Abstract
Background:
Medical natural language processing (NLP) systems have been developed to identify, extract and encode information within clinical narrative text. However, the role of NLP in clinical research and patient care remains limited. Pancreatic cysts are common. Some pancreatic cysts, such as intraductal papillary mucinous neoplasms (IPMNs), have malignant potential and require extended periods of surveillance. We seek to develop a novel NLP system that could be applied in our clinical network to develop a functional registry of IPMN patients.
Objectives:
This study aims to validate the accuracy of our novel NLP system in the identification of surgical patients with pathologically confirmed IPMN in comparison with our pre-existing manually created surgical database (standard reference).
Methods:
The Regenstrief EXtraction Tool (REX) was used to extract pancreatic cyst patient data from medical text files from Indiana University Health. The system was assessed periodically by direct sampling and review of medical records. Results were compared with the standard reference.
Results:
Natural language processing detected 5694 unique patients with pancreas cysts, in 215 of whom surgical pathology had confirmed IPMN. The NLP software identified all but seven patients present in the surgical database and identified an additional 37 IPMN patients not previously included in the surgical database. Using the standard reference, the sensitivity of the NLP program was 97.5% (95% confidence interval [CI] 94.8–98.9%) and its positive predictive value was 95.5% (95% CI 92.3–97.5%).
Conclusions:
Natural language processing is a reliable and accurate method for identifying selected patient cohorts and may facilitate the identification and follow-up of patients with IPMN.
Keywords: Intraductal papillary mucinous neoplasm, pancreatic cancer, prevention, cystic neoplasm, precancerous, natural language processing, data mining
Introduction
The use of prospective clinical databases has increased significantly in the last decade as a result of technological advancements in data entry and processing.1,2 Although clinical databases facilitate the tracking of clinical outcomes and allow for better-quality research, their longterm impact on patient care remains unclear.3 Despite recent software development, personnel time and effort are consumed in the creation of the framework, data entry, storage and maintenance of databases.
Pancreatic cysts are becoming increasingly recognized. This is in part a result of increasing resolution and the enhanced sensitivity of cross-sectional imaging. Radiographic and autopsy studies suggest that the incidence of pancreatic cysts may range from 0.4% to 24% depending upon size threshold.4,5 Some pancreatic cysts demonstrate premalignant behaviour and some will ultimately progress to pancreatic cancer. Because of their variable malignant potential, the management of pancreatic cysts may be surgical, observational or both.
One of the most common pancreatic cysts with malignant potential is the intraductal papillary mucinous neoplasm (IPMN).6 These neoplasms may occur anywhere in the pancreatic ductal system and often present first on cross-sectional imaging in an asymptomatic patient.7 When removed, these lesions may harbour invasive cancer in up to 50% of cases depending on patient symptoms, signs, radiographic features of the cyst and cyst fluid analysis.8 After their removal, the remaining pancreas must be observed in patients with IPMN because there is a significant incidence of multifocality and new lesion formation. Thus, the accurate identification, surveillance and treatment of IPMN combine to represent an opportunity to prevent pancreatic cancer. As the natural history of pancreatic cysts is quite variable and often poorly understood in mainstream medicine, patients with IPMN may not always be identified, tracked and treated optimally. At present there is no statewide or nationwide standard programme or system in place to optimize the identification, tracking and treatment of patients with pancreatic cysts for the purposes of either patient care or clinical investigation. The institution of such a programme could have tremendous potential from both a clinical and investigational standpoint. Current practice relies heavily on individual practitioners and their use of manually entered patient databases accrued retrospectively. Although these can be easily queried for retrospective studies, they have two main drawbacks. Firstly, they require a significant amount of time and manpower to create and maintain. Secondly, whether they are sufficiently sensitive to capture all cases of interest has not been validated.
Natural language processing (NLP) describes the computerized approach to analysing text. It is based on both a set of theories and a set of technologies and is best defined as a range of computational techniques for analysing and representing naturally occurring text at one or more levels of linguistic analysis to achieve human-like language processing for a range of applications. These computational techniques can be used to analyse naturally occurring texts in any language, mode or genre. The texts can be oral or written. In addition, the text to be analysed should ideally be gathered from actual usage, and should not be modified or altered for NLP.
In medicine, a wealth of patient data are gathered as narrative free text; this text may be written or transcribed by doctors, nurses and other health care professionals. With the increasing adoption of electronic medical record (EMR) systems, more of these text data are becoming electronic and therefore available for computer processing. However, by contrast with numerical data (such as laboratory test results or blood pressure readings), the data in medical narrative documents are unstructured and cannot be utilized in computerized applications. Two basic approaches have been used in attempts to provide structure to the text reports a clinician creates. One of these involves avoiding the creation of unstructured data altogether through the use of a form-based user interface so that the clinician chooses to mark appropriate concepts and values from a list of possible entries. This approach has been criticized by clinicians because of its frequent inability to capture many of the nuances represented in free text documentation, as well as the increased time it requires. The other approach has been to use NLP to perform information retrieval on narrative medical documents and to convert the data in documents into a form suitable for various applications. A medical NLP system is one that is applied to processing clinical documents (such as radiology or pathology reports, or discharge summaries) that are produced during the actual clinical care of a patient.
One of the oldest and most studied medical NLP systems is the Medical Language Extraction and Encoding System (MedLEE), developed by Carol Friedman et al. at Columbia University in the mid-1990s.9 MedLEE was developed to address a need to transform narrative text in patient reports to structured and encoded data so that the data could be stored in the clinical repository and accessed by other applications.
MedLEE has been studied extensively. In 1996, Jain et al.10 used it to identify suspected tuberculosis patients by processing chest radiograph reports. MedLEE and the reference standard (human review) agreed on nearly 89% of the reports. In 1997, Jain and Friedman11 used it to identify mammogram findings suspicious for breast cancer and found it to accurately identify such findings and to compare favourably with the reference standard. In 2005, Melton and Hripcsak12 described a study in which MedLEE was tasked to identify adverse events using discharge summaries.
The Regenstrief Institute in Indianapolis, IN, USA has developed an NLP program called the Regenstrief EXtraction Tool (REX). REX is a rule-based NLP system written in Java and has successfully extracted patient data from radiology reports,13 admission notes,14 microbiology results15 and pathology reports. REX uses a combination of regular expressions and algorithms to detect where in text keywords or phrases related to a concept are found. It then examines ‘windows’ of words before and after the concept phrase to determine context (i.e. positive, negated, historical, family history, related, etc.).
In this particular application of REX, the primary goal was to identify all patients with a confirmed surgical pathological diagnosis of IPMN within a large multi-hospital clinical network. Identifying IPMN patients in this setting is more difficult than identifying patients with other medical conditions for several reasons. The method often used by researchers to identify patients with medical conditions involves using ICD-9 billing codes. Although research shows that using ICD-9 codes to identify patients can be inaccurate,16,17 it is frequently used because it is a relatively quick and easy method for identifying large cohorts of patients. Unfortunately, this method is not useful for identifying patients with IPMN because this condition is rarely coded as a billable diagnosis with ICD-9 codes. Medication data, sometimes used as a surrogate for a diagnosis and occasionally used to identify patients, are not useful to identify IPMN patients because no medications are given to treat this condition and the medications used to mitigate its symptoms are not exclusively used in IPMN. Often the only evidence that a patient has an IPMN is a statement in a narrative medical report, such as a radiology, endoscopy or pathology report. Given the large number of patients and text reports in our network, it would be extremely time-consuming and labour-intensive to manually review text reports to detect patients with IPMN. In addition, the above effort would be applied only to existing reports; additional manpower would be needed on an ongoing basis to manually review newly created reports to detect new patients with IPMN.
REX NLP software was used to identify IPMN patients and accurately extract key elements from their text reports. The aim of our study was to compare the results of an NLP-guided search for patients with a surgical pathology-confirmed diagnosis of IPMN with a standard manually maintained surgical database. We performed a validation study of the system's accuracy in identifying these patients and of its ability to build a database of IPMN patient data. We hypothesized that the NLP software would prove superior to the standard surgical database in its ability to identify patients with IPMN. We also hypothesized that NLP would more readily provide additional data, including date of diagnosis, method of detection and context.
Materials and methods
This study was approved by the Indiana University Health Institutional Review Board (IRB) and conducted in compliance with its regulations. During 1985–2009, a manually created surgical database was compiled for all patients undergoing resection for IPMN at Indiana University Hospital. The methodology for the creation of this database involved the prospective collection and accrual of patients identified as undergoing pancreatic resection for IPMN by the operating surgeon under IRB approval. An up-to-date registry of each of these patients was maintained in database format. At quarterly intervals, data regarding patient demographics, radiologic, pathologic and operative characteristics, and outcomes were added in a retrospective fashion to populate the remaining fields of the surgical database. All of this process of data search and accrual was performed manually by clinical staff. Subsequently, these data are intermittently re-reviewed and validated by manual chart review prior to use in any clinical research or investigation. We consider this dataset the most comprehensive for patients with resected IPMN at our institution and used it for the purposes of comparison with the newly created NLP database.
Evolution of REX to suit pancreas cyst research
REX was modified to enable it to accurately identify patients with pancreatic cysts. Towards that goal, a knowledge base for the concept of pancreatic cysts had to be created. Firstly, a group of concept words that could be used to identify pancreatic cysts were assembled by our team of experts in pancreas disease. In addition, a literature search and a Unified Medical Language System (UMLS)18 review of pancreatic cysts were conducted to identify additional keywords or phrases that express the concept of pancreatic cyst. Finally, a training set of clinical text reports were collected to identify additional keywords or phrases that clinicians use to express the concept of pancreatic cyst. The same training set was used to identify any contextual markers specific to pancreatic cyst that needed to be added to the knowledge base. Several meetings were held among the authors to discuss which data elements would be of value to researchers and clinicians and should be extracted from the text reports. Based on the group's consensus, we added fields to the pancreatic cyst knowledge base, as well as additional algorithmic rules to REX that enabled the extraction of these data. Specifically, REX was reprogrammed to identify a total of 11 pancreatic concepts (Table 1). These included 10 classes of pancreatic cysts/tumours, as well as pancreatic duct dilatation/ectasia.
Table 1.
The various ‘cyst concepts’ utilized by the natural language processing software to identify patients with pancreatic cysts
| Pancreatic concept |
|---|
| Pancreatic cyst |
| Pseudocyst |
| Intraductal papillary mucinous neoplasm |
| Serous cyst |
| Retention cyst |
| Cystic neuroendocrine tumour |
| Cystic degeneration |
| Mucinous cystic neoplasm |
| Mucinous cystadenoma |
| Islet cell tumour |
| Pancreatic duct dilatation/ectasia |
REX and pancreatic cysts: search details
The NLP research conducted in our study is innovative. Our interest group identified four abilities required to identify and categorize study cohorts. We enhanced REX to provide these capabilities. They are: (i) the ability to prioritize by pancreatic cyst type; (ii) the ability to prioritize by report type; (iii) the ability to prioritize by report date, and (iv) the ability to prioritize by context.
We will discuss each of these enhancements in terms of their details, the rationale for adding them to REX and the methods we used to add them to the existing REX software. We will also give examples of how these enhancements might be of benefit to researchers.
Prioritizing by pancreatic cyst type
This prioritization is important because there may be several citations of pancreatic cyst in a patient's medical record and it is vital to isolate any instances when more detailed or specific citations of pancreatic cyst are made. Prior to such a modification, REX would identify the first citation of pancreatic cyst in a positive context chronologically. This is suboptimal because several pathological types of pancreatic cyst exist, such as pseudocysts, serous cystic neoplasms (SCNs), intraductal papillary mucinous neoplasms (IPMNs) and mucinous cystic neoplasms (MCNs), with variable malignant potential. Therefore, it is important to isolate the report that specifies the type of cyst. For example, a patient's medical record may include reports that contain both non-specific statements about pancreatic cysts (e.g. ‘A pancreatic cyst was seen’), as well as more specific statements (e.g. ‘Pathology demonstrated a mucinous cystic neoplasm of the pancreas’). In this case, ideally, the search system would disregard the non-specific citation in favour of isolating the report that contains the more specific statement. Conversely, if a patient's record only contained non-specific citation(s) of pancreatic cyst, this patient would still be captured, but in a different, more general cohort. As REX was unable to perform this prioritization/categorization function, we added this enhancement by developing algorithmic rules to achieve this prioritization.
Prioritizing by report type
This is important when there are several types of report that contain citations of pancreatic cyst, but one report type is considered more definitive. For example, a report on an abdominal computed tomography (CT) scan may state that a pancreatic cyst appears to be an MCN. Although a radiologist may suspect the cyst is an MCN based on radiological appearance, this can only be definitively determined by a pathologist during cyst wall biopsy. Therefore, if a patient's medical record has both a radiology and a pathology report that contain statements affirming the presence of an MCN, ideally the pathology report is prioritized over the radiology report because it is more definitive. Conversely, if a patient's medical record contains only a radiology report that cites an MCN, we should capture it as well, albeit in a category that is less certain of the MCN designation. REX was previously unable to perform this prioritization/categorization by report type. REX could identify patients with pancreatic cysts on CT scans as well as patients with pancreatic cysts on pathology reports, but was unable to categorize and filter the two sets by detecting duplicate patients (i.e. REX was able only to place the patients into two groups and a cross-referencing of the two groups to identify all patients in the CT scan group but not also in the pathology group required a separate processing step by researchers). To add this in conjunction with the first enhancement, we created algorithms to allow for this prioritization/categorization and integrated them into REX.
Prioritizing by report date
Pancreatic cyst researchers at our institution have identified that the date of diagnosis is an important element in identifying patients. Because the patient data in our database extend back as far as the mid-1970s, it is valuable to know the date of the report that indicates pancreatic cyst. This information is helpful in prioritizing which patients should be contacted for future follow-up if needed. Prior to this project, REX did not have the capability to extract dates from reports and could not perform temporal reasoning tasks (i.e. date comparisons). Therefore, it had no ability to identify patients with positive results reported before or after a given date. To add this enhancement, temporal data fields were added to the pancreatic cyst knowledge base and additional programming was added to REX to enable it to extract dates from reports and compare and calculate intervals between dates in the report and the target date in the knowledge base.
Prioritizing by context
As stated previously, REX can identify several contexts that could be linked to the citation of a concept. For the pancreatic cyst study, the highest priority context is positive. However, in contexts other than positive, researchers desire the ability to prioritize based on context. For example, if in a patient's medical record there are citations of pancreatic cyst in both the historical and negated contexts, REX would be expected to favour the report in the historical context because this is greater evidence that the patient has pancreatic cyst, at least in the past. In this instance, the citation of pancreatic cyst in a negated context does not necessarily indicate that the patient does not have the condition, only that a specific study (such as an abdominal X-ray) was negative for pancreatic cyst. Prior to this project, REX had no ability to favour one context over another except for the positive context. REX programming was updated to enable awareness of the current context of a concept within a given patient's record to give the software the capability to favour one context over another.
In addition, REX was modified and extended to enable the automated creation of a research database populated with data extracted from free text reports. One of the goals of this project was to accurately and automatically extract important data from text reports of pancreatic cyst cohorts and place those data into a database in order to relieve researchers of the time, expense and effort required to do this manually. This enhancement required major revisions of the REX code because previously REX could only output a text file of the positive report with the concept that was identified appended to the report. This enhancement consisted of two parts: (i) the creation of methods to identify and extract key data elements from positive reports, and (ii) the creation of mechanisms whereby REX inputs the extracted data into the appropriate fields of the database.
After these initial modifications and enhancements of the REX software had been completed, a training set of reports was collected. These were used to test and further refine the software. This training set consisted of all text reports that contained the terms ‘pancreas’ and ‘cyst’ or all patients in our database produced during the 1-year period from January to December 2007 inclusive. This training set was processed through REX and then analysed to detect false positive and false negative errors. Analysis of the cause of these errors led to further modification and enhancement of the REX code (algorithmic rules) to correct them. A series of modifications were made, the training set was rerun through REX and the output was analysed again for errors. This modification–process–analysis cycle was repeated several times until REX achieved sufficient accuracy. Examples of revisions made to the REX software during this phase are shown in Table 2.
Table 2.
Examples of revisions made to the natural language processing software during the project
| Date | Category | Revision |
|---|---|---|
| 08/08 | Cyst type | Added ‘sphenoid’, ‘ethmoid’, ‘ovarian’ and ‘sinus’ to report keyword exceptions to prevent false positive identification of pancreatic cyst |
| 09/08 | Context window size | Changed length of ambiguous context window from +/− 8 words to +/− 12 words |
| 09/08 | Context phrases | Added ‘nor’ to negation context phrase list and ‘inconclusive’ to ambiguous context phase list |
| 10/08 | Report type priority | Placed endoscopy/radiology reports at second highest level of report priority behind pathology reports |
| 12/08 | Cyst type | Added islet cell tumour to cyst types identified |
| 01/09 | Cyst type | Added cyst size/duct size extraction capability |
| 02/09 | Report frequency | Added calculation of number of endoscopy/radiology tests for each patient |
| 06/09 | Context phrases | Removed ‘borderline’ from ambiguous context phrase list |
| 06/09 | Cyst type priority | Moved ‘pancreatic cyst’ to a lower priority than ‘pseudocyst’ |
Evaluation of REX
After REX achieved sufficient accuracy in processing the training set, the program was applied to process all 165 000 reports collected for this study. A validation study was then performed to assess the accuracy of REX in identifying and extracting data for IPMN patients. Patients with a pathological diagnosis of IPMN identified by REX were cross-checked across the surgical IPMN database. Patient charts were reviewed and particular attention was paid to those identified by one system but not the other.
Statistical methods
The validation process was performed using a ‘universe’ of true positive cases of IPMN patients who underwent surgical resection. Patients in this cohort were identified by either NLP or the standard cyst surgical database or both. All cases in this cohort were individually validated by careful review of existing medical records. The sensitivity of each system was calculated as the proportion of patients with surgical pathology of IPMN correctly identified by the particular system compared with the total number of true positive cases in the cohort. Similar criteria were applied to calculate positive predictive value. The specificity of either system could not be assessed as a result of our inability to confidently identify all surgical IPMN patients missed by both systems. Confidence intervals (95% CIs) were calculated.
Results
Natural language processing identified 5694 unique patients with pancreatic cysts. Among these, 852 were in the mucinous category based on pathological (surgical pathology, tissue biopsy or fine needle aspiration) and non-pathological (mainly imaging) text reports. The rest of the patients identified by NLP either had a dilated pancreatic duct suggestive of main duct IPMN (n = 1375) or had pancreatic cysts cited in a non-mucinous context (n = 3467). Natural language processing was able to further categorize patients in these groups based on the availability of surgical pathology (Fig. 1).
Figure 1.
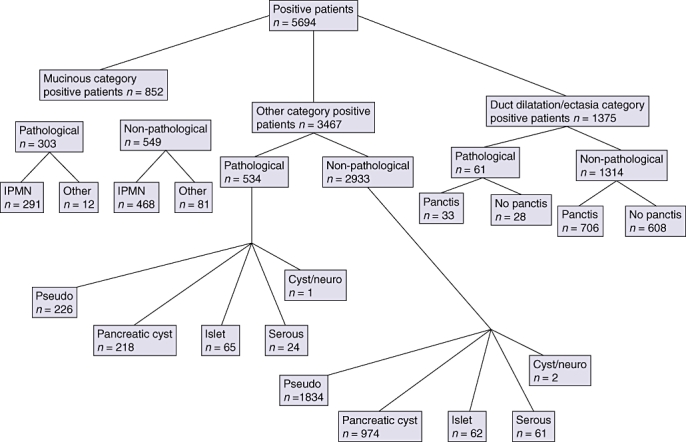
Diagram demonstrating the total number of patients identified by natural language processing and their breakdown into the various subcategories. IPMN, intraductal papillary mucinous neoplasm
The number of pathologically confirmed IPMN patients identified using both systems was 319. Of these, 21 patients (in the surgical database) did not have identifiable medical record numbers by NLP and were excluded, leaving 298 patients who constituted the ‘universe’ for this study. In this group, 205 patients were correctly identified by both systems. The number of patients identified by one system and not the other were 86 and seven for NLP and the surgical database, respectively (Fig. 2). The relatively large group of 86 patients identified by NLP included 36 patients with cytopathological diagnosis of IPMN obtained during gastrointestinal procedures (endoscopic ultrasound [EUS] or endoscopic retrograde cholangiopancreatography [ERCP]) (Fig. 3). As patients in this group either did not undergo surgery and were clinically monitored, or underwent surgery at a different institution, they were not part of our surgical IPMN database. Nonetheless, even accounting for this, 37 patients were identified only by the NLP system and were not picked up by the surgical database. The difference in sensitivity between the two systems was statistically significant (P < 0.001) (Table 3).
Figure 2.
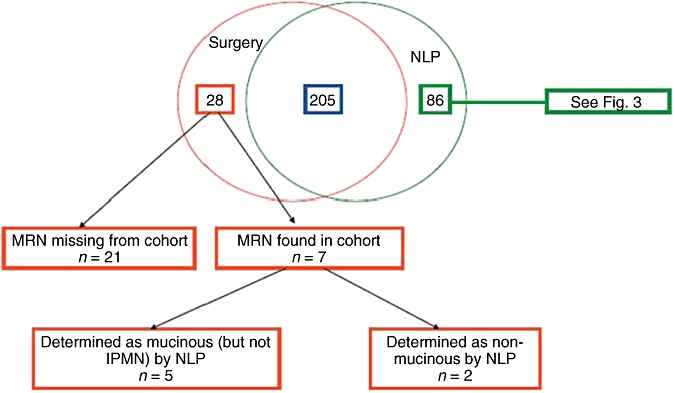
Venn diagram showing patients identified by both systems (the natural language processing [NLP] platform and the surgical database) and each system separately. MRN, medical record number; IPMN, intraductal papillary mucinous neoplasm
Figure 3.
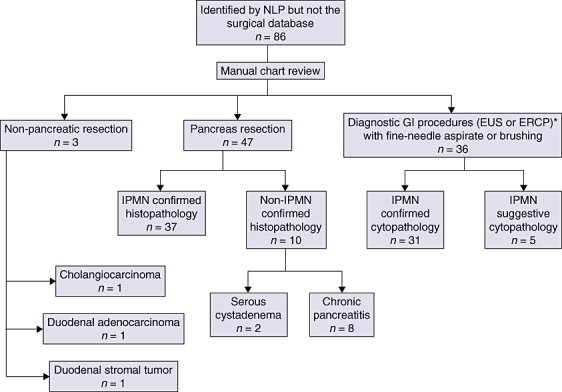
Flow diagram showing details of the subgroup of patients identified by natural language processing (NLP) but not the surgical database. GI, gastrointestinal; EUS, endoscopic ultrasound; ERCP, endoscopic retrograde cholangiopancreatography; IPMN, intraductal papillary mucinous neoplasm
Table 3.
Comparison of the sensitivity and positive predictive values of the natural language processing (NLP) platform and the surgical database
| System used | Sensitivity, % (95% CI) | Positive predictive value, % (95% CI) |
|---|---|---|
| NLP platform | 97.5 (94.8–98.9) | 95.5 (92.3–97.5) |
| Surgical database | 85.1 (80.0–89.2) | 100 (97.8–100) |
95% CI, 95% confidence interval
Discussion
The use of EMRs has revolutionized the delivery of medical care in numerous ways. One potential area, which is only beginning to be explored, is the ability to identify and track patients based on a particular disease or clinical characteristic. This role has important implications for both clinical research and patient care. The current standard tool remains the prospectively accrued clinical registry. At the outset, the aim of this study was to compare the results of an NLP-guided search for patients with a surgical diagnosis of IPMN with a standard manually maintained surgical registry. We find that NLP provides a more accurate method for identification of IPMN patients within a single health network. Although manual medical record review and analysis remain important elements in clinical and outcomes research, patient capture is more accurate when employing an NLP system for this purpose.
To our knowledge, this is the first study comparing the performance of a smart search engine like NLP with a human-constructed database to be reported in the literature. The NLP program demonstrated outstanding sensitivity of 97.5% compared with the surgical database (P < 0.001) and a positive predictive value of 95.5%. This demonstrates the feasibility and reliability of NLP in creating a clinical database using a limited number of entry criteria. Indeed, the NLP program uniquely identified 37 patients with IPMN, representing an increase of approximately 20% in the size of our existing surgical database. Although the majority of patients in this group were not suspected to have IPMN at the time of surgical resection and IPMN was subsequently identified in the surgical pathology specimens, the implications of this finding for the subsequent management of patients are significant. This cohort of patients is at risk for IPMN recurrence and therefore should be carefully screened longitudinally. Although the optimal method and interval of surveillance remain controversial, current consensus guidelines indicate the importance of at least annual cross-sectional imaging of the pancreatic remnant in patients with surgically resected IPMN.8
As electronic patient data become more prevalent through the increased adoption of EMRs, using NLP to assist with the automated creation of patient cohorts for research will become an increasingly valuable methodology. Several studies have shown that the traditional method of identifying patients, namely, by using administrative claims databases, can be inaccurate. In a study similar to ours, in which patients with precancerous conditions needed to be identified, Jacobson and Gerson found that the method of using ICD-9 codes to identify patients with Barrett's oesophagus was inaccurate.19 The NLP system used in this study could be modified and applied to identify patients with other precancerous lesions, including Barrett's oesophagus, as well as other conditions not routinely coded with ICD-9 codes, such as cardiomegaly or pleural effusion.
A major limitation of this comparative study relates to our inability to determine the total number of patients with IPMN within the health network captured by neither system. In other words, the specificity of the two systems could not be determined or compared. The fact that a significant number of patients (n = 37) were not included in the surgical database and that seven were missed by the NLP system implies that neither of the two systems in isolation represents a suitable reference standard.
We envision several future directions and uses for the REX NLP system. The system used in this study has the potential to perform data mining of medical text reports beyond simply identifying patients with certain conditions. With enhancements to the core system, REX can be programmed to extract specific data elements from the text report related to the condition of interest. For example, enhancements to REX can be performed in the future in order to automatically identify and extract data on pancreatic cyst location and size, as well as pancreatic duct diameter, from pathology reports. In addition, REX could be modified to recognize concomitant conditions or diagnoses, such as in patients with pancreatic cysts who develop new or worsening diabetes mellitus. Our next step will include implementing a surveillance version of REX within our electronic patient network in order to automatically detect new pancreatic cyst patients as they are discovered.
In summary, the Regenstrief NLP REX platform provides an accurate method for identifying IPMN patients within a single health network. This platform has the potential for broad application to other medical conditions. Furthermore, we believe the NLP REX platform could be applied by other similar regional health organization networks to impact care on a regional or national level.
Acknowledgments
This study was supported by the Indiana Genomics Initiative (INGEN) of Indiana University. INGEN is supported in part by Lilly Endowment, Inc., Indianapolis, IN, USA.
Conflicts of interest
None declared.
References
- 1.Tierney WM, McDonald CJ. Practice databases and their uses in clinical research. Stat Med. 1991;10:541–557. doi: 10.1002/sim.4780100406. [DOI] [PubMed] [Google Scholar]
- 2.Black N, Payne M. Improving the use of clinical databases. BMJ. 2002;324:1194. doi: 10.1136/bmj.324.7347.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harvey S, Rowan K, Harrison D, Black N. Using clinical databases to evaluate healthcare interventions. Int J Technol Assess Health Care. 2010;26:86–94. doi: 10.1017/S0266462309990572. [DOI] [PubMed] [Google Scholar]
- 4.Kimura W, Nagai H, Kuroda A, Muto T, Esaki Y. Analysis of small cystic lesions of the pancreas. Int J Pancreatol. 1995;18:197–206. doi: 10.1007/BF02784942. [DOI] [PubMed] [Google Scholar]
- 5.Laffan TA, Horton KM, Klein AP, Berlanstein B, Siegelman SS, Kawamoto S, et al. Prevalence of unsuspected pancreatic cysts on MDCT. AJR Am J Roentgenol. 2008;191:802–807. doi: 10.2214/AJR.07.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohashi K, Murakami Y, Marayama M. Four cases of mucus-secreting pancreatic cancer. Prog Dig Endoscopy. 1982;20:348–351. [Google Scholar]
- 7.Waters JA, Schmidt CM. Intraductal papillary mucinous neoplasm – when to resect. Adv Surg. 2008;42:87–108. doi: 10.1016/j.yasu.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka M, Chari S, Adsay V, Fernandez-del Castillo C, Falconi M, Shimizu M, et al. International consensus guidelines for management of intraductal papillary mucinous neoplasms and mucinous cystic neoplasms of the pancreas. Pancreatology. 2006;6:17–32. doi: 10.1159/000090023. [DOI] [PubMed] [Google Scholar]
- 9.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1:161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain NL, Knirsch CA, Friedman C, Hripcsak G. Identification of suspected tuberculosis patients based on natural language processing of chest radiograph reports. Proc AMIA Annu Fall Symp. 1996;1996:542–546. [PMC free article] [PubMed] [Google Scholar]
- 11.Jain NL, Friedman C. Identification of findings suspicious for breast cancer based on natural language processing of mammogram reports. Proc AMIA Annu Fall Symp. 1997;1997:829–833. [PMC free article] [PubMed] [Google Scholar]
- 12.Melton GB, Hripcsak G. Automated detection of adverse events using natural language processing of discharge summaries. J Am Med Inform Assoc. 2005;12:448–457. doi: 10.1197/jamia.M1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedlin J, McDonald CJ. A natural language processing system to extract and code concepts relating to congestive heart failure from chest radiology reports. AMIA Annu Symp Proc. 2006;2006:269–273. [PMC free article] [PubMed] [Google Scholar]
- 14.Friedlin J, McDonald CJ. Using a natural language processing system to extract and code family history data from admission reports. AMIA Annu Symp Proc. 2006;2006:925. [PMC free article] [PubMed] [Google Scholar]
- 15.Friedlin J, Grannis S, Overhage JM. Using natural language processing to improve accuracy of automated notifiable disease reporting. AMIA Annu Symp Proc. 2008;2008:207–211. [PMC free article] [PubMed] [Google Scholar]
- 16.Miller ML, Wang MC. Accuracy of ICD-9-CM coding of cervical spine fractures: implications for research using administrative databases. Annu Proc Assoc Adv Automot Med. 2008;52:101–105. [PMC free article] [PubMed] [Google Scholar]
- 17.Singh JA, Holmgren AR, Noorbaloochi S. Accuracy of Veterans Administration databases for a diagnosis of rheumatoid arthritis. Arthritis Rheum. 2004;51:952–957. doi: 10.1002/art.20827. [DOI] [PubMed] [Google Scholar]
- 18.National Institutes of Health, National Library of Medicine. UMLS (Unified Medical Language System) http://www.nlm.nih.gov/research/umls/. [Accessed 1 September 2010.
- 19.Jacobson BC, Gerson LB. The inaccuracy of ICD-9-CM Code 530.2 for identifying patients with Barrett's esophagus. Dis Esophagus. 2008;21:452–456. doi: 10.1111/j.1442-2050.2007.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


