Abstract
Signal transduction along the Ras/MAPK pathway has been generally thought to take place at the plasma membrane. It is now evident that the plasma membrane is not the only platform capable of Ras/MAPK signal induction. Fusion of Ras with green fluorescent protein and the development of genetically encoded fluorescent probes for Ras activation have revealed signaling events on a variety of intracellular membranes including endosomes, the Golgi apparatus and the endoplasmic reticulum. Thus, the Ras/MAPK pathway is spatially compartmentalized within cells and this may afford greater complexity of signal output.
Keywords: Ras, Endosomes, Golgi, Farnesylation, Compartmentalization
1. Introduction
The field of signal transduction concerns itself with the molecular mechanisms whereby biochemical signals are transduced across the plasma membrane and propagated within the cell in order to orchestrate a desired cellular response. More than any other field, signal transduction holds the promise of informing the process of drug discovery. Among the myriad of signaling molecules that have received great scrutiny in recent decades, perhaps none has been more intensely studied than Ras. For more than three decades great effort has been made to understand the intrinsic signaling properties of Ras proteins and the signaling networks regulated by them. The hope that the study of Ras signaling will lead to novel anti‐cancer therapies has in no small part fueled the intense interest. For many years the study of Ras involved the “what and when” of signaling as investigators catalogued the upstream activators, negative regulators and downstream effectors of the GTPase and studied the kinetics of the Ras/MAPK pathway. Following from the discovery that Ras is expressed on several subcellular compartments and seeking to help explain the diversity of signal outputs emanating from a biochemically simple binary switch, Ras biologists have more recently focused on the “where” of signaling.
The plasma membrane (PM) is often considered the primary signaling platform because signaling complexes are assembled here when transmembrane receptors are engaged by extracellular ligands. Several sets of discoveries contributed to the initial assignment of Ras exclusively to the PM. First was the discovery that Ras proteins are peripheral membrane proteins localized on the inner leaflet of the plasma membrane (Willingham et al., 1980). Second was the discovery that Ras is associated with membranes by virtue of post‐translational modification with lipids (Hancock et al., 1990). Finally, in genetic studies in flies, Ras was placed immediately downstream of growth factor receptors (Schlessinger, 2000). The revolution in cell biology ushered in by the age of green fluorescent protein (GFP) provoked a reassessment of the spatiotemporal aspects of Ras signaling. Using genetically encoded fluorescent probes, Ras signaling has been observed on intracellular membranes. In addition to the PM, Ras and/or MAPK signaling has now been observed on endosomes, the endoplasmic reticulum (ER), the Golgi apparatus, and mitochondria. Ras signaling from each of these platforms plays a role in the control of a wide variety of cellular processes, including growth, survival and differentiation. Subcellular compartmentalization of signaling, such as that regulated by Ras, provides one explanation for the apparent complexity of signaling outputs elaborated by individual signaling molecules and, in the case of Ras, forms a framework for understanding the evolution of four isoforms that differ primarily in the way they are targeted to cellular membranes. In this review, we give an overview of the current understanding of compartmentalized signaling focusing on the Ras/MAPK pathway.
2. Ras biology – the GTPase, the oncogene
Ras proteins are prototypical members of the superfamily of small GTPases. They transmit signals from cell surface receptors to a variety of effectors and thereby regulate pathways governing cell proliferation, differentiation, and programmed cell death (Karnoub and Weinberg, 2008). Ras proteins act as molecular switches. Signal‐induced conversion of the inactive to active state is mediated by guanine nucleotide‐exchange factors (GEFs) that stimulate the exchange of GDP for GTP. This is accomplished by catalyzing the release of GDP from the guanine nucleotide binding pocket. Once nucleotide free, Ras next binds GTP because it is tenfold more abundant in cytosol than is GDP. A marked conformational change caused by GTP binding leads to activation of Ras (Vetter and Wittinghofer, 2001). The effector domain engages downstream signaling molecules only when the protein is in the GTP‐bound state. The activation state of Ras is self‐limited by the intrinsic GTPase activity of the protein. However, Ras, like most signaling GTPases, is a poor enzyme. The catalytic activity of Ras is greatly enhanced by GTPase activating proteins (GAPs). GEFs and GAPs thus cooperate to generate a critical level of regulation, allowing the signal to turn on and off and to persist for a relatively short but variable period of time. The availability of constitutively active as well as dominant‐negative forms of Ras have made it possible to characterize its biological function (Feig, 1999). The dominant‐active forms are constitutively GTP‐bound and mimic the oncogenic forms of the protein.
Ras genes encoded by rat sarcoma viruses, v‐H‐ras and v‐K‐ras, were among the first oncogenes to be recognized (Harvey, 1964; Kirsten and Mayer, 1969). These viral genes are mutant forms of cellular protooncogenes (DeFeo et al., 1981; Ellis et al., 1981; Ruta et al., 1986). Mutations that render the GTPase insensitive to the action of GAP and therefore lock Ras in the GTP‐bound state account for its oncogenic activity (Barbacid, 1987; Lowy and Willumsen, 1993). Activated Ras alleles are the oncogene most frequently associated with human carcinomas (Barbacid, 1987), accounting for the great impact of Ras on human health. The mammalian genome encodes three ras genes that give rise to four gene products, N‐Ras, H‐Ras, K‐Ras4A, and K‐Ras4B. All isoforms are ubiquitously expressed, although isoform ratios vary from tissue to tissue. K‐ras4A and K‐Ras4B are splice variants of the K‐Ras gene that use alternative fourth exons. Mutations in ras genes are found in thirty percent of all human cancers. Tumors differ both in the isoforms associated with the disease and in the incidence of mutations of that isoform. For example, whereas 90% of pancreatic adenocarcinomas are associated with an oncogenic Ras mutation that is invariably in the K‐Ras gene, only 10% of bladder carcinomas harbor Ras mutations, and these occur in the H‐Ras gene (Bos, 1989).
3. Ras processing and trafficking
Although distinct in trafficking and steady‐state localization, one universal feature common to all Ras isoforms is that their localization on the cytosolic leaflet of cellular membranes is required for biological function. Evidence for Ras isoform‐specific signaling has been provided from tumor profiling, knockout mice, and overexpression studies (Hancock, 2003). The subcellular compartmentalization of different Ras isoforms is partially overlapping, yet distinct, and accounts for their biological differences. Ras proteins begin their lives in the cytosol as globular hydrophilic proteins that display a C‐terminal CAAX sequence (Figure 1). This sequence is the signal for a series of post‐translational modifications that include farnesylation, AAX proteolysis and carboxyl methylation (reviewed in (Mor and Philips, 2006)). The enzymes that catalyze AAX removal, Rce1, and carboxyl methylation, Icmt, are restricted to the ER, which serves as a way station for nascent Ras (Boyartchuk et al., 1997; Dai et al., 1998). H‐Ras, N‐Ras and K‐Ras4A are further modified with one or two palmitates (Hancock et al., 1989). The enzyme that palmitoylates Ras resides on the Golgi apparatus (Swarthout et al., 2005). In contrast to the other isoforms, K‐Ras4B is not palmitoylated and requires no further modification for full membrane affinity. Instead, K‐Ras4B localization depends on a polybasic region immediately upstream of its C‐terminal farnesyl cysteine (Hancock et al., 1990). Thus, K‐Ras4B, hereafter referred to simply as K‐Ras, is unique among the Ras isoforms and its subcellular trafficking is distinct. K‐Ras is also the isoform most often associated with human cancer. Cancer biologists hope to exploit the unique features of K‐Ras trafficking to develop new anti‐Ras drugs.
Figure 1.
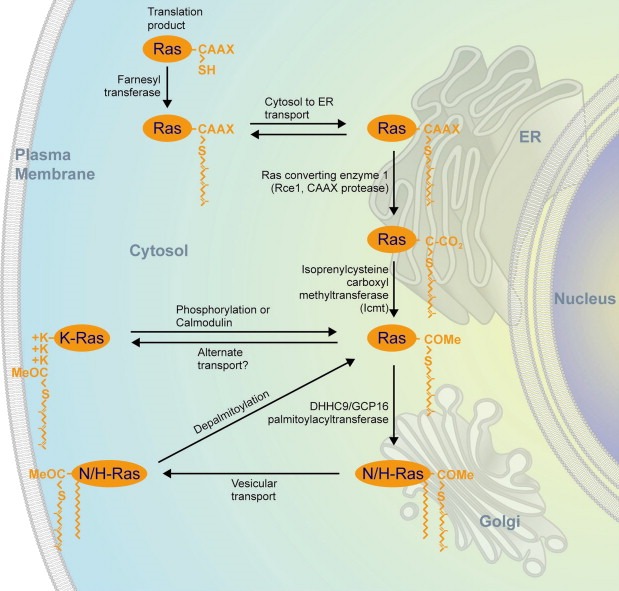
Post‐translational modification and trafficking of Ras. Ras proteins are synthesized in the cytosol where their CAAX C‐terminal sequence is recognized by farnesyltransferase. The farnesylated proteins are sent to the endoplasmic reticulum (ER) where they encounter Ras converting enzyme (Rce1) and isoprenylcysteine carboxyl methyltransferase (Icmt) that subsequently remove the AAX amino acids and methyl esterify the α carboxyl group. From here K‐Ras is sent directly to the plasma membrane (PM) but N‐Ras and H‐Ras travel to the Golgi where they are modified with one or two palmitates before being sent on to the PM. K‐Ras recycles back to endomembranes as a consequence of phosphorylation by PKC on serine 181 or through the action of calmodulin. N‐Ras and H‐Ras recycle back to the Golgi as a consequence of depalmitoylation at the PM. Reprinted, with permission, from the Annual Review of Immunology, Volume 24 ©2006 by Annual Reviews, www.annualreviews.org.
As peripheral membrane proteins, Ras proteins have two ways by which they can move from one membrane compartment to another. First, they can travel like intrinsic membrane proteins that are transferred from compartment to compartment via vesicular transport. Second, they can detach from the donor membrane and move through the aqueous phase of the cytosol, with or without a chaperone to shield their farnesyl chain, to the acceptor membrane. Significant evidence exists for each mode of transport. GFP‐tagged Ras proteins can be readily observed on highly motile vesicles, some of which travel along microtubules in a linear, saltatory fashion (Choy et al., 1999). N‐Ras and H‐Ras have been found to undergo a palmitoylaton/depalmitoylation cycle whereby depalmitoylation favors release of the GTPases from the inner leaflet of the PM from whence they travel retrograde, via diffusion through the cytosol, to the Golgi apparatus. Upon arrival at the Golgi, Ras proteins are replamitloylated and thereby once again affinity trapped in the membrane and sent back to the PM by vesicular transport (Goodwin et al., 2005; Rocks et al., 2005). Because K‐Ras associates with the PM via an intrinsic polybasic sequence rather than a labile palmitate modification, on first principles one might assume that its membrane association is constitutive. Elegant studies by Silvius showed this is not the case. Rather, PM K‐Ras is in a dynamic equilibrium with a pool in the cytosol (Silvius et al., 2006). More recently, the association of K‐Ras with the inner leaflet of the PM has been shown to be regulated by calmodulin binding to the polybasic region (Fivaz and Meyer, 2005) and by phosphorylation by PKC of serine 181 within the polybasic region (see below) (Bivona et al., 2006).
Compartmentalization of Ras isoforms does not end with delivery of the mature proteins to the PM. A large amount of evidence has been generated over the past decade that demonstrates that H‐Ras and K‐Ras reside in distinct microdomains within the PM, the former partitioning into lipid ordered domains high in cholesterol and sometimes referred to as lipid‐rafts and the latter restricted to disordered domains (Hancock and Parton, 2005). Remarkably, the partition of Ras proteins into microdomains, referred to as nanoclusters, is regulated by the GTP/GDP state of the protein suggesting that either the G domain participates in membrane association or this region controls the conformation of the hypervariable C‐terminus in such a way as to affect microdomain partition (Prior et al., 2001). The influence of PM microdomain localization on Ras signaling may be as profound as that of organelle localization (Inder et al., 2008). Indeed, Hancock et al. have put forth elegant models demonstrating how nanoclusters of Ras can transform analog signal input into digital output (Tian et al., 2007). However, this area has been extensively reviewed in recent years (Henis et al., 2009) and we will focus here on subcellular localization at the level of the organelle.
GFP‐tagged Ras proteins have been visualized on PM, Golgi apparatus, ER, mitochondria and a variety of endosomes. Ras isoforms display different degrees of association with endomembranes; N≥H>K‐Ras (Choy et al., 1999). These observations raise the obvious question of whether endomembrane associated Ras is capable of signaling and, perhaps more important, whether the signal output varies depending on the subcellular platform? Fluorescent probes of Ras activity have been used to address this question and the answer appears to be, yes; Ras can become activated on endomembrane and send a signal down various pathways and the outcome and duration of signaling depends, to some extent, on location (Chiu et al., 2002).
4. Ras signaling
The best‐characterized signaling pathway regulated by Ras is the MAPK pathway that proceeds through Erk1 and Erk2. After growth factors bind to their cognate protein tyrosine kinase receptors (PTKRs), these receptors dimerize, which in turn allows cross‐phosphorylation of tyrosine residues in their cytosolic domains catalyzed by the intrinsic kinase domain (Schlessinger, 2000). Among the signaling molecules that bind to phosphotyrosines on the cytosolic domains of these receptors is the adaptor Grb2 that binds SOS, which in turn acts as a GEF for Ras proteins. Thus, phosphorylation of PTKRs leads to the recruitment of SOS to the PM where it can encounter Ras. GTP‐bound Ras then recruits the Raf‐1 kinase. The molecular details of the kinase activation of Raf‐1 remains poorly understood but require membrane association of Ras (Morrison and Cutler, 1997). Raf‐1 subsequently phosphorylates and activates MEK (MAPK/Erk kinase), a dual specificity tyrosine/threonine kinase, that, in turn, phosphorylates and activates Erk1 and Erk2. The latter are serine/threonine kinases with numerous substrates, both nuclear and cytoplasmic. Phospho‐Erk forms dimers that translocate to the nucleus, where they phosphorylate the Ets family of transcription factors, including Elk‐1. In this way a signal originating from an extracellular stimulus is transmitted from the cell surface to the nucleus where gene transcription is modulated.
The Raf‐1/Erk pathway is not the only one regulated by Ras. Ras effectors can be defined as those proteins that bind to Ras via its effector domain only when Ras is GTP‐bound. More stringently defined, the function of the effector must be modulated by the binding to GTP‐Ras. Today, more than ten types of proteins are characterized as putative Ras effectors (Repasky et al., 2004). Apart from Raf‐1, the best‐characterized effectors are phosphatidylinositol 3‐kinase (PI3K) and members of a family of exchange factors for the small GTPase Ral, e.g. RalGDS. Although the Raf‐1/MAPK pathway has been defined as sufficient for Ras‐mediated transformation of mouse fibroblasts, recent data have suggested that in human cells the RalGDS pathway is the most important for transformation (Hamad et al., 2002). In addition, PI3K activates a pro‐survival kinase is Akt/PKB, which in some circumstances may be essential for oncogenesis (Gupta et al., 2007).
5. Ras signaling on the ER and Golgi
GFP fusions with Ras proteins established that Ras transits the ER en route to the PM and that at steady‐state palmitoylated isoforms of Ras are expressed on the Golgi apparatus as well as the PM (Choy et al., 1999; Apolloni et al., 2000). The first indication that intracellular Ras was capable of activation downstream of growth factor signaling came from studies using GFP fused to the Ras binding domain (RBD) of Raf‐1 (Chiu et al., 2002). At about the same time, Matsuda designed innovative FRET activation probes for Ras family proteins that he designated RAICHU. Using RAICHU‐Ras, Matsuda reported that activation was limited to the PM (Mochizuki et al., 2001). However, RAICHU‐Ras was targeted to membranes using the C‐terminus of K‐Ras4B such that it bypassed the Golgi and therefore did not accurately report spatiotemporal signaling of palmitoylated Ras isoforms. Using GFP‐RBD probes that are spatially unbiased it is now well established that GTP‐bound Ras accumulates on the Golgi as well as the PM in response to growth factor signaling in fibroblasts and TCR signaling in lymphocytes (Rocks et al., 2005; Chiu et al., 2002; Bivona et al., 2003; Caloca et al., 2003; Mor et al., 2007).
GFP‐RBD probes report Ras activation by simple translocation. Their biggest drawback is that they are not sensitive enough to report activation of endogenous Ras and co‐expression of wild‐type Ras isoforms is required in live cell assays. However, using FRET based analysis, Chiu et al. confirmed that the results observed with GFP‐RBD expressed with untagged, wild‐type H‐Ras are identical to those seen with endogenous Ras (Chiu et al., 2002; Bivona et al., 2003). Augsten et al. have recently introduced a trimeric GFP‐RBD probe that contains three tandem, doubly mutated RBD domains because the equivalent probe with native domains proved to be highly toxic. These investigators claim that their probe is sensitive enough to report activation of endogenous Ras, which they detect only on the PM of Jurkat T cells stimulated with PMA and ionomycin (Augsten et al., 2006). Physiologic signaling through the TCR was not reported. Thus, although there is some controversy in the field, there remains a preponderance of evidence supporting the idea that GTP‐bound palmitoylated Ras accumulates on the Golgi downstream of receptor signaling.
Although Ras activation on the Golgi has been reported in a number of cell types it has been most extensively studied in T lymphocytes downstream of TCR signaling. This is due to several features specific to T cells. First, although T cells express both N‐Ras and K‐Ras it appears to be N‐Ras that is most important for T cell activation (Pérez de Castro et al., 2004) and, when hematopoetic malignancies are associated with a mutated ras gene, it is most often nras. Second, whereas GFP‐RBD recruitment to the Golgi in fibroblasts can take more than 20min of growth factor signaling (Chiu et al., 2002), the same phenomenon in T cells stimulated through the TCR can be detected in under 2min (Bivona et al., 2003). Finally, RasGRP1, the exchange factor associated with Ras activation on the Golgi is highly expressed in T cells. Spatiotemporal Ras signaling has proven to be very interesting in T lymphocytes. When the TCR alone was engaged there was a rapid and robust accumulation of GTP‐Ras on the Golgi without any detectable signal at the PM (Bivona et al., 2003; Mor et al., 2007). However, when more physiologic signaling was initiated by simultaneous ligation of both the TCR and the LFA‐1 co‐receptor, robust accumulation of GTP‐Ras on the PM was observed in addition to a pool activated on the Golgi. Both pools of GTP‐bound Ras were controlled by RasGRP1 that gained access to the PM following LFA‐1 engagement because the integrin co‐receptor activated phospholipase D2 to generate phosphatidic acid which was then converted to DAG (Mor et al., 2007). Thus, the combination of receptors engaged on T lymphocytes determines the spatiotemporal features of Ras signaling.
If GTP‐bound Ras accumulates on the Golgi there are two mechanisms that may drive this process. First, Ras could be activated at the PM and traffic to the Golgi apparatus in a retrograde pathway following depalmitoylation at the PM. This model is favored by Bastiaens et al. who showed, using photoactivatable GFP, that H‐Ras and N‐Ras traffic retrograde from PM to Golgi via a non‐vesicular, diffusion limited pathway (Rocks et al., 2005), a conclusion supported by the FRAP studies of Kenworthy (Goodwin et al., 2005). This model requires a relatively long half‐life of GTP binding suggesting a lack of access to GAPs during depalmitoylation at the PM, cytosolic transport and accumulation on the Golgi. An alternative, model holds that H‐Ras and N‐Ras can be activated in situ on the Golgi. This model is made quite plausible in lymphocytes where RasGRP1 is regulated by diacylglycerol and calcium and has affinity for the Golgi following TCR engagement (Bivona et al., 2003; Mor et al., 2007). Of course, these two models are not mutually exclusive and GTP‐bound Ras on the Golgi may derive from both pathways. In support of the idea that Ras exchange factors preferentially activate spatially restricted pools of Ras, Arozarena et al. reported that RasGRF activated H‐Ras on the ER (Arozarena et al., 2004).
Perhaps more relevant that GTP‐bound Ras on the Golgi or ER is the question of whether activated Ras so situated can signal to downstream effectors and, if so, are these signals qualitatively different than those emanating from the PM? To address this question Chiu et al. stringently targeted constitutively activated Ras proteins to various subcellular membranes using a variety of transmembrane segments fused to the N‐terminus of Ras. H‐Ras61L potently transformed rodent fibroblasts when restricted to either the ER or Golgi (Chiu et al., 2002; Bivona et al., 2003). Importantly, when signaling output to three biochemical pathways was compared following expression of endomembrane‐tethered Ras, Erk and Akt were as efficiently activated from the Golgi as from the PM but Jnk was not. In contrast, activated Ras restricted to the ER was very efficient in activating Jnk providing a compelling demonstration of compartmentalized signaling (Chiu et al., 2002). Crespo et al. took a similar approach and found preferential signaling down the RalGDS pathway from Golgi‐tethered Ras and robust signaling to Erk, Akt and Jnk from ER restricted Ras (Matallanas et al., 2006). Although these investigators found little Erk phosphorylation induced by Golgi‐tethered Ras, Elk‐1 phosphorylation and luciferase activity was nevertheless stimulated equally well from this compartment (Casar et al., 2009). Thus, although some results differ, the idea of compartmentalized signaling to specific effectors is supported by both studies.
Artificially targeting Ras to various organelles has firmly established that signaling is possible from these locations but does not establish that such compartmentalized signaling occurs in vivo. One advance toward establishing such a model in vivo comes from genetic studies of fission yeast. In Schizosaccharomyces pombe a single Ras protein, Ras1p, controls at least two distinct pathways, that regulating cell morphology and that regulating mating. Onken et al. showed that these distinct pathways are regulated by Ras1p in different locations; signals emanating from the ER controlled cell morphology and those originating at the PM drove mating (Onken et al., 2006). The most compelling data to date for a biological role for compartmentalized Ras signaling comes from the study of T lymphocytes. Palmer et al. have explained a long baffling paradox of lymphocyte signaling by spatiotemporal signaling of Ras. Positive versus negative selection of thymocytes represent diametrically opposed processes leading, respectively, to proliferation or programmed cell death. Yet the Ras/MAPK pathway is required for both biological outcomes. Daniels et al. showed that whereas strong antigens that stimulate negative selection activate the Ras/MAPK pathway at the PM, weaker antigens that induce positive selection initiate signaling from the Golgi (Daniels et al., 2006). Thus the biological outcome of Ras signaling can depend on the subcellular platform from which signaling is initiated.
6. Ras on mitochondria
All three Ras isoforms have been found associated with subcellular fractions enriched in mitochondria (Rebollo et al., 1999; Wolfman et al., 2006). In contrast, numerous studies utilizing live cell imaging with GFP‐tagged Ras proteins failed to detect Ras on mitochondria despite unambiguous expression on ER, Golgi and a variety of vesicles. One explanation for this discrepancy is that localization of Ras by subcellular fractionation is complicated by ex vivo release of Ras from membranes decorated with the protein in intact cells and subsequent non‐specific adsorption of the released Ras onto membranes and organelles in the cellular homogenate, a phenomenon that is particularly problematic with K‐Ras that undergoes non‐specific electrostatic interactions via its C‐terminal polybasic region. More compelling evidence for mitochondrial association of Ras was provided by Bivona et al. who reported GFP‐K‐Ras translocation to the mitochondrial outer membrane in living cells treated with the PKC agonist bryostatin‐1 (Bivona et al., 2006). A farnesyl‐electrostatic switch involving phosphorylation of serine 181 in the polybasic region serves to partially neutralize the electrostatic charge and promote release from the PM. Surprisingly, phospho‐K‐Ras was associated with programmed cell death and Bcl‐Xl was required for induction of apoptosis. Like phospho‐K‐Ras, Bcl‐Xl resides on the mitochondrial outer membrane where it interacts with K‐Ras. Thus, K‐Ras signaling from mitochondria may not only engage distinct effectors but may have a biological outcome (cell death) diametrically opposed to Ras signaling from other membranes (proliferation and survival). It may be possible to exploit this feature of K‐Ras signaling to develop anti‐cancer drugs.
7. Ras signaling on endosomes
Because endosomes derive from the PM and are well known to internalize PTKRs it is not surprising that Ras signaling has been reported on endosomes. Indeed, endosomes serve as the most diverse and dynamic endomembrane compartment upon which Ras has been found to reside and signal. Originally conceived of as a process that limits signaling by removing receptors from the surface, several groups have subsequently shown that, in many contexts, efficient growth factor signaling requires endocytosis (Di Fiore and De Camilli, 2001). The first evidence for endosomal signaling came from subcellular fractionation studies in which Shc, Grb2, mSOS, and phospho‐Raf‐1 were differentially observed on endosomes following EGF or insulin stimulation (Di Guglielmo et al., 1994). Subsequently inhibition of clathrin‐mediated endocytosis with agents such as dominant‐negative dynamin (K44A) proved to inhibit rather than enhance Ras/MAPK signaling (Kranenburg et al., 1999; Vieira et al., 1996). Endosomal signaling may affect the kinetics of signaling: growth factor stimulated MAPK activity has been reported to be transient from the PM and sustained from endosomes (Oksvold et al., 2001; Taub et al., 2007).
Several methods have been used to demonstrate that PTKRs remain active upon internalization on endosomes. EGF remains bound to its receptor in early endosomes and the EGF receptor (EGFR) itself is phosphorylated after internalization (Burke et al., 2001). In addition to phosphorylated, active PTKRs, other upstream components of Ras signaling have been localized on endosomes, including Shc, Grb2, SOS, and PLCγ1 (Chiu et al., 2002; Di Guglielmo et al., 1994; Oksvold et al., 2001; Burke et al., 2001; Jiang and Sorkin, 2002; Matsuda et al., 2001; Sorkin et al., 2000; Sorkin and Von Zastrow, 2002; Wang et al., 2001). Ras itself was observed on endosomes using subcellular fractionation (Pol et al., 1998). Using GFP fusion proteins and live cell imaging, Ras has been localized to vesicles, including endosomes (Choy et al., 1999; Jiang and Sorkin, 2002). Sorkin et al. have used YFP‐RBD to reveal GTP‐bound Ras on vesicles decorated with internalized EGF (Jiang and Sorkin, 2002). Roy et al. showed that H‐Ras but not K‐Ras signaling requires endocytosis (Roy et al., 2002) and this result was confirmed by Omerovic et al. who found that N‐Ras signaling also required endocytosis (Omerovic et al., 2008).
Piertro De Camilli et al. recently provided a major advance in endosomal signaling when they showed that signaling endosomes, characterized by association of the adaptor protein APPL1, mature into EEA1 positive early endosomes by accumulation of PI3P and can be reverted by hydrolysis of the phosphate at the 3 position (Zoncu et al., 2009), (Figure 2). Importantly, reversion of early endosomes enhanced growth factor signaling demonstrating that the APPL1 positive compartment is particularly adapted for signaling. Thus, the complexity of endosomal signaling is greater than previously appreciated. If one takes into account that signaling has been observed from both clathrin‐dependent and independent endosomes, the complexity of the system increases even more.
Figure 2.

Ras signaling from endomembranes. N‐Ras and H‐Ras decorate the cytoplasmic surface of EEA1+/Rab5+/PI3P+ early endosomes (EE) as a consequence of endocytosis and, when diubiquitinated (Ub), are retained on this compartment. Alternatively they can be recycled to the plasma membrane (PM), perhaps via Rab11+ recycling endosomes (RE). APPL1+/PI3P‐ signaling endosomes (SE) that derive from and can be returned to the bulk pool of EEs have recently been described that are competent for MAPK signaling but have not yet been examined for Ras. N‐Ras and H‐Ras also cycle between the PM and Golgi as a consequence of a palmitoylation/depalmitoylation cycle traveling anterograde on secretory vesicles (SV) and retrograde via diffusion. K‐Ras also decorates various endosomes, including Lamp1/2+/Rab7+ late endosomes (LE) and multivesicular bodies (MVB), although it remains unclear whether this is a consequence of endocytosis as opposed to transport to the organelle from the PM via the cytosol. K‐Ras phosphorylated at the PM by PKC translocates to the outer surface of mitochondria as well as to Golgi and ER. Solid black tails on Ras proteins depict the farnesyl modification, green tails represent palmitates and +++ designates the polybasic region of K‐Ras.
Internalized receptors are either further trafficked to lysosomes for degradation or recycled back to the PM (Carpenter, 2000). A recent study by Lu et al. adds late endosomes and lysosomes to the pantheon of membrane platforms from which K‐Ras signals (Figure 2). In this study EGFR signaling was associated with the progressive accumulation of GFP‐K‐Ras but not N‐Ras or H‐Ras on early EEA1/Rab5 bearing endosomes, Rab7‐marked endosomes, LAMP1/2‐ marked lysosomes and multivesicular bodies (MVBs) (Lu et al., 2009). Furthermore, late endosomes, lysosomes and MVBs were shown to serve not only as platforms for Ras signaling but also as sites for K‐Ras degradation. K‐Ras was stabilized by inhibitors of lysosomal degradation but not by proteasome inhibitors (Lu et al., 2009). This latter observation is somewhat surprising given the disposition of K‐Ras and other Ras proteins to the cytosol. The occurrence of large multivesicular structures decorated with GFP‐Kras, as seen by Lu et al., could be the result of the long‐term incubation with protease inhibitors employed in these studies. Protease inhibitors are known to induce autophagosome formation, perhaps through a lysosomal stress pathway (Ostenfeld et al., 2008). Furthermore, because protease inhibitors upregulate the lysosomal membrane proteins, LAMP‐1/2 (Fehrenbacher et al., 2008), the increased co‐localization of K‐Ras with LAMP‐1/2 observed by Lu et al. may also be a consequence of protease inhibition.
It remains to be clarified whether K‐Ras association with endosomes is a result of clathrin‐mediated endocytosis of membranes carrying K‐Ras, or if K‐Ras arrives on these compartments by translocation through the cytosol, or both. Regardless of the mechanism of trafficking to late endosomes, the results of Lu et al. contradict earlier studies from several groups that found that, whereas N‐Ras and H‐Ras trafficked stably associate with endosomes, K‐Ras does not (Roy et al., 2002; Omerovic et al., 2008; Jura et al., 2006). This differential trafficking pattern has been attributed to the preferential modification of H‐ and N‐Ras by non‐degradative mono and Lys 63‐linked di ubiquitination. The selective targeting of H‐ and N‐Ras for ubiquitination is determined by their C‐terminal membrane targeting regions likely reflecting the confinement of the enzymatic machinery that controls Ras ubiquitination to specific membrane compartments either at the level of organelles or membrane nanodomains. Similarly to other membrane cargo proteins, the ubiquitination of H‐ and N‐Ras promotes their association with endosomes (Figure 2). As a consequence, the pool of PM‐associated Ras molecules that is available for the recruitment and activation of Raf‐1 is reduced and ERK activation is attenuated (Jura et al., 2006). Consistent with these findings, it has been recently shown that that in Drosophila, maintaining a threshold of Ras ubiquitination is critical to prevent inappropriate Ras‐ERK activation in vivo (Yan et al., 2009). Thus, rather than functioning as signaling endosomes, the pool of endosomes bearing ubiquitinated Ras may not be capable of signaling and may thereby afford a mechanism for downregulation of Ras/MAPK signaling.
Other recent findings shed some doubt on the idea of endosomes as fully competent MAPK signaling platforms. MEK2‐GFP was observed both on the PM and endosomes but the activated form of the kinase was detected only on the PM (Galperin and Sorkin, 2008). Moreover, the population of endosomes decorated with MEK2‐GFP was distinct from those that carried activated EGFR. Interestingly, in this study silencing of clathrin heavy chain augmented EGF stimulation of Erk, a result inconsistent with studies using dominant‐negative dynamin as a way of blocking endocytosis (Di Fiore and De Camilli, 2001). Thus it is clear that the complexity and physiologic relevance of signaling from endosomes remains to be fully elucidated.
8. MAPK scaffolds
Compartmentalized Ras signaling is facilitated in part by the differential subcellular trafficking of Ras isoforms. However, Ras localization is not the only mechanism for compartmentalization. An increasingly recognized component of MAPK and other signaling pathways are scaffold proteins that play no direct role in catalysis but serve as platforms upon which signaling complexes can assemble. Because many scaffolds are spatially restricted within cells this class of molecule can contribute to and, in some cases define, compartmentalized signaling. Scaffolding molecules in the MAPK pathways serve to organize the various MAPK modules, such as Erk, Jnk and p38 (Karandikar and Cobb, 1999). A well‐studied Erk scaffold is the kinase suppressor of Ras (KSR), a protein poorly named since it is not a kinase and was first identified in screens in flies (Therrien et al., 1995) and worms (Kornfeld et al., 1995; Sundaram and Han, 1995) as a positive regulator of the Ras/MAPK pathway. KSR is a multidomain protein that binds Raf‐1, MEK and Erk, as well as other proteins. Like Raf‐1, KSR is sequestered in the cytosol by 14‐3‐3 proteins in resting cells. Upon mitogenic stimulation KSR translocates to the PM when it becomes dephosphorylated at serine 392 and looses its affinity for 14‐3‐3 (Cacace et al., 1999; Muller et al., 2001). Thus, KSR serves as inducible scaffold for Ras/MAPK signaling with specificity for the PM. Recently the specificity of KSR has been further resolved to the nanoscale by a study that reports that the scaffolding activity of KSR1 is specific to lipid microdomains within the PM (Casar et al., 2009). Interestingly, in this study IQGAP1 also served as a lipid‐raft specific MAPK scaffold that promoted phosphorylation of EGFR (Casar et al., 2009).
MEK partner 1 (MP1) was first identified in a yeast two‐hybrid screen as a binding partner of MEK1 (Schaeffer et al., 1998). It preferentially binds to MEK1 and Erk1, not MEK2 and Erk2. Binding facilitated the phosphorylation of Erk1 by MEK1, thus fulfilling the criteria for a scaffold (Schaeffer et al., 1998). Interestingly, MP1 was also found to interact with p14, a highly conserved protein that resides on the cytoplasmic face of early endosomes (Teis et al., 2002). Overexpression studies revealed that MP1 increases Erk signaling, but only when overexpressed with p14. Using the C‐terminus of K‐Ras, the MP1/p14 complex was ectopically targeted to the PM. This construct failed to augment Erk activation, indicating that the endosomal location is essential for the scaffolding function of MP1/p14 (Teis et al., 2002). Thus, MP1 is an endosome‐specific scaffold for MEK and Erk. Interestingly, while MP1/p14 was not required for early activation at the PM, it was required for the activation seen on endosomes 10–30min after EGF stimulation (Teis et al., 2002).
MP1 is not the only MAPK scaffold on endosomes; β‐arrestin also provides this function in GPCR signaling. This multifunctional protein regulates internalization of GPCRs into clathrin‐coated vesicles (Goodman et al., 1996; Laporte et al., 1999) and serves as a scaffold for both the Erk (Luttrell et al., 2001) and Jnk (McDonald et al., 2000) MAPK modules. Several studies place β‐arrestin functionally in the transmission of signals from Raf‐1 to MEK and Erk on endosomes. Raf‐1 overexpression increased MEK and Erk binding to β‐arrestin (Luttrell et al., 2001) and a dominant‐negative form of β‐arrestin blocked Erk activation downstream of a GPCR (Crosby, 1953; Daaka et al., 1998; DeFea et al., 2000). Thus, endosomes are a subcellular site that supports both PTKR and GPCR signaling to Erk, and each system uses distinct scaffolds on this organelle.
Sef is yet another MEK/Erk scaffold that affords spatial specificity. Sef resides on the Golgi apparatus (Torii et al., 2004). Sef was originally identified as a negative regulator of fibroblast growth factor (Furthauer et al., 2002; Tsang et al., 2002). Sef binds MEK only when the kinase is phosphorylated and activated. In the canonical MAPK cascade Erk dissociates from MEK once it is phosphorylated forms dimers that subsequently enters the nucleus. However, activated Erk remains associated with MEK on Sef, preventing Erk's translocation into the nucleus and preventing is interaction with nuclear substrates such as Elk‐1. Nevertheless, active Erk associated with Sef on the Golgi is capable of phosphorylating cytosolic substrates such as RSK2 (Torii et al., 2004). When Elk‐1 was tagged with a nuclear export signal, it became artificially localized to the cytoplasm and became a substrate for Sef‐associated phospho‐Erk (Torii et al., 2004). This makes Sef a compartment‐specific scaffold protein that directs Erk activity to one set of substrates over another (Philips, 2004). Recently, Sef was reported to be required for Ras signaling from the ER (Casar et al., 2009). Like Sef, β‐arrestin 1 can also shift the substrate specificity of Erk during GPCR signaling such that cytosolic substrates are favored and cell proliferation is not triggered (DeFea et al., 2000). Similarly, β‐arrestin 2 acts to sequester Jnk3 in the cytosol (McDonald et al., 2000). Thus the spatiotemporal controls of Ras/MAPK signaling imparted by the various scaffolds translate into substrate specificity and therefore pathway selection.
9. Concluding remarks
One of the central paradoxes of signal transduction is how a signaling molecule such as Ras, which appears from a biochemical point of view to be a simple binary switch, can independently regulate so many different pathways. One way to enhance the complexity of Ras signaling is to by compartmentalization within cells. The nature of Ras as a peripheral membrane protein conditionally associated with the cytoplasmic leaflet of cellular membranes allows for a diversity of localizations on various organelles and, taken to the nanoscale, in a variety of membrane microdomains. The peripatetic nature of Ras allows for compartmentalized signaling. Indeed, Ras/MAPK signaling has now been established on PM, endosomes, Golgi and ER and K‐Ras signals from the surface of mitochondria. In the case of T lymphocytes it appears that the location of Ras/MAPK signaling dictates the biological outcome. As new tools are developed to allow measurement of compartmentalized Ras/MAPK signaling in live cells and in vivo we are bound to learn more about how location dictates function.
Acknowledgements
This work was supported by NIH Grants GM055279, CA116034 and CA118495 to M.R.P and CA055360 to D.B‐S.
Fehrenbacher Nicole, Bar-Sagi Dafna, Philips Mark, (2009), Ras/MAPK signaling from endomembranes, Molecular Oncology, 3, doi: 10.1016/j.molonc.2009.06.004.
References
- Apolloni, A. , Prior, I.A. , Lindsay, M. , Parton, R.G. , Hancock, J.F. , 2000. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol.. 20, 2475–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arozarena, I. , Matallanas, D. , Berciano, M.T. , Sanz-Moreno, V. , Calvo, F. , Munoz, M.T. , Egea, G. , Lafarga, M. , Crespo, P. , 2004. Activation of H-Ras in the endoplasmic reticulum by the RasGRF family guanine nucleotide exchange factors. Mol. Cell. Biol.. 24, 1516–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augsten, M. , Pusch, R. , Biskup, C. , Rennert, K. , Wittig, U. , Beyer, K. , Blume, A. , Wetzker, R. , Friedrich, K. , Rubio, I. , 2006. Live-cell imaging of endogenous Ras-GTP illustrates predominant Ras activation at the plasma membrane. EMBO Rep.. 7, 46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacid, M. , 1987. ras Genes. Ann. Rev. Biochem.. 56, 779–827. [DOI] [PubMed] [Google Scholar]
- Bivona, T.G. , Perez De Castro, I. , Ahearn, I.M. , Grana, T.M. , Chiu, V.K. , Lockyer, P.J. , Cullen, P.J. , Pellicer, A. , Cox, A.D. , Philips, M.R. , 2003. Phospholipase Cγ activates Ras on the Golgi apparatus by means of RasGRP1. Nature. 424, 694–698. [DOI] [PubMed] [Google Scholar]
- Bivona, T.G. , Quatela, S.E. , Bodemann, B.O. , Ahearn, I.O. , Soskis, M.J. , Mor, A. , Miura, J. , Wiener, H.H. , Wright, L. , Saba, S.G. , 2006. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol. Cell.. 21, 481–493. [DOI] [PubMed] [Google Scholar]
- Bos, J.L. , 1989. ras Oncogenes in human cancer: a review. Cancer Res.. 49, 4682–4689. [PubMed] [Google Scholar]
- Boyartchuk, V.L. , Ashby, M.N. , Rine, J. , 1997. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 275, 1796–1800. [DOI] [PubMed] [Google Scholar]
- Burke, P. , Schooler, K. , Wiley, H.S. , 2001. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol. Biol. Cell.. 12, 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacace, A.M. , Michaud, N.R. , Therrien, M. , Mathes, K. , Copeland, T. , Rubin, G.M. , Morrison, D.K. , 1999. Identification of constitutive and ras-inducible phosphorylation sites of KSR: implications for 14-3-3 binding, mitogen-activated protein kinase binding, and KSR overexpression. Mol. Cell. Biol.. 19, 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caloca, M.J. , Zugaza, J.L. , Matallanas, D. , Crespo, P. , Bustelo, X.R. , 2003. Vav mediates Ras stimulation by direct activation of the GDP/GTP exchange factor Ras GRP1. EMBO J.. 22, 3326–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, G. , 2000. The EGF receptor: a nexus for trafficking and signaling. Bioessays. 22, 697–707. [DOI] [PubMed] [Google Scholar]
- Casar, B. , Arozarena, I. , Sanz-Moreno, V. , Pinto, A. , Agudo-Ibanez, L. , Marais, R. , Lewis, R.E. , Berciano, M.T. , Crespo, P. , 2009. Ras subcellular localization defines extracellular signal-regulated kinase 1 and 2 substrate specificity through distinct utilization of scaffold proteins. Mol. Cell. Biol.. 29, 1338–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, V.K. , Bivona, T. , Hach, A. , Sajous, J.B. , Silletti, J. , Wiener, H. , Johnson, R.L. , Cox, A.D. , Philips, M.R. , 2002. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell. Biol.. 4, 343–350. [DOI] [PubMed] [Google Scholar]
- Choy, E. , Chiu, V.K. , Silletti, J. , Feoktistov, M. , Morimoto, T. , Michaelson, D. , Ivanov, I.E. , Philips, M.R. , 1999. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 98, 69–80. [DOI] [PubMed] [Google Scholar]
- Crosby, W.H. , 1953. Blood. 8, 769–812. [PubMed] [Google Scholar]
- Daaka, Y. , Luttrell, L.M. , Ahn, S. , Della Rocca, G.J. , Ferguson, S.S. , Caron, M.G. , Lefkowitz, R.J. , 1998. Essential role for G protein-coupled receptor endocytosis in the activation of mitogen-activated protein kinase. J. Biol. Chem.. 273, 685–688. [DOI] [PubMed] [Google Scholar]
- Dai, Q. , Choy, E. , Chiu, V. , Romano, J. , Slivka, S. , Steitz, S. , Michaelis, S. , Philips, M.R. , 1998. Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J. Biol. Chem.. 273, 15030–15034. [DOI] [PubMed] [Google Scholar]
- Daniels, M.A. , Teixeiro, E. , Gill, J. , Hausmann, B. , Roubaty, D. , Holmberg, K. , Werlen, G. , Hollander, G.A. , Gascoigne, N.R. , Palmer, E. , 2006. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 444, 724–729. [DOI] [PubMed] [Google Scholar]
- DeFea, K.A. , Zalevsky, J. , Thoma, M.S. , Dery, O. , Mullins, R.D. , Bunnett, N.W. , 2000. Beta-arrestin-dependent endocytosis of proteinase-activated receptor 2 is required for intracellular targeting of activated ERK1/2. J. Cell. Biol.. 148, 1267–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFeo, D. , Gonda, M.A. , Young, H.A. , Chang, E.H. , Lowy, D.R. , Scolnick, E.M. , Ellis, R.W. , 1981. Analysis of two divergent rat genomic clones homologous to the transforming gene of Harvey murine sarcoma virus. Proc. Natl Acad. Sci. U S A. 78, 3328–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore, P.P. , De Camilli, P. , 2001. Endocytosis and signaling. An inseparable partnership. Cell. 106, 1–4. [DOI] [PubMed] [Google Scholar]
- Di Guglielmo, G.M. , Baass, P.C. , Ou, W.J. , Posner, B.I. , Bergeron, J.J. , 1994. Compartmentalization of SHC, GRB2 and mSOS, and hyperphosphorylation of Raf-1 by EGF but not insulin in liver parenchyma. EMBO J.. 13, 4269–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, R.W. , DeFeo, D. , Shih, T.Y. , Gonda, M.A. , Young, H.A. , Tsuchida, N. , Lowy, D.R.S. , 1981. The p21 src genes of Harvey and Kirsten sarcoma viruses originate from divergent members of a family of normal vertebrate genes. Nature. 292, 506–511. [DOI] [PubMed] [Google Scholar]
- Fehrenbacher, N. , Bastholm, L. , Kirkegaard-Sorensen, T. , Rafn, B. , Bottzauw, T. , Nielsen, C. , Weber, E. , Shirasawa, S. , Kallunki, T. , Jaattela, M. , 2008. Sensitization to the lysosomal cell death pathway by oncogene-induced down-regulation of lysosome-associated membrane proteins 1 and 2. Cancer Res.. 68, 6623–6633. [DOI] [PubMed] [Google Scholar]
- Feig, L.A. , 1999. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat. Cell. Biol.. 1, E25–27. [DOI] [PubMed] [Google Scholar]
- Fivaz, M. , Meyer, T. , 2005. Reversible intracellular translocation of KRas but not HRas in hippocampal neurons regulated by Ca2+/calmodulin. J. Cell. Biol.. 170, 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furthauer, M. , Lin, W. , Ang, S.L. , Thisse, B. , Thisse, C. , 2002. Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nat. Cell. Biol.. 4, 170–174. [DOI] [PubMed] [Google Scholar]
- Galperin, E. , Sorkin, A. , 2008. Endosomal targeting of MEK2 requires RAF, MEK kinase activity and clathrin-dependent endocytosis. Traffic. 9, 1776–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, O.B. , Krupnick, J.G. , Santini, F. , Gurevich, V.V. , Penn, R.B. , Gagnon, A.W. , Keen, J.H. , Benovic, J.L. , 1996. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 383, 447–450. [DOI] [PubMed] [Google Scholar]
- Goodwin, J.S. , Drake, K.R. , Rogers, C. , Wright, L. , Lippincott-Schwartz, J. , Philips, M.R. , Kenworthy, A.K. , 2005. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J. Cell. Biol.. 170, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S. , Ramjaun, A.R. , Haiko, P. , Wang, Y. , Warne, P.H. , Nicke, B. , Nye, E. , Stamp, G. , Alitalo, K. , Downward, J. , 2007. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 129, 957–968. [DOI] [PubMed] [Google Scholar]
- Hamad, N.M. , Elconin, J.H. , Karnoub, A.E. , Bai, W. , Rich, J.N. , Abraham, R.T. , Der, C.J. , Counter, C.M. , 2002. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev.. 16, 2045–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, J.F. , Parton, R.G. , 2005. Ras plasma membrane signalling platforms. Biochem. J.. 389, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, J.F. , Magee, A.I. , Childs, J.E. , Marshall, C.J. , 1989. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 57, 1167–1177. [DOI] [PubMed] [Google Scholar]
- Hancock, J.F. , Paterson, H. , Marshall, C.J. , 1990. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 63, 133–139. [DOI] [PubMed] [Google Scholar]
- Hancock, J.F. , 2003. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell. Biol.. 4, 373–384. [DOI] [PubMed] [Google Scholar]
- Harvey, J.J. , 1964. An unidentified virus which causes the rapid production of tumours in mice. Nature. 204, 1104–1105. [DOI] [PubMed] [Google Scholar]
- Henis, Y.I. , Hancock, J.F. , Prior, I.A. , 2009. Ras acylation, compartmentalization and signaling nanoclusters. (Review) Mol. Membr. Biol.. 26, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inder, K. , Harding, A. , Plowman, S.J. , Philips, M.R. , Parton, R.G. , Hancock, J.F. , 2008. Activation of the MAPK module from different spatial locations generates distinct system outputs. Mol. Biol. Cell.. 19, 4776–4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X. , Sorkin, A. , 2002. Coordinated traffic of Grb2 and Ras during epidermal growth factor receptor endocytosis visualized in living cells. Mol. Biol. Cell.. 13, 1522–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jura, N. , Scotto-Lavino, E. , Sobczyk, A. , Bar-Sagi, D. , 2006. Differential modification of Ras proteins by ubiquitination. Mol. Cell.. 21, 679–687. [DOI] [PubMed] [Google Scholar]
- Karandikar, M. , Cobb, M.H. , 1999. Scaffolding and protein interactions in MAP kinase modules. Cell. Calcium. 26, 219–226. [DOI] [PubMed] [Google Scholar]
- Karnoub, A.E. , Weinberg, R.A. , 2008. Ras oncogenes: split personalities. Nat. Rev. Mol. Cell. Biol.. 9, 517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsten, W.H. , Mayer, L.A. , 1969. Malignant lymphomas of extrathymic origin induced in rats by murine erythroblastosis virus. J. Natl Cancer Inst.. 43, 735–746. [PubMed] [Google Scholar]
- Kornfeld, K. , Hom, D.B. , Horvitz, H.R. , 1995. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans . Cell. 83, 903–913. [DOI] [PubMed] [Google Scholar]
- Kranenburg, O. , Verlaan, I. , Moolenaar, W.H. , 1999. Dynamin is required for the activation of mitogen-activated protein (MAP) kinase by MAP kinase kinase. J. Biol. Chem.. 274, 35301–35304. [DOI] [PubMed] [Google Scholar]
- Laporte, S.A. , Oakley, R.H. , Zhang, J. , Holt, J.A. , Ferguson, S.S. , Caron, M.G. , Barak, L.S. , 1999. The beta2-adrenergic receptor/betaarrestin complex recruits the clathrin adaptor AP-2 during endocytosis. Proc. Natl Acad. Sci. U S A. 96, 3712–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy, D.R. , Willumsen, B.M. , 1993. Function and regulation of ras. Ann. Rev. Biochem.. 62, 851–891. [DOI] [PubMed] [Google Scholar]
- Lu, A. , Tebar, F. , Alvarez-Moya, B. , Lopez-Alcala, C. , Calvo, M. , Enrich, C. , Agell, N. , Nakamura, T. , Matsuda, M. , Bachs, O. , 2009. A clathrin-dependent pathway leads to KRas signaling on late endosomes en route to lysosomes. J. Cell. Biol.. 184, 863–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttrell, L.M. , Roudabush, F.L. , Choy, E.W. , Miller, W.E. , Field, M.E. , Pierce, K.L. , Lefkowitz, R.J. , 2001. Activation and targeting of extracellular signal-regulated kinases by beta-arrestin scaffolds. Proc. Natl Acad. Sci. U S A. 98, 2449–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matallanas, D. , Sanz-Moreno, V. , Arozarena, I. , Calvo, F. , Agudo-Ibanez, L. , Santos, E. , Berciano, M.T. , Crespo, P. , 2006. Distinct utilization of effectors and biological outcomes resulting from site-specific Ras activation: Ras functions in lipid rafts and golgi complex are dispensable for proliferation and transformation. Mol. Cell. Biol.. 26, 100–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, M. , Paterson, H.F. , Rodriguez, R. , Fensome, A.C. , Ellis, M.V. , Swann, K. , Katan, M. , 2001. Real time fluorescence imaging of PLC gamma translocation and its interaction with the epidermal growth factor receptor. J. Cell. Biol.. 153, 599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, P.H. , Chow, C.W. , Miller, W.E. , Laporte, S.A. , Field, M.E. , Lin, F.T. , Davis, R.J. , Lefkowitz, R.J. , 2000. Beta-arrestin 2: a receptor-regulated MAPK scaffold for the activation of JNK3. Science. 290, 1574–1577. [DOI] [PubMed] [Google Scholar]
- Mochizuki, N. , Yamashita, S. , Kurokawa, K. , Ohba, Y. , Nagai, T. , Miyawaki, A. , Matsuda, M. , 2001. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 411, 1065–1068. [DOI] [PubMed] [Google Scholar]
- Mor, A. , Philips, M.R. , 2006. Compartmentalized Ras/MAPK signaling. Ann. Rev. Immunol.. [DOI] [PubMed] [Google Scholar]
- Mor, A. , Campi, G. , Du, G. , Zheng, Y. , Foster, D.A. , Dustin, M.L. , Philips, M.R. , 2007. The lymphocyte function-associated antigen-1 receptor costimulates plasma membrane Ras via phospholipase D2. Nat. Cell. Biol.. 9, 713–719. [DOI] [PubMed] [Google Scholar]
- Morrison, D.K. , Cutler, R.E. , 1997. The complexity of Raf-1 regulation. Curr. Opin. Cell. Biol.. 9, 174–179. [DOI] [PubMed] [Google Scholar]
- Muller, J. , Ory, S. , Copeland, T. , Piwnica-Worms, H. , Morrison, D.K. , 2001. C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol. Cell.. 8, 983–993. [DOI] [PubMed] [Google Scholar]
- Oksvold, M.P. , Skarpen, E. , Wierod, L. , Paulsen, R.E. , Huitfeldt, H.S. , 2001. Re-localization of activated EGF receptor and its signal transducers to multivesicular compartments downstream of early endosomes in response to EGF. Eur. J. Cell. Biol.. 80, 285–294. [DOI] [PubMed] [Google Scholar]
- Omerovic, J. , Hammond, D.E. , Clague, M.J. , Prior, I.A. , 2008. Ras isoform abundance and signalling in human cancer cell lines. Oncogene. 27, 2754–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onken, B. , Wiener, H. , Philips, M. , Chang, E.C. , 2006. Compartmentalized signaling of Ras in fission yeast. Proc. Natl Acad. Sci. U S A. 103, 9045–9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostenfeld, M.S. , Hoyer-Hansen, M. , Bastholm, L. , Fehrenbacher, N. , Olsen, O.D. , Groth-Pedersen, L. , Puustinen, P. , Kirkegaard-Sorensen, T. , Nylandsted, J. , Farkas, T. , 2008. Anti-cancer agent siramesine is a lysosomotropic detergent that induces cytoprotective autophagosome accumulation. Autophagy. 4, 487–499. [DOI] [PubMed] [Google Scholar]
- Pérez de Castro, I. , Bivona, T. , Philips, M. , Pellicer, A. , 2004. Ras activation in Jurkat T cells following low-grade stimulation of the T-cell receptor is specific to N-Ras and occurs only on the Golgi. Mol. Cell. Biol.. 24, 3485–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips, M.R. , 2004. Sef: a MEK/ERK catcher on the Golgi. Mol. Cell.. 15, 168–169. [DOI] [PubMed] [Google Scholar]
- Pol, A. , Calvo, M. , Enrich, C. , 1998. Isolated endosomes from quiescent rat liver contain the signal transduction machinery. Differential distribution of activated Raf-1 and Mek in the endocytic compartment. FEBS Lett.. 441, 34–38. [DOI] [PubMed] [Google Scholar]
- Prior, I.A. , Harding, A. , Yan, J. , Sluimer, J. , Parton, R.G. , Hancock, J.F. , 2001. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat. Cell. Biol.. 3, 368–375. [DOI] [PubMed] [Google Scholar]
- Rebollo, A. , Perez-Sala, D. , Martinez, A.C. , 1999. Bcl-2 differentially targets K-, N-, and H-Ras to mitochondria in IL-2 supplemented or deprived cells: implications in prevention of apoptosis. Oncogene. 18, 4930–4939. [DOI] [PubMed] [Google Scholar]
- Repasky, G.A. , Chenette, E.J. , Der, C.J. , 2004. Renewing the conspiracy theory debate: does Raf function alone to mediate Ras oncogenesis?. Trends Cell. Biol.. 14, 639–647. [DOI] [PubMed] [Google Scholar]
- Rocks, O. , Peyker, A. , Kahms, M. , Verveer, P.J. , Koerner, C. , Lumbierres, M. , Kuhlmann, J. , Waldmann, H. , Wittinghofer, A. , Bastiaens, P.I. , 2005. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 307, 1746–1752. [DOI] [PubMed] [Google Scholar]
- Roy, S. , Wyse, B. , Hancock, J.F. , 2002. H-Ras signaling and K-Ras signaling are differentially dependent on endocytosis. Mol. Cell. Biol.. 22, 5128–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruta, M. , Wolford, R. , Dhar, R. , Defeo-Jones, D. , Ellis, R.W. , Scolnick, E.M. , 1986. Nucleotide sequence of the two rat cellular rasH genes. Mol. Cell. Biol.. 6, 1706–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer, H.J. , Catling, A.D. , Eblen, S.T. , Collier, L.S. , Krauss, A. , Weber, M.J. , 1998. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 281, 1668–1671. [DOI] [PubMed] [Google Scholar]
- Schlessinger, J. , 2000. Cell signaling by receptor tyrosine kinases. Cell. 103, 211–225. [DOI] [PubMed] [Google Scholar]
- Silvius, J.R. , Bhagatji, P. , Leventis, R. , Terrone, D. , 2006. K-ras4B and prenylated proteins lacking “second signals” associate dynamically with cellular membranes. Mol. Biol. Cell.. 17, 192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin, A. , Von Zastrow, M. , 2002. Signal transduction and endocytosis: close encounters of many kinds. Nat. Rev. Mol. Cell. Biol.. 3, 600–614. [DOI] [PubMed] [Google Scholar]
- Sorkin, A. , McClure, M. , Huang, F. , Carter, R. , 2000. Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr. Biol.. 10, 1395–1398. [DOI] [PubMed] [Google Scholar]
- Sundaram, M. , Han, M. , 1995. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell. 83, 889–901. [DOI] [PubMed] [Google Scholar]
- Swarthout, J.T. , Lobo, S. , Farh, L. , Croke, M.R. , Greentree, W.K. , Deschenes, R.J. , Linder, M.E. , 2005. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J. Biol. Chem.. 280, 31141–31148. [DOI] [PubMed] [Google Scholar]
- Taub, N. , Teis, D. , Ebner, H.L. , Hess, M.W. , Huber, L.A. , 2007. Late endosomal traffic of the epidermal growth factor receptor ensures spatial and temporal fidelity of mitogen-activated protein kinase signaling. Mol. Biol. Cell.. 18, 4698–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis, D. , Wunderlich, W. , Huber, L.A. , 2002. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev. Cell.. 3, 803–814. [DOI] [PubMed] [Google Scholar]
- Therrien, M. , Chang, H.C. , Solomon, N.M. , Karim, F.D. , Wassarman, D.A. , Rubin, G.M. , 1995. KSR, a novel protein kinase required for RAS signal transduction. Cell. 83, 879–888. [DOI] [PubMed] [Google Scholar]
- Tian, T. , Harding, A. , Inder, K. , Plowman, S. , Parton, R.G. , Hancock, J.F. , 2007. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat. Cell. Biol.. 9, 905–914. [DOI] [PubMed] [Google Scholar]
- Torii, S. , Kusakabe, M. , Yamamoto, T. , Maekawa, M. , Nishida, E. , 2004. Sef is a spatial regulator for Ras/MAP kinase signaling. Dev. Cell.. 7, 33–44. [DOI] [PubMed] [Google Scholar]
- Tsang, M. , Friesel, R. , Kudoh, T. , Dawid, I.B. , 2002. Identification of Sef, a novel modulator of FGF signalling. Nat. Cell. Biol.. 4, 165–169. [DOI] [PubMed] [Google Scholar]
- Vetter, I.R. , Wittinghofer, A. , 2001. The guanine nucleotide-binding switch in three dimensions. Science. 294, 1299–1304. [DOI] [PubMed] [Google Scholar]
- Vieira, A.V. , Lamaze, C. , Schmid, S.L. , 1996. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 274, 2086–2089. [DOI] [PubMed] [Google Scholar]
- Wang, X.J. , Liao, H.J. , Chattopadhyay, A. , Carpenter, G. , 2001. EGF-dependent translocation of green fluorescent protein-tagged PLC-gamma1 to the plasma membrane and endosomes. Exp. Cell. Res.. 267, 28–36. [DOI] [PubMed] [Google Scholar]
- Willingham, M.C. , Pastan, I. , Shih, T.Y. , Scolnick, E.M. , 1980. Localization of the src gene product of the Harvey strain of MSV to plasma membrane of transformed cells by electron microscopic immunocytochemistry. Cell. 19, 1005–1014. [DOI] [PubMed] [Google Scholar]
- Wolfman, J.C. , Planchon, S.M. , Liao, J. , Wolfman, A. , 2006. Structural and functional consequences of c-N-Ras constitutively associated with intact mitochondria. Biochim. Biophys. Acta. 1763, 1108–1124. [DOI] [PubMed] [Google Scholar]
- Yan, H. , Chin, M.L. , Horvath, E.A. , Kane, E.A. , Pfleger, C.M. , 2009. Impairment of ubiquitylation by mutation in Drosophila E1 promotes both cell-autonomous and non-cell-autonomous Ras-ERK activation in vivo. J. Cell. Sci.. 122, 1461–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu, R. , Perera, R.M. , Balkin, D.M. , Pirruccello, M. , Toomre, D. , De Camilli, P. , 2009. A phosphoinositide switch controls the maturation and signaling properties of APPL endosomes. Cell. 136, 1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]


