Abstract
During evolution the average chain length of polyisoprenoid glycosyl carrier lipids increased from C55 (prokaryotes) to C75 (yeast) to C95 (mammalian cells). In this study, the ability of the E. coli enzyme, undecaprenyl pyrophosphate synthase (UPPS), to complement the loss of the yeast cis-isoprenyltransferase in the rer2Δ mutant was tested to determine if (55)dolichyl phosphate (Dol-P) could functionally substitute in the protein N-glycosylation pathway for (75)Dol-P, the normal isoprenologue synthesized in S. cerevisiae. First, expression of UPPS in the yeast mutant was found to complement the growth and the hypoglycosylation of carboxypeptidase Y defects suggesting that the (55)polyprenyl-P-P intermediate was converted to (55)Dol-P and that (55)Dol-P could effectively substitute for (75)Dol-P in the biosynthesis and function of Man-P-Dol, Glc-P-Dol and Glc3Man9GlcNAc2-P-P-Dol (mature DLO) in the protein N-glycosylation pathway and glycosylphosphatidylinositol anchor assembly. In support of this conclusion, mutant cells expressing UPPS (1) synthesized (55)Dol-P based on MS analysis, (2) utilized (55)Dol-P to form Man-P-(55)Dol in vitro and in vivo, and (3) synthesized N-linked glycoproteins at virtually normal rates as assessed by metabolic labeling with [3H]mannose. In addition, an N-terminal GFP-tagged construct of UPPS was shown to localize to the endoplasmic reticulum of Chinese hamster ovary cells. Consistent with the synthesis of (55)Dol-P by the transfected cells, microsomes from the transfected cells synthesized the [14C](55)polyprenyl-P-P intermediate when incubated with [14C]isopentenyl pyrophosphate and [3H]Man-P-(55)Dol when incubated with GDP-[3H]Man. These results indicate that (C55)polyisoprenoid chains, significantly shorter than the natural glycosyl carrier lipid, can function in the transbilayer movement of DLOs in the endoplasmic reticulum of yeast and mammalian cells, and that conserved sequences in the cis-isoprenyltransferases are recognized by, yet to be identified, binding partners in the endoplasmic reticulum of mammalian cells.
Keywords: CHO cells, cis-isoprenyltransferase, dolichyl phosphate, undecaprenyl pyrophosphate synthase, yeast mutant
Introduction
Polyisoprenoid glycosyl carrier lipids play key roles in the assembly of cell wall components in bacteria (Lennarz and Scher 1972; Bugg and Brandish 1994) and in protein N-glycosylation, O- and C-mannosylation of glycoproteins and glycosylphosphatidylinositol (GPI) anchors in yeast and mammalian cells (Schenk, Fernandez et al. 2001; Helenius and Aebi 2004). Although the functional significance is not understood, it is well established that the chain length of the polyisoprenoid moieties of the glycosyl carrier lipids has been elongated during evolution (Schenk, Fernandez et al. 2001). The chain length of the polyisoprenoid carrier lipids is determined by the specificity of a family of conserved cis-isoprenyltransferases (cis-IPTases) enzymatically elongating farnesyl pyrophosphate (F-P-P) in prokaryotes (C55) and eukaryotes (C90–100) (Schenk, Fernandez et al. 2001). cDNAs encoding these enzymes in M. luteus (Shimizu et al. 1998), E. coli (Apfel et al. 1999), S. cerevisiae (Sato et al. 1999; Sato et al. 2001; Schenk, Fernandez et al. 2001; Schenk, Rush et al. 2001), Arabidopsis (Oh et al. 2000), Homo sapiens (Endo et al. 2003; Shridas et al. 2003) and Giardia (Grabinska et al. 2010) have been cloned.
A key molecular distinction between the prokaryotic and eukaryotic carrier lipids is that the 2-3 double bond of the α-isoprene unit of the eukaryotic carrier lipid, dolichol, is reduced. In bacteria, undecaprenyl pyrophosphate (Und-P-P), the end-product of the undecaprenyl pyrophosphate synthase (UPPS) reaction sequence, must be cleaved to Und-P to be utilized for lipid intermediate synthesis (Goldman and Strominger 1972). In eukaryotes, the current view is that the (95)polyprenyl pyrophosphate (Poly-P-P) product of the cis-IPTase reactions is dephosphorylated by currently unknown phosphatases, and dolichol is formed when the α-isoprene unit is reduced by a microsomal reductase (Sagami et al. 1993). Dolichol is then converted to dolichyl phosphate (Dol-P), the “activated” form of the carrier lipid, by dolichol kinase (Allen et al. 1978; Burton et al. 1979; Schenk, Fernandez et al. 2001; Fernandez et al. 2002).
Although hydropathy plots of the human cis-IPTase (hCIT) do not predict any membrane-spanning α-helices, the enzyme is localized to the endoplasmic reticulum (ER) (Shridas et al. 2003), raising the possibility that binding partners play a role in the ER association of the mammalian enzyme. Furthermore, the observations that the level of cis-IPTase activity is a rate-controlling factor in Dol-P and lipid intermediate biosynthesis (Crick and Waechter 1994; Crick et al. 1994; Konrad and Merz 1996) emphasize the importance of understanding how the cis-IPTase is regulated and the nature of its association with the ER.
In this study, when the ability of the bacterial uppS gene from E. coli to complement the rer2Δ mutant in S. cerevisiae was tested, the growth and hypoglycosylation defects in the yeast mutant were restored, indicating that (55)Dol-P could functionally substitute for the natural (75)Dol-P in the lipid-mediated protein N-glycosylation pathway. In another related aspect of this investigation, when an N-terminal GFP-tagged construct of bacterial UPPS was expressed in Chinese hamster ovary (CHO) cells, the bacterial enzyme, containing conserved domains present in the mammalian cis-IPTase, was found to localize to the ER in a functional form. The functional significance of shorter chain Dol-Ps being utilized for lipid intermediate biosynthesis by eukaryotic enzymes, the failure of CHO cells to elongate the (55)Poly-P-P end-product formed by UPPS and the possibility that conserved sequences in the bacterial and mammalian cis-IPTases are involved in the recognition of the isoprenyltransferase by potential binding partners in the ER are discussed.
Results
Expression of E. coli uppS complements growth and protein N-glycosylation defects in the yeast mutant, rer2Δ
The uppS gene was expressed in the S. cerevisiae rer2Δ mutant to determine if the bacterial enzyme could correct the growth and hypoglycosylation defects. From the results illustrated in Figure 1, it can be seen that expression of the bacterial cis-IPTase complements the defects in growth (panel A) and the hypoglycosylation of carboxypeptidase Y (CPY) (panel B). Transformation with the empty plasmid did not correct these defects. As shown previously, transformation with the hCIT and the RER2 gene restores normal N-glycosylation of CPY (Figure 1, panel B). These results indicated that S. cerevisiae was capable of utilizing a C55-Poly-P-P for the synthesis of (55)Dol-P and that the shorter polyisoprenoid carrier lipid was utilized for the lipid intermediate pathway for protein N-glycosylation.
Fig. 1.
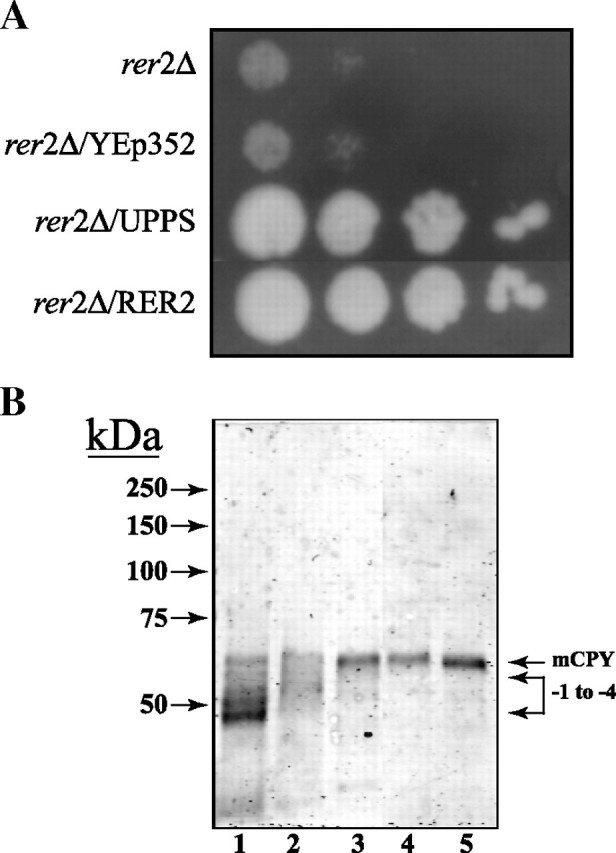
Expression of E. coli UPPS in S. cerevisiae rer2Δ mutant complements defects in growth and protein N-glycosylation. (A) S. cerevisiae rer2Δ cells were transformed with the empty plasmid YEp352, UPPS, or RER2, under transcriptional regulation of the putative yeast RER2 promoter, plated onto YPD agar and cultured for 3 days at 30°C, as described in “Materials and methods”. (B) Extracts from S. cerevisiae cells were separated by SDS-PAGE (8% acrylamide) and analyzed by Western blot with anti-CPY antibody (Zufferey et al. 1995). The positions of mature CPY (mCPY) and hypoglycosylated glycoforms lacking 1-4 N-linked oligosaccharide chains are indicated (-1 to -4). rer2Δ cells (lane 1) were transformed with YEp352 containing either empty vector (lane 2), UPPS (lane 3), hCIT (lane 4), or RER2 (lane 5), under transcriptional regulation of the putative yeast RER2 promoter, cultured in either YPD or ura selection medium (transformed cell lines).
To extend the evidence that expression of UPPS restored protein N-glycosylation, cells expressing the bacterial enzyme were assayed by metabolic labeling with [3H]mannose. The results in Table I show that the rate of incorporation of [3H]mannose into yeast glycoproteins in the rer2Δ mutant is reduced by a factor of 10 relative to control wild-type cells. However, the rate of [3H]mannose incorporation into the glycoprotein fraction of cells expressing UPPS was 62% relative to wild type cells and 82% of the rate calculated for mutant cells complemented with the RER2 gene (Table I). Similar increases in [3H]mannose incorporation into dolichol-linked oligosaccharides (DLOs) were also observed (data not included).
Table I.
Incorporation of [3H]mannose into [3H]Man-glycoprotein by metabolic labeling of various yeast strains
| Yeast strain | [3H-Man]glycoprotein
(dpm × 10 − 6/mg) |
|---|---|
| rer2Δ | 4.3 |
| rer2Δ/YEp352 | 8.6 |
| Wild type (SS328) | 42.6 |
| rer2Δ/UPPS | 26.9 |
| rer2Δ/RER2 | 32.7 |
Cells from the indicated yeast strains were labeled metabolically for 15 min with [3H]mannose and analyzed for incorporation of [3H]mannose into the glycoprotein fraction as described in “Materials and methods”. The data are average values of duplicate analyses and are representative of several experiments.
Expression of bacterial UPPS activity in S. cerevisiae
To confirm that rer2Δ mutant cells expressing the UPPS gene are synthesizing the bacterial length (C55) isoprenol, membrane fractions were prepared from rer2Δ mutant cells following transformation with YEp352 containing various cis-IPTase genes and analyzed for cis-IPTase activity. Table II shows that there is a measurable increase in cis-IPTase activity in membrane fractions from cells expressing the E. coli UPPS gene and the authentic yeast RER2 gene when compared with either untransformed cells or cells transformed with empty plasmid (YEp352). To determine whether the additional cis-IPTase activity is directing the synthesis of a C55 isoprenol, the enzymatic products of the cis-IPTase activities were dephosphorylated and analyzed by thin layer chromatography (TLC) on Baker Si-C18 reverse-phase silica plates as shown in Figure 2. Very low levels of polyprenols were synthesized in membrane fractions from rer2Δ mutant cells (Figure 2, panel A) or rer2Δ mutant cells transformed with an empty vector (Figure 2, panel B), but microsomes from rer2Δ mutant cells transformed with the uppS gene (Figure 2, panel C) synthesize a C55-polyprenol. The rer2Δ mutant transformed with RER2 (Figure 2, panel D) synthesizes the natural C75-polyprenol.
Table II.
cis-IPTase activity in microsomal vesicles from various yeast strains
| Yeast strain |
cis-IPTase activity
(pmol/min/mg) |
|---|---|
| rer2Δ | 3.6 |
| rer2Δ/YEp352 | 3.4 |
| rer2Δ/UPPS | 12.7 |
| rer2Δ/RER2 | 20.8 |
Microsomal fractions were prepared from the indicated yeast strains and assayed for cis-IPTase activity as described in “Materials and methods”. The data are average values of duplicate analyses and are representative of three separate experiments.
Fig. 2.
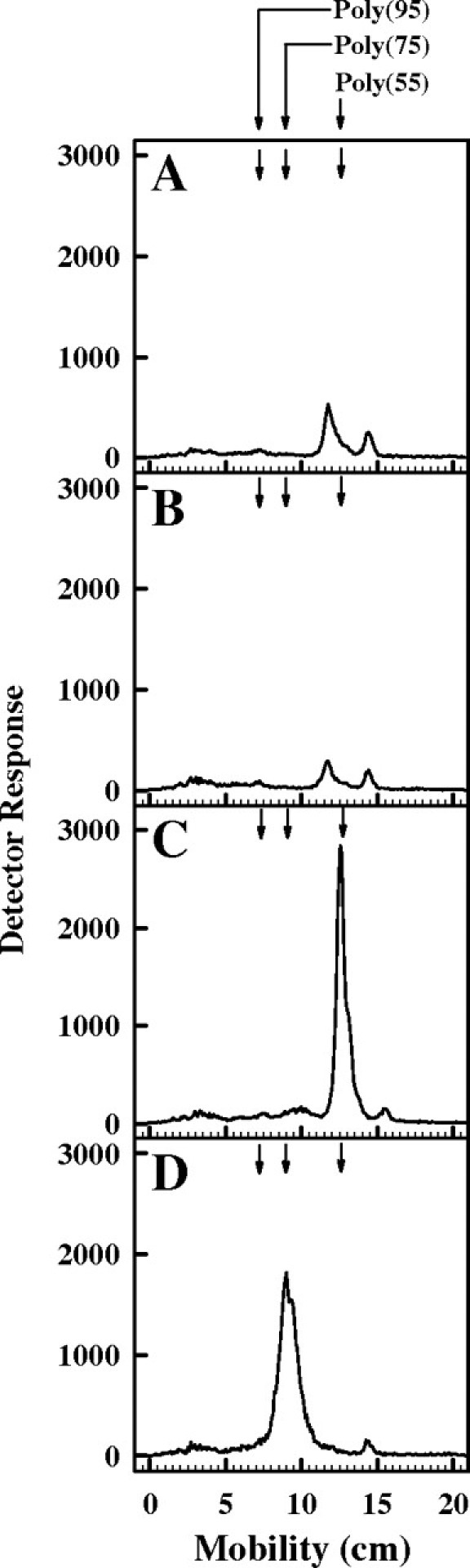
Characterization of products of cis-IPTase activity in various yeast strains. The [14C]Poly-P-P products of the cis-IPTase assays described in Table II from rer2Δ cells (A), cells transformed with the empty plasmid YEp352 (B), UPPS (C), or RER2 (D) were dephosphorylated, resolved by TLC on Baker Si-C18 reverse-phase silica plates developed with acetone/H2O (98:2) and the radioactive zones detected by radiochromatoscanning with a Bioscan AR2000 Imaging Scanner (Bioscan, Washington, DC). The positions of standard polyprenols (C55, C75, and C95; Warszawa, Warsaw, Poland) are indicated by the arrows.
Biosynthesis of Man-P-(55)Dol by microsomes from rer2Δ mutant cells transformed with bacterial UPPS
To determine if rer2Δ mutant cells transformed with UPPS, produced Dol-P and if it was utilized as substrate by Man-P-Dol synthase (MPDS), microsomes were isolated and assayed for MPDS activity. From Table III, it can be seen that the initial rate of Man-P-Dol synthesis in microsomes from rer2Δ mutant cells is substantially reduced when compared to rer2Δ cells expressing the yeast RER2 gene. Transformation with UPPS increased the initial rate of Man-P-Dol synthesis in microsomes when dependent on endogenous Dol-P to slightly less than 50% of the rate in mutant cells transformed with the natural cis-IPTase. When MPDS was assayed in the presence of saturating levels of exogenous Dol-P, comparable levels of MPDS activity were observed for all strains, indicating that the different rates were due to a deficiency of Dol-P rather than MPDS protein.
Table III.
Transfer of [3H]mannose from GDP-[3H]Man into [3H]Man-P-Dol in microsomal fractions from various yeast strains
| Yeast strain | [3H]Man-P-Dol
No addition (pmol/mg) |
[3H]Man-P-Dol
Plus Dol-P (pmol/mg) |
|---|---|---|
| rer2Δ | 1.9 | 218.6 |
| rer2Δ/YEp352 | 1.9 | 197.9 |
| rer2Δ/UPPS | 6.9 | 179 |
| rer2Δ/RER2 | 15.8 | 233.8 |
Microsomal fractions were prepared from the indicated yeast strains and assayed for the transfer of [3H]mannose from GDP-[3H]Man into [3H]Man-P-Dol in the presence and absence of saturating levels of exogenously added Dol-P as described in “Materials and methods”. The results are representative of three separate experiments.
To investigate whether mutant cells transformed with UPPS synthesized (55)Dol-P or the natural substrate, (75)Dol-P, the mannolipid products synthesized in vitro were examined by TLC. The analysis illustrated in Figure 3 shows that microsomes from the rer2Δ and mutant cells transformed with the empty plasmid synthesized virtually no Man-P-Dol (panels A and B). However, cells transformed with UPPS synthesized a mannolipid product that comigrated with the Man-P-(55)Dol standard (panel C). As expected when mutant cells were transformed with RER2, Man-P-(75)Dol was synthesized by microsomal fractions (panel D).
Fig. 3.
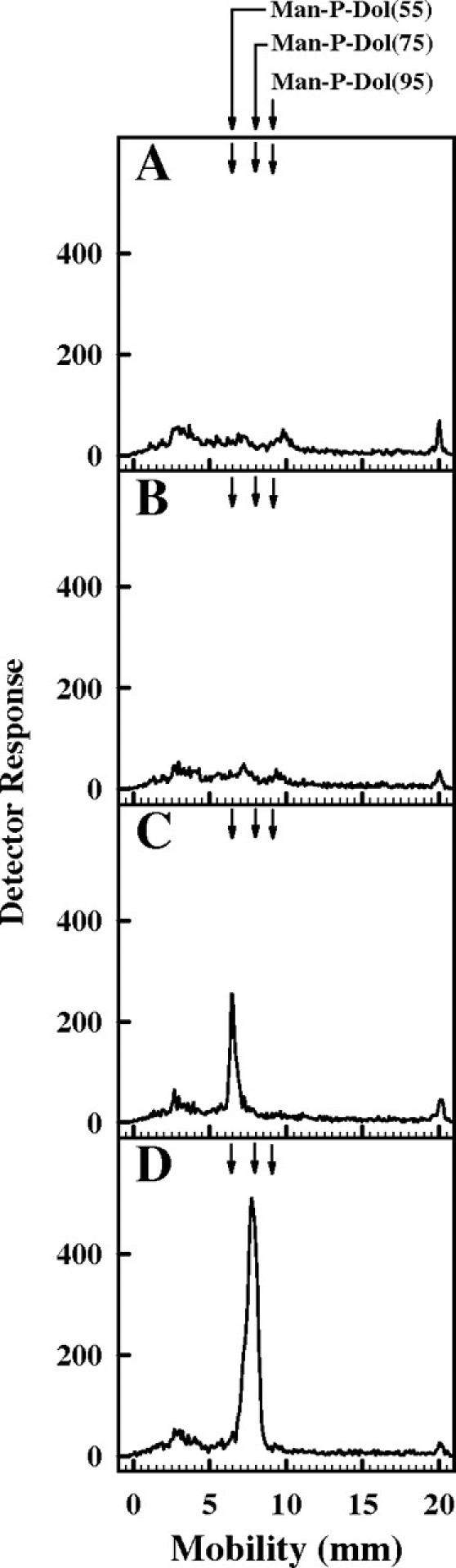
Characterization of products of Man-P-Dol synthase reactions by thin layer chromatography. The [3H]mannolipid products of the Man-P-Dol synthase activities from rer2Δ cells (A), cells transformed with the empty plasmid (B), uppS (C), or RER2 (D) were resolved by TLC on Baker Si-250 silica plates developed in CHCl3/CH3OH/H2O (75:25:4) and detected by a Bioscan AR2000 Imaging Scanner (Bioscan, Washington, DC). The migration of standard Man-P-(55)Dol, Man-P-(75)Dol, and Man-P-(95)Dol is indicated by the arrows.
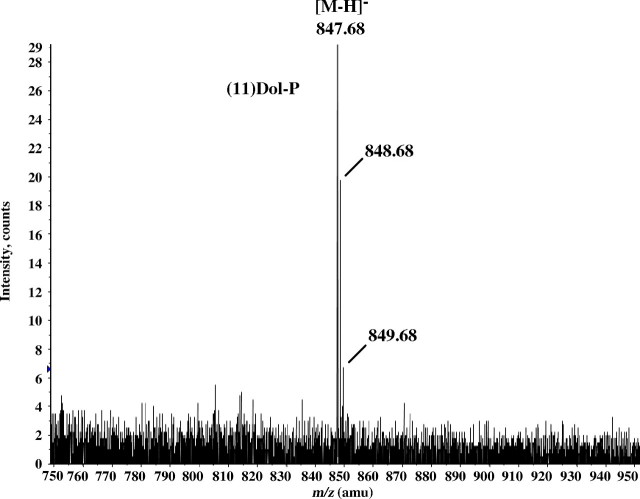
To demonstrate directly that rer2Δ mutant cells expressing E. coli UPPS are forming Dol(55)-P, membrane fractions from the pertinent yeast strains were analyzed by LC-MS. From Figure 4, it can be seen that the only prominent high molecular weight ion eluting in the chromatographic region containing the polyisoprenyl phosphates is m/z = 847.68, corresponding to the molecular ion for Dol(55)-P after loss of a proton. A similar analysis of the unphosphorylated polyprenols in this extract showed primarily m/z = 827.753, arising from [(55)Dol + acetate]−, and a minor peak at m/z = 895.814, corresponding to [(60)Dol + acetate]− (data not shown). Similar analyses from membrane fractions derived from rer2Δ mutant cells or mutant cells transformed with the empty plasmid revealed only molecular ions indicative of very small amounts of Dol-Ps in the (100–110)Dol-P range, presumably due to derepression of endogenous SRT1 activity (Sato et al. 2001; Schenk, Rush et al. 2001). However, these low levels were not sufficient to complement the growth and hypoglycosylation defects.
Fig. 4.
Mass spectrum of (55)Dol-P isolated from microsomes of rer2Δ cells expressing UPPS. Microsomes (1 mg membrane protein) from rer2Δ cells expressing UPPS were incubated in 1 mL of 40% CH3OH containing 3 N KOH at 100°C for 1 h. Following incubation, the reaction mixtures were neutralized with acetic acid and diluted with CHCl3 and CH3OH to a final ratio of CHCl3/CH3OH/H2O (3:2:1) and partitioned. The aqueous phase was discarded, and the organic phase was washed two times with 1/3 vol of CHCl3/CH3OH/0.9% NaCl (3:48:47). The washed organic phase was dried, dissolved in mobile phase A, and analyzed by negative ion LC-MS as described in “Materials and methods”. M/Z = 847.684, corresponding to the molecular ion of (55)Dol-P after loss of a proton [M-1], was the only molecular ion attributable to a polyprenyl phosphate detected in the sample.
Localization of functional GFP-UPPS in the ER of CHO cells
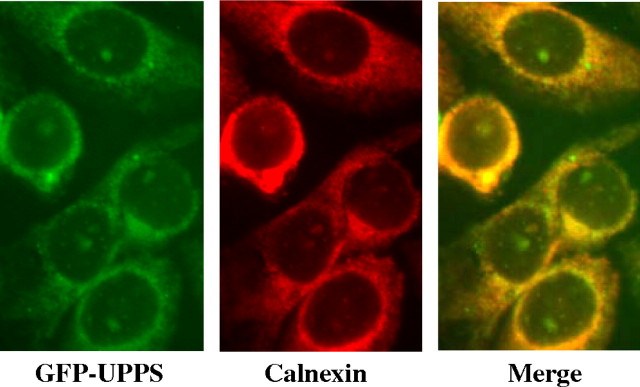
A GFP-tagged construct of E. coli UPPS was also expressed in CHO cells to see if the bacterial protein, containing domains conserved in the hCIT, would localize to the ER in a functional form. From Figure 5 (left panel), it can be seen that when the GFP-tagged UPPS was transfected into CHO cells and detected by immunofluorescence, a reticular pattern characteristic of an ER/nuclear envelope localization was observed. Consistent with an ER localization, this pattern overlapped with the ER marker calnexin (middle and right panels).
Fig. 5.
ER localization of GFP-UPPS expressed in CHO cells by immunofluorescence. CHO cells expressing E. coli UPPS with an N-terminal GFP tag (left, green) were viewed, with and without treatment with rabbit anticalnexin antibody, followed by Texas Red conjugated antirabbit antibody (center, red), with a Nikon Eclipse E600 microscope, as described in “Materials and methods”. The overlap image (right, merge) indicates colocalization of GFP-UPPS with calnexin.
Biosynthesis of Man-P-Dol(55) in microsomes of CHO cells transfected with UPPS
Two enzymological approaches were taken to establish that the bacterial cis-IPTase was associated with the CHO ER in a functional form. First, when microsomal fractions from CHO cells expressing UPPS were assayed for cis-IPTase activity, there was an approximately 20-fold increase in activity relative to microsomal fractions from CHO-K1 cells (Table IV). When the enzymatic products formed by microsomes from CHO-K1 cells were analyzed after strong alkali treatment, as expected (95)Poly-P was the major product. In contrast to this result, when the products formed by microsomes from transfected cells were similarly characterized, Poly-Ps ranging in size from 6 to 11 isoprene units were the major products. Very low levels of (95)Poly-P, if any, were detected. This result suggests that the GFP-tagged UPPS may have displaced a large number of the endogenous cis-IPTases from the ER. It is also plausible that the endogenous cis-IPTase activity is extremely low due to competing for the common substrate, F-P-P, with a relatively high level of UPPS expressed in the transfectants.
Table IV.
cis-IPTase activity in microsomal fractions from control and CHO cells expressing UPPS
| Microsomes from |
cis-IPTase activity
(pmol/min/mg) |
|---|---|
| CHO-K1 | 1.4 |
| CHO/UPPS | 37.7 |
Microsomal fractions were prepared from CHO-K1 and CHO/UPPS cells and assayed for cis-IPTase activity as described in “Materials and methods”. The data are average values of duplicate analyses and are representative of three separate experiments.
Further evidence for the synthesis of (55)Dol-P by the transfected cells was obtained by showing that Man-P-(55)Dol and Man-P-(95)Dol were formed by microsomes in the MPDS reactions, dependent on endogenous Dol-P (Table V). Only [3H]Man-P-(95)Dol was formed by microsomes from untransfected CHO-K1 cells. The presence of a saturated α-isoprene unit in the Man-P-(55/95)Dol products was confirmed by demonstrating that the [3H]mannolipids were stable to treatment with 50% phenol at 68°C for 1 h as expected for Man-P-Dols (Parodi and Quesada-Allue 1982) (Table VI). Under these conditions, > 80% of a Man-P-undecaprenol standard was hydrolyzed.
Table V.
Transfer of [3H]mannose from GDP-[3H]Man into [3H]Man-P-(55/95)Dol in microsomal fractions from CHO-K1 and CHO cells expressing UPPS
| Microsomes from | [3H]Man-P-(55)Dol
(pmol/mg/min) |
[3H]Man-P-(95)Dol
(pmol/mg/min) |
|---|---|---|
| CHO-K1 | n.d. | 1.4 |
| CHO/UPPS | 1.9 | 1.8 |
Microsomal fractions were prepared from either CHO-K1 or CHO/UPPS cells and assayed for the transfer of [3H]mannose from GDP-[3H]Man into [3H]Man-P-(55/95)Dol described in “Materials and methods”. The two mannolipids were resolved as in Figure 3. The data are average values of duplicate analyses and are representative of at least two separate experiments. n. d., not detected.
Table VI.
Effect of aqueous phenol on [3H]mannolipid product synthesized from GDP-[3H]Man in microsomal fractions from CHO-K1 and CHO cells expressing UPPS
| Mannolipid synthesized by | [3H]Mannolipid hydrolyzed
(%) |
|---|---|
| CHO-K1 microsomes | 8.1 |
| CHO/UPPS microsomes | 9.4 |
| Man-P-Dol standard | 7.8 |
| Man-P-Und standard | 81.5 |
[3H]Mannose-labeled mannolipid products were prepared from microsomal fractions from either CHO-K1 or CHO/UPPS cells and incubated at 68°C in 50% phenol as described by Parodi and Quesada-Allue (1982). The data are average values of duplicate analyses.
Discussion
Evolutionary changes in the cis-IPTases catalyzing chain elongation and the corresponding length of polyisoprenoid glycosyl carrier lipids are now well established. To address the question of whether the relatively shorter, C55-carrier lipid in prokaryotic cells, could functionally substitute in S. cerevisiae for the longer chain C75-dolichols, the ability of the E. coli UPPS to complement defects in the yeast rer2Δ mutant was investigated. The results presented in this paper demonstrate that the expression of the bacterial UPPS corrected the defects in growth, protein N-glycosylation, and presumably GPI anchor assembly. In a recent related study, Grabinska et al. (2010) have shown that the cis-IPTase from the protist Giardia lamblia, which synthesizes (55/60)polyprenols, is also capable of complementing the defects in growth, protein N-glycosylation and GPI anchor assembly in the rer2Δ mutant.
Several experimental approaches in this study establish that (55)Dol-P could effectively substitute for the natural (75)Dol-P for the synthesis of Man-P-(55)Dol and the mature DLO. The identification of (55)Dol-P was confirmed by mass spectrometry, and the formation of Man-P-(55)Dol is based on chemical and chromatographic criteria. An interesting enzymological implication from these results is that the end-product, Und-P-P, of the bacterial cis-IPTase reaction sequence is further processed by the putative phosphatases and reductase that convert the natural (75)Poly-P-P to (75)Dol-P in yeast. There is also evidence that there is a relaxed specificity for the chain length of the polyprenyl pyrophosphate intermediates since the C19–22 intermediates formed by the alternative cis-IPTase, SRT1, are converted to (95–110)Dol-Ps (Sato et al. 2001; Schenk, Rush et al. 2001). Induction of SRT1 evidently plays no role in the complementation of the rer2Δ mutant in our studies since no (95–110)Dol-Ps were observed when the mutant was transformed with UPPS. Sato et al. (2001) have previously shown that SRT1 is localized in lipid bodies and induced in the stationary phase. The precise function of the C95–110 dolichols and Dol-Ps remains to be established.
In another aspect of this study, it has been shown that the bacterial UPPS localizes to the ER when transfected into CHO cells in a functional form and is not directed there as unfolded polypeptides. The evidence that UPPS is functional is based on the finding that cis-IPTase activity was elevated in the transfectants relative to control cells and MPDS in microsomes from the transfected cells produced Man-P-(55)Dol. The formation of (55)Dol-P in the cells transfected with UPPS indicates that the Und-P-P is processed by the phosphatases/reductase proposed to convert Poly-P-P to Dol-P. It is possible that the end-product of the chain-elongation stage, in this case (55)Poly-P-P, is “channeled” directly to the putative phosphatase/reductase and then the cytidine triphosphate-mediated kinase to complete Dol-P synthesis, and that the fully unsaturated intermediate cannot diffuse to the active site of the endogenous cis-IPTase to be further elongated.
All of these results raise the intriguing question of why the chain length of polyisoprenoid glycosyl carrier lipids has been elongated from bacterial (C55) to eurkaryotic cells (C75–95) during evolution. Although the width of the bilayer in bacterial cytoplasmic membranes is apparently not substantially larger than the ER in yeast and mammalian cells, it is possible that some biophysical aspects of the eukaryotic ER require longer chains in order for the lipid-linked “activated” glycosyl units to traverse the hydrophobic core of the respective bilayers.
The observation that bacterial UPPS is localized to the ER in CHO cells is also very interesting in view of the fact that the hCIT is localized in the ER even though hydropathy plots do not predict the presence of any membrane-spanning α-helices. Thus, these results suggest that domains in the mammalian cis-IPTases conserved from the bacterial enzymes are involved in the recognition of the mammalian enzymes by potential binding partners in the ER. This proposal is supported by the observation that the GFP-tagged UPPS apparently displaces a large amount of the functional endogenous cis-IPTase from the ER when transfected into CHO-K1 cells. Studies are now in progress to identify potential binding partners and to define the conserved domains recognized by the ER binding partners.
Materials and methods
Materials
[2-3H]Mannose (60 Ci/mmol) and [14C]isopentenyl pyrophosphate (I-P-P) (55 mCi/mmol) were obtained from American Radiolabeled Chemicals (St. Louis, MO). GDP-[3H]mannose was prepared enzymatically as described previously (Rush et al. 1993). YEp352 (yeast shuttle vector), strain YG932 (yeast rer2Δ mutant), and the cDNA for yeast RER2 were gifts from Drs. M. Aebi (ETH, Zurich, Switzerland) and F. Fernandez (GlycoVaxyn, Schlieren, Switzerland). Vector PEX-T21 containing the E. coli uppS sequence was a generous gift from Steffi Balada (GlycoVaxyn, Schlieren, Switzerland). Anti-CPY antiserum was purchased from Molecular Probes (Invitrogen, Eugene, OR). Undecaprenol and purified individual isoprenologues of (55)Dol, (75)Dol, and (95)Dol were obtained from Warzawa (Warsaw, Poland) and phosphorylated chemically as described by Danilov et al. (1989) and Danilov and Chojnacki (1981). [3H]Man-P-(55)Dol, [3H]Man-P-(75)Dol, and [3H]Man-P-(95)Dol were prepared enzymatically using partially purified Man-P-Und synthase from M. luteus and purified as described previously (Rush et al. 1993).
Molecular cloning of cis-IPTases, construction of YEp352 shuttle vectors and expression in S. cerevisiae
The coding sequence of the E. coli uppS gene was amplified by polymerase chain reaction (PCR) using the two primers (forward) 5′-TTCCCGGG ATGTTGTCTGCTACTCAACCAC-3′ and (reverse) 5′-TTCAAGCTTCAA TGATGATGATGATGATGGGCTGTTTCATCACCGGC-3′ with Xma1 and HindIII restriction sites (underlined) and E. coli genomic DNA as template. The reverse primer is designed to introduce a His6-tag at the carboxy terminus of the protein. The PCR product was digested with Xma1 and HindIII and ligated into similarly prepared YEp352, containing the putative yeast RER2 promoter (the 250 bp region immediately upstream of the yeast RER2 gene) described previously (Shridas et al. 2003). The structure of the resulting plasmid with E. coli uppS downstream of the putative yeast promoter sequence was verified by sequence analysis (Eurofins MWG Biotech, info@Eurofins.com). Yeast strains were transformed as described by Schiestl and Gietz (1989) and transformants were identified by uracil selection.
Preparation of pEGFP-C1:UPPS
The DNA sequence encoding the E. coli UPPS gene was amplified by PCR using primers (forward) 5′-TTGAATTCGATGTTGTCTGCTACTCAACCA-3′ and (reverse) 5′-TTGTCGACTAGGCTGTTTCATCACCGGGC-3′ containing EcoR1 and SalI restriction sites (underlined). Following digestion with the appropriate restriction enzymes, the insert was purified and ligated into similarly digested pEGFP-C1 to produce an expression plasmid encoding the bacterial cis-IPTase, UPPS, containing an N-terminal GFP tag. The structure and sequence of the expected plasmid were verified by direct sequencing by Davis Sequencing (Davis, CA).
Yeast culture and preparation of yeast microsomes
Yeast strains SS328 (MATα ade2-101 ura3-52 his3Δ200 lys2-801) and YG932 (rer2Δ mutant) (MATα rer2Δ::kanMX4 ade2-101 ura3-52 his3Δ200 lys2-801) were obtained from Dr. M. Aebi (ETH, Zurich, Switzerland). The strains were cultured at 30°C in 1% yeast extract (Becton, Dickinson and Co., Sparks, MD), 2% Bacto-Peptone (Becton, Dickinson and Co., Sparks, MD), and 2% glucose (yeast peptone dextrose, YPD). Yeast strains transformed with YEp352 (Hill et al. 1986) and its derivatives were grown in 0.67% yeast nitrogen base, 50 mM sodium succinate pH 5.0, 2% glucose and all necessary auxotrophic requirements, except for uracil.
Yeast microsomes were prepared from logarithmically growing cell cultures by homogenization (tight-fitting Dounce homogenizer) following Lyticase (Sigma-Aldrich, St. Louis, MO) treatment as described previously (Fernandez et al. 2001).
CHO cell culture, transfection with pEGFP-C1/uppS and preparation of crude microsomal fractions
CHO cells were maintained at 37°C in 5% CO2 in DMEM/F-12 50/50 with glutamine and 15 mM HEPES (Mediatech, Manassas, VA), supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 μg/mL streptomycin. CHO cells were transfected at 95% confluence using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) following the manufacturer’s protocol. Twenty-four hours following transfection, cell monolayers were scraped into 0.01 M Tris–HCl, pH 7.4, 0.25 M sucrose, 1 mM ethylenediaminetetraacetic acid (EDTA) (buffer A) and homogenized with 20 passes with a tight-fitting Dounce homogenizer. The homogenates were centrifuged at 1000 × g, 10 min to remove unbroken cells and debris. Crude microsomes were collected by sedimentation at 100,000 × g, 10 min in a TL-100A tabletop ultracentrifuge and resuspended in buffer A.
Detection of UPPS in CHO cells by immunofluorescence
To localize GFP-tagged UPPS by immunofluorescence, CHO cells were grown on 12-mm coverslips to 30–45% confluence and transfected with pEGFP-C1/UPPS, as described above. After 24 h in culture, the coverslips were washed in PBS and fixed with cold (− 60°C) methanol for 6 min, followed by blocking with 5% FBS in Tris-buffered saline for 20 min at room temperature. The fixed coverslips were incubated with rabbit anticalnexin antibody (0.050 mL of 1:100 diluted) for 1 h and washed 3 × with blocking solution. After washing, the coverslips were probed with secondary antibody (Texas-Red conjugated sheep antirabbit IgG, Amersham) for 1 h and washed 3 × with blocking solution followed by PBS. Coverslips were mounted on glass slides with Vectashield mounting solution (Vector Laboratories, Burlingame, CA). The slides were viewed with a Nikon Eclipse E600 microscope.
In vitro assays of MPDS activity
Reaction mixtures for MPDS contained microsomes from yeast or CHO cells (0.25 mg protein), 50 mM Tris–HCl, pH 8, 5 mM AMP, 12.5 mM MgCl2, 1 mM sodium orthovanadate, and 10 μM GDP-[3H]Man (1000 dpm/pmol) in a total volume of 0.05 mL. Samples were incubated with GDP-[3H]Man for 1 min, following a 5-min pre-incubation, at 30°C and the reaction terminated by the addition of 20 vols CHCl3/CH3OH (2:1). The incorporation of [3H]mannose into [3H]Man-P-Dol was assayed as described previously (Waechter et al. 1976). Where indicated, Dol-P (100 μM) was added as a dispersion in 1% CHAPS, with a final CHAPS concentration of 0.1%.
Metabolic labeling of S. cerevisiae
Cell cultures were labeled metabolically by incubation with [3H]mannose (5 μCi/mL) at a cell density of 200 OD600 units/mL in ura selection medium containing 0.1% glucose and 25 mg/mL uracil. Following incubation for 15 min at 30°C, the cell cultures were quickly chilled by the addition of crushed ice (0.5 gm/mL). The cells were recovered by centrifugation (1000 × g, 10 min), resuspended in 5 mL ice-cold PBS, and sedimented again. Incorporation of [3H]mannose into [3H-Man]glycoprotein was determined as described previously (Waechter et al. 1983).
Assays for cis-IPTase activity in yeast and CHO microsomal fractions
Reaction mixtures for the assay of yeast cis-IPTase contained 25 mM HEPES-NaOH, pH 8.5, 5 mM MgCl2, 0.5 mM sodium orthovanadate, 0.1 mM F-P-P, microsomes (0.1–0.5 mg membrane protein), and 20 μM [14C]I-P-P (121 dpm/pmol) in a total volume of 0.05 mL. Following incubation for 15 min at 30°C, the reactions were terminated by the addition of 20 vols CHCl3/CH3OH (2:1). The enzymatic synthesis of labeled polyprenyl-P-P products was assayed basically as described previously (Shridas et al. 2003).
Reaction mixtures assaying cis-IPTase activity in CHO microsomes contained crude microsomes (30–40 μg protein), 25 mM Tris–HCl (pH 8.5), 5 mM MgCl2, 1.25 mM DTT, 2.5 mM sodium orthovanadate, 10 μM squalestatin, 100 μM F-P-P, 0.35% Triton X-100, and 45 μM [1-14C]I-P-P (55 mCi/mmol) in a total volume of 0.050 mL. After incubation at 37°C for 1 h, the reaction was terminated by the addition of 1 mL CHCl3/CH3OH (2:1). The enzymatic synthesis of labeled polyprenyl-P-P products was then assayed basically as described previously (Crick et al. 1991).
Characterization of products formed by cis-IPtase activities in microsomal fractions from yeast and CHO cells
To determine the chain length of the products formed in the yeast cis-IPTase reactions in vitro, the [14C]-labeled enzymatic products were dispersed by brief sonication (Kontes Micro Ultrasonic Cell Disruptor at 40% full power) in 1% Triton X-100 and dephosphorylated by overnight incubation (37°C) with calf brain microsomes (0.2 mg, in 50 mM Tris–Cl, pH 7.4, 2.5 mM EDTA, 0.5% Triton X-100 in a total volume of 0.1 mL). Following incubation, the [14C]-labeled products were recovered by organic extraction and Folch partition. The organic phase was dried under nitrogen, redissolved in hexane and chromatographed on a 1 gm column of Bio-Sil HA (Bio-Rad Laboratories, Richmond, CA) equilibrated in hexane. The Bio-Sil HA column was washed with 5 column volumes of hexane and then eluted with 1 mL portions of hexane/diethyl ether (9:1). The dephosphorylated [14C]polyprenols eluting in fractions 2–4 were pooled, dried under nitrogen, redissolved in a small volume of hexane, and resolved by chromatography on Baker Si-C18 TLC plates in acetone/water (98:2). The [14C]-polyprenols were detected by radiochromatoscanning with a Bioscan AR2000 Imaging Scanner (Bioscan, Washington, DC). The zones corresponding to polyprenol standards (obtained from Warszawa, Warsaw, Poland) were located by exposure to iodine vapors.
To determine the chain length of the products formed in the cis-IPTase reactions with CHO microsomes in vitro, the [14C]Poly-P-P products were converted to Poly-P by strong alkaline hydrolysis (70% methanol containing 3.6 M KOH, 2 h, 100°C). Following alkaline hydrolysis, the reactions were diluted with CHCl3 and H2O and partitioned, as described previously (Crick et al. 1991), and analyzed by TLC on Silica gel G TLC plates in CHCl3/CH3OH/H2O (75:25:4). The [14C]Poly-Ps and Poly-P standards were detected as described above.
Analysis of yeast cells and microsomal fractions for dolichol and Dol-P by NP-LC/MS methods
Normal phase LC-ESI/MS of lipids was performed using an Agilent 1200 Quaternary LC system coupled to a QSTAR XL quadrupole time-of-flight tandem mass spectrometer (Applied Biosystems, Foster City, CA). An Ascentis® Si HPLC column (5 μm, 25 cm × 2.1 mm) was used. Mobile phase A consisted of chloroform/methanol/aqueous ammonium hydroxide (800:195:5, v/v/v). Mobile phase B consisted of chloroform/methanol/water/ aqueous ammonium hydroxide (60:34:5:0.5, v/v/v/v). Mobile phase C consisted of chloroform/methanol/water/aqueous ammonium hydroxide (45:45:9.5:0.5, v/v/v/v). The elution program consisted of the following: 100% mobile phase A was held isocratically for 2 min and then linearly increased to 100% mobile phase B over 14 min and held at 100% B for 11 min. The LC gradient was then changed to 100% mobile phase C over 3 min and held at 100% C for 3 min, and finally returned to 100% A over 0.5 min and held at 100% A for 5 min. The total LC flow rate was 300 μL/min. The postcolumn splitter diverted ~ 10% of the LC flow to the ESI source of the Q-Star XL mass spectrometer, with MS settings as follows: IS = − 4500 V, CUR = 20 psi, GS1 = 20 psi, DP = − 55 V, and FP = − 150 V. Nitrogen was used as the collision gas for MS/MS experiments. Data acquisition and analysis were performed using the instrument’s Analyst QS software.
Analytical procedures
Protein concentrations were determined using the bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL) following precipitation of membrane proteins with deoxycholate and trichloroacetic acid according to the Pierce Biotechnology bulletin, “Eliminate interfering substances from samples for BCA protein assay.” Samples were analyzed for radioactivity by scintillation spectrometry in a Packard Tri-Carb 2100TR liquid scintillation spectrometer following the addition of 0.5 mL 1% SDS and 4 mL Econosafe Economical Biodegradable Counting Cocktail (Research Products International, Corp., Mount Prospect, IL).
Funding
This work was supported by National Institutes of Health (NIH) grant GM36035 (CJW), and the mass spectrometry facility in the Department of Biochemistry of the Duke University Medical Center and Dr. Ziqiang Guan are supported by the LIPID MAPS Large Scale Collaborative grant number GM-069338 from NIH.
Acknowledgments
The authors thank Steffi Balada (GlycoVaxyn, Schlieren, Switzerland) for providing the plasmid containing the E. coli UPPS.
Conflict of interest statement
None declared.
Abbreviations
- BCA
bicinchoninic acid
- CHO
Chinese hamster ovary
- cis-IPTase
cis-isoprenyltransferase
- CPY
carboxypeptidase Y
- DLO
dolichol-linked oligosaccharides, Glc3Man9GlcNAc2-P-P-dolichol, mature DLO
- Dol-P
dolichyl monophosphate
- EDTA
ethylenediaminetetraacetic acid
- ER
endoplasmic reticulum
- FBS
fetal bovine serum
- F-P-P
farnesyl pyrophosphate
- GPI
glycosylphosphatidylinositol
- hCIT
human cis-isoprenyltransferase
- I-P-P
isopentenyl pyrophosphate
- MPDS
Man-P-Dol synthase
- PCR
polymerase chain reaction
- Poly-P-P
fully unsaturated polyprenyl pyrophosphate
- Und-P
undecaprenyl monophosphate
- Und-P-P,
undecaprenyl pyrophosphate
- UPPS
undecaprenyl pyrophosphate synthase
- YPD
yeast peptone dextrose
References
- Allen CM, Jr, Kalin JR, Sack J, Verizzo D. CTP-dependent dolichol phosphorylation by mammalian cell homogenates. Biochemistry. 1978;17:5020–5026. doi: 10.1021/bi00616a025. [DOI] [PubMed] [Google Scholar]
- Apfel CM, Takacs M, Fountoulakis M, Stieger M, Keck W. Use of genomics to identify bacterial undecaprenyl pyrophosphate synthetase: Cloning, expression, and characterization of the essential uppS gene. J Bacteriol. 1999;181:483–492. doi: 10.1128/jb.181.2.483-492.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugg TD, Brandish PE. From peptidoglycan to glycoproteins: Common features of lipid-linked oligosaccharide biosynthesis. FEMS Microbiol Lett. 1994;119:255–262. doi: 10.1111/j.1574-6968.1994.tb06898.x. [DOI] [PubMed] [Google Scholar]
- Burda P, Aebi M. The dolichol pathway of N-linked glycosylation. Biochim Biophys Acta. 1999;1426:239–257. doi: 10.1016/s0304-4165(98)00127-5. [DOI] [PubMed] [Google Scholar]
- Burton WA, Scher MG, Waechter CJ. Enzymatic phosphorylation of dolichol in central nervous tissue. J Biol Chem. 1979;254:7129–7136. [PubMed] [Google Scholar]
- Crick DC, Waechter CJ. Long-chain cis-isoprenyltransferase activity is induced early in the developmental program for protein N-glycosylation in embryonic rat brain cells. J Neurochem. 1994;62:247–256. doi: 10.1046/j.1471-4159.1994.62010247.x. [DOI] [PubMed] [Google Scholar]
- Crick DC, Rush JS, Waechter CJ. Characterization and localization of a long-chain isoprenyltransferase activity in porcine brain: Proposed role in the biosynthesis of dolichyl phosphate. J Neurochem. 1991;57:1354–1362. doi: 10.1111/j.1471-4159.1991.tb08301.x. [DOI] [PubMed] [Google Scholar]
- Crick DC, Scocca JR, Rush JS, Frank DW, Krag SS, Waechter CJ. Induction of dolichyl-saccharide intermediate biosynthesis corresponds to increased long chain cis-isoprenyltransferase activity during the mitogenic response in mouse B cells. J Biol Chem. 1994;269:10559–10565. [PubMed] [Google Scholar]
- Danilov LL, Chojnacki T. A simple procedure for preparing Dolichyl monophosphate by the use of POCl3. FEBS Lett. 1981;131:310–312. [Google Scholar]
- Danilov LL, Druzhinina TN, Nalinchuk NA, Maltev SD, Shibaev VN. Polyprenyl phosphates: Synthesis and structure-activity relationship for a biosynthetic system of Salmonella anatum O-specific polysaccharide. Chem Phys Lipids. 1989;51:191–203. doi: 10.1016/0009-3084(89)90006-6. [DOI] [PubMed] [Google Scholar]
- Endo S, Zhang YW, Takahashi S, Koyama T. Identification of human dehydrodolichyl diphosphate synthase gene. Biochim Biophys Acta. 2003;1625:291–295. doi: 10.1016/s0167-4781(02)00628-0. [DOI] [PubMed] [Google Scholar]
- Fernandez F, Rush JS, Toke DA, Han G-S, Quinn J, Carman GMY, Voelker DR, Aebi M, Waechter CJ. The CWH8 gene encodes a dolichyl pyrophosphatase with a luminally oriented active site in the endoplasmic reticulum of Sacchar\omyces cerevisiae. J Biol Chem. 2001;276:31344–41464. doi: 10.1074/jbc.M105544200. [DOI] [PubMed] [Google Scholar]
- Fernandez F, Shridas P, Aebi M, Jiang S-M, Waechter CJ. Cloning and expression of a cDNA that encodes dolichol kinase in human brain. FASEB J. 2002 doi: 10.1093/glycob/cwf068. 16 Abstract 674.4. [DOI] [PubMed] [Google Scholar]
- Fujihashi M, Zhang Y-W, Higuchi Y, Li X-Y, Koyama T, Miki K. Crystal structure of cis-prenyl chain elongating enzyme, undecaprenyl diphosphate synthase. Proc Natl Acad Sci USA. 2001;98:4337–4342. doi: 10.1073/pnas.071514398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman R, Strominger JL. Purification and properties of C 55-isoprenylpyrophosphate phosphatase from Micrococcus lysodeikticus. J Biol Chem. 1972;247:5116–5122. [PubMed] [Google Scholar]
- Grabinska KA, Cui J, Chatterjee A, Guan Z, Raetz CR, Robbins PW, Samuelson J. Molecular characterization of the cis-prenyltransferase of Giardia lamblia. Glycobiology. 2010;20(7):824–832. doi: 10.1093/glycob/cwq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Herscovics A, Orlean P. Glycoprotein biosynthesis in yeast. FASEB J. 1993;7:540–550. doi: 10.1096/fasebj.7.6.8472892. [DOI] [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner TJ, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Konrad M, Merz WE. Long-term effect of cyclic AMP on N-glycosylation is caused by an increase in the activity of the cis-prenyltransferase. Biochem J. 1996;316:575–581. doi: 10.1042/bj3160575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennarz WJ, Scher MG. Metabolism and function of polyisoprenol sugar intermediates in membrane-associated reactions. Biochim Biophys Acta. 1972;265:417–441. doi: 10.1016/0304-4157(72)90015-9. [DOI] [PubMed] [Google Scholar]
- Ogura K, Koyama T. Enzymatic aspects of isoprenoid chain elongation. Chem Rev. 1998;98:1263–1276. doi: 10.1021/cr9600464. [DOI] [PubMed] [Google Scholar]
- Oh SK, Han KH, Ryu SB, Kang H. Molecular cloning, expression, and functional analysis of a cis-prenyltransferase from Arabidopsis thaliana. Implications in rubber biosynthesis. J Biol Chem. 2000;275:18482–18488. doi: 10.1074/jbc.M002000200. [DOI] [PubMed] [Google Scholar]
- Parodi AJ, Quesada-Allue LA. Protein glycosylation in Trypanosoma cruzi. 1. Characterization of dolichol-bound monosaccharides and oligosaccharides synthesized “in vivo”. J Biol Chem. 1982;257:7637–7640. [PubMed] [Google Scholar]
- Rush JS, Shelling JG, Zingg NS, Ray PH, Waechter CJ. Mannosylphosphoryldolichol-mediated reactions in oligosaccharide-P-P-dolichol biosynthesis. Recognition of the saturated alpha-isoprene unit of the mannosyl donor by pig brain mannosyltransferases. J Biol Chem. 1993;268:13110–13117. [PubMed] [Google Scholar]
- Sagami H, Kurisake A, Ogura K. Formation of dolichol from dehydrodolichol is catalyzed by NADPH-dependent reductase localised in microsomes of rat liver. J Biol Chem. 1993;268:10109–10113. [PubMed] [Google Scholar]
- Sato M, Sato K, Nishikawa S, Hirata A, Kato J, Nakano A. The yeast RER2 gene, identified by endoplasmic reticulum protein localization mutations, encodes cis-prenyltransferase, a key enzyme in dolichol synthesis. Mol Cell Biol. 1999;19:471–483. doi: 10.1128/mcb.19.1.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Fujisaki S, Sato K, Nishimura Y, Nakano A. Yeast Saccharomyces cerevisiae has two cis-prenyltransferases with different properties and localizations. Implication for their distinct physiological roles in dolichol synthesis. Genes Cells. 2001;6:495–506. doi: 10.1046/j.1365-2443.2001.00438.x. [DOI] [PubMed] [Google Scholar]
- Schenk B, Fernandez F, Waechter CJ. The ins(ide) and outs(ide) of dolichyl phosphate biosynthesis and recycling in the endoplasmic reticulum. Glycobiology. 2001;11:R61–R70. doi: 10.1093/glycob/11.5.61r. [DOI] [PubMed] [Google Scholar]
- Schenk B, Rush JS, Waechter CJ, Aebi M. An alternative cis-isoprenyltransferase activity in yeast that produces polyisoprenols with chain lengths similar to mammalian dolichols. Glycobiology. 2001;11:89–98. doi: 10.1093/glycob/11.1.89. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Shimizu N, Koyama T, Ogura K. Molecular cloning, expression, and purification of undecaprenyl diphosphate synthase. No sequence similarity between E- and Z-prenyl diphosphate synthases. J Biol Chem. 1998;273:19476–19481. doi: 10.1074/jbc.273.31.19476. [DOI] [PubMed] [Google Scholar]
- Shridas P, Rush JS, Waechter CJ. Identification and characterization of a cDNA encoding a long-chain cis-isoprenyltranferase involved in dolichyl monophosphate biosynthesis in the ER of brain cells. Biochem Biophys Res Commun. 2003;312:1349–1356. doi: 10.1016/j.bbrc.2003.11.065. [DOI] [PubMed] [Google Scholar]
- Waechter CJ, Kennedy JL, Harford JB. Lipid intermediates involved in the assembly of membrane-associated glycoproteins in calf brain white matter. Arch Biochem Biophys. 1976;174:726–737. doi: 10.1016/0003-9861(76)90403-3. [DOI] [PubMed] [Google Scholar]
- Waechter CJ, Schmidt JW, Catterall WA. Glycosylation is required for maintenance of functional sodium channels in neuroblastoma cells. J Biol Chem. 1983;258:5117–5123. [PubMed] [Google Scholar]
- Zufferey R, Knauer R, Burda P, Stagljar I, te Heesen S, Lehle L, Aebi M. STT3, a highly conserved protein required for yeast oligosaccharyl transferase activity in vivo. EMBO J. 1995;14:4949–4960. doi: 10.1002/j.1460-2075.1995.tb00178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]