Abstract
Myriad genetic and epigenetic alterations are required to drive normal cells toward malignant transformation. These somatic events commandeer many signaling pathways that cooperate to endow aspiring cancer cells with a full range of biological capabilities needed to grow, disseminate and ultimately kill its host. Cancer genomes are highly rearranged and are characterized by complex translocations and regional copy number alterations that target loci harboring cancer-relevant genes. Efforts to uncover the underlying mechanisms driving genome instability in cancer have revealed a prominent role for telomeres. Telomeres are nucleoprotein structures that protect the ends of eukaryotic chromosomes and are particularly vulnerable due to progressive shortening during each round of DNA replication and, thus, a lifetime of tissue renewal places the organism at risk for increasing chromosomal instability. Indeed, telomere erosion has been documented in aging tissues and hyperproliferative disease states—conditions strongly associated with increased cancer risk. Telomere dysfunction can produce the opposing pathophysiological states of degenerative aging or cancer with the specific outcome dictated by the integrity of DNA damage checkpoint responses. In most advanced cancers, telomerase is reactivated and serves to maintain telomere length and emerging data have also documented the capacity of telomerase to directly regulate cancer-promoting pathways. This review covers the role of telomeres and telomerase in the biology of normal tissue stem/progenitor cells and in the development of cancer.
Telomeres protect chromosome ends
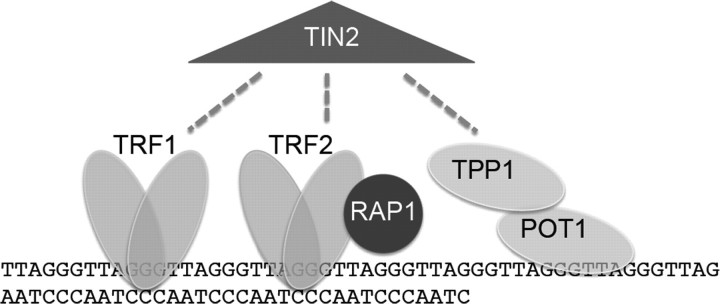
Telomeres are comprised of tracts of G-rich nucleotide repeats that serve as binding sites for a large array of proteins (Figure 1). In vertebrates, telomeres are composed of the sequence TTAGGG and include a double-stranded tract of repeats many kilobases long and an obligate single-stranded 3′ overhang, measuring a few hundred nucleotides (1). The single-stranded overhang can fold back on the double-stranded telomere in a lariat structure termed the t-loop, which serves to sequester the chromosome terminus (2). Double-stranded telomere sequences are bound directly by two sequence-specific DNA binding proteins telomeric repeat binding factor 1 (TRF1) and telomeric repeat binding factor 2 (TRF2), which in turn interact with a larger number of proteins. TRF2 is critical for telomere end protection and can facilitate formation of the t-loop conformation. Disruption of TRF2 through overexpression of a dominant-negative form or through deletion in TRF2-knockout mouse embryo fibroblasts leads to loss of the protective capped structure, characterized by processing of the 3′ overhang and ligation of chromosome ends (3,4). TRF1 can serve to modulate telomere length, but it also serves to facilitate DNA replication through the telomere repeats, which acts as fragile DNA sites (5–7).
Fig. 1.
Telomeres protect chromosome ends. Telomeres are TTAGGG double-stranded DNA repeats culminating in a single-stranded overhang. The shelterin protein complex protects telomeres, facilitates replication and controls access of telomerase. TRF1 and TRF2 bind the double-stranded telomere. POT1 and TPP1 are OB-fold containing proteins associated with the single stranded overhang. RAP1 binds TRF2, and TIN2 is a central component of the complex interacting with TRF1, TRF2 and TPP1. Telomeres can exist in a looped conformation, termed the t-loop, in which the single-stranded overhang folds back on the double-stranded telomere to sequester the end. The 3′ hydroxyl group represents the substrate for telomere addition by telomerase.
TRF1 and TRF2 each interact with a common factor TRF1-interacting nuclear factor (TIN2) (8), which nucleates a six-member complex, termed shelterin, that also includes telomeric repeat binding factor 2-interacting protein, protection of telomeres 1 (POT1) and TPP1 (9). Telomeric repeat binding factor 2-interacting protein is a TRF2-interacting factor, related to Rap1p, a critical telomere-binding protein in Saccharomyces cerevisiae (10). Unlike yeast Rap1p, mammalian telomeric repeat binding factor 2-interacting protein does not bind telomeric DNA directly but is instead recruited to telomeres through its interaction with TRF2, where it may act to aid in repression of non-homologous end joining (11,12). TRF1-interacting nuclear factor 2 also interacts with the subcomplex of shelterin that binds the single-stranded overhang—TPP1 and POT1, two oligonucleotide/oligosaccharide binding (OB)-fold containing proteins (13–15). POT1 directly binds the single-stranded telomere sequences and interacts directly with TPP1. POT1 and TPP1 serve a role in protecting the single-stranded portion of the telomere because loss of POT1 impairs telomere capping (16–19). In addition, POT1 and TPP1 can control telomerase action at telomeres. Overexpression of POT1 leads to telomere shortening by inhibiting telomerase action at the telomere (20). In contrast, POT1 and TPP1 in vitro serve as potent enhancers of telomerase processivity (21,22), thus this single-stranded telomere complex serves an important role in regulating telomerase at the telomere.
Replicative senescence and crisis in human fibroblasts
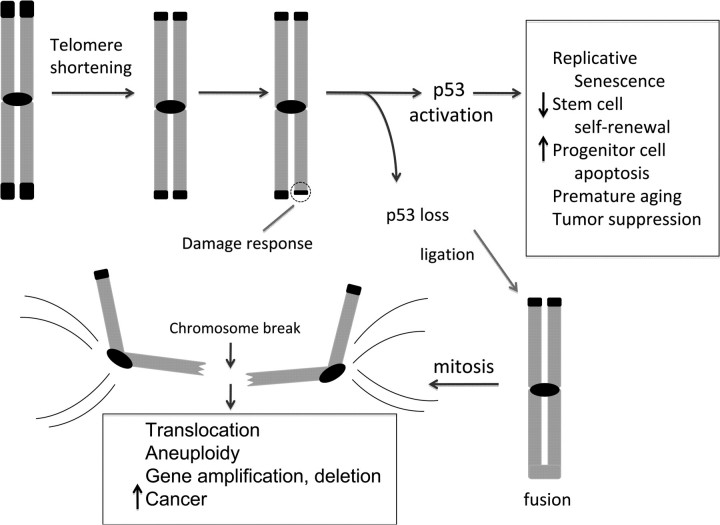
One of the earliest connections between telomeres and cancer was inferred from the study of primary human fibroblasts grown in cell culture (Figure 2). These primary human cells exhibited limited replicative potential, dividing ∼60–80 population doublings before entry into a senescent state (23). In contrast, established cancer lines divided indefinitely with passage in culture. A first critical clue of the mechanistic basis for this distinction came with the observation that telomeres shorten progressively during division of normal human fibroblasts in culture, yet are maintained in cancer cells (24). These shortened telomeres activate a ‘senescence’ program, and this growth arrest phenotype can be neutralized through inactivation of p53 and Rb (the Retinoblastoma tumor suppressor protein) (25,26). However, as these human fibroblasts undergo additional cell divisions, telomere erosion continues and the loss of telomere capping function produces rampant chromosomal instability and widespread apoptosis, a cellular state termed crisis (27). Senescence and crisis in these fibroblasts are effectively averted by overexpresssion of telomerase reverse transcriptase (TERT), the catalytic protein subunit of telomerase, an enzyme complex that synthesizes telomere repeats (28,29). Immortalization of primary human cells by TERT overexpression has been repeated in many different cell types. This remarkable ability of telomerase to endow primary human cells, which are destined to senesce, with immortal growth potential provoked widespread speculation that telomerase reactivation plays a near obligate role in human cancer development, whereas the lack of such activity promotes aging and degenerative disorders.
Fig. 2.
Telomere shortening activates p53 and drives formation of epithelial cancers through gene amplification and deletion. Telomeres shorten progressively with cell division due to the end-replication problem in settings of insufficient telomerase, including in human fibroblasts, aging tissues, early cancers and diseases of high cellular turnover. Critical telomere shortening compromises the telomere cap and results in a DNA damage response that activates the p53 tumor suppressor protein. This activation of p53 induces replicative senescence in cultured human fibroblasts, impairs stem cell self-renewal, induces apoptosis in tissue progenitor cells, causes premature aging and strongly suppresses tumor formation. If p53 is mutated or deleted, these responses to telomere dysfunction are mitigated and chromosomal fusions are tolerated. The generation of fused chromosomes results in dicentric chromosomes (chromosomes with two centromeres) and when these attach to opposite spindle poles, chromosome breakage occurs. These broken ends serve as potent catalysts for translocations, focal amplifications and focal deletions. Such CNAs drive development of carcinomas and explain the widespread gene copy number changes seen in human cancers.
Human telomerase components and telomerase trafficking in cancer cells
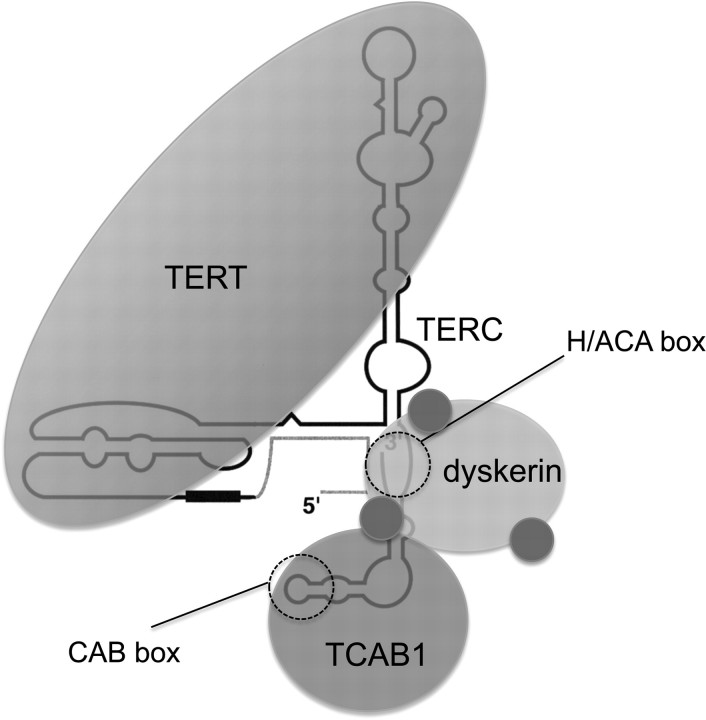
The telomerase ribonucleoprotein enzyme is composed of a minimal catalytic core, which includes TERT protein and the telomerase RNA component (TERC) (Figure 3) (30–33). TERC functions as the template for the enzyme to add telomere repeats in a reverse transcriptase reaction at the chromosome end. Although TERT and TERC are sufficient to generate telomerase activity in vitro in rabbit reticulocyte lysates (34), additional protein components of the enzyme are required in human cells. The 3′ end of TERC in vertebrates contains sequence motifs lacking in TERC from single-cell eukaryotes. In this region, mouse and human TERC possess an H/ACA sequence motif that defines a specific class of non-coding RNAs (35,36). These H/ACA RNAs act as guides for the modification of other cellular RNAs and they can be divided into two groups. H/ACA small nucleolar RNAs (snoRNAs) accumulate in the nucleolus and are involved in modification of ribosomal RNAs. A related group of RNAs, H/ACA small Cajal body-specific RNA (scaRNAs), accumulate in Cajal bodies and direct the modification of splicing RNAs (37). The difference in cellular trafficking between the H/ACA snoRNAs and the H/ACA scaRNAs is attributable to the presence of another sequence motif, termed the Cajal body box or CAB box. Cajal bodies are subnuclear sites of ribonucleoprotein assembly and modification (38). TERC has not only an H/ACA sequence but also a CAB box and therefore resembles a scaRNA. In fact, TERC accumulates in human cancer cells specifically within the Cajal body by RNA fluorescence in situ hybridization (39–42).
Fig. 3.
Telomerase is a large multisubunit RNP. Telomerase is a reverse transcriptase that adds telomere repeats to chromosome ends. The minimal catalytic core is composed of TERT, the telomerase reverse transcriptase, and TERC, the telomerase RNA, which acts as the template for telomere addition. The 5′ end of TERC contains the template region (black box) and 3′ end of TERC contains two sequences that act as binding sites for additional telomerase protein factors. The H/ACA box represents the binding site for dyskerin, a protein critical for telomerase assembly and for stability of TERC. Dyskerin has three small associated proteins—NHP2, NOP10 and GAR1—shown as blue spheres. TCAB1 is a WD40 repeat protein that recognizes the CAB box in TERC. TCAB1 interacts with dyskerin and is crucial for facilitating telomerase trafficking to Cajal bodies and for telomere maintenance.
H/ACA RNAs depend upon a specific assembly pathway that requires the protein dyskerin. Dyskerin is an RNA-binding protein and an enzyme itself—a pseudouridine synthetase. Together with several small associated proteins including NHP2, NOP10 and GAR1, dyskerin binds H/ACA RNAs and is required for their stability. The H/ACA snoRNAs and scaRNAs direct this enzyme complex to their complementary RNAs—ribosomal RNAs and splicing RNAs, respectively—where the dyskerin complex catalyzes the isomerization of uridines to pseudouridines. These modifications are important for the function of these target RNAs. Although the function of dyskerin as a pseudouridine synthetase is not thought to be involved in telomerase, dyskerin is essential for telomerase. Dyskerin is associated with all active telomerase in human cancer cell extracts (43–45). Our understanding of how telomerase, and other scaRNA ribonucleoproteins, traffic specifically to Cajal bodies, rather than to nucleoli, was enhanced by the recent finding of a new protein telomerase Cajal body protein 1 (TCAB1) in telomerase complexes. TCAB1 (also known as WDR79) is a WD40-repeat containing protein that associates with dyskerin and binds the CAB box in scaRNAs (46,47). TCAB1 associates with a majority of both telomerase activity and telomerase RNA in human cancer cell extracts. In addition, TCAB1 specifically associates with H/ACA scaRNAs but not H/ACA snoRNAs or other small RNAs in Cajal bodies or nucleoli. Depletion of TCAB1 disrupts telomerase localization to Cajal bodies and leads to progressive telomere shortening. Therefore, telomerase requires additional proteins that are stable components of the holoenzyme, for proper assembly, trafficking and function in human cancer cells.
Expression of telomerase in mouse and human tissues
Early studies cataloging TERT expression and telomerase activity reported potent suppression in human somatic tissues, yet robust expression and activity in germ cells and cancer cells (48). However, more sensitive analysis of primary human samples has since revealed modest levels of telomerase activity in proliferative tissues with high renewal potential such as the bone marrow, skin, gastrointestinal tract and testis as well as in activated lymphocytes (49–51). Within these renewing tissues, telomerase is most active in the resident stem/progenitor cell compartments—particularly in early hematopoietic progenitor cells (49), in the gastrointestinal crypt epithelium (52) and in hair follicle progenitor cells (53). Telomerase expression in stem and progenitor cells appears to be insufficient to maintain telomeres, particularly with the stresses of bone marrow transplantation or with advancing age. Telomerase is reactivated during the reprogramming of human fibroblasts to induced pluripotent stem cells and is expressed robustly in human embryonic stem cells (54).
Telomerase activity is regulated in large part at the level of TERT gene transcription and, accordingly, a TERT-green fluorescent protein transgenic reporter mouse showed green fluorescent protein expression in hematopoietic stem/progenitor cells and in crypt cells of the gastrointestinal tract (55). The TERT promoter is controlled in some contexts by the c-Myc oncogene, which recognizes E-boxes within the promoter (56,57), and by estrogen receptor and Sp1 (58). Beyond TERT regulation, the telomerase holoenzyme is probably subjected to posttranscriptional regulation as evidenced by modulation of telomerase activity by the ubiquitin ligase MKRN1 (59). At present, it remains unclear to what extent the other components of the telomerase holoenzyme are regulated, how assembly of telomerase is controlled, whether and how posttranslational modifications impact on enzyme access and activity and how many other pathways impinge on control of TERT protein. Additionally, while telomerase is clearly an important factor in progenitor cell biology, uncertainty surrounds the extent to which telomerase is differentially regulated in progenitor cell compartments and the full extent of biological functions of telomerase in processes other than its classical role in telomere maintenance (see below).
Telomerase-knockout mice reveal a critical role for telomeres in progenitor cell function
The identification of mammalian TERC, and later mammalian TERT, allowed the generation of telomerase-knockout mice to test the function of telomerase in development and tissue biology. Surprisingly, each telomerase component was found to be dispensable for life and these telomerase-knockout mice were remarkably normal (60). The survival of the mouse was attributed to the much longer telomeres of laboratory mouse strains. Indeed, continued breeding of first generation (G1) TERC−/− mice to yield successive generations of TERC−/− mice resulted in progressive loss of telomere reserves and ultimately provoked severe degeneration of highly proliferative tissues. G5–G6 TERC mice became infertile, showed high rates of apoptosis in testis and germ cell depletion, exhibited impaired bone marrow function and diminished proliferation of lymphocytes (61). The onset and severity of these defects tracked specifically with the degree of telomere dysfunction. Early generation TERC−/− mice with intact telomeres were grossly unaffected and healthy, whereas late generation TERC−/− mice showed a shortened lifespan and suffered from multi-organ degenerative decline. In the G5–G6 TERC−/− tissues, there was clear cytogenetic evidence of progressive telomere erosion including ‘signal-free’ ends, chromosomal end-to-end fusions and anaphase bridging at mitosis (60–62).
The severe degenerative tissue defects in the late-generation telomerase-knockout mice appear linked to impaired proliferation and increased apoptosis in cycling progenitor cells and to a self-renewal defect in tissue stem cells. These defects are particularly evident in the very high rates of progenitor cell apoptosis in the intestinal crypts with resultant villus atrophy (63,64) as well as in the diminished transplant potential of hematopoietic stem cells from telomerase-deficient mice. Specifically, G1 TERT−/− or G1 TERC−/− hematopoietic stem cells were reduced in their ability to serially reconstitute the blood of irradiated mice compared with wild-type controls (65) and this defect was more severe in late-generation TERC−/− hematopoietic stem cells (66). The defects in proliferation and survival seen in late-generation telomerase-knockout mice appear to be caused by a subset of critically short telomeres, rather than a reduction in mean telomere length. Intercrossing late-generation TERC−/− mice with short telomeres together with wild-type mice with long telomeres yields TERC−/− animals that possess an increased mean telomere length and half the number of short dysfunctional telomeres. These mice exhibit comparable apoptotic and proliferative defects as the original late-generation TERC−/− mice consistent with the view that a subset of dysfunctional telomeres is a key driver of the progenitor cell defects in TERC−/− mice (66–68).
Telomeres, the p53 pathway and aging
Critically short telomeres induce cellular and tissue defects through checkpoint signaling pathways that converge on the p53 tumor suppressor protein, a well-known cellular stress sensor that enforces cell cycle arrest or cell death in response to diverse stimuli including activated oncogenes, DNA damage and hypoxia (69) (Figure 2). In late-generation TERC−/− mice, p53 protein is stabilized and loss of p53 significantly attenuates the cell cycle arrest and cell death phenotypes in TERC−/− tissues (70). Telomere uncapping through disruption of TRF2 also strongly activates p53 in cell culture (71). Indeed, the signaling from uncapped telomeres has been dissected in cultured cells to reveal that as a telomere becomes critically short, or when it becomes acutely uncapped, telomere structure is disrupted in a way that enables its recognition as a classic DNA double-strand break. Telomeres in human fibroblasts approaching senescence, as well as telomeres in cells in which TRF2 function has been disrupted, acquire foci comprised of DNA damage proteins typical of those present at internal double-strand breaks in chromosomes. These telomere dysfunction-induced foci include the histone variant gamma-H2AX, and DNA damage protein p53BP1, the Mre11 complex and phosphorylated ataxia telangiectasia mutated (ATM) (72,73).
The signaling network linked to p53 has been the focus of several studies. Signaling from uncapped telomeres can be mediated by either of the key DNA damage-signaling kinases, ATM or ataxia telangiectasia and Rad3 related. When the double-stranded portion of the telomere complex is disrupted, for example through inhibition of TRF2, the ATM kinase is activated. In contrast, impairing the single-stranded telomere overhang through deletion of POT1 leads to activation of the ataxia telangiectasia and Rad3 related kinase (74,75). The genes downstream of p53 that are responsible for executing the responses to dysfunctional telomeres have been less well studied. However, the cyclin-dependent kinase inhibitor p21 has been shown to participate in the p53-dependent telomere checkpoint response in both human fibroblasts and in TERC−/− mice (66,76).
Late-generation telomerase-knockout mice exhibit a premature aging syndrome characterized by hair graying, shortened survival and impaired responses to acute and chronic stress (63,77). These findings support the hypothesis that telomere shortening detected in aging humans may contribute to many aging phenotypes. From this perspective, an eroded telomere may represent an important, genotoxic signal that can activate DNA damage-signaling pathways capable of accelerating aging (78). Indeed, it is worth noting that reactive oxygen species, which can induce DNA damage, are capable of accelerating telomere erosion that may further amplify checkpoint responses (79). Furthermore, many of the aging phenotypes observed in the telomerase-knockout mouse were also encountered in mice harboring a hyperactivated p53 germline allele (80). Together, these data establish the importance of p53-dependent DNA damage signaling in the aging process. This model implies that factors affecting genome integrity (such as telomeres, reactive oxygen species and DNA repair) and/or modulating p53-dependent responses (such as sirtuins) will impact on the aging phenotype, a view that is clearly supported by the dramatic attenuation of degenerative aging in telomerase-deficient mice null for p53.
Telomere shortening inhibits tumorigenesis in models with intact p53 pathways
To determine whether telomere attrition influences the malignant transformation process, telomerase-knockout mice have been intercrossed with various engineered mouse models of cancer. These model systems have afforded the opportunity to study in vivo the effect of telomere shortening on cancer development in different tissue types and in different genotypic contexts. In one series of studies, the impact of telomerase deficiency was assayed in mice null for the Ink4a/Arf gene, which encodes two distinct tumor suppressor proteins: the cyclin-dependent kinase inhibitor p16 and the p53 activator p19Arf (the Alternative Reading Frame protein encoded within the CDKN2A locus) (81,82). It is worth noting that, although p19ARF regulates p53, ARF’s prime mission is to sense aberrant cell cycle entry or hyperactivated oncogenes by blocking MDM2 (transformed mouse 3T3 double minute 2)-induced degradation of p53, resulting in p53 stabilization (83,84). ARF plays no discernable role in sensing DNA damage (85). Ink4a/Arf mutant mice develop lymphomas and sarcomas with short latency and high penetrance (86); however, in the setting of telomere dysfunction, G4/G5 TERC−/− Ink4a/Arf−/− mice show reduced tumor incidence and increased latency (87,88) and no change in the tumor spectrum. Notably, these G4/G5 TERC−/− Ink4a/Arf−/− tumors showed activation of alternative lengthening of telomeres and frequent mutation of p53, underscoring the importance of telomere maintenance and p53-dependent telomere checkpoint responses in tumorigenesis.
Quantitative analysis of mice with multiple intestinal neoplasia (min) provided further evidence that telomere shortening can dramatically impair tumor development in epithelial cells at specific stages in the malignant transformation process. The adenomatous polyposis coli (APC) tumor suppressor gene is mutated in patients with familial adenomatous polyposis, a disease characterized by hundreds of colonic polyps, and in 80% of sporadic human colon cancers. Mice heterozygous for the APC min mutation (APCmin) develop numerous gastrointestinal adenomas, in similar fashion to patients with APC mutations. However, the lesions in mice do not progress at high frequency to invasive adenocarcinoma, presumably due to the shortened lifespan of APCmin mice, caused by anemia secondary to bleeding from their gastrointestinal adenomas. In this APCmin model, the addition of telomere dysfunction resulted in an increase in early neoplastic lesions probably due to chromosome instability resulting in increased loss of the remaining wild-type APC allele. However, these G4 TERC−/− APCmin mice exhibited a profound suppression of more advanced adenomatous lesions in association with significant intratumoral growth arrest and apoptosis (89). The antitumor effects of telomere dysfunction resulted in dramatically increased survival in G4 TERC−/− APCmin mice by preventing blood loss caused by gastrointestinal tract adenomas. In a transgenic breast cancer model driven by Polyoma Middle T antigen, telomere dysfunction significantly impaired not only tumor latency and penetrance but also reduced metastatic potential (90). In a classical chemical carcinogenesis model, application of 7,12-dimethylbenz(a)anthracene (DMBA) and phorbol esters 12-O-tetradecanoylphorbol 13-acetate induces high rates of skin papilloma formation by inducing activating mutations in the H-RAS gene (91). DMBA and phorbol ester treatment of G5 TERC−/− mice resulted in a 20-fold reduction in papilloma frequency compared with mice with intact telomere function, demonstrating profound impairment of keratinocyte transformation in the setting of telomere dysfunction (92).
Attempts to understand the relative contributions of senescence and apoptosis in tumor suppression by dysfunctional telomeres have found evidence for both. Grieder and colleagues studied the impact of telomere dysfunction in the classical Eμ-Myc B-cell lymphoma model (93). In that setting, dysfunctional telomeres induced a strong cellular senescence phenotype in the emerging lymphomas resulting in potent suppression of lymphomagenesis. In studies employing a p53 allele (R172P) capable of enforcing senescence but defective in apoptosis, dysfunctional telomeres strongly inhibited the spontaneous lymphomas and sarcomas to which p53-mutant mice normally succumb. However, the R172P allele was not as efficient as wild-type p53 in inhibiting chemically induced papillomas, suggesting that senescence was insufficient to block transformation of keratinocytes in this model (94). Together, these findings indicate that senescence and apoptosis responses induced by telomere dysfunction and p53 activation contribute to tumor suppression and that their relative roles probably vary based on cell type and the genetic context of the incipient cancer.
The role of telomerase in the transformation of cultured human cells has also been explored in great detail. In the 1980s, the introduction of oncogenes and transforming viral oncoproteins into primary rodent cells established the requirement of combinations of these cancer genes (95). However, similar cellular transformation experiments with potent oncogene combinations proved to be highly inefficient in the malignant transformation of cultured human cells. However, upon cotransfection with the human TERT gene, defined sets of oncogenes, specifically activated H-RAS, SV40 large T antigen and SV40 small t antigen, resulted in the efficient malignant transformation of human fibroblasts (96). Similar approaches have been used to transform human mammary cells (97), melanocytes (98) and many diverse cell types. In the vast majority of cases, enforced expression of exogenous TERT was required for transformation, although there are some exceptions such as human keratinocytes wherein Inhibitor NFκB α (IκBα) and activated H-RAS can drive transformation to squamous cell carcinoma (98). More recently, lentiviral oncogene transduction of human mammary epithelia cells directly from reduction mammoplasty results in breast adenocarcinoma following short-term culture and immediate orthotopic implantation (99). Importantly, in both the squamous cell carcinoma model and the breast adenocarcinoma model, the emergent cancers show activation of endogenous telomerase activity. These cell culture-based systems are consistent with the seminal observation that the vast majority of spontaneous human cancers express robust levels of telomerase activity (48).
Collectively, the aforementioned mouse models and human cell systems highlight the importance of telomeres in the transformation process. In all four mouse models, telomere dysfunction and the retention of a p53-dependent DNA damage response resulted in impaired growth of tumors. Those tumors that did emerge showed activation of alternative lengthening of telomeres (ALT), mutation of p53 and/or a diminished capacity to achieve a fully malignant state (88,100). Similarly, the data in human cells also strongly support the view that telomerase reactivation is required for malignant transformation and, if TERT is not exogenously supplied, endogenous telomerase is expressed through selection, through direct regulation by the cancer genes employed or through transformation of a rare progenitor cell population expressing telomerase in the culture of human cells.
Telomere dysfunction is a key mutator mechanism driving epithelial carcinogenesis
While the above studies established the importance of telomere maintenance in the acquisition of a fully transformed state, the analysis of mice deficient for both telomerase and p53 expanded our view of how telomere dysfunction impacts on the genesis of malignancies. Mice deficient for p53 have been instrumental in dissecting many functions of this critical tumor suppressor protein in processes of malignant transformation. However, while p53-mutant mice are cancer prone, we noted that their predilection for mesenchymal and lymphoid malignancies, rather than epithelial cancers, did not represent the tumor types most commonly possessing p53 mutations in humans (101,102). Genetic studies designed to assess the interaction of p53 and telomere dysfunction yielded unanticipated insights into the genesis of epithelial cancers, which are the most common malignancies affecting the human population.
The p53 response is known to be critical in sensing DNA damage and executing cellular checkpoint responses to repair or eliminate such cells (103). Reasoning that an eroded telomere might be similar to a DNA damage signal, we assessed the impact of p53 loss on the degenerative phenotypes of mice with dysfunctional telomeres. In G5/G6 TERC−/− p53−/− mice, we showed dramatic and widespread restoration of cellularity and cellular proliferation and reduction in apoptosis across many tissues—findings consistent with deactivation of a p53-dependent DNA damage response (70). However, this cell survival and continued cycling resulted in the accumulation of cells with strikingly abnormal cytogenetic profiles consistent with ongoing telomere dysfunction and its associated chromosomal fusion-breakage cycles upon cell division. In notable contrast to the aforementioned models (intact for DNA damage signaling), G5/G6 TERC−/− p53−/− (or p53+/−) mice showed significant acceleration of the rate of tumor formation and altered the spectrum of tumor types (104). In contrast to the typical tumor spectrum of p53+/− mice with intact telomeres, G5/G6 TERC−/− p53+/− mice developed epithelial cancers, particularly those of the skin, breast and gastrointestinal tract. In fact, by one year of age, all mice in this cohort harbored neoplastic lesions of the colonic epithelium. Thus, telomere shortening and dysfunction that proceeds unchecked in the setting of deactivated p53 serves to accelerate, rather than limit, carcinogenesis and promotes tumor formation in epithelial compartments.
The G5/G6 TERC−/− p53+/− epithelial tumors sustained loss of the wild-type p53 allele and showed frequent anaphase bridge formation consistent with the formation of dicentric chromosomes. On the cytogenetic level, spectral karyotype analysis showed chromosomal end fusions and numerous translocations between non-homologous chromosomes. These translocations were of the non-reciprocal type and were not seen in cancers from p53-deficient mice with long telomeres. Unlike classical balanced translocations, non-reciprocal translocations lead to copy number changes of the genes residing near the translocation breakage-rearrangement point (105). High-resolution copy number analysis, coupled with spectral karyotyping, have since demonstrated that these non-reciprocal translocations are a major driver of regional amplification or deletion of cancer-relevant genes (106,107).
In line with telomere dysfunction as a driver of such genome instability, disruption of TRF2 in primary human fibroblasts has been shown to promote the formation of non-reciprocal translocations (108). Thus, either progressive telomere shortening or acute telomere uncapping can lead to non-reciprocal translocations through a fusion-breakage mechanism. In addition, mice mutant for the telomere-binding protein TPP1 develop adrenocortical dysplasia (acd) and other developmental abnormalities that are largely rescued by p53 deficiency (109,110). Similar to the G5/G6 TERC−/− p53+/− model, TPP1acd/acd p53+/− mice are highly prone to epithelial carcinomas of the skin, which possess non-reciprocal translocations (111).
Thus, impairment of telomere capping function, either through progressive shortening or through immediate uncapping, can drive carcinoma development by destabilizing chromosomes (Figure 2). Importantly, this telomere-related chromosome instability provides a mechanism for regional amplification or deletion, which under biological pressure can select for changes in oncogenes and tumor suppressor genes. Epithelial cancers, as opposed to hematopoietic or mesenchymal malignancies, have long been recognized to require a larger number of mutations to achieve a malignant state (112). In this light, our studies suggest that a major basis for the cross-species difference in tumor spectrum between humans and mice relates to an absence of a key mutator mechanism in mice that enables epithelial carcinogenesis. But what is the evidence that telomere dysfunction is a relevant mechanism driving epithelial cancers in humans?
Telomere dysfunction drives focal amplifications and deletions of cancer-relevant loci: a model for generation of complex human cancer genomes
Analysis of cancer genomes has been aided by advances in array comparative genome hybridization (aCGH), which allows a global view of DNA copy number changes with high resolution. Extensive aCGH studies on human cancers using a variety of platforms have revealed widespread focal amplifications and deletions in human cancers which are now known to harbor oncogenes and tumor suppressor genes, respectively (113). At the same time, for the vast majority of such amplifications and deletions, the relevant cancer genes is not yet known for these loci so the question remains as to whether such loci contain driver cancer genes or are simply the by-product of genome instability and fragile sites. The argument for the cancer relevance for these copy number alterations (CNAs) comes from aCGH analyses of tumors from telomerase-deficient mice. These copy number profiles have revealed genomic events remarkably similar to those of human cancers. Carcinomas from G4–G7 TERC−/− p53+/− mice showed a marked increase in focal CNAs by aCGH, compared with early generation TERC−/− p53+/− tumors with long telomeres (106). Similarly, skin carcinomas from TPP1acd/acd p53+/− tumors exhibited focal CNAs by aCGH (111). Importantly, these abundant focal CNAs occur specifically in tumors driven by dysfunctional telomeres. Conventional mouse models driven by activated oncogenes or loss of tumor suppressor genes exhibited fewer focal CNAs, instead showing genomic profiles characterized by gain or loss of whole chromosomes (114–118). The paucity of focal CNAs in these conventional models is due, in fact, to the long telomeres of mice, which preclude critical telomere shortening and prevent telomere-based crisis in would-be cancer cells.
The dramatic accumulation of focal CNAs in G4–G7 TERC−/− p53+/− tumors prompted us to speculate that chromosome breakage process associated with telomere dysfunction might provide a major mechanism driving amplifications and deletions in human cancer genomes (Figure 2). The fundamental importance of chromosomal breakage is reinforced by analogous genomic changes in cancers from mice deficient for non-homologous end joining and p53. Cells from these mice also sustain breakage events that are associated with CNA accumulation (119). Our hypothesis that telomere-based crisis and deactivation of DNA damage signaling are a major mechanism driving epithelial carcinogenesis in humans, particularly the aged, is consistent with multiple observations including (i) telomere shortening in epithelial compartments with advancing age, (ii) short telomeres in established tumors indicating a period of telomere erosion at some point in tumor development, (iii) massive genome instability at very early stages of the malignant transformation process, yet quelling of instability with further progression (consistent with telomerase reactivation) and (iv) presence of anaphase bridging at a time when genome instability and inactivation of p53 are observed in evolving epithelial cancers.
More specifically, the multiple lines of evidence supporting the importance of telomere-based crisis as a prime mechanism shaping cancer genomes and driving epithelial carcinogenesis in aged humans are as follows. Early experiments relying on Southern blot analysis showed significantly shorter telomeres in breast and colon cancers compared with adjacent normal tissue samples (120–122). The development of fluorescence in situ hybridization-based techniques for telomere length analysis has been a substantial technical advance in this area, allowing analysis of individual cell populations within a cancer. Telomere fluorescence in situ hybridization analysis has confirmed the fact that telomeres are markedly shorter in cancerous epithelium compared with pathologically normal epithelium or stromal cell compartments within the same sample. With this in situ technique, telomeres were shown to be shorter in invasive cancers of the breast, prostate and pancreas, as well as in their pre-invasive counterparts (123–126). Pre-invasive cancers of the bladder, cervix, colon, esophagus and oral cavity were also shown to have very short telomeres in 89% of cases studied. These data indicate that telomeres shorten during epithelial cancer development in humans and that short telomeres are evident at the earliest stages of human carcinogenesis (127,128).
Concomitant with telomere shortening in early stages of cancer, there is evidence that telomeres become dysfunctional during human tumorigenesis. Anaphase bridges, a hallmark of dysfunctional telomeres, are low in adenomas but increase substantially at the adenoma–carcinoma transition. Anaphase bridges become significantly more abundant at the pre-invasive and invasive colon cancer stages (89). In human breast cancer, rates of chromosomal instability peaked at the ductal carcinoma in situ stage. As in the case of colon cancer, anaphase bridges were seen in pre-invasive stages and this evidence for chromosomal instability occurred during a period a dramatic telomere shortening (126). Ductal carcinoma in situ is the stage at which telomerase is upregulated, which is consistent with the idea that telomere stabilization then reduces the rate of chromosomal changes at the invasive and metastatic stages (129). Similarly, upregulation of telomerase occurs at the dysplastic adenoma to carcinoma transition in human colon cancer, facilitating enhanced telomere maintenance and fostering relative stability late in tumor progression (120). These findings suggest that extended proliferation in early stages of tumorigenesis leads to telomere shortening in part because telomerase levels are inadequate at these stages. In fact, diseases of high turnover are associated with precisely this type of telomere shortening, telomere dysfunction and cancer predisposition. Inflammatory bowel disease results in chronic inflammation and increased proliferation in the gastrointestinal tract. Ulcerative colitis is a form of inflammatory bowel disease that affects the colon and is associated with telomere shortening, increased anaphase bridges and a high rate of colon cancer (130). Similarly, the chronic cellular damage that leads to liver cirrhosis results in shortened telomeres, anaphase bridges and a dramatically increased risk of liver cancer (131–134). Thus, conditions of enhanced proliferation and tissue damage in humans accelerate telomere shortening and destabilize chromosomes in a manner similar to that seen in many human carcinomas.
The incorporation of telomere dysfunction in mouse cancer models changes their genomic profile such that focal amplifications and deletions, typically rare in mouse tumors, become abundant. These findings suggest that mouse cancers arising due to dysfunctional telomeres may serve as a model for cancer gene discovery. Furthermore, these mouse tumors provide a powerful platform for sifting through the complexity of the human cancer genome. Thymic lymphomas driven by severe telomere dysfunction in G5 TERC−/− ATM−/ p53+/− mice were analyzed by aCGH and showed abundant focal gene amplifications and deletions. These CNAs from the mouse lymphomas were compared with CNAs in a broad variety of human cancers (107). Remarkably, the majority of recurrent CNAs in these murine lymphomas were syntenic to amplified/deleted regions in human cancers of diverse types. These syntenic changes included alterations in known cancer genes such as NOTCH1, MYB, FBXW7 and PTEN. However, and more strikingly, the gene target of these CNAs is not known in the vast majority of loci indicating that a large number of cancer genes remain to be discovered. Such data are consistent with recent deep multidimensional analysis of human cancers (135). Together, these data indicate that the human genome is widely altered through changes in gene copy number and that telomere shortening during tumorigenesis can serve as a key mutator mechanism.
Extracurricular activities of telomerase in cancer and stem cell biology
These data from telomerase-deficient mice provide important insights into the early stages of tumor development when telomerase levels are low. Together, the data from humans and mice are consistent with a model in which a period of genomic instability early in tumor development is followed by a period of relative stability after telomerase is upregulated. In addition to stabilizing chromosomes, upregulation of telomerase clearly serves to enhance proliferation of cancer cells by maintaining telomere sequences, thereby reducing intolerable levels of chromosomal instability and preventing the DNA damage checkpoint responses that would slow proliferation, induce senescence or trigger cell death.
While mouse and human systems have clearly established a critical role for telomerase, in particular enforced TERT expression, in telomere maintenance and immortalization, mounting evidence has revealed additional activities of TERT that are independent of telomere synthesis. Overexpression experiments in transgenic mice and in human cells have shown that TERT exhibits activities in cellular transformation (136–138), proliferation (139), stem cell biology (140–142), cell survival (143,144) and chromatin regulation (145). An active role for TERT in tissue progenitor cells was revealed in a conditional gain-of-function system in transgenic mice. Expression of TERT under control of a tetracycline-regulated promoter in mouse skin led to a rapid developmental transition in mouse hair follicles, from the resting phase, telogen, to the active phase, anagen. TERT drove this dramatic change by activating quiescent hair follicle stem cells in their niche, the bulge region. These effects of TERT occurred in mice null for TERC and therefore are independent of the canonical role of telomerase in adding telomere repeats (146). Stable overexpression of TERT in mouse skin enhanced the ability of bulge stem cells to enter cell cycle when stimulated with phorbol esters (147). Conditional overexpression of a TERT point mutant lacking reverse transcriptase function led to an identical phenotype of hair growth and bulge stem cell proliferation (148).
These observations indicated that TERT possesses properties of a developmental regulator and indeed TERT exerts these potent developmental effects as a modulator in the Wnt signaling pathway (149). TERT interacts with the chromatin remodeling protein Brg-1, which binds β-catenin, the central transactivator in the Wnt pathway. TERT is recruited to Wnt target gene chromatin in cells stimulated by Wnts and serves to enhance the transcriptional output of the Wnt program in this context (148,149). The ability of TERT to enhance Wnt signaling explains how TERT activates bulge stem cells since overexpression of β-catenin in mouse skin causes a very similar stem cell-activation phenotype (150–152). In addition, TERT was shown to interact with RNA component of mitochondrial RNA processing endoribonuclease, the RNA component of ribonuclease P, and in this context TERT is able to act as an RNA-dependent RNA polymerase. These findings suggest that TERT may amplify small non-coding RNAs and exert other activities through this mechanism (153). Together, these findings indicate that the near universal reactivation of TERT in human cancers may promote tumor progression, proliferation or survival through multiple mechanisms. Upregulation of TERT may yield enhanced telomerase activity and therefore stabilize short telomeres, supporting unlimited cell division. In addition, by enhanceing Wnt signals in human tumors, TERT may support proliferation and survival of cancer cells through more direct mechanisms.
Germline mutations of telomerase components underlie the human genetic disease dyskeratosis congenita
The roles of telomere dysfunction in inhibiting tissue progenitor cell function and in promoting cancer are further supported by findings in a rare genetic disorder dyskeratosis congenita (DC). DC is characterized by a triad of cutaneous findings including oral leukoplakia, skin hyperpigmentation and nail dystrophy. However, the major morbidity of the disease is due to a high incidence of aplastic anemia and pulmonary fibrosis (154,155). In its X-linked form, DC is caused by mutations in dyskerin, which leads to reduced levels of TERC and telomerase activity (43). Autosomal dominant forms are caused by mutations in TERT, TERC or the telomere binding protein TRF1-interacting nuclear factor 2 (156–159). Autosomal recessive forms can be caused by mutations in the dyskerin-associated proteins NHP2 (160) and NOP10 (161). In all cases, telomeres in DC patients are very short, typically shorter than the first percentile of age-matched controls. The severe defects in maintenance of blood, which results in life-threatening aplastic anemia, implicate a defect at the level of the hematopoietic stem cell. Other aspects of the disease including the oral leukoplakia, nail dystrophy and pulmonary disease suggest the possibility that dysfunctional telomeres in this disorder broadly impair function of stem cells and progenitor cells, findings in close agreement with those in telomerase-knockout mice. Patients with DC are prone to myelodysplastic syndrome, a preleukemic condition, and the oral leukoplakia is a precursor lesion for invasive squamous cell carcinoma of the oral mucosa. Thus, telomere shortening syndromes in humans recapitulate the defects in stem cell function, impaired maintenance of proliferative tissues and the tumor predisposition seen in telomerase-knockout mice.
Conclusions and perspectives
Data from mouse and human systems indicate that intact telomere function is crucial for cell proliferation and survival and that telomere shortening can profoundly impact regenerative tissues, particularly in progenitor cell compartments. In the absence of p53—the key mediator of the response to dysfunctional telomeres—cell death and cell cycle arrest responses in progenitor cells are mitigated. This checkpoint defect allows survival in the face of continued telomere shortening, which now leads to rampant chromosomal instability through chromosome fusion-bridge-breakage cycles. Data from mouse and human cancers support the hypothesis that dysfunctional telomeres are a key driver of focal gene amplification and deletion in epithelial tumors. Several factors conspire to enhance telomere shortening in incipient human tumors, including advancing age, proliferation in preneoplastic lesions and diseases of high cellular turnover. Telomerase-knockout mice offer the opportunity to enable cancer gene discovery through analyses of syntenic regions in mouse and human tumors, which will facilitate the discovery of driver mutations, and therefore novel therapeutic targets, in human cancers. Understanding why telomeres shorten so significantly in tumorigenesis and other disease states, despite telomerase expression, is an important goal for the field. Possibilities include insufficient telomerase levels, impaired regulation or the possibility that a subset of telomerase components is limiting. An improved understanding of telomerase function may suggest specific strategies for stabilizing telomeres in aging tissues to prevent cancer and other diseases, and alternatively, more sophisticated approaches for inhibiting telomerase in mature human cancers. The clear requirement for telomere maintenance by telomerase in renewing tissues, coupled with the emerging telomere-independent role for TERT in tissue progenitor cells, indicates that manipulating TERT may indeed fulfill its early promise in regenerative medicine. Continued investigation in the field of telomeres and telomerase will yield new and important insights into how progenitor cells are regulated, how tissues and organisms age and how cancer genomes evolve.
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- acd
adrenocortical dysplasia
- aCGH
array comparative genome hybridization
- APC
adenomatous polyposis coli
- ATM
ataxia telangiectasia mutated
- CNA
copy number alterations
- DC
dyskeratosis congenita
- min
multiple intestinal neoplasia
- POT1
protection of telomeres 1
- scaRNA
small Cajal body-specific RNA
- snoRNA
small nucleolar RNA
- TCAB1
telomerase Cajal body protein 1
- TERC
telomerase RNA component
- TERT
telomerase reverse transcriptase
- TRF1
telomeric repeat binding factor 1
- TRF2
telomeric repeat binding factor 2
References
- 1.Palm W, et al. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 2.Griffith JD, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 3.van Steensel B, et al. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 4.Celli GB, et al. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 5.van Steensel B, et al. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 6.Smogorzewska A, et al. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sfeir A, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim SH, et al. TIN2, a new regulator of telomere length in human cells. Nat. Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 10.Li B, et al. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 11.Bae NS, et al. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol. Cell. 2007;26:323–334. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 12.Sarthy J, et al. Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J. 2009;28:3390–3399. doi: 10.1038/emboj.2009.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baumann P, et al. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 14.Ye JZ, et al. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu D, et al. PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 16.Hockemeyer D, et al. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24:2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hockemeyer D, et al. Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 18.Wu L, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 19.Hockemeyer D, et al. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat. Struct. Mol. Biol. 2007;14:754–761. doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- 20.Loayza D, et al. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;424:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 21.Wang F, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 22.Xin H, et al. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 23.Hayflick L, et al. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 24.Harley CB, et al. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 25.Shay JW, et al. A role for both RB and p53 in the regulation of human cellular senescence. Exp. Cell Res. 1991;196:33–39. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- 26.Hara E, et al. Cooperative effect of antisense-Rb and antisense-p53 oligomers on the extension of life span in human diploid fibroblasts, TIG-1. Biochem. Biophys. Res. Commun. 1991;179:528–534. doi: 10.1016/0006-291x(91)91403-y. [DOI] [PubMed] [Google Scholar]
- 27.Counter CM, et al. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 29.Counter CM, et al. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc. Natl Acad. Sci. USA. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greider CW, et al. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 31.Lingner J, et al. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura TM, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 33.Meyerson M, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 34.Weinrich SL, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell JR, et al. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3’ end. Mol. Cell. Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen JL, et al. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 37.Matera AG, et al. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 38.Cioce M, et al. Cajal bodies: a long history of discovery. Annu. Rev. Cell Dev. Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- 39.Zhu Y, et al. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol. Biol. Cell. 2004;15:81–90. doi: 10.1091/mbc.E03-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jady BE, et al. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol. 2004;164:647–652. doi: 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomlinson RL, et al. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell. 2006;17:955–965. doi: 10.1091/mbc.E05-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cristofari G, et al. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol. Cell. 2007;27:882–889. doi: 10.1016/j.molcel.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 43.Mitchell JR, et al. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 44.Cohen SB, et al. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315:1850–1853. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- 45.Venteicher AS, et al. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Venteicher AS, et al. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tycowski KT, et al. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol. Cell. 2009;34:47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim NW, et al. Specific association of human telomerase activity 965 with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 49.Yui J, et al. Telomerase activity in candidate stem cells from fetal liver and adult bone marrow. Blood. 1998;91:3255–3262. [PubMed] [Google Scholar]
- 50.Liu K, et al. Constitutive and regulated expression of telomerase reverse transcriptase (hTERT) in human lymphocytes. Proc. Natl Acad. Sci. USA. 1999;96:5147–5152. doi: 10.1073/pnas.96.9.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forsyth NR, et al. Telomerase and differentiation in multicellular organisms: turn it off, turn it on, and turn it off again. Differentiation. 2002;69:188–197. doi: 10.1046/j.1432-0436.2002.690412.x. [DOI] [PubMed] [Google Scholar]
- 52.Bachor C, et al. Telomerase is active in normal gastrointestinal mucosa and not up-regulated in precancerous lesions. J. Cancer Res. Clin. Oncol. 1999;125:453–460. doi: 10.1007/s004320050302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramirez RD, et al. Telomerase activity concentrates in the mitotically active segments of human hair follicles. J. Invest. Dermatol. 1997;108:113–117. doi: 10.1111/1523-1747.ep12285654. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 55.Breault DT, et al. Generation of mTert-GFP mice as a model to identify and study tissue progenitor cells. Proc. Natl Acad. Sci. USA. 2008;105:10420–10425. doi: 10.1073/pnas.0804800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greenberg RA, et al. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene. 1999;18:1219–1226. doi: 10.1038/sj.onc.1202669. [DOI] [PubMed] [Google Scholar]
- 57.Wu KJ, et al. Direct activation of TERT transcription by c-MYC. Nat. Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 58.Kyo S, et al. Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci. 2008;99:1528–1538. doi: 10.1111/j.1349-7006.2008.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim JH, et al. Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev. 2005;19:776–781. doi: 10.1101/gad.1289405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blasco MA, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 61.Lee HW, et al. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 62.Hande MP, et al. Telomere length dynamics and chromosomal instability in cells derived from telomerase null mice. J. Cell Biol. 1999;144:589–601. doi: 10.1083/jcb.144.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rudolph KL, et al. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- 64.Wong KK, et al. Telomere dysfunction impairs DNA repair and enhances sensitivity to ionizing radiation. Nat. Genet. 2000;26:85–88. doi: 10.1038/79232. [DOI] [PubMed] [Google Scholar]
- 65.Allsopp RC, et al. Telomerase is required to slow telomere shortening and extend replicative lifespan of HSCs during serial transplantation. Blood. 2003;102:517–520. doi: 10.1182/blood-2002-07-2334. [DOI] [PubMed] [Google Scholar]
- 66.Choudhury AR, et al. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat. Genet. 2007;39:99–105. doi: 10.1038/ng1937. [DOI] [PubMed] [Google Scholar]
- 67.Hemann MT, et al. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 68.Samper E, et al. Restoration of telomerase activity rescues chromosomal instability and premature aging in Terc-/- mice with short telomeres. EMBO Rep. 2001;2:800–807. doi: 10.1093/embo-reports/kve174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krizhanovsky V, et al. Implications of cellular senescence in tissue damage response, tumor suppression, and stem cell biology. Cold Spring Harb. Symp. Quant.Biol. 2008;73:513–522. doi: 10.1101/sqb.2008.73.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chin L, et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 71.Karlseder J, et al. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 72.Takai H, et al. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 73.d'Adda di Fagagna F, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 74.Denchi EL, et al. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 75.Guo X, et al. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 2007;26:4709–4719. doi: 10.1038/sj.emboj.7601893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brown JP, et al. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 77.Herrera E, et al. Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J. 1999;18:2950–2960. doi: 10.1093/emboj/18.11.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garinis GA, et al. DNA damage and ageing: new-age ideas for an age-old problem. Nat. Cell Biol. 2008;10:1241–1247. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ayouaz A, et al. Telomeres: hallmarks of radiosensitivity. Biochimie. 2008;90:60–72. doi: 10.1016/j.biochi.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 80.Tyner SD, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 81.Quelle DE, et al. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 82.Kim WY, et al. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 83.Pomerantz J, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2′s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 84.Kamijo T, et al. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl Acad. Sci. USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kamijo T, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 86.Serrano M, et al. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 87.Greenberg RA, et al. Short dysfunctional telomeres impair tumorigenesis in the INK4a(delta2/3) cancer-prone mouse. Cell. 1999;97:515–525. doi: 10.1016/s0092-8674(00)80761-8. [DOI] [PubMed] [Google Scholar]
- 88.Khoo CM, et al. Ink4a/Arf tumor suppressor does not modulate the degenerative conditions or tumor spectrum of the telomerase-deficient mouse. Proc. Natl Acad. Sci. USA. 2007;104:3931–3936. doi: 10.1073/pnas.0700093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rudolph KL, et al. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat. Genet. 2001;28:155–159. doi: 10.1038/88871. [DOI] [PubMed] [Google Scholar]
- 90.Jaskelioff M, et al. Telomerase deficiency and telomere dysfunction inhibit mammary tumors induced by polyomavirus middle T oncogene. Oncogene. 2009 doi: 10.1038/onc.2009.268. [DOI] [PubMed] [Google Scholar]
- 91.Balmain A, et al. Activation of the mouse cellular Harvey-ras gene in chemically induced benign skin papillomas. Nature. 1984;307:658–660. doi: 10.1038/307658a0. [DOI] [PubMed] [Google Scholar]
- 92.Gonzalez-Suarez E, et al. Telomerase-deficient mice with short telomeres are resistant to skin tumorigenesis. Nat. Genet. 2000;26:114–117. doi: 10.1038/79089. [DOI] [PubMed] [Google Scholar]
- 93.Feldser DM, et al. Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell. 2007;11:461–469. doi: 10.1016/j.ccr.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cosme-Blanco W, et al. Telomere dysfunction suppresses spontaneous tumorigenesis in vivo by initiating p53-dependent cellular senescence. EMBO Rep. 2007;8:497–503. doi: 10.1038/sj.embor.7400937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Land H, et al. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 96.Hahn WC, et al. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 97.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dajee M, et al. NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421:639–643. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- 99.Wu M, et al. Dissecting genetic requirements of human breast tumorigenesis in a tissue transgenic model of human breast cancer in mice. Proc. Natl Acad. Sci. USA. 2009;106:7022–7027. doi: 10.1073/pnas.0811785106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chang S, et al. Telomere-based crisis: functional differences between telomerase activation and ALT in tumor progression. Genes Dev. 2003;17:88–100. doi: 10.1101/gad.1029903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Harvey M, et al. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat. Genet. 1993;5:225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 102.Jacks T, et al. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 103.Kastan MB, et al. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 104.Artandi SE, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 105.Windle B, et al. A central role for chromosome breakage in gene amplification, deletion formation, and amplicon integration. Genes Dev. 1991;5:160–174. doi: 10.1101/gad.5.2.160. [DOI] [PubMed] [Google Scholar]
- 106.O'Hagan RC, et al. Telomere dysfunction provokes regional amplification and deletion in cancer genomes. Cancer Cell. 2002;2:149–155. doi: 10.1016/s1535-6108(02)00094-6. [DOI] [PubMed] [Google Scholar]
- 107.Maser RS, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smogorzewska A, et al. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol. 2005;12:1635–44. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- 109.Keegan CE, et al. Urogenital and caudal dysgenesis in adrenocortical dysplasia (acd) mice is caused by a splicing mutation in a novel telomeric regulator. Hum. Mol. Genet. 2005;14:113–123. doi: 10.1093/hmg/ddi011. [DOI] [PubMed] [Google Scholar]
- 110.Vlangos CN, et al. Caudal regression in adrenocortical dysplasia (acd) mice is caused by telomere dysfunction with subsequent p53-dependent apoptosis. Dev. Biol. 2009;334:418–428. doi: 10.1016/j.ydbio.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Else T, et al. Genetic p53deficiency partially rescues the adrenocortical dysplasia (acd) phenotype at the expense of increased tumorigenesis. Cancer Cell. 2009;15:465–476. doi: 10.1016/j.ccr.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Armitage P, et al. The age distribution of cancer and a multi-stage theory of carcinogensis. Br. J. Cancer. 1954;8:1–12. doi: 10.1038/bjc.1954.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pinkel D, et al. Array comparative genomic hybridization and its applications in cancer. Nat. Genet. 2005;37(suppl.):S11–S17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- 114.Bardeesy N, et al. Dual inactivation of RB and p53 pathways in RAS-induced melanomas. Mol. Cell. Biol. 2001;21:2144–2153. doi: 10.1128/MCB.21.6.2144-2153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Donehower LA, et al. Effects of genetic background on tumorigenesis in p53-deficient mice. Mol. Carcinog. 1995;14:16–22. doi: 10.1002/mc.2940140105. [DOI] [PubMed] [Google Scholar]
- 116.Hodgson G, et al. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat. Genet. 2001;29:459–464. doi: 10.1038/ng771. [DOI] [PubMed] [Google Scholar]
- 117.Albertson DG, et al. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat. Genet. 2000;25:144–146. doi: 10.1038/75985. [DOI] [PubMed] [Google Scholar]
- 118.Sweet-Cordero A, et al. Comparison of gene expression and DNA copy number changes in a murine model of lung cancer. Genes, Chromosomes Cancer. 2006;45:338–348. doi: 10.1002/gcc.20296. [DOI] [PubMed] [Google Scholar]
- 119.Zhu C, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 120.Chadeneau C, et al. Telomerase activity associated with acquisition of malignancy in human colorectal cancer. Cancer Res. 1995;55:2533–2536. [PubMed] [Google Scholar]
- 121.Engelhardt M, et al. Telomerase and telomere length in the development and progression of premalignant lesions to colorectal cancer. Clin. Cancer Res. 1997;3:1931–1941. [PubMed] [Google Scholar]
- 122.Odagiri E, et al. Reduction of telomeric length and c-erbB-2 gene amplification in human breast cancer, fibroadenoma, and gynecomastia. Relationship to histologic grade and clinical parameters. Cancer. 1994;73:2978–2984. doi: 10.1002/1097-0142(19940615)73:12<2978::aid-cncr2820731215>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 123.Meeker AK, et al. Telomere length assessment in human archival tissues: combined telomere fluorescence in situ hybridization and immunostaining. Am. J. Pathol. 2002;160:1259–1268. doi: 10.1016/S0002-9440(10)62553-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Meeker AK, et al. Telomere shortening occurs in subsets of normal breast epithelium as well as in situ and invasive carcinoma. Am. J. Pathol. 2004;164:925–935. doi: 10.1016/S0002-9440(10)63180-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.van Heek NT, et al. Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am. J. Pathol. 2002;161:1541–1547. doi: 10.1016/S0002-9440(10)64432-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chin K, et al. In situ analyses of genome instability in breast cancer. Nat. Genet. 2004;36:984–988. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- 127.Meeker AK, et al. Telomere length abnormalities occur early in the initiation of epithelial carcinogenesis. Clin. Cancer Res. 2004;10:3317–3326. doi: 10.1158/1078-0432.CCR-0984-03. [DOI] [PubMed] [Google Scholar]
- 128.Meeker AK, et al. Recent advances in telomere biology: implications for human cancer. Curr. Opin. Oncol. 2004;16:32–38. doi: 10.1097/00001622-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 129.Shpitz B, et al. Telomerase activity in ductal carcinoma in situ of the breast. Breast Cancer Res. Treat. 1999;58:65–69. doi: 10.1023/a:1006394209922. [DOI] [PubMed] [Google Scholar]
- 130.O'Sullivan JN, et al. Chromosomal instability in ulcerative colitis is related to telomere shortening. Nat. Genet. 2002;32:280–284. doi: 10.1038/ng989. [DOI] [PubMed] [Google Scholar]
- 131.Rudolph KL, et al. Inhibition of experimental liver cirrhosis in mice by telomerase gene delivery. Science. 2000;287:1253–1258. doi: 10.1126/science.287.5456.1253. [DOI] [PubMed] [Google Scholar]
- 132.Miura N, et al. Progressive telomere shortening and telomerase reactivation during hepatocellular carcinogenesis. Cancer Genet. Cytogenet. 1997;93:56–62. doi: 10.1016/s0165-4608(96)00329-9. [DOI] [PubMed] [Google Scholar]
- 133.Urabe Y, et al. Telomere length in human liver diseases. Liver. 1996;16:293–297. doi: 10.1111/j.1600-0676.1996.tb00748.x. [DOI] [PubMed] [Google Scholar]
- 134.Kitada T, et al. Telomere shortening in chronic liver diseases. Biochem. Biophys. Res. Commun. 1995;211:33–39. doi: 10.1006/bbrc.1995.1774. [DOI] [PubMed] [Google Scholar]
- 135.Zheng H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Artandi SE. Telomere shortening and cell fates in mouse models of neoplasia. Trends Mol. Med. 2002;8:44–47. doi: 10.1016/s1471-4914(01)02222-5. [DOI] [PubMed] [Google Scholar]
- 137.Gonzalez-Suarez E, et al. Increased epidermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. EMBO J. 2001;20:2619–2630. doi: 10.1093/emboj/20.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stewart SA, et al. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc. Natl Acad. Sci. USA. 2002;99:12606–12611. doi: 10.1073/pnas.182407599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Smith LL, et al. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat. Cell Biol. 2003;5:474–479. doi: 10.1038/ncb985. [DOI] [PubMed] [Google Scholar]
- 140.Armstrong L, et al. Overexpression of telomerase confers growth advantage, stress resistance, and enhanced differentiation of ESCs toward the hematopoietic lineage. Stem Cells. 2005;23:516–529. doi: 10.1634/stemcells.2004-0269. [DOI] [PubMed] [Google Scholar]
- 141.Imamura S, et al. A non-canonical function of zebrafish telomerase reverse transcriptase is required for developmental hematopoiesis. PLoS One. 2008;3:e3364. doi: 10.1371/journal.pone.0003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang C, et al. A key role for telomerase reverse transcriptase unit in modulating human embryonic stem cell proliferation, cell cycle dynamics, and in vitro differentiation. Stem Cells. 2008;26:850–863. doi: 10.1634/stemcells.2007-0677. [DOI] [PubMed] [Google Scholar]
- 143.Fu W, et al. Anti-apoptotic role of telomerase in pheochromocytoma cells. J. Biol. Chem. 1999;274:7264–7271. doi: 10.1074/jbc.274.11.7264. [DOI] [PubMed] [Google Scholar]
- 144.Lee J, et al. TERT promotes cellular and organismal survival independently of telomerase activity. Oncogene. 2008;27:3754–3760. doi: 10.1038/sj.onc.1211037. [DOI] [PubMed] [Google Scholar]
- 145.Masutomi K, et al. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl Acad. Sci. USA. 2005;102:8222–8227. doi: 10.1073/pnas.0503095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sarin KY, et al. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature. 2005;436:1048–1052. doi: 10.1038/nature03836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Flores I, et al. Effects of telomerase and telomere length on epidermal stem cell behavior. Science. 2005;309:1253–1256. doi: 10.1126/science.1115025. [DOI] [PubMed] [Google Scholar]
- 148.Choi J, et al. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008;4:e10. doi: 10.1371/journal.pgen.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Park JI, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460:66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gat U, et al. De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 151.Lo Celso C, et al. Transient activation of beta-catenin signalling in adult mouse epidermis is sufficient to induce new hair follicles but continuous activation is required to maintain hair follicle tumours. Development. 2004;131:1787–1799. doi: 10.1242/dev.01052. [DOI] [PubMed] [Google Scholar]
- 152.Van Mater D, et al. Transient activation of beta -catenin signaling in cutaneous keratinocytes is sufficient to trigger the active growth phase of the hair cycle in mice. Genes Dev. 2003;17:1219–1224. doi: 10.1101/gad.1076103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Maida Y, et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Savage SA, et al. Dyskeratosis congenita. Hematol. Oncol. Clin. North Am. 2009;23:215–231. doi: 10.1016/j.hoc.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Armanios M. Syndromes of telomere shortening. Annu. Rev. Genomics Hum. Genet. 2009;10:45–61. doi: 10.1146/annurev-genom-082908-150046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vulliamy T, et al. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 157.Armanios M, et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc. Natl Acad. Sci. USA. 2005;102:15960–15964. doi: 10.1073/pnas.0508124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Savage SA, et al. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am. J. Hum. Genet. 2008;82:501–509. doi: 10.1016/j.ajhg.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Walne AJ, et al. TINF2 mutations result in very short telomeres: analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood. 2008;112:3594–3600. doi: 10.1182/blood-2008-05-153445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Vulliamy T, et al. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc. Natl Acad. Sci. USA. 2008;105:8073–8078. doi: 10.1073/pnas.0800042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Walne AJ, et al. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein NOP10. Hum. Mol. Genet. 2007;16:1619–1629. doi: 10.1093/hmg/ddm111. [DOI] [PMC free article] [PubMed] [Google Scholar]