Abstract
Pre-mRNA processing, including 5′-end capping, splicing, editing, and polyadenylation, consists of a series of orchestrated and primarily cotranscriptional steps that ensure both the high fidelity and extreme diversity characteristic of eukaryotic gene expression. Alternative splicing and editing allow relatively small genomes to encode vast proteomic arrays while alternative 3′-end formation enables variations in mRNA localization, translation, and stability. Of course, this mechanistic complexity comes at a high price. Mutations in the myriad of RNA sequence elements that regulate mRNA biogenesis, as well as the trans-acting factors that act upon these sequences, underlie a number of human diseases. In this review, we focus on one of these key RNA processing steps, splicing, to highlight recent studies that describe both conventional and novel pathogenic mechanisms that underlie muscle and neurological diseases.
Muscle cells and neurons rely heavily on alternative splicing to generate variants of proteins such as dystrophin. Mutations in splicing factors consequently underlie several muscular and neurological diseases.
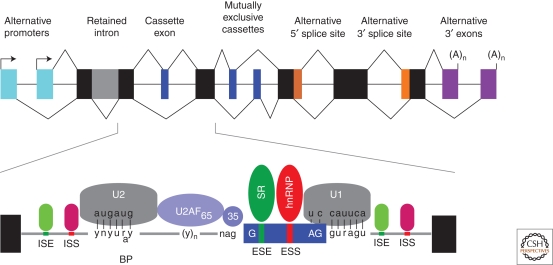
Deep sequencing and microarray analyses of the human transcriptome have revealed that >90% of our genes undergo alternative splicing, which permits a limited genome to encode the vast proteomic repertoire required for the human interactome (Blencowe 2006; Wang and Burge 2008; Wang et al. 2008). From a biochemical viewpoint, the RNA splicing reaction is a relatively simple process that consists of two transesterification reactions (Black and Grabowski 2003; Konarska and Query 2005; House and Lynch 2008). However, this apparent simplicity is compounded by the fact that exons are generally buried within much larger introns and both authentic, and many more false, splice sites exist in genes. In addition, multiple alternative splicing modes are utilized in vivo (Fig. 1). Finding the majority of these sites is the function of the major spliceosome, which is composed of five small nuclear ribonucleoprotein complexes (snRNPs) and ∼170 proteins (Deckert et al. 2006; Behzadnia et al. 2007; Bessonov et al. 2008; Wahl et al. 2009). Conserved sequence elements at exon-intron boundaries, the 5′ and 3′ splice sites of each exon, and the branchpoint facilitate spliceosome recruitment via base-pairing interactions with five spliceosome-associated snRNAs (U1, U2, U4/6, U5) (Fig. 2). A minor spliceosome, which recognizes variant splice sites and contains a unique set of snRNAs (U11, U12, U4atac, U6atac) in addition to U5, also exists in the nucleus (Will and Luhrmann 2005; Steitz et al. 2008). Spliceosomal recognition of these core elements is modulated by a myriad of additional sequence elements in both exons and introns that either activate (exonic splicing enhancer, ESE; intronic splicing enhancer, ISE) or repress (exonic splicing silencer, ESS; intronic splicing silencer, ISS) spliceosome recruitment by interacting with a variety of splicing factors, including serine-arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) (Fig. 1) (Martinez-Contreras et al. 2007; Long and Caceres 2009). The density of these sites is astonishing, and current estimates suggest that splicing regulatory motifs comprise >75% of the nucleotides in an average exon (Chasin 2007).
Figure 1.
RNA sequences and factors involved in the regulation of alternative splicing. The upper diagram shows various patterns of alternative splicing using a 12-exon (black and colored boxes) gene that contains alternative promoters (arrows adjacent to turquoise boxes), constitutive exons (black), a retained intron (gray), a cassette and mutual exclusion exons (blue), alternative 5′ and 3′ splice sites (orange), and alternative 3′ exons with polyadenylation sites (A)n. The lower diagram highlights the conserved RNA sequence elements at the branch point (BP, U2 binding site) and the 5′ (U1 binding site) and 3′ (U2AF heterodimer binding site) splice sites of an alternatively spliced cassette exon (blue box). Also shown are the less conserved exonic and intronic splicing enhancer (ESE, ISE) and silencer (ESS, ISS) elements that are recognized by multiple factors including the SR and hnRNP proteins.
Figure 2.
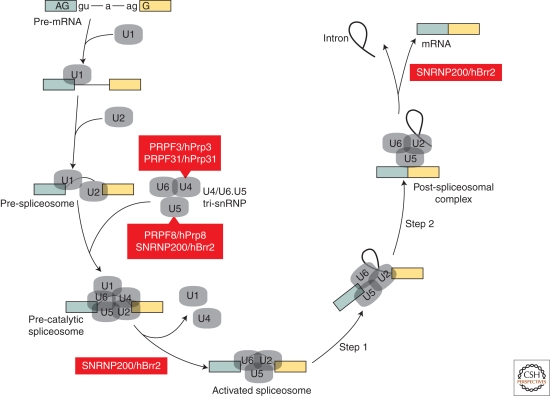
Spliceosome dynamics and retinopathy. The assembly/disassembly cycle of the spliceosome (adapted with permission from Wahl et al. 2009) during splicing with only the pre-mRNA (blue and yellow boxes, exons; thick black line, intron) and snRNP (grey) complexes illustrated to highlight the roles of U4-associated (PRPF3/hPrp3, PRPF31/hPrp31) and U5-associated (PRPF8/hPrp8, SNRP200/hBrr2) proteins as well as several other steps that require the DExD/H-type RNA-dependent ATPase/helicase. Steps 1 and 2 refer to the first and second catalytic steps of splicing.
Both the importance and complexity of splicing regulation is typified by the expression of dystrophin, a 427 kDa protein encoded by the DMD gene composed of 79 exons and 8 promoters, in skeletal muscle (Ervasti 2007; Gurvich et al. 2008). Dystrophin plays an essential role in skeletal muscle cells, or myofibers, since it binds to both actin and β-dystroglycan and thus provides a vital link between the actin-myosin contractile apparatus and the extracellular matrix (ECM). A number of single nucleotide mutations result in skeletal muscle degenerative diseases, such as the severe Duchenne (DMD) and the milder Becker (BMD) muscular dystrophies, as well as X-linked dilated cardiomyopathy. For Duchenne, mutations often disrupt the DMD open reading frame, sometimes by altering splicing enhancers and silencers, while the reading frame, and some dystrophin function, is mostly preserved in BMD. Although every tissue requires flexibility in gene expression, the significance of alternative splicing to human function is particularly striking in the central nervous system. For example, CACNA1H, which encodes a T-type low-voltage-activated calcium channel and is a candidate gene for idiopathic generalized epilepsies, is extensively alternatively spliced to potentially generate >4,000 mRNA variants (Zhong et al. 2006). Therefore, it is not surprising that errors in splicing regulation have been linked to a number muscle and neurologic diseases and this number is likely to increase in the future.
Since a number of excellent reviews have been published recently that survey how defects in splicing are associated with a broad range of human diseases (Licatalosi and Darnell 2006; Cooper et al. 2009; Tazi et al. 2009), this review focuses on only a few hereditary diseases in which mutations result in unusual pathogenic mechanisms, which have led to some intriguing insights into splicing regulatory mechanisms. Given the essential nature of the splicing reaction in all tissues, we begin by examining the paradox of why mutations in select spliceosomal components cause retinal degeneration while loss of a snRNP assembly factor leads to a lower motor neuron disease and proximal muscle atrophy. In addition to the core spliceosome, numerous trans-acting factors recognize pre-mRNA sequence elements to orchestrate spliceosome recruitment and alternative splicing. Thus, we next review the roles of both tandem repeats and dispersed repetitive elements in modulating splicing regulation and the emerging roles of repeat polymorphisms and reiterative element mutations on the pathogenesis of specific diseases.
GHOSTS IN THE MACHINE
The spliceosome is a remarkably dynamic macromolecular machine that depends on the recognition of loose consensus sequence elements at splice sites and the branch point (Fig. 2) (Wahl et al. 2009). This splice site detection process is facilitated by a number of other splicing factors that promote snRNP recruitment (Wang and Burge 2008; Licatalosi and Darnell 2010). For splicing to occur, snRNPs and the pre-mRNA undergo a series of association-dissociation steps and conformational transitions and, although the splicing reaction involves only two catalytic steps, intron removal must be accomplished with great precision in all tissues. While splicing errors underlie a large number of human diseases, it is puzzling that some diseases, which are characterized by the degeneration of only certain cell types, are caused by mutations either in ubiquitously expressed spliceosome components or depletion of factors important for snRNP assembly.
The Spliceosome and Retinopathies
Autosomal dominant retinitis pigmentosa (adRP) is characterized by progressive degeneration of predominantly rod photoreceptor neurons resulting in visual impairment. Some forms of adRP are caused by mutations in four pre-mRNA processing factors (PRPF3, PRPF8, PRPF31, SNRNP200) that are components of the ubiquitously expressed U4/U6.U5 tri-snRNP, which is recruited as a preassembled complex to form the functional spliceosome (Fig. 2) (Mordes et al. 2006; Wahl et al. 2009; Zhao et al. 2009). Another gene, PIM1-associated protein (PAP1), which encodes a protein that interacts with PRPF3 to regulate splicing, also causes adRP (Maita et al. 2004; Maita et al. 2005). Why do mutations in these specific genes cause a specific eye disease? One possible explanation relates to the unusual metabolic requirements of photoreceptor cells, including circadian oscillation of transcript levels (von Schantz et al. 1999), and the importance of tri-snRNP dynamics during splicing. Although remodeling of spliceosomal subunits is common during splicing, the U4/U6.U5 tri-snRNP presents an extreme case since catalytic activation of the spliceosome requires disruption of the extensively base-paired U4/U6 duplex to allow U6 snRNA interactions with U2 and the pre-mRNA (Wahl et al. 2009). Evidence that tri-snRNP remodeling is particularly important for photoreceptors comes from recent reports that SNRNP200 mutations cause adRP (Li et al. 2009; Zhao et al. 2009). SNRNP200, also known as hBrr2, is a DExD/H-box protein that is required for both unwinding of the U4/U6 duplex and spliceosome disassembly. Interestingly, the SNRNP200 mutation associated with more severe adRP causes a larger defect in U4/U6 unwinding activity (Zhao et al. 2009). Of course, it is possible that PRPF3, PRPF8, PRPF31, SNRNP200, and PAP1 have nonsplicing functions that are particularly critical for retinal function, but this possibility seems less likely given their common association with the tri-snRNP.
Both loss-of-function and gain-of-function models have been proposed to explain why mutations in multiple components of the tri-snRNP result in retinal disease (Mordes et al. 2006). For loss-of-function models, tri-snRNP-associated mutations result in haploinsufficiency that only cause overt disease in photoreceptors because these neurons are unusually sensitive to decrements in splicing function due to the high demand for specific proteins, such as rhodopsin (RHO mutations also cause adRP). For example, more than 40 PRPF31 mutations have been reported and the majority of these create a premature termination codon (PTC), transcript degradation via the nonsense-mediated decay (NMD) pathway, and reduced mutant allele expression (Rio Frio et al. 2008). Additionally, the incomplete penetrance noted for PRPF31 mutations may reflect variations in expression from the normal allele (Vithana et al. 2003; Rivolta et al. 2006). A potential problem with this haploinsufficiency model is that other neurons (e.g., Purkinje cells) and other tissues (e.g., regenerating muscle) also place considerable demands on transcription and RNA processing. Indeed, a recent study generated mice carrying either a human ADRP mutation in Prpf31 (Prpf31A261P/+ knockin) or Prpf31+/− heterozygous knockout mice. Although the mouse retina is enriched in rod photoreceptors compared to humans, these lines fail to model adRP and do not develop retinal degeneration, although the corresponding homozygous knockin and knockout mice are embryonic lethal (Bujakowska et al. 2009). In contrast, heterozygous Rho+/− mice, which express lower levels of rhodopsin, develop retinal degeneration (Lem et al. 1999). While this is a single study and may reflect functional differences in the rodent versus human retina, the failure to model adRP in mice will impede further testing of the haploinsufficiency model. A caveat is that Prpf31A261P/+ knockin mice were generated on a mixed genetic background, so it is possible that mutant Prpf31 alleles will show a retinal phenotype when placed on a different background (Buchner et al. 2003). Additionally, mouse models should be generated for other adRP mutations, particularly Snrnp200. For gain-of-function models, mutant tri-snRNP proteins might alter the spliceosome so that it generates, or fails to efficiently proofread, incorrectly spliced mRNAs (Mordes et al. 2006; Zhao et al. 2009). Another pathogenic mechanism, characteristic of some neurodegenerative diseases, is that U4/U6.U5 tri-snRNP mutations lead to the synthesis of aggregation-prone proteins, which are particularly toxic to photoreceptors.
snRNP Biogenesis and Motor Neuron Disease
The spliceosome requires the functions of multiple snRNPs, which contain a seven-member ring of Sm proteins that assemble onto the Sm site of each snRNA in the cytoplasm with the exception of U6. A critical factor in this assembly process in vivo is the survival of motor neurons gene 1 (SMN1), which is ubiquitously expressed in human tissues (Monani 2005; Burghes and Beattie 2009; Chari et al. 2009). In addition to SMN, this ATP-dependent SMN “assemblyosome” complex consists of several GEMINS (SIP1/GEMIN2, DDX20/GEMIN3, GEMINS4-8) and STRAP/UNRIP (Paushkin et al. 2002; Burghes and Beattie 2009). Loss of SMN1 expression, due to either mutation or deletion, causes the autosomal recessive disease spinal muscular atrophy (SMA), a leading cause of infantile mortality. SMA is associated with the degeneration of lower motor neurons in the ventral horn of the spinal cord and muscle atrophy, which ranges from severe Type I disease, characterized by an early (<6 months) age of onset and death (<2 years of age), to Type III disease, which manifests later during development (>18 months) and patients may have a normal lifespan (Monani 2005). In a cruel twist of fate, the human genome contains the nearly identical gene SMN2, but this copy fails to compensate for SMN1 loss due to a C → T transition in SMN2 exon 7. This single nucleotide change blocks inclusion of exon 7 and results in inhibition of SMN protein oligomerization, and/or the generation of a C-terminal degradation signal, leading to low levels of functional protein (Monani 2005; Burghes and Beattie 2009; Cho and Dreyfuss 2010).
Similar to adRP, both splicing and non-splicing models have been proposed for SMA pathogenesis to explain how loss-of-function mutations in a ubiquitously expressed gene result in selective effects on specific cell populations. Splicing models emphasize the possibility that lower motor neurons may be particularly sensitive to SMA-associated declines in SMN activity and relative snRNP abundance, which causes mis-splicing of critical pre-mRNAs. In support of this possibility, there is a strong correlation between phenotypic severity, SMN levels, and snRNP assembly activity in mouse models of SMA (Gabanella et al. 2007). Loss of SMN in these models also causes cell-type specific changes in snRNA levels, with minor spliceosome snRNAs particularly affected in brain and spinal cord (Gabanella et al. 2007; Zhang et al. 2008). Exon microarrays have been used to detect widespread splicing abnormalities in the Smn−/−; SMN2; SMNΔ7 mouse SMA model, with a maximum lifespan of 14 days, at end-stage disease (postnatal day 11, P11), which led to the suggestion that SMA is a pan-splicing disease (Zhang et al. 2008). This analysis has been challenged since symptomatic mice were analyzed, suggesting that alternative splicing changes may represent secondary effects due to disease progression. Indeed, a comparison of presymptomatic (postnatal day 1, P1), early-symptomatic (P7), and late-symptomatic (P13) mice showed that the majority of splicing changes in spinal cord were only observed in late-symptomatic mice (Baumer et al. 2009). Nevertheless, it is possible that many gene expression and RNA splicing changes have been missed in presymptomatic mice using microarrays so it will be important to re-examine these mice using RNA-Seq technologies.
Efforts to develop effective SMA therapies have employed several elegant splicing-related strategies designed to enhance SMN2 exon 7 splicing levels (Khoo and Krainer 2009; Sendtner 2010). Prominent among these are approaches based on bifunctional antisense oligonucleotides (ASOs) or U7 snRNAs composed of sequences complementary to SMN2 exon 7 for targeting coupled to splicing enhancer (ASO-ESE, U7-ESE), or minimal RS domain peptide, motifs to increase exon 7 inclusion. Evidence that this strategy is effective in vivo has been provided recently. SMA model mice (Smn−/−; hSMN2+/+), which also express a U7-ESE transgene, show a dramatic improvement in survival over Smn−/−; hSMN2+/+ mice alone (Meyer et al. 2009). Importantly, extensive ASO tiling analyses across exon 7 and flanking introns 6 and 7 revealed additional splicing silencers, and ASO-mediated antisense masking of these sequence motifs increases exon 7 inclusion in several tissues of human SMN2 transgenic mice (Hua et al. 2007; Hua et al. 2008).
Non-splicing functions of SMN have also been reported. Indeed, this protein has been proposed to be a master assembler of RNP complexes (Terns and Terns 2001). For example, the Sm-like (LSm) proteins also form ring structures that function in cytoplasmic RNA turnover and RNA–RNA interactions, but it is not known if SMN activity is essential for this assembly process (Wilusz and Wilusz 2005). Other proposed SMN functions include transcriptional regulation and axonal mRNP transport and translation (Monani 2005; Burghes and Beattie 2009).
REPETITIVE ELEMENTS AND SPLICING
Studies on trans-acting factor mutations have expanded our understanding of spliceosome assembly and function. However, many other diseases are caused by mutations in pre-mRNA sequence elements, and there is a growing appreciation for the importance of reiterative sequences in splicing regulation. Repetitive sequence elements comprise >50% of the human genome and are broadly divided into two categories, dispersed and tandem repeats (Richard et al. 2008). Dispersed repeats include tRNA genes, gene paralogs, and transposon/transposon-derived elements such as long- and short-interspersed elements (LINEs, SINEs) and LTR retrotransposons. Among these, transposable elements (TEs) may cause disease either by insertional mutagenesis or nonallelic homologous recombination between TEs (Belancio et al. 2009). An intriguing example of how these elements can affect splicing regulation is provided by the short interspersed element (SINE) Alu. Alu is a small (∼300 nt) primate-specific non-LTR retrotransposon that is the most abundant (>106 copies) repetitive element in the human genome (Keren et al. 2010). Alu elements are dimeric structures composed of left and right arms joined by an A-rich linker region and are preferentially located in introns in gene-rich regions often in the antisense orientation. During evolution, Alu mutations have resulted in the appearance of regulatory sequences that are recognized by the spliceosome so that these elements are sometimes “exonized” or included in the mRNA product as internal alternative exons (Lev-Maor et al. 2003; Keren et al. 2010). Other mutations can affect Alu-associated splicing events. For example, a C→T transition in intron 6 of the CTDP1 gene, which encodes a phosphatase associated with the C-terminal domain in RNA polymerase II, generates a 5′ splice site while also activating an upstream cryptic 3′ splice site in an antisense Alu element, leading to its inclusion and the autosomal recessive disorder congenital cataracts facial dysmorphism neuropathy (CCFDN) (Varon et al. 2003).
Tandem repeats include gene tandems, rDNA arrays, and satellite DNAs. The latter group, specifically polymorphic microsatellites, plays a prominent role in hereditary neurological disease and splicing dysregulation. Aside from short repeats, the contraction of a larger repeat, the D4Z4 3.3 kb macrosatellite on chromosome 4q, also appears to underlie at least one muscular dystrophy, and recent studies suggest that contraction-induced alterations in chromatin structure and RNA processing may also be pathogenic.
Microsatellite-induced Disruption of Alternative Splicing
The human genome contains ∼253,000 di-/tri-/tetra-nucleotide microsatellite repeats (Richard et al. 2008). These simple sequence repeats may contract or expand due to errors in DNA replication, recombination, and repair. When the latter occurs, transcription of some expanded repeats results in the synthesis of RNA hairpin structures, which are toxic because they affect the activities of several splicing factors. For the neuromuscular disease myotonic dystrophy (DM), the DMPK and CNBP/ZNF9 genes contain CTG and CCTG expansions, respectively. DM type 1 (DM1) is associated with DMPK (CTG)37–>3,500, while type 2 disease (DM2) is caused by CNBP (CCTG)75–∼11,000, expansions. DM is an autosomal dominant disease that affects multiple systems including skeletal (myotonia, weakness/wasting) and heart (arrhythmias) muscles, the eye (subcapsular cataracts), and the brain (mental retardation, cerebral atrophy). Moreover, DM is both a congenital (CDM) and adult-onset degenerative disease. Curiously, CDM is only associated with CTGexp mutations in the DMPK gene. While the molecular basis for this unique association has not been delineated, one possibility that has not been examined is the potential for problematic transmission of very large CNBP CCTGexp (e.g., >1,000) alleles through the germline, so disease-associated CNBP mutant alleles might only be generated postnatally.
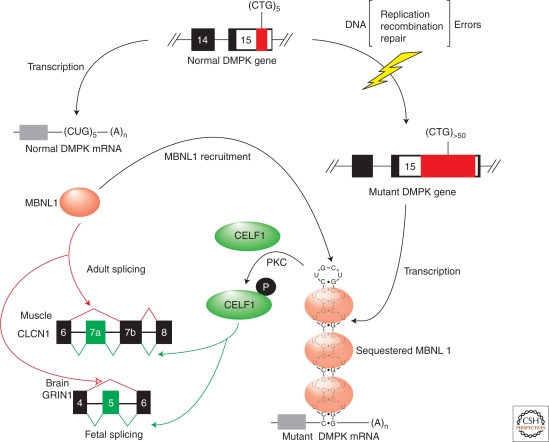
Upon transcription of the mutant DMPK and CNBP/ZNF9 genes, the corresponding CUG and CCUG expansion (CUGexp, CCUGexp) RNAs accumulate in nuclear, or ribonuclear, foci and inhibit the muscleblind-like (MBNL) proteins while also increasing CUGBP1/ETR-3-like factor (CELF) levels (Fig. 3). Coordinate MBNL loss and CELF1 (formerly CUGBP1) up-regulation occurs because (C)CUGexp RNAs form RNA hairpins that sequester MBNL proteins while also activating protein kinase C (PKC), which results in elevated CELF1 steady state levels due to hyperphosphorylation. The MBNL and CELF proteins act antagonistically to control alternative splicing during development. MBNL1 promotes adult, while CELF1 increases fetal, splicing patterns for specific exons during the fetal to adult transition, so fetal isoform expression persists in adult DM tissues and causes characteristic disease manifestations. The timing of disease onset in adults could reflect the continued somatic expansion of these repeats, particularly in tissues with postmitotic cell populations (e.g., muscle, brain), so that a pathogenic threshold is crossed. Genome-wide expression and splice junction microarray analyses, as well as a crystal structure of two MBNL1 zinc finger-like domains (Zn3/4) bound to r(CGCUGU), indicate that MBNL1 preferentially recognizes a core YGCY motif consistent with its high affinity binding to CUGexp and CCUGexp RNAs (Castle et al. 2008; Teplova and Patel 2008; Du et al. 2010; Goers et al. 2010). In agreement with studies on the brain-specific Nova family of splicing regulators and the importance of binding site position relative to the regulated exon (Ule et al. 2006; Ule and Darnell 2007), binding of MBNL1 upstream of the 3′ splice site promotes skipping of that alternative exon while binding downstream of the 5′ splice site favors inclusion (Du et al. 2010). A similar scenario has been described for CELF proteins that recognize a UGUGU motif (Faustino and Cooper 2005; Castle et al. 2008; Daughters et al. 2009). CELF proteins contain three RNA recognition motifs (RRM1-3) and the C-terminal RRM of CELF1 forms specific contacts with the UGU trinucleotide (Tsuda et al. 2009). Both Mbnl1 knockout and CELF1 transgenic mouse models have been generated, which recapitulate some features of DM disease (Kanadia et al. 2003; Timchenko et al. 2004; Ho et al. 2005). Nevertheless, the relative contributions of MBNL1 and CELF1 misregulation to pathogenesis are unclear, and the molecular etiology of congenital DM remains a mystery.
Figure 3.
RNA-mediated disease mechanism in myotonic dystrophy. Errors in DNA replication, recombination, and repair may lead to an increase in a normal length (CTG)5 (small red box) repeat located in DMPK exon 15 to (CTG)>50 expansions (large red box). Following transcription, these mutant mRNAs with CUGexp RNA hairpins sequester the MBNL proteins (red ovals) while also triggering activation of protein kinase C (PKC), which results in hyperphosphorylation (white P in black circle) of CELF1 (green oval). MBNL proteins are required during postnatal development to activate adult splicing patterns (red arrows) while CELF1 promotes fetal exon splicing (green arrows). Only specific exons are regulated by these two antagonistic splicing factors. For example, MBNL induces adult-specific skipping of CLCN1 exon 7a in muscle and GRIN1/NMDAR1 exon 5 in brain, while CELF1 promotes inclusion of these fetal exons. Open red and green arrowheads indicate that the precise roles of MBNL and CELF1 in GRIN1 pre-mRNA splicing have not been determined.
The aberrant sequestration of splicing factors by another CG-rich microsatellite has been reported for the late-onset neurological disease fragile X-associated tremor ataxia syndrome (FXTAS). The primary features of FXTAS are action tremor and gait ataxia but cognitive decline, executive function deficits, and mild parkinsonism have also been noted (Hagerman et al. 2001; Grigsby et al. 2007). This neurodegenerative disorder, which was originally identified in older adult carriers of premutation alleles of the fragile X syndrome (FRAXA) mental retardation gene (FMR1), is caused by (CGG)55–200 expansions in the FMR1 5′ UTR that are shorter than those observed for FRAXA-associated (CGG)>230 repeats (Hagerman and Hagerman 2007). In neurons and astrocytes, transcripts from the mutant FMR1 gene accumulate in large ubiquitin-positive intranuclear inclusions, together with a number of other proteins. Interestingly, these toxic CGGexp RNAs are expressed in other cell types, including fibroblasts, and during embryonic development, which might explain the neurodevelopmental abnormalities observed in some premutation children (Garcia-Arocena et al. 2010). Several RNA-binding proteins, including CELF1 and hnRNP A2/B1, have been implicated in FXTAS pathogenesis, but a recent study indicates that the splicing factor Sam68, encoded by the KHDRBS1 gene, is particularly prone to sequestration by CGGexp RNAs (Sellier et al. 2010). Although this is a surprising finding because previous studies have indicated that Sam68 preferentially recognizes A- and U-rich sequences, the Sam68 CGGexp interaction region does not include the central KH RNA-binding domain. Instead, Sam68 interacts with CGGexp RNA via an N-terminal region important for protein–protein interactions, so recruitment to nuclear rCGGexp inclusions is probably mediated by another uncharacterized factor. In support of the hypothesis that Sam68 plays an important role in FXTAS, expression of this protein is required for neuronal differentiation of P19 cells and Khdrbs1−/– null mice develop motor coordination deficiencies, as indicated by rotarod and hindlimb beam walking tests, although cerebellar degeneration has not been reported (Lukong and Richard 2008; Chawla et al. 2009). Significantly, Sam68 depletion, either by sequestration in FXTAS brains or by shRNA-mediated knockdowns, results in the alternative splicing of some Sam68 target exons. Unlike DM, where specific mis-splicing events have been linked to distinct pathological consequences (e.g., myotonia due to aberrant inclusion of CLCN1 exon 7a), it is not clear if FXTAS is a developmental splicing disease. To address this concern, splicing-sensitive microarray and RNA-Seq analyses should be performed to determine if a FXTAS-specific splicing signature emerges.
Microsatellite expansions and the disruption of developmentally regulated splicing transitions have been implicated in other neurological diseases. The autosomal dominant spinocerebellar ataxias (SCAs) are a group of late-onset and progressive neurodegenerative diseases that are characterized by cerebellar atrophy resulting in ataxia (abnormal limb movement/coordination), tremor, and premature death (Carlson et al. 2009). SCA type 8 (SCA8) is particularly interesting since it is transmitted with reduced penetrance and caused by CTG•CAG repeat expansions in both coding (ATXN8 gene) and non-coding (ATXN8OS) regions since the locus is bidirectionally transcribed (Moseley et al. 2006). For ATXN8OS, CUGexp RNAs accumulate in ribonuclear foci that are detectable in the human SCA8 cerebellum, particularly in molecular layer interneurons and Bergmann glia. Ribonuclear foci are also detectable in a BAC transgenic mouse model for SCA8. These mice express a human (CTG•CAG)116 expansion and develop characteristic motor deficits possibly due to loss of cerebellar GABAergic inhibition. A recent study confirmed that expression of ATXN8OS CUGexp RNAs affects cerebellar inhibitory pathways by altering the activities of developmentally regulated splicing events controlled by CELF1 and MBNL1. Using the crosslinking/immunoprecipation protocol (CLIP), a number of brain RNA targets for mouse Cugbp1 were revealed including the GABA-A transporter 4, GAT4/GABT4A. Expression of CUGexp RNA, which was shown to be coordinately regulated by MBNL1 and CUGBP1, leads to persistence of the fetal GAT4/GABT4 splicing pattern, chronically elevated GAT4/GABT4 levels, and loss of GABAergic inhibition in the granular layer of the cerebellum (Daughters et al. 2009).
SCA type 31 (SCA31) has also been reported to be associated with a TGGAAexp insertion in newly indentified introns of the TK2 and BEAN genes, which partially overlap and are transcribed in opposite directions on chromosome 16q22.1 (Sato et al. 2009). Similar to DM, mutant SCA31 RNAs accumulate in ribonuclear foci, so this SCA may also be an RNA-mediated disease, although these foci are detectable in Purkinje cells in SCA31. However, the splicing factors involved may be different since UGGAA repeats are not predicted to form stem-loop structures. A clue to the identity of these factors has emerged from studies on repetitive satellite III (SatIII) DNA repeats, which are composed of (GGAAT)n CAAC(C/A)CGAGT repeats (Valgardsdottir et al. 2008). Thermal and chemical stresses trigger SatIII transcription and these RNAs concentrate in nuclear stress bodies (nSBs), which also contain splicing factors including SFRS1 (ASF/SF2). Interestingly, SFRS1 and SFRS9 (SRp30c) bind to (UGGAA)8 RNAs in vitro, so it is possible that sequestration of these splicing factors causes misregulation of alternative splicing in Purkinje cells and degeneration (Sato et al. 2009).
In contrast to DM and SCA31, Friedreich ataxia (FRDA) is an autosomal recessive disease characterized by atrophy of primary sensory neurons in the dorsal root ganglia as well as hypertrophic cardiomyopathy (Pandolfo 2008). FRDA is caused by noncoding GAA expansions in the first intron of the FXN gene, which encodes the frataxin protein that is associated with the mitochondrial matrix and involved in iron homeostasis. Normal FXN alleles have up to ∼38 GAA repeats that expand to ∼70 to >1,000 repeats in FRDA. These GAA expansions are currently believed to be pathogenic because they inhibit transcription by altering DNA structure, by forming non-B DNA triplexes and “sticky” DNA, and by promoting chromatin condensation (Sakamoto et al. 1999; Saveliev et al. 2003). Recently, GAA expansions have also been shown to influence alternative splicing regulation. Insertion of a (GAA)100 expansion downstream of an alternative exon in a hybrid minigene splicing reporter resulted in position-dependent effects on exon skipping, flanking intron retention, and splicing of a cryptic exon containing the (GAA)100 repeat (Baralle et al. 2008). More significantly, insertion of (GAA)100, but not a complementary (TTC)100 repeat, into a frataxin minigene reporter did not alter nascent transcript levels but did inhibit splicing of FXN intron 1. Thus, mis-splicing of FXN pre-mRNA may be a primary event that leads to decreased FXN mRNA levels.
Satellite Contractions: Potential Effects on RNA Processing
While loss-of-function mutations alter cis-acting splicing regulatory sequences in the DMD gene to cause exon skipping and frame-shifting in DMD/BMD, toxic C(C)UGexp RNAs expressed in DM alter the activities of trans-acting splicing factors that control the expression of developmentally regulated isoforms. A third mechanism that affects splicing regulation may be important in the molecular etiology of another muscular dystrophy, facioscapulohumeral muscular dystrophy (FSHD). FSHD is caused by the contraction of a D4Z4 macrosatellite repeat array in the subtelomeric region of chromosome 4qter (van der Maarel et al. 2007; Tawil 2008). This 3.3 kb repeat is normally 11–100 D4Z4 units in length but contracts to 1–10 repeats in affected individuals and is preferentially associated with the 4qA161 haplotype. Current position-effect models propose that repeat contraction leads to a decrease in D4Z4 epigenetic modifications resulting in increased expression of genes that are either centromeric, including the spliceosome-associated protein FRG1, or within the repeat, such as the toxic double homeobox protein DUX4. In support of the former proposal, transgenic mice overexpressing FRG1 10- to 50-fold develop several FSHD-associated muscle features including kyphosis and RNA splicing changes (Gabellini et al. 2006). However, microarray expression studies have challenged the possibility that FRG1 is overexpressed in FSHD muscle (Osborne et al. 2007; Klooster et al. 2009). Alternatively, contraction-related changes in chromatin structure, or hypomethylation in the absence of contractions also known as phenotypic FSHD, may lead to changes in transcript abundance and/or processing from specific D4Z4 repeats that are toxic to muscle cells. Recent studies have focused on the most telomeric D4Z4 repeat, which together with allele-specific flanking sequences, produces transcripts that are processed into si/miRNA-sized fragments as well as alternatively processed DUX4 RNAs that potentially antagonize myogenesis (Dixit et al. 2007; Bosnakovski et al. 2008; Bosnakovski et al. 2009; Snider et al. 2009). Future studies should determine if D4Z4, and perhaps other repetitive element, contractions activate transcription of locus-specific repeat arrays while coordinately altering transcript processing to generate pathogenic RNAs and/or proteins.
Concluding Remarks
Studies designed to clarify the molecular basis of some puzzling hereditary neurological disorders have provided both surprising results and fertile territory for further exploration. The key question of why mutations in genes encoding broadly expressed factors target specific cell types, such as tri-snRNP-associated proteins in adRP and SMN1 in SMA, remains unresolved, but the link to splicing misregulation has been reinforced by recent reports. Dynamic mutations associated with several muscular dystrophies and ataxias have also highlighted a novel disease mechanism, RNA-mediated pathogenesis, and the critical roles of novel alternative splicing factors during development. Future developments should yield more surprises and strengthen the connection between aberrant RNA processing and disease.
ACKNOWLEDGMENTS
Research in our lab is funded by the NIH (AR046799, NS048843, NS058901) and the MDA.
Footnotes
Editors: Tom Misteli and David L. Spector
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Baralle M, Pastor T, Bussani E, Pagani F 2008. Influence of Friedreich ataxia GAA noncoding repeat expansions on pre-mRNA processing. Am J Hum Genet 83: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer D, Lee S, Nicholson G, Davies JL, Parkinson NJ, Murray LM, Gillingwater TH, Ansorge O, Davies KE, Talbot K 2009. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet 5: e1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, Will CL, Urlaub H, Stark H, et al. 2007. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J 26: 1737–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Deininger PL, Roy-Engel AM 2009. LINE dancing in the human genome: transposable elements and disease. Genome Med 1: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R 2008. Isolation of an active step I spliceosome and composition of its RNP core. Nature 452: 846–850 [DOI] [PubMed] [Google Scholar]
- Black DL, Grabowski PJ 2003. Alternative pre-mRNA splicing and neuronal function. Prog Mol Subcell Biol 31: 187–216 [DOI] [PubMed] [Google Scholar]
- Blencowe BJ 2006. Alternative splicing: new insights from global analyses. Cell 126: 37–47 [DOI] [PubMed] [Google Scholar]
- Bosnakovski D, Daughters RS, Xu Z, Slack JM, Kyba M 2009. Biphasic myopathic phenotype of mouse DUX, an ORF within conserved FSHD-related repeats. PLoS One 4: e7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosnakovski D, Xu Z, Gang EJ, Galindo CL, Liu M, Simsek T, Garner HR, Agha-Mohammadi S, Tassin A, Coppee F, et al. 2008. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J 27: 2766–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner DA, Trudeau M, Meisler MH 2003. SCNM1, a putative RNA splicing factor that modifies disease severity in mice. Science 301: 967–969 [DOI] [PubMed] [Google Scholar]
- Bujakowska KM, Maubaret C, Chakarova CF, Tanimoto N, Beck SC, Fahl E, Humphries MM, Kenna P, Makarov E, Makarova O, et al. 2009. Study of gene targeted mouse models of splicing factor gene Prpf31 implicated in human autosomal dominant retinitis pigmentosa (RP). Invest Ophthalmol Vis Sci. 50: 5927–5933 [DOI] [PubMed] [Google Scholar]
- Burghes AH, Beattie CE 2009. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci 10: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson KM, Andresen JM, Orr HT 2009. Emerging pathogenic pathways in the spinocerebellar ataxias. Curr Opin Genet Dev 19: 247–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM 2008. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet 40: 1416–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari A, Paknia E, Fischer U 2009. The role of RNP biogenesis in spinal muscular atrophy. Curr Opin Cell Biol 21: 387–393 [DOI] [PubMed] [Google Scholar]
- Chasin LA 2007. Searching for splicing motifs. Adv Exp Med Biol 623: 85–106 [DOI] [PubMed] [Google Scholar]
- Chawla G, Lin CH, Han A, Shiue L, Ares M Jr, Black DL 2009. Sam68 regulates a set of alternatively spliced exons during neurogenesis. Mol Cell Biol 29: 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Dreyfuss G 2010. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev 24: 438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G 2009. RNA and disease. Cell 136: 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LP 2009. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet 5: e1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Luhrmann R 2006. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol 26: 5528–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit M, Ansseau E, Tassin A, Winokur S, Shi R, Qian H, Sauvage S, Matteotti C, van Acker AM, Leo O, et al. 2007. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci U S A 104: 18157–18162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, et al. 2010. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol 17: 187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ervasti JM 2007. Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim Biophys Acta 1772: 108–117 [DOI] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA 2005. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol Cell Biol 25: 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L 2007. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One 2: e921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini D, D'Antona G, Moggio M, Prelle A, Zecca C, Adami R, Angeletti B, Ciscato P, Pellegrino MA, Bottinelli R, et al. 2006. Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature 439: 973–977 [DOI] [PubMed] [Google Scholar]
- Garcia-Arocena D, Yang JE, Brouwer JR, Tassone F, Iwahashi C, Berry-Kravis EM, Goetz CG, Sumis AM, Zhou L, Nguyen DV, et al. 2010. Fibroblast phenotype in male carriers of FMR1 premutation alleles. Hum Mol Genet 19: 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goers ES, Purcell J, Voelker RB, Gates DP, Berglund JA 2010. MBNL1 binds GC motifs embedded in pyrimidines to regulate alternative splicing. Nucleic Acids Res 38: 2467–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigsby J, Brega AG, Leehey MA, Goodrich GK, Jacquemont S, Loesch DZ, Cogswell JB, Epstein J, Wilson R, Jardini T, et al. 2007. Impairment of executive cognitive functioning in males with fragile X-associated tremor/ataxia syndrome. Mov Disord 22: 645–650 [DOI] [PubMed] [Google Scholar]
- Gurvich OL, Tuohy TM, Howard MT, Finkel RS, Medne L, Anderson CB, Weiss RB, Wilton SD, Flanigan KM 2008. DMD pseudoexon mutations: splicing efficiency, phenotype, and potential therapy. Ann Neurol 63: 81–89 [DOI] [PubMed] [Google Scholar]
- Hagerman PJ, Hagerman RJ 2007. Fragile X-associated tremor/ataxia syndrome–an older face of the fragile X gene. Nat Clin Pract Neurol 3: 107–112 [DOI] [PubMed] [Google Scholar]
- Hagerman RJ, Leehey M, Heinrichs W, Tassone F, Wilson R, Hills J, Grigsby J, Gage B, Hagerman PJ 2001. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology 57: 127–130 [DOI] [PubMed] [Google Scholar]
- Ho TH, Bundman D, Armstrong DL, Cooper TA 2005. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet 14: 1539–1547 [DOI] [PubMed] [Google Scholar]
- House AE, Lynch KW 2008. Regulation of alternative splicing: more than just the ABCs. J Biol Chem 283: 1217–1221 [DOI] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR 2007. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol 5: e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR 2008. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet 82: 834–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS 2003. A muscleblind knockout model for myotonic dystrophy. Science 302: 1978–1980 [DOI] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G 2010. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11: 345–355 [DOI] [PubMed] [Google Scholar]
- Khoo B, Krainer AR 2009. Splicing therapeutics in SMN2 and APOB. Curr Opin Mol Ther 11: 108–115 [PMC free article] [PubMed] [Google Scholar]
- Klooster R, Straasheijm K, Shah B, Sowden J, Frants R, Thornton C, Tawil R, van der Maarel S 2009. Comprehensive expression analysis of FSHD candidate genes at the mRNA and protein level. Eur J Hum Genet 17: 1615–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Query CC 2005. Insights into the mechanisms of splicing: more lessons from the ribosome. Genes Dev 19: 2255–2260 [DOI] [PubMed] [Google Scholar]
- Lem J, Krasnoperova NV, Calvert PD, Kosaras B, Cameron DA, Nicolo M, Makino CL, Sidman RL 1999. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc Natl Acad Sci U S A 96: 736–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev-Maor G, Sorek R, Shomron N, Ast G 2003. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science 300: 1288–1291 [DOI] [PubMed] [Google Scholar]
- Li N, Mei H, Macdonald IM, Jiao X, Hejtmancik F 2009. Mutations in ASCC3L1 on chromosome 2q11.2 are associated with autosomal dominant retinitis pigmentosa in a Chinese Family. Invest Ophthalmol Vis Sci 51: 1036–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB 2006. Splicing regulation in neurologic disease. Neuron 52: 93–101 [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB 2010. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet 11: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF 2009. The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417: 15–27 [DOI] [PubMed] [Google Scholar]
- Lukong KE, Richard S 2008. Motor coordination defects in mice deficient for the Sam68 RNA-binding protein. Behav Brain Res 189: 357–363 [DOI] [PubMed] [Google Scholar]
- Maita H, Kitaura H, Ariga H, Iguchi-Ariga SM 2005. Association of PAP-1 and Prp3p, the products of causative genes of dominant retinitis pigmentosa, in the tri-snRNP complex. Exp Cell Res 302: 61–68 [DOI] [PubMed] [Google Scholar]
- Maita H, Kitaura H, Keen TJ, Inglehearn CF, Ariga H, Iguchi-Ariga SM 2004. PAP-1, the mutated gene underlying the RP9 form of dominant retinitis pigmentosa, is a splicing factor. Exp Cell Res 300: 283–296 [DOI] [PubMed] [Google Scholar]
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B 2007. hnRNP proteins and splicing control. Adv Exp Med Biol 623: 123–147 [DOI] [PubMed] [Google Scholar]
- Meyer K, Marquis J, Trüb J, Nlend RN, Verp S, Ruepp M-D, Imboden H, Barde I, Trono D, Schümperli D 2009. Rescue of a severe mouse model for spinal muscular atrophy by U7 snRNA-mediated splicing modulation. Hum Mol Genet 18: 546–555 [DOI] [PubMed] [Google Scholar]
- Monani UR 2005. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron 48: 885–896 [DOI] [PubMed] [Google Scholar]
- Mordes D, Luo X, Kar A, Kuo D, Xu L, Fushimi K, Yu G, Sternberg P Jr, Wu JY 2006. Pre-mRNA splicing and retinitis pigmentosa. Mol Vis 12: 1259–1271 [PMC free article] [PubMed] [Google Scholar]
- Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, Chen G, Weatherspoon MR, Clark HB, Ebner TJ, et al. 2006. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet 38: 758–769 [DOI] [PubMed] [Google Scholar]
- Osborne RJ, Welle S, Venance SL, Thornton CA, Tawil R 2007. Expression profile of FSHD supports a link between retinal vasculopathy and muscular dystrophy. Neurology 68: 569–577 [DOI] [PubMed] [Google Scholar]
- Pandolfo M 2008. Friedreich ataxia. Arch Neurol 65: 1296–1303 [DOI] [PubMed] [Google Scholar]
- Paushkin S, Gubitz AK, Massenet S, Dreyfuss G 2002. The SMN complex, an assemblyosome of ribonucleoproteins. Curr Opin Cell Biol 14: 305–312 [DOI] [PubMed] [Google Scholar]
- Richard GF, Kerrest A, Dujon B 2008. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev 72: 686–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio Frio T, Wade NM, Ransijn A, Berson EL, Beckmann JS, Rivolta C 2008. Premature termination codons in PRPF31 cause retinitis pigmentosa via haploinsufficiency due to nonsense-mediated mRNA decay. J Clin Invest 118: 1519–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivolta C, McGee TL, Rio Frio T, Jensen RV, Berson EL, Dryja TP 2006. Variation in retinitis pigmentosa-11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum Mutat 27: 644–653 [DOI] [PubMed] [Google Scholar]
- Sakamoto N, Chastain PD, Parniewski P, Ohshima K, Pandolfo M, Griffith JD, Wells RD 1999. Sticky DNA: self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedreich's ataxia. Mol Cell 3: 465–475 [DOI] [PubMed] [Google Scholar]
- Sato N, Amino T, Kobayashi K, Asakawa S, Ishiguro T, Tsunemi T, Takahashi M, Matsuura T, Flanigan KM, Iwasaki S, et al. 2009. Spinocerebellar Ataxia Type 31 Is Associated with “Inserted” Penta-Nucleotide Repeats Containing (TGGAA)(n). Am J Hum Genet 85: 544–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveliev A, Everett C, Sharpe T, Webster Z, Festenstein R 2003. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature 422: 909–913 [DOI] [PubMed] [Google Scholar]
- Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, et al. 2010. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J 29: 1248–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendtner M 2010. Therapy development in spinal muscular atrophy. Nat Neurosci 13: 795–799 [DOI] [PubMed] [Google Scholar]
- Snider L, Asawachaicharn A, Tyler AE, Geng LN, Petek LM, Maves L, Miller DG, Lemmers RJ, Winokur ST, Tawil R, et al. 2009. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum Mol Genet 18: 2414–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz JA, Dreyfuss G, Krainer AR, Lamond AI, Matera AG, Padgett RA 2008. Where in the cell is the minor spliceosome? Proc Natl Acad Sci U S A 105: 8485–8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawil R 2008. Facioscapulohumeral muscular dystrophy. Neurotherapeutics 5: 601–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J, Bakkour N, Stamm S 2009. Alternative splicing and disease. Biochim Biophys Acta 1792: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Patel DJ 2008. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol 15: 1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns MP, Terns RM 2001. Macromolecular complexes: SMN–the master assembler. Curr Biol 11: R862–864 [DOI] [PubMed] [Google Scholar]
- Timchenko NA, Patel R, Iakova P, Cai ZJ, Quan L, Timchenko LT 2004. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J Biol Chem 279: 13129–13139 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Kuwasako K, Takahashi M, Someya T, Inoue M, Terada T, Kobayashi N, Shirouzu M, Kigawa T, Tanaka A, et al. 2009. Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res 37: 5151–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Darnell RB 2007. Functional and mechanistic insights from genome-wide studies of splicing regulation in the brain. Adv Exp Med Biol 623: 148–160 [DOI] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB 2006. An RNA map predicting Nova-dependent splicing regulation. Nature 444: 580–586 [DOI] [PubMed] [Google Scholar]
- Valgardsdottir R, Chiodi I, Giordano M, Rossi A, Bazzini S, Ghigna C, Riva S, Biamonti G 2008. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res 36: 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maarel SM, Frants RR, Padberg GW 2007. Facioscapulohumeral muscular dystrophy. Biochim Biophys Acta 1772: 186–194 [DOI] [PubMed] [Google Scholar]
- Varon R, Gooding R, Steglich C, Marns L, Angelicheva D, Yong KK, Ambrugger P, Reinhold A, Morar B, Baas F, et al. Partial deficiency of the C-terminal-domain phosphatase of RNA polymerase II is associated with congenital cataracts facial dysmorphism neuropathy syndrome. Nat Genet 35: 185–189 [DOI] [PubMed] [Google Scholar]
- Vithana EN, Abu-Safieh L, Pelosini L, Winchester E, Hornan D, Bird AC, Hunt DM, Bustin SA, Bhattacharya SS 2003. Expression of PRPF31 mRNA in patients with autosomal dominant retinitis pigmentosa: a molecular clue for incomplete penetrance? Invest Ophthalmol Vis Sci 44: 4204–4209 [DOI] [PubMed] [Google Scholar]
- von Schantz M, Lucas RJ, Foster RG 1999. Circadian oscillation of photopigment transcript levels in the mouse retina. Brain Res Mol Brain Res 72: 108–114 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Burge CB 2008. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R 2005. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol Chem 386: 713–724 [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J 2005. Eukaryotic Lsm proteins: lessons from bacteria. Nat Struct Mol Biol 12: 1031–1036 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G 2008. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133: 585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Bellur DL, Lu S, Zhao F, Grassi MA, Bowne SJ, Sullivan LS, Daiger SP, Chen LJ, Pang CP, et al. 2009. Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. Am J Hum Genet 85: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Liu JR, Kyle JW, Hanck DA, Agnew WS 2006. A profile of alternative RNA splicing and transcript variation of CACNA1H, a human T-channel gene candidate for idiopathic generalized epilepsies. Hum Mol Genet 15: 1497–1512 [DOI] [PubMed] [Google Scholar]