activating ARFs, either on the promoter (L) or as free heterodimers (D,J).
Auxin Regulation of the Aux/IAA-ARF System
How then does auxin affect events at the AuxRE? The key to understanding this and so how information is carried in this system lies in the dynamic changes in the abundance of Aux/IAA repressors in response to auxin stimulation. With a few exceptions, Aux/IAAs are very unstable proteins with recorded half-lives as short as six minutes (Abel et al. 1994) and crucially, their instability is enhanced by auxin (Gray et al. 2001; Zenser et al. 2001). The degradation of Aux/IAAs is mediated by the ubiquitin-proteasome system in which target proteins are marked for degradation in the 26S proteasome by their covalent modification with the peptide modifier ubiquitin (Gray et al. 2001; Moon et al. 2004; Maraschin et al. 2009) (Fig. 2F). The conjugation of ubiquitin to Aux/IAA proteins is catalysed by an E3 ubiquitin-ligase complex called SCFTIR1/AFB (Gray et al. 1999; Gray et al. 2001; Dharmasiri et al. 2005b). SCF-type E3s are so called because they consist of, among other proteins, a SKP subunit (termed ASK proteins in Arabidopsis), a CULLIN subunit and an F-box protein (Moon et al. 2004). It is the F-box protein component that is responsible recruiting specific target proteins for ubiquitination by the core ASK and CULLIN subunits and associated catalytic factors. In the case of Aux/IAAs, the cognate F-box protein is TIR1, or one of 5 other related F-box proteins called Auxin F-box proteins (AFBs) (Ruegger et al. 1998; Dharmasiri et al. 2005b). The specific selection of Aux/IAAs is mediated by an interaction between a set of leucine-rich repeats (LRRs) in TIR1 and a short, so-called, “degron” motif within domain II of the Aux/IAA (Fig. 1B) (Ramos et al. 2001; Tan et al. 2007). Consistent with the auxin-induced instability of Aux/IAAs, auxin enhances this interaction and does so in a startlingly short signal transduction chain: auxin itself is bound directly between TIR1 and the Aux/IAA, thereby increasing the affinity of the ternary complex and thus the opportunity for ubiquitination of the bound Aux/IAA through the action associated E2 ubiquitin conjugating enzymes (Dharmasiri et al. 2005a; Kepinski and Leyser 2005; Tan et al. 2007) (see Calderon-Villalobos et al. 2010).
These dynamic changes in Aux/IAA protein abundance are the primary drivers of auxin-induced transcriptional change and together, the AFB-Aux/IAA-ARF system represents a core minimal mechanism in auxin response (Fig. 1C, orange box). Although likely incomplete, this network of physically interacting components forms a contiguous mechanistic link between the arrival of auxin in a cell and outputs in terms of gene expression control: when auxin levels are low, Aux/IAAs are relatively stable and are able to interact with DNA-bound ARFs, thereby inducing a TPL-mediated repressive chromatin state at those loci. As auxin levels rise, increased SCFTIR1/AFB-mediated ubiquitination prompts a rapid drop in Aux/IAA levels, relieving repression of a range of ARFA target genes (Figs. 1C, 2). Because most Aux/IAA genes are themselves subject to regulation by the Aux/IAA-ARF system (Abel et al. 1995), a crucial negative feedback loop is formed that allows repression to be returned to the system and, as will be discussed later, leads to a complex control of the steady-state levels of individual Aux/IAA proteins. Additionally, auxin can affect the topology of the Aux/IAA-ARF system through the induction of certain ARF genes, most notably ARF19 (Okushima et al. 2005). Importantly, ARF19 seems to regulate its own auxin-induced expression thereby forming a direct and positive feedback loop (Okushima et al. 2005).
Clearly, this is a generic and rather abstract view of the system's operation and one that takes no account of the numbers of different Aux/IAAs and ARFs involved. In particular, the established corepressor activity of the Aux/IAAs raises the question of the extent to which interactions between Aux/IAAs and repressing ARFR are of consequence and indeed, whether they occur at all in vivo. The mechanism of ARFR-mediated repression is yet to be fully established but unless quantitative differences in the level and/or type of repression at a particular locus are meaningful in a signaling context, the formation of Aux/IAA-ARFR dimers on an AuxRE would appear not to be significant. Although Aux/IAA-ARFR interactions have been shown by yeast-two-hybrid analysis (Ouellet et al. 2001), there are reports that interactions between Aux/IAAs and ARFR are weaker than those with ARFA (Tiwari et al. 2003). It seems more likely that ARFR contribute to auxin response by competing with ARFA for AuxRE binding sites, thereby offering a mechanism to alter the sensitivity of those targets to auxin according to the relative levels of relevant ARFA and ARFR in a particular tissue or cell type. Nevertheless it is still possible that Aux/IAAs could affect ARFR activity by interfering with the formation of ARFR dimers through blocking or sequestration as noted earlier.
It is also the case that not all members of the AFB, Aux/IAA and ARF families in Arabidopsis conform to the typical structure and biology described earlier. For example, two AFB proteins, AFB4 and AFB5 can be distinguished from the rest of the AFB family both structurally by a long amino-terminal extension and genetically (in the case of AFB5) because in contrast to loss-of-function in TIR1, AFB1, AFB2, and AFB3, which all confer resistance to exogenous auxin, afb5 mutants are slightly hypersensitive to IAA (Dharmasiri et al. 2005b; Walsh et al. 2006). In the case of the Aux/IAAs, several members (IAAs 20, 30, 32, 33, and 34) lack a recognizable domain II degron motif suggesting that these Aux/IAAs might be long-lived proteins, something that has been confirmed in the case of IAA20 (Dreher et al. 2006). Lastly, of the 23 ARFs in Arabidopsis ARFs 3/ETTIN, 13, 17 and 23 lack carboxy-terminal dimerization domains (CTDDs) (Guilfoyle and Hagen 2007). ARF23 contains a stop codon in its DNA-binding domain and may be a pseudogene. The particular significance (if any) of the lack of CTDDs in these ARFs is not clear, although the striking floral phenotypes associated with loss-of-function of ARF3/ETTIN show that CTDDs are not absolutely required for some ARF functions (Sessions et al. 1997). It is worth noting that some ARFs have been shown in in vitro assays to bind more stably to AuxREs as ARF-ARF dimers and that this more stable binding requires the presence of the CTDDs (Ulmasov et al. 1999b).
SPECIFICITY AND INFORMATION PROCESSING AT THE CELLULAR LEVEL
The complex mechanisms underlying auxin distribution set up a landscape of auxin concentration throughout the plant to which individual cells respond appropriately. How this happens is still far from clear. Importantly, it is not known how or if a field of cells experiencing a gradient of auxin can interpret that gradient in a quantitative way or the extent to which outputs can be linear and continuous or more stepped and threshold-dependent or indeed, if more than one mode of control can operate simultaneously. The core AFB-Aux/IAA-ARF mechanism provides a means to turn genes on and off in response to auxin but to understand the capacity of this system to respond differently to different kinds of auxin stimulation it is useful to break the system down into the key sets of interactions among the different family members. These are: the interaction of the AFB receptors with auxin and Aux/IAA proteins (the perception event); the Aux/IAA-ARF event; the Aux/IAA-corepressor event; and finally, the ARF-promoter event. At several steps in this network of physical interactions specificities and binding preferences, either predicted or proven to exist, provide a means to generate specific responses to auxin pulses of differing length and amplitude in the cell.
Specific Interactions in the AFB-Aux/IAA-ARF System
At the level of auxin perception, genetic and biochemical analysis points to significant functional specificity among the six TIR1/AFB receptors (Dharmasiri et al. 2005b; Parry et al. 2009). This is perhaps not surprising given that among the 24 or so Aux/IAAs with recognizable domain II degron motifs there is significant variation at more than half of the degron positions (Dreher et al. 2006). Thus it is highly likely that this structural variation among both receptors and Aux/IAAs will be reflected in a range of affinities for each receptor-auxin-Aux/IAA complex. In turn, such differences in affinity would be predicted to translate to differences in Aux/IAA stability and indeed the half-lives of Aux/IAA proteins range from extremely short at 6 min to much longer at 80 min (Dreher et al. 2006). Therefore this specificity in the very first step of TIR1/AFB-mediated auxin signal transduction has the potential to affect profoundly the balance of downstream interactions between Aux/IAAs and ARFs and so, how the auxin signal in that particular cell is interpreted.
Next, specific interactions among Aux/IAAs and ARFs represent the class of interactions with perhaps the greatest potential to encode information in the system. Because different ARFA proteins seem to regulate distinct asz well as overlapping subsets of auxin-regulated genes, binding preferences among Aux/IAAs and ARFs represent a way to direct repression to particular sets of loci. Several lines of evidence support the existence of such binding preferences but most convincing are data from the analysis of semi-dominant stabilizing mutations in several Arabidopsis Aux/IAA genes. Although there are very few phenotypes associated with loss of Aux/IAA function (Overvoorde et al. 2005), gain-of-function mutations in different Aux/IAAs give rise to a range of striking phenotypes, some common and some entirely opposite (Liscum and Reed 2002). It is the latter group that are informative here because although some differences can be attributed to the expression pattern of the stabilized proteins, several of these opposite phenotypes persist when the mutant proteins are expressed in the same cell type and/or from the same promoter (Knox et al. 2003; Weijers et al. 2005a; Muto et al. 2007). A good example is the effect of stabilized SHY2/IAA3 and AXR3/IAA17 proteins on the production of root hairs. Driven by the same inducible promoter, too much IAA3 protein results in longer root hairs whereas too much IAA17 protein stops root hair production all together (Knox et al. 2003). Whatever the details of the molecular basis of these phenotypes, it is clear that these mutant proteins can do different things and although there are other possibilities, the most parsimonious explanation is that they are interacting predominantly with different ARFs, which are in turn targeting distinct subsets of auxin-regulated loci. This idea is consistent with recent domain-swapping experiments in which the carboxy-terminal dimerization domains of mutant axr3-1 and shy2-2 proteins were exchanged. This work showed that when driven by the AXR3 promoter, only chimeric proteins with the carboxy-terminal dimerization domains of AXR3 were able to bring about the dramatic reduction in root hair production characteristic of the axr3-1 mutant (A. Readshaw, L. Armitage, S. Kepinski, and O. Leyser, unpubl.). Thus, in this context at least, the bulk of the distinct function of AXR3 can be accounted for by specific interactions with other ARFs and/or Aux/IAAs (or indeed, as yet unknown auxiliary factors). In addition it is possible that in other contexts or for other Aux/IAAs, specific interactions between Aux/IAAs and TPL/TPR proteins could also be significant, especially in the event of highly tissue-specific patterns of TPL/TPR expression.
A corollary of the apparent existence of binding preferences among Aux/IAA and ARF proteins is that there must also be specificity in the binding of ARF proteins to different cis-regulatory elements. This idea that individual ARFs or subsets of ARFs regulate particular sets of genes is consistent with a number of studies but has not been shown unequivocally (Hardtke et al. 2004; Okushima et al. 2005). Indeed, very little is known about the nature of the interactions between ARFs and the cis-regulatory elements of their target genes. A number of these elements which contribute to the auxin responsiveness of various promoters have been characterized but only the TGTCTC-containing AuxRE motifs have been associated with ARF binding (Ulmasov et al. 1997a; Ulmasov et al. 1999b). Gel shift experiments using various ARFs identified different tolerances for substitutions of residues at +5 and +6 in the consensus TGTCTC AuxRE providing some experimental support for selectivity in ARF-DNA interactions (Ulmasov et al. 1999b). It is highly likely that coupling and auxiliary cis elements and associated transcription factors will also have a significant impact on binding specificity and further that entirely non-TGTCNC-based AuxREs will exist (Guilfoyle et al. 1998). All of these ideas require a rigorous and systematic analysis.
Complex Regulation Within the AFB-Aux/IAA-ARF System
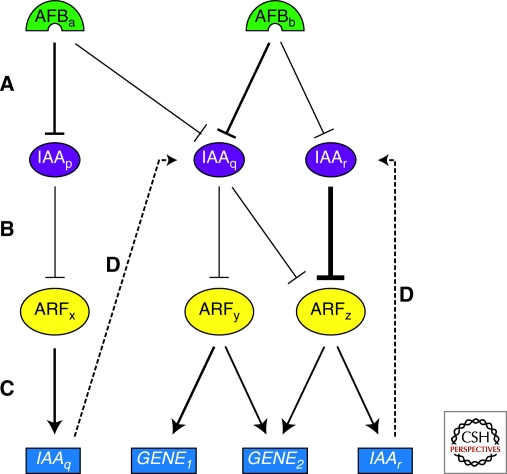
The significance of binding specificities within the AFB-Aux/IAA-ARF system in terms of information processing becomes clear when we think about the temporal changes in topology of the Aux/IAA-ARF mechanism in response to auxin and other stimuli. The principal mechanism of change here is the modulation of Aux/IAA levels and the fact that the rate of change of concentrations of different Aux/IAAs in response to auxin varies considerably across the family (Dreher et al. 2006). As well as variation in Aux/IAA half-lives, Aux/IAA genes are induced by exogenous auxin over a range of time and dose response profiles, some gene transcripts taking minutes to reach maximal levels with others taking hours (Abel et al. 1995). These two phenomena are of course linked by multiple individual Aux/IAA feedback loops which, because of the binding preferences at the level of Aux/IAAs and ARFs, and ARFs and target genes (including Aux/IAAs), makes the control of Aux/IAA levels across the family likely to be highly nonlinear. Further complexity arises from the fact that several ARF genes are also induced by auxin over relatively short time frames (Okushima et al. 2005; M. Kieffer and S. Kepinski, unpubl.), introducing the probability of amplifying positive feedback loops within the networks. Together this all means that information is carried principally in the coming and going of Aux/IAAs, with the output of the system at any given point being determined by the relative abundance of Aux/IAAs and ARFs. It is therefore easy to see how the duration and magnitude of differences in the abundance of any two Aux/IAAs of distinct function in a responding cell can represent a “piece” of information (Fig. 3). The challenge is to understand precisely how Aux/IAA (and ARF) abundance changes in response to auxin and other signals, and to know which downstream gene sets are regulated by which ARFs.
Figure 3.
Schematic representation of the complexity generated by differential interaction in the auxin pathway machinery. Hypothetical interaction network of the AFB receptors with Aux/IAA proteins (A), Aux/IAAs with ARFs (B), and ARFs with different target genes (C) are shown as arrows. The thickness of lines indicates the affinity of the interaction. Further complexity is generated by feedback loops of Aux/IAA (D) and ARF (not drawn) expression. As shown, Aux/IAAs can directly repress their own transcription (right) or that of other Aux/IAAs (left). Note that the tissue-specific lack of one or more factors would change auxin responsiveness and target gene expression.
This is a non-trivial problem and indeed the complexity of the Aux/IAA-ARF system with its highly looped feedback architecture means that there are a wide range of possible gene expression control behaviors, which are not immediately apparent outside of a computational framework. Modeling of simple Aux/IAA-ARF systems containing only one or two species of ARF and Aux/IAA has shown that as well as straightforward graded outputs in response to graded changes in auxin concentration, these simple systems can show bistability, that is, the system can output in one of two steady-states but not in any intermediate states (Middleton et al. 2010; G. Mirams, A. Middleton, L. Bridge, M. Kieffer, J. King and S. Kepinski, unpubl.). Such bistable (or even multistable) switches are common throughout prokaryotic and eukaryotic biological systems and are important in terms of development because they can convert graded inputs into switch-like “on/off” outputs (Brandman and Meyer 2008). This allows the generation of sharp transcriptional borders, and hence sharp transitions between developmental phases. In extreme cases these switches can be nonreversible but more commonly, bistable control systems are able to return to previous states but show hysteresis, meaning that the system can maintain a given state for some time after the loss of input signal required to achieve that state in the first place (Ferrell 2002; Ninfa and Mayo 2004). Another important and related feature of such bistable switches is that their response to a given level of input signal is dependent on the history of the cell, so that a low level of input signal that is insufficient to induce a response in a cell in an inactive state can be sufficient to maintain an pre-existing activated state. It is worth pointing out that the type of gene expression control at work, graded or switchlike, would depend on the connectivity in the AFB-Aux/IAA-ARF networks and importantly in the “wiring” to the downstream target genes. This means that it is perfectly possible for more than one type of behavior to originate from overlapping and largely similar Aux/IAA-ARF topologies in a single cell.
AUXIN GRADIENTS, AUXIN RESPONSE, AND DEVELOPMENTAL CONTROL
These ideas about the possible modes of auxin's control of gene expression are directly relevant to a number of auxin-regulated phenomena, which in all cases appear to be dependent on thresholds of auxin response. To explore these it is useful to think about auxin signaling in the context of the auxin gradients and accumulations that correlate with developmental events and transitions. This is less straightforward than it might at first appear because of the extreme difficulty measuring auxin levels in vivo. Therefore most of what is known about auxin distribution in planta has been inferred indirectly from a combination of auxin response, for the most part using the synthetic auxin responsive-promoter DR5 (Ulmasov et al. 1997b), and the occurrence and polarity of PIN auxin efflux carriers to indicate the vector of auxin flux (Benkova et al. 2003; Friml et al. 2003; Tanaka et al. 2006). This pragmatic approach to the difficulty of tracking auxin distribution is a reasonable one as long as it is remembered that expression driven by DR5 elements, itself mediated by Aux/IAAs and ARFs, is not necessarily a linear readout of auxin levels in a given tissue and is likely to also be influenced by tissue-specific variations in auxin responsiveness. Nevertheless, accepting these caveats, there does seem to be a credible case to support the existence of at least two broad classes of auxin gradient; those that are shallow, in which there appear to be relatively modest changes in concentration across a field of cells, and those that are very steep, in which adjacent cells seem to have dramatically different levels of auxin (Tanaka et al. 2006; Grieneisen et al. 2007). The latter are more usefully described as auxin maxima (or minima) and a prime example is of the induction of lateral root primordia. Here, PIN-mediated auxin accumulation in one or two cells of the xylem-pole pericycle is sufficient to initiate a lateral root primordium and subsequent patterns of PIN protein expression and polarity sustain an auxin distribution that drives the entire lateral root developmental program (Casimiro et al. 2001; Benkova et al. 2003; De Smet et al. 2006). Interestingly, the regular spacing of lateral roots in Arabidopsis seems to be based on a transient oscillatory increase in AFB-Aux/IAA-ARF activity in pericycle cells that are transiting the basal root meristem, several millimeters distal to the eventual site of lateral root emergence (De Smet et al. 2007). This suggests that this early auxin response might prime subsets of xylem-pole pericycle cells to respond differently (more readily) to later auxin activation of primordium initiation in the older root (De Smet et al. 2007). It is therefore possible that this priming of otherwise identical pericycle cells represents a hysteretic presensitization of the system.
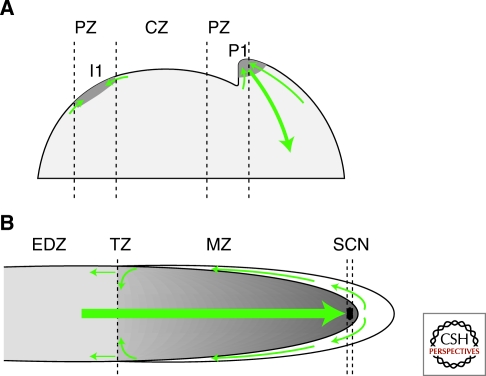
The fact that all xylem-pole pericycle cells can form lateral root founder cells in response to high levels of auxin suggests that initiating a lateral root is merely a case of exceeding a threshold auxin concentration (Benkova et al. 2003). This idea of thresholds of responsiveness is played out most elegantly in the shoot apical meristem (SAM) where complex PIN-mediated patterns of auxin accumulation and ARF-mediated response drive the induction of lateral organs such as leaves and flowers from the meristem flanks (Reinhardt et al. 2003; Heisler et al. 2005). The single layer of epidermis (L1) in the peripheral zone (PZ) of the SAM represents a field of cells within which existing primordia act as auxin sinks (Fig. 4A). The consequent depletion of auxin from adjacent PZ tissue means that new primordia are formed only at a certain minimal distance from existing primordia, giving rise to a regular arrangement of lateral organs known as phyllotaxis (Reinhardt et al. 2003). PIN1 expression and polarity in the L1 layer is responsible for the generation of auxin maxima via a so-called “up the gradient” model for PIN protein polarization, where as the name suggests, PIN proteins are orientated so that auxin is pumped out toward to adjacent cells with the highest auxin concentration (Jonsson et al. 2006; Smith et al. 2006). This is one of the many examples of self-organization in auxin biology. As well as putative non-genomic effects of auxin on PIN protein retention at the plasma membrane that could contribute to an up-the-gradient flow (Paciorek et al. 2005), the fact that PIN1 is induced by auxin via the AFB-Aux/IAA-ARF system is another positive feedback component of this mechanism (Vieten et al. 2005). In this way a convergence point is formed that marks precisely the incipient primordium and once a threshold concentration of auxin is reached, primordium initiation is begun (Reinhardt et al. 2003; Bayer et al. 2009). In addition to this, experimental and mathematical modeling approaches have identified the possibility of a second auxin-dependent developmental transition in the primordium program, triggered once an even higher auxin concentration threshold is reached at the convergence point (Bayer et al. 2009): Midway through primordium development auxin flow switches from predominantly toward the convergence point in the L1 layer to away from it and down into the underlying tissues as part of the specification of the vasculature that will form the midvein of the developing leaf (Sachs 2000; Reinhardt et al. 2003) (Fig. 4A). This demands an opposite “with-the-flux” polarization of PIN proteins which is part of a more general concept of self-organization in auxin biology known as the canalization hypothesis which proposes that increasingly narrow paths or “canals” for auxin transport can be formed by the action of positive feedback loops acting at the level of individual cells (Sachs 2000; Sauer et al. 2006). Recent modeling work evaluating these seemingly incompatible arrangements of PINs has raised the possibility that both mechanisms could operate simultaneously in a given cell with the balance of activity between them determined by thresholds of auxin concentration (Bayer et al. 2009).
Figure 4.
Auxin distribution and flux at the root and shoot apical meristems. Predicted auxin distribution is represented by shades of grey, where darker means higher concentration. Green arrows represent flux with arrowhead representing the direction of the flow and thickness is proportional to the flux of auxin. (A) In the shoot apical meristem, PIN-mediated auxin transport generates a discrete auxin maximum in the peripheral zone (PZ), known as the convergence point, which marks the site of the incipient primordium (I1). Above a certain threshold concentration, a lateral organ is initiated (in this case P1). Once the developing primordium reaches a certain stage, a new flux of auxin down and away from the convergence point is initiated, which guides the specification of vascular tissue. The distribution of auxin in the central zone (CZ) is still unknown and therefore has not be represented. (B) In the root meristem PIN-mediated polar transport generates an auxin maximum at the root tip that positions and maintains the stem-cell niche (SCN), and an auxin gradient throughout the meristematic zone (MZ) that sustains cell proliferation. Auxin is transported to the root tip through the central vascular tissue and then from the SCN is moved laterally and up and away from the tip. Importantly, a significant proportion of the basipetal auxin flow is refluxed back into the central acropetal flow at the transition zone (TZ). This system of polar auxin transport is predicted to generate a gradient of auxin that is very high in the SCN, moderately high in the meristematic zone (MZ) and much lower in the elongation/differentiation zone (EDZ).
Gradients of Auxin Concentration and Response in the Root Tip
These examples of very defined and highly asymmetric localizations of auxin and auxin response are taken from developmental contexts in which an existing field of cells is subsequently patterned by local auxin accumulation or flux. The second class of auxin gradient, that of the shallower, graded distributions occurs in more complicated developmental scenarios in which auxin is tightly linked with the production of the cells that it is subsequently proposed also to regulate. The two principal examples here are of the root apex and the vascular cambium in the wood-forming tissues of trees. In the root apex, the stem cell niche (SCN), containing the quiescent center and stem cells, gives rise to all cell types of the root (Sabatini et al. 1999). Cells laid down behind the SCN as the root apex advances remain mitotically active for a period before beginning to elongate and differentiate (Beemster and Baskin 1998). Thus the proximo-distal axis of the root tip can be divided into at least two broad zones; the meristematic zone in which cells divide and the elongation/differentiation zone in which proliferation gives way to cell elongation and differentiation (Beemster and Baskin 1998; Moubayidin et al. 2009).
Mathematical modeling, supported as far as possible by experimental observation, suggests that this longitudinal pattern of developmental transitions can be correlated with an overall gradient of auxin, which shifts from high in the root apex to low in the elongation and differentiation zones (Grieneisen et al. 2007; Petersson et al. 2009) (Fig. 4B). A dynamic and coordinated system of PIN protein expression and orientation moves auxin around the growing root tip creating a distal auxin maximum that, with other inputs, positions and maintains the SCN (Blilou et al. 2005; Xu et al. 2006). Proximal to this, an accompanying diminishing gradient of auxin appears to play a role in regulating the extent of proliferation in the meristem and thus the transition to elongation and differentiation (Blilou et al. 2005; Grieneisen et al. 2007). PINs are arranged so that the main flux of auxin is down the central vascular tissues into the SCN auxin maximum (Fig. 4B). From here, auxin is moved laterally outward through the lateral root cap and back up and away from the tip through the lateral root cap and epidermis. Importantly, at the transition zone between the meristematic and elongation zones, PIN proteins are orientated so that a significant portion of the auxin flowing away from the tip is refluxed laterally back into the central acropetal flow (Blilou et al. 2005). This reflux loop contributes to the maintenance of both the discrete distal maximum and overall to the high levels of auxin throughout the meristem relative to the adjacent elongation zone, creating a steeper drop in auxin concentration across the MZ/EZ boundary that has been proposed to prompt the transition to elongation and differentiation (Grieneisen et al. 2007). Importantly, mathematical modeling of the PIN system in the root tip has shown that it possesses impressive self-organizing properties and is predicted to be capable of generating the observed patterns of auxin distribution, even in the absence of feedbacks with auxin response (Grieneisen et al. 2007).
The extent to which auxin responses along the proximo-distal root axis are mediated by the AFB-Aux/IAA-ARF system is not entirely clear. It is certainly involved the positioning of the SCN by regulating indirectly the expression of members of the PLETHORA (PLT) family of AP2-domain transcription factors which are required for stem cell maintenance (Sabatini et al. 1999; Aida et al. 2004). In turn, PLT expression is required for the proper expression of the PIN proteins that mediate both the formation of the distal auxin maximum and the root tip auxin reflux loop more generally (Aida et al. 2004; Blilou et al. 2005). This positive feedback loop is thus another stabilizing input into self-organizing auxin distribution in the root tip.
Although PLT expression and other evidence of high levels of AFB-Aux/IAA-ARF activity in the SCN could be considered to be a straightforward case of response to an auxin maximum, PLTs may also have other dose-dependent functions in addition to the maintenance of the SCN that could be related to the interpretation of a proximal-distal auxin gradient. The expression of the four PLT family proteins with demonstrated functions in the root tip (PLT1, PLT2, PLT3, and BABY BOOM [BBM]) is highest in the SCN, diminishing to varying extents into the proximal meristem (Galinha et al. 2007). In addition to the loss of stem cells, plt mutant combinations also have a short meristem phenotype. The targeted misexpression of PLT2 in only the proximal meristem of plt1 plt2 mutants is sufficient to suppress this meristem phenotype without rescuing distal stem cell defects suggesting that PLT2 can have an input into proliferation in the meristem spatially and temporally separate from that which controls stem cell maintenance (Galinha et al. 2007). What is less clear is the extent to which the lower levels of PLT in the meristem represent a fundamentally distinct regulation of cellular activity from that observed in the SCN. Certainly, without further genetic intervention to release the effects of parallel inputs into SCN specification (via the down-regulation of the RETINOBLASTOMA (RBR) pathway), inducible overexpression of PLT by itself is not sufficient to increase the size of the stem cell pool (Galinha et al. 2007). Thus it is not possible to know whether differences in higher and lower doses of PLT direct truly different aspects stemness and meristemness respectively. Further, because of the feedback relationships between PLTs and auxin distribution it is difficult to assess the extent to which PLT expression and action represents a functional readout of a proximo-distal auxin gradient. For example it is also possible that PLT2 misexpression in the proximal meristem affects auxin homeostasis in the MZ and thus has indirect effects on proliferation through parallel AFB-Aux/IAA-ARF-independent mechanisms.
The Morphogen Question
Regardless of whether or not gradients of PLT are a readout of auxin levels in the root that mediate changes in cellular activity as cells transit away from the SCN, there seems little doubt that high concentrations of auxin are required for the specification of the SCN, moderately high concentrations are required to sustain division in the root meristem and as a corollary of this, elongation and differentiation are only possible at much lower levels still. This being the case the question of whether auxin can be considered a morphogen has often been raised, the answer to which has invariably been no owing to the rather strict nature of the modern definition of a morphogen (Sabatini et al. 1999; Bhalerao and Bennett 2003; Benkova et al. 2009). This subject has been covered extensively elsewhere and so comment here will be restricted to pointing out how very different and more complex these developmental scenarios are from animal embryo context in which the modern morphogen concept was constructed (Wolpert 1969). Although the central idea of Wolpert's iconic French flag hypothesis can be grafted to these putative plant examples of morphogen action it is important to remember that these are dynamic growing structures and so every cell in the morphogenetic field has arisen in the same peak of auxin concentration. What follows from this is that the potential of a cell to respond to auxin in the next unit of developmental space is the product of auxin concentration and the prevailing auxin signaling topology at that point or in other words, each cell's response will be dependent on the history of previous responses to higher levels of auxin further up the gradient. In its requirements of the cell's ability to distinguish different levels of stimulation this then is distinct from the situation where a field of cells derives positional information from a morphogenetic gradient. Here, it is a temporal as much as a spatial question because the question is of how a single cell behaves during a descent down an auxin concentration gradient formed as the auxin maximum in the root tip moves farther away.
Auxin as a Self-Organizer
There are of course examples in plant development in which an existing field of cells is patterned by auxin. Again, the inter-primordium L1 layer of the SAM is a good example, but here cells in the PZ field will do only one of two things; make a leaf or a flower primordium if auxin concentrations are high enough, or continue with a default stem tissue program if they are not (Reinhardt et al. 2003). Thus strictly speaking auxin is simply a permissive signal for lateral organ initiation in the SAM. If not a morphogen then, can we at least say whether there are circumstances in which auxin can be an instructive signal? Put another way, is there any example in normal development in which different concentrations of auxin within a field of cells can induce more than two fates or states (i.e., a prestimulation fate and a poststimulation fate)? Indeed it is difficult to find any example in which this condition is met. Despite this, the argument for an instructive role for auxin in development has been advanced (Reinhardt et al. 2003) and can be supported if auxin's capacity to self-organize is considered. Several examples of self-organization have already been highlighted here and in all of these cases the underlying mechanism is the regulatory feedback between auxin transport and auxin response (e.g., Fig. 1C). From this point of view, the auxin transport-auxin response “module” that underlies phyllotactic patterning possesses a high level of instructive capacity; without the requirement for signals other than auxin, responses at the level of individual cells lead to the emergence of pattern across tissues.
Self-Organization in Time as Well as Space
As well as the spatial control generated by feedback between auxin transport and response, the many feedback loops between individual components of the AFB-Aux/IAA-ARF system provide another layer of self-organizational capacity that can operate over time within individual cells and their descendants. An example is of the priming of xylem-pole pericycle cells for later activation as lateral root founder cells as described above (De Smet et al. 2007). The idea that one specific AFB-Aux/IAA-ARF topology can give rise to another with a different potential to respond to auxin is a powerful one because it provides a means to generate a more complex and multilayered response to a simple auxin input signal. Instances of evolving auxin signaling topologies are beginning to come to light. For example, for some time it has been known that a regulatory module consisting of the Aux/IAA SOLITARY ROOT (SLR/IAA14) and the ARFs NPH4/ARF7 and ARF19 is required for lateral root initiation and organogenesis (Fukaki et al. 2002; Vanneste et al. 2005). More recently a second module involving the Aux/IAA BODENLOS (BDL)/IAA12 and the ARF MP/ARF5 has been shown to act after SLR-ARF7/19 and indeed the over-expression of MP was able to suppress the lateral rootless phenotype of slr (De Smet et al. 2010b). Although it is not clear whether the induction of this BDL/MP module is directly dependent on ARF7/19 regulation, BDL is certainly an auxin-inducible gene and there is evidence that MP expression is also moderately induced by auxin treatment (Abel et al. 1995; Wenzel et al. 2007).
This concept of auxin signaling topologies self-organizing over time is also relevant to the robustness of auxin-regulated development. This is because a system that depends on a sequence of auxin-triggered events can be buffered against fluctuation and noise in auxin concentration so that quantitative differences in auxin input need not affect the qualitative output. For example, in response to treatment with high levels of auxins such as IAA or NAA, the xylem-pole pericycle responds by initiating many LR primordia, each developing as a more or less perfectly-formed lateral root (Benkova et al. 2003). Thus, across a range of auxin input signals, the integrity of the individual LR developmental program is maintained. Drilling down further into the LR development program reveals yet more examples of highly feedback-regulated downstream modules that contribute to the robustness and integrity of development. For example, the transcription factor NAC1, a positive regulator of LR development is induced by auxin in an ARF-dependent manner but this induction is also accompanied by the expression of both the micro RNA miR164 which negatively regulates NAC1 transcript levels and the ubiquitin ligase SINAT5 which targets the NAC1 protein for degradation, suggesting that only a brief input from NAC1 is required for normal lateral root development (Xie et al. 2000; Xie et al. 2002; Guo et al. 2005).
DEVELOPMENTAL CONTEXT AND AUXIN RESPONSE
Whatever the self-organizing capacity of auxin to pattern developmental events in time and space, the nature of those events is constrained by developmental context as evidenced by the unmistakably root- and shoot-specific outcomes of auxin accumulation in the xylem-pole pericycle and PZ of the shoot apex respectively. How then are these auxin response contexts set? Considering the rather fundamental nature of this question, surprisingly little is known of how variation in auxin responsiveness is specified. There are two principal levels at which a specific auxin response context can be established: spatial variation in either the core AFB-Aux/IAA-ARF system or the connections of downstream effectors to different context-specific AFB-Aux/IAA-ARF modules. It is extremely likely that, as a general rule for auxin action, variation at both of these levels would be required to allow the flexibility necessary for appropriate developmental outputs to be generated in each distinct developmental context. The process of organogenesis at the SAM highlights a possible exception in which spatial variation in the AFB-Aux/IAA-ARF system alone may be sufficient to account for specification of a distinct auxin response context. This is because it is only the PZ of the SAM, and not the adjacent central zone (CZ), that can respond to auxin with the development of a lateral organ (Reinhardt et al. 2000; Reinhardt et al. 2003). Analysis of DR5::GFP across the entire shoot apex after treatment with exogenous auxin shows that AFB-Aux/IAA-ARF action in the CZ is suppressed in some way, pointing to differences in AFB-Aux/IAA-ARF topology between these meristem zones (de Reuille et al. 2006). It is also entirely possible that other factors outside of the known AFB-Aux/IAA-ARF system might contribute to context-specific responses: the identification of novel ARF interactors such as the Myb transcription factor MYB77 (Shin et al. 2007) has highlighted the possibility that auxiliary factors might bind to ARFs or other components of the AFB-Aux/IAA-ARF system to modulate their activity. From this viewpoint, the core AFB-Aux/IAA-ARF module would provide a basic scaffold of auxin regulation into which auxiliary (transcription) factors are integrated to generate distinct auxin response contexts.
Patterning Spatial Variation in Auxin Responsiveness
The evidence for tissue- and cell-type-specific variation in Aux/IAA and ARF expression in particular is abundant (Abel et al. 1995; Birnbaum et al. 2003; Weijers et al. 2006; Brady et al. 2007) but for the most part little is known of the functional significance of this variation and in no case has a mechanistic link to an upstream patterning mechanism been made. Perhaps the best understood developmental context in this respect is the Arabidopsis embryo (for review see Möller and Weijers 2009). Here, MP/ARF5 and BDL/IAA12 contribute to the initiation of the embryonic root by specifying the uppermost cell of the suspensor (the extraemryonic column of cells that supports the embryo proper) to become the hypophysis (Weijers et al. 2006; Schlereth et al. 2010). Importantly MP and BDL expression is confined to the central cells of the proembryo immediately above the suspensor cells. From here they act to promote the transport of auxin to the incipient hypophysis by regulating the expression of the PIN protein PIN1 (Weijers et al. 2006). This generates a maximum of AFB-Aux/IAA-ARF-mediated response that is essential for hypohysis specification. Interestingly the the Aux/IAAs and ARFs that mediate the hyphophyseal response to auxin are distinct from MP and BDL: although loss-of-function mp mutants and stabilized gain-of-function bdl mutants both fail to specify the hypophysis and consequently lack a root, the expression of mutant bdl protein in the hypophysis has no effect on embryonic root development (Weijers et al. 2006). Recent work has identified a basic helix-loop-helix transcription factor, itself a target of MP (TARGET OF MONOPTEROS 7, TMO7), that is also required for hypophysis specification. Importantly, TMO7 moves from its site of synthesis in the central cells to the hypophysis providing another MP-mediated input into embryonic root development (Schlereth et al. 2010).
This BDL-MP activity is part of the elaboration of an initial apical-basal asymmetry that exists in the egg cell even before fertilization (Friml et al. 2003; De Smet et al. 2010a). An asymmetric zygotic division creates a smaller apical cell that divides to form the proembryo and a larger basal cell that is the origin of the suspensor tissue (De Smet et al. 2010a). This suggests that proembryo and suspensor-specific AFB-Aux/IAA-ARF topologies may be traced back to the inherent asymmetry of the egg cell. Both the mechanism of patterning these distinct auxin response contexts and the origin of the initial asymmetry in the egg cell are unknown. In the case of the latter it would not be entirely unexpected if auxin was found to be a (maternal) polarizing signal here too. Also, the fact that DR5 activity precedes that of BDL/MP expression in the proembryo might suggest that yet another AFB-Aux/IAA-ARF topology is required for this initial response (Friml et al. 2003), again hinting at the possibility of evolving Aux/IAA-ARF regimes. Indeed, despite the existence of very dramatic fluxes of auxin initially up to the apical cells and then down to the hypophysis, the fundamental apical-basal axis appears remarkably resistant to perturbation of auxin homeostasis (Friml et al. 2003; Weijers et al. 2005b). This could be yet another example of sequential, temporally-damped, feedback-regulated, self-organization of AFB-Aux/IAA-ARF activity and auxin movement conferring developmental robustness across a range of auxin concentrations.
Contextual Inputs—Environmental Regulation of Auxin Response
So far auxin has been discussed in isolation, principally to draw attention to its remarkable capacity for self-organization and developmental control. Of course, the auxin response system operates as part of a much larger system and is connected at many levels, or nodes, to other signaling subsystems within the plant, including those for the other major plant hormones (Kieffer et al. 2010; (Stewart and Nemhauser 2010). The integration of intrinsic developmental and extrinsic environmental signals in the auxin response system is a crucial component of the postembryonic plasticity of plant development. Although there are many examples of signal integration via the modulation of auxin distribution, responses to light and gravity being obvious cases, most relevant here are environmental signals whose primary effect is to alter development by altering the sensitivity of tissues to auxin (Perez-Torres et al. 2008). A good example is of the root response to phosphate starvation, which includes an increase in lateral rooting and root hair length and a decrease in the growth of the primary root. These phenotypes are reminiscent of auxin treatment and a recent study has revealed at least one component of this similarity in response by demonstrating that Pi starvation promotes the expression of the auxin receptor TIR1, rendering the root hypersensitive to IAA and promoting auxin response (Perez-Torres et al. 2008).
The modulation of auxin sensitivity via TIR1/AFB regulation has also been implicated in the defense response to the pathogen P. syringae. Bacterial infection induces the expression of the micro-RNA miR393 which negatively regulates TIR1, AFB2, and AFB3 mRNA, thereby diminishing auxin response and restricting pathogen growth (Navarro et al. 2006). These examples of signal-induced changes in auxin sensitivity represent another dimension of contextual control in which intrinsic auxin-regulated developmental processes can be modulated to optimize the fitness of the plant.
CONCLUSIONS
The remarkable versatility of auxin as a signaling molecule has led to a search for unifying concepts that may explain its repeated use as a developmental regulator. This article has focused on the capacity of the AFB-Aux/IAA-ARF system both alone and in combination with feedback regulation of auxin transport, to interpret and effect responses to variation in auxin distribution by forming context-specific and often self-organizing response modules. It is important to point out that other auxin signaling systems, such as those involving ABP1, have received little attention here (see Tromas et al. 2010b for detailed review). These additional systems of course add further layers of control and complexity to auxin response. Recent work also suggests ABP1-mediated signaling is also linked by feedback loops to both AFB-Aux/IAA-ARF-mediated response (Tromas et al. 2010a,b). It is also the case that even in this extended discussion on the topic, simplifications have been made for sake of clarity. Perhaps the most obvious is the idea of pairs of Aux/IAAs and ARFs regulating particular processes. While it is true that in the case of BDL and MP for example, these proteins have been shown to work together, this does not mean that other coexpressed ARFs and Aux/IAAs (especially the latter) do not contribute to MP-mediated responses. It is more likely that the affinities between any given Aux/IAA and ARF pair will range from very low to very high and therefore outputs in any given cell at a given time will be the product of a complex set of interactions.
The picture that emerges is one of almost unfathomable complexity and certainly sufficient flexibility to regulate different processes simultaneously in time and space, and with constant reference to environmental signals. At the same time, the ability now to address auxin response at a mechanistic level, taking a rational and where possible quantitative and mathematical approach to understanding the genetic and biophysical interactions of the core AFB-Aux/IAA-ARF components and their downstream targets in each developmental context, provides an extremely powerful framework within which fundamentally new understanding of plant biology can be generated. Because auxin impacts on so much of development, it is reasonable to begin to think of how multiscale computational models of auxin distribution and response could form the fabric of organ- and organism-level models of plant development. Such an approach, while presently extremely challenging both experimentally and mathematically, offers perhaps the greatest opportunity to capture the plasticity of development in all its glorious complexity.
ACKNOWLEDGMENTS
We would like to apologize to those authors whose valuable contributions could not be included because of space constraints. Work in the authors' laboratory is supported by the Biotechnology and Biological Research Council (BB/F007418/1 and BB/F013981/1), the Engineering and Physical Sciences Research Council (EP/G039496/1), and by a White Rose Studentship to M.D.B.
Footnotes
Editors: Mark Estelle, Dolf Weijers, Ottoline Leyser, and Karin Ljung
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
- Abel S, Oeller PW, Theologis A 1994. Early auxin-induced genes encode short-lived nuclear proteins. Proc Natl Acad Sci 91: 326–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Nguyen MD, Theologis A 1995. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol 251: 533–549 [DOI] [PubMed] [Google Scholar]
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, Blakeslee JJ, Lee OR, Mravec J, Sauer M, Titapiwatanakun B, Makam SN, Bouchard R, Geisler M, Martinoia E, Friml J, Peer WA, Murphy AS 2007. Interactions of PIN and PGP auxin transport mechanisms. Biochem Soc Trans 35: 137–141 [DOI] [PubMed] [Google Scholar]
- Bayer EM, Smith RS, Mandel T, Nakayama N, Sauer M, Prusinkiewicz P, Kuhlemeier C 2009. Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev 23: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GT, Baskin TI 1998. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol 116: 1515–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkova E, Ivanchenko MG, Friml J, Shishkova S, Dubrovsky JG 2009. A morphogenetic trigger: Is there an emerging concept in plant developmental biology? Trends Plant Sci 14: 189–193 [DOI] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA 1996. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 16: 948–950 [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Bennett MJ 2003. The case for morphogens in plants. Nat Cell Biol 5: 939–943 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN 2003. A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN 2007. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brandman O, Meyer T 2008. Feedback loops shape cellular signals in space and time. Science 322: 390–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Villalobos LI, Tan X, Zheng N, Estelle M 2010. Auxin Perception—Structural Insights. Cold Spring Harb Perspect Biol 2: a005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao R, Beeckman T, Dhooge S, Swarup R, Graham N, Inze D, Sandberg G, Casero P, et al. 2001. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Reuille PB, Bohn-Courseau I, Ljung K, Morin H, Carraro N, Godin C, Traas J 2006. Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc Natl Acad Sci 103: 1627–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Lau S, Mayer U, Jurgens G 2010a. Embryogenesis—the humble beginnings of plant life. Plant J 61: 959–970 [DOI] [PubMed] [Google Scholar]
- De Smet I, Lau S, Voss U, Vanneste S, Benjamins R, Rademacher E, Schlereth A, De Rybel B, Vassileva V, Grunewald W, et al. 2010b. Bimodular auxin response controls organogenesis in Arabidopsis. Proc Natl Acad Sci 1072705–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frey N, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. 2007. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690 [DOI] [PubMed] [Google Scholar]
- De Smet I, Vanneste S, Inze D, Beeckman T 2006. Lateral root initiation or the birth of a new meristem. Plant Mol Biol 60: 871–887 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M 2005a. The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M 2005b. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J 2006. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE Jr 2002. Self-perpetuating states in signal transduction: Positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol 14: 140–148 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jurgens G 2003. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M 2002. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B 2007. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M 1999. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13: 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M 2001. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Maree AF, Hogeweg P, Scheres B 2007. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013 [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G 2007. Auxin response factors. Curr Opin Plant Biol 10: 453–460 [DOI] [PubMed] [Google Scholar]
- Guilfoyle T, Hagen G, Ulmasov T, Murfett J 1998. How does auxin turn on genes? Plant Physiol 118: 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HS, Xie Q, Fei JF, Chua NH 2005. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17: 1376–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen G, Guilfoyle TJ, Berleth T 2004. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131: 1089–1100 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM 2005. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Jonsson H, Heisler MG, Shapiro BE, Meyerowitz EM, Mjolsness E 2006. An auxin-driven polarized transport model for phyllotaxis. Proc Natl Acad Sci 103: 1633–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O 2002. Ubiquitination and auxin signaling: A degrading story. Plant Cell 14 Suppl: S81–S95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kieffer M, Neve J, Kepinski S 2010. Defining auxin response contexts in plant development. Curr Opin Plant Biol 13: 12–20 [DOI] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A 1997. Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci 94: 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox K, Grierson CS, Leyser O 2003. AXR3 and SHY2 interact to regulate root hair development. Development 130: 5769–5777 [DOI] [PubMed] [Google Scholar]
- Leyser O 2005. Auxin distribution and plant pattern formation: How many angels can dance on the point of PIN? Cell 121: 819–822 [DOI] [PubMed] [Google Scholar]
- Liscum E, Reed JW 2002. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49: 387–400 [PubMed] [Google Scholar]
- Lokerse AS, Weijers D 2009. Auxin enters the matrix–assembly of response machineries for specific outputs. Curr Opin Plant Biol 12: 520–526 [DOI] [PubMed] [Google Scholar]
- Maraschin S, Memelink J, Offringa R 2009. Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J 59: 100–109 [DOI] [PubMed] [Google Scholar]
- Middleton AM, King JR, Bennett MJ, Owen MR 2010. Mathematical modelling of the Aux/IAA negative feedback loop. Bull Math Biol. [DOI] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M 2004. The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L, Di Mambro R, Sabatini S 2009. Cytokinin-auxin crosstalk. Trends Plant Sci 14: 557–562 [DOI] [PubMed] [Google Scholar]
- Möller B, Weijers D 2009. Auxin control of embryo patterning. Cold Spring Harb Perspect Biol 1: a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto H, Watahiki MK, Nakamoto D, Kinjo M, Yamamoto KT 2007. Specificity and similarity of functions of the Aux/IAA genes in auxin signaling of Arabidopsis revealed by promoter-exchange experiments among MSG2/IAA19, AXR2/IAA7, and SLR/IAA14. Plant Physiol 144: 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD 2006. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Ninfa AJ, Mayo AE 2004. Hysteresis vs. graded responses: The connections make all the difference. Sci STKE 2004: e20. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde P, Arima K, Alonso J, Chan A, Chang C, Ecker J, Hughes B, Lui A, Nguyen D, et al. 2005. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A 2001. IAA17/AXR3: biochemical insight into an auxin mutant phenotype. Plant Cell 13: 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde P, Okushima Y, Alonso J, Chan A, Chang C, Ecker J, Hughes B, Liu A, Onodera C, Quach H, et al. 2005. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17: 3282–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T, Zazimalova E, Ruthardt N, Petrasek J, Stierhof Y, Kleine-Vehn J, Morris D, Emans N, Jurgens G, Geldner N, et al. 2005. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256 [DOI] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M 2009. Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci 106: 22540–22545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Torres CA, Lopez-Bucio J, Cruz-Ramirez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L 2008. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20: 3258–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K 2009. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J 2001. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13: 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Mandel T, Kuhlemeier C 2000. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C 2003. Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M 1998. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev 12: 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. 1999. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Sachs T 2000. Integrating cellular and organismic aspects of vascular differentiation. Plant Cell Physiol 41: 649–656 [DOI] [PubMed] [Google Scholar]
- Salmon J, Ramos J, Callis J 2008. Degradation of the auxin response factor ARF1. Plant J 54: 118–128 [DOI] [PubMed] [Google Scholar]
- Sauer M, Balla J, Luschnig C, Wisniewska J, Reinohl V, Friml J, Benkova E 2006. Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev 20: 2902–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A, Moller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jurgens G, Weijers D 2010. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916 [DOI] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC 1997. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124: 4481–4491 [DOI] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP 2007. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19: 2440–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RS, Guyomarc'h S, Mandel T, Reinhardt D, Kuhlemeier C, Prusinkiewicz P 2006. A plausible model of phyllotaxis. Proc Natl Acad Sci 103: 1301–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JL, Nemhauser JL 2010. Do trees grow on money? Auxin as the currency of the cellular economy. Cold Spring Harb Perspect Biol 2: a001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA 2008. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Dhonukshe P, Brewer PB, Friml J 2006. Spatiotemporal asymmetric auxin distribution: A means to coordinate plant development. Cell Mol Life Sci 63: 2738–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T 2003. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ 2004. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ 2001. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas A, Braun N, Muller P, Khodus T, Paponov IA, Palme K, Ljung K, Lee JY, Benfey P, Murray JA, Scheres B, Perrot-Rechenmann C 2010a. The AUXIN BINDING PROTEIN 1 is required for differential auxin responses mediating root growth. PLoS ONE 4: e6648 doi:10.1371/journal.pone.0006648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas A, Paponov I, Perrot-Rechenmann C 2010b. AUXIN BINDING PROTEIN 1: functional and evolutionary aspects. Trends Plant Sci 15: 436–446 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ 1997a. ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ 1999a. Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci 96: 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ 1999b. Dimerization and DNA binding of auxin response factors. Plant J 19: 309–319 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ 1997b. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, De Rybel B, Beemster G, Ljung K, De Smet I, Van Isterdael G, Naudts M, Iida R, Gruissem W, Tasaka M, et al. 2005. Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17: 3035–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wisniewska J, Benkova E, Benjamins R, Beeckman T, Luschnig C, Friml J 2005. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531 [DOI] [PubMed] [Google Scholar]
- Walsh TA, Neal R, Merlo AO, Honma M, Hicks GR, Wolff K, Matsumura W, Davies JP 2006. Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol 142: 542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jager KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jurgens G 2005a. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. Embo J 24: 1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Sauer M, Meurette O, Friml J, Ljung K, Sandberg G, Hooykaas P, Offringa R 2005b. Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell 17: 2517–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Schlereth A, Ehrismann JS, Schwank G, Kientz M, Jurgens G 2006. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell 10: 265–270 [DOI] [PubMed] [Google Scholar]
- Wenzel CL, Schuetz M, Yu Q, Mattsson J 2007. Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in Arabidopsis thaliana. Plant J 49: 387–398 [DOI] [PubMed] [Google Scholar]
- Wolpert L 1969. Positional information and the spatial pattern of cellular differentiation. J Theor Biol 25: 1–47 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B 2005. Auxin: Regulation, action, and interaction. Ann Bot 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua NH 2000. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14: 3024–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH 2002. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170 [DOI] [PubMed] [Google Scholar]
- Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, Scheres B 2006. A molecular framework for plant regeneration. Science 311: 385–388 [DOI] [PubMed] [Google Scholar]
- Zenser N, Ellsmore A, Leasure C, Callis J 2001. Auxin modulates the degradation rate of Aux/IAA proteins. Proc Natl Acad Sci 98: 11795–11800 [DOI] [PMC free article] [PubMed] [Google Scholar]