SYNOPSIS
The eukaryotic translation initiation factor 5A (eIF5A) is the only cellular protein containing hypusine, [Nε-(4-amino-2-hydroxybutyl)lysine]. eIF5A is activated by the posttranslational synthesis of hypusine and the hypusine modification is essential for cell proliferation. In this study, we report selective acetylation of the hypusine and/or deoxyhypusine residue of eIF5A by a key polyamine catabolic enzyme, spermidine/spermine- N1-acetyltransferase 1 (SSAT1). This enzyme normally catalyzes the N1-acetylation of spermine and spermidine to form acetyl-derivatives, which in turn are degraded to lower polyamines. Although SSAT1 has been reported to exert other effects in cells by its interaction with other cellular proteins, eIF5A is the first target protein specifically acetylated by SSAT1. Hypusine or deoxyhypusine, as the free amino acid, does not act as a substrate for SSAT1, suggesting a macromolecular interaction between eIF5A and SSAT1. Indeed, the binding of eIF5A and SSAT1 was confirmed by pull-down assays. The effect of the acetylation of hypusine on eIF5A activity was assessed by comparison of acetylated vs non-acetylated bovine testis eIF5A in the methionyl-puromycin synthesis assay. The loss of eIF5A activity by this SSAT1-mediated acetylation confirms the strict structural requirement for the hypusine side chain and suggests a possible regulation of eIF5A by hypusine acetylation/deacetylation.
Keywords: eIF5A, hypusine, SSAT1, posttranslational modification, polyamine metabolism, acetylation
INTRODUCTION
Spermidine/spermine-N1-acetyltransferase 1 (SSAT1) is the key enzyme in the metabolism of polyamines and catalyzes acetylation of spermidine or spermine at the N1 of the aminopropyl moiety (reviews [1, 2]). The monoacetylated spermidine or spermine is oxidatively degraded by N1-acetylpolyamine oxidase N-acetylaminopropanal and a smaller polyamine, or the acetylated polyamines may be excreted from cells leading to a decrease in cellular polyamine levels. The level of SSAT1 is normally very low, but can be rapidly induced by a variety of stimuli, including polyamines, polyamine analogs, toxic chemicals, certain drugs and growth factors. Induction of SSAT1 results in an increase in metabolic flux, calorie consumption and fatty acid metabolism [3-6] and SSAT1 has been implicated in pathological conditions such as pancreatitis [7], obesity and diabetes [3, 5, 6]. SSAT1 has also been implicated in cancer, for its growth suppressive effects through depletion of polyamines, and also for its transforming effects due to increased tissue putrescine or due to reactive oxygen species generated from oxidation of acetyl polyamines [8, 9].
In addition to its role in polyamine catabolism, SSAT1 has been proposed to interact with certain cellular proteins. It has been reported to be involved in integrin-mediated cell migration by its binding to the α9β1 integrin [10], which functions in conjunction with the potassium channel Kir4.2 modulated by local polyamine concentrations [11]. SSAT1 and its closely related enzyme, SSAT2 (TLAT), were implicated in the regulation of the HIF-1α by their binding to HIF-1α [12, 13]. The binding of SSAT1 to the diamine exporter protein SLC3A2 suggests a concerted mechanism of polyamine acetylation and its export. Although these effects appear to be dependent on SSAT1 enzyme activity, it is unclear whether they are mediated by acetylation of polyamines or by acetylation of these SSAT1 binding partners or other substrates. The autoacetylation of one specific Lys (K26) and the open active site structure of SSAT1 [14] leave open the possible acetylation of other substrates beside polyamines.
The putative eukaryotic translation initiation factor 5A (eIF5A)5 contains a polyamine-lysine conjugated amino acid, hypusine, [Nε-(4-amino-2-hydroxybutyl)lysine] (at one site, 1 mol per mol) (see reviews [15-17]), which is essential for its activity. The long side chain of the hypusine/deoxyhypusine resembles the polyamine spermidine/homospermidine. Hypusine is formed post-translationally in the eIF5A precursor, eIF5A(Lys), by two consecutive enzymatic steps. In the first step, deoxyhypusine synthase (DHS) catalyzes the transfer of the aminobutyl moiety from the polyamine spermidine to a specific lysine residue (Lys50 for the human eIF5A) to form the deoxyhypusine [Nε-(4-aminobutyl)lysine] intermediate [18]. This deoxyhypusine residue is subsequently hydroxylated by deoxyhypusine hydroxylase (DOHH) [19] to form the biologically active eIF5A. The vital role of the hypusine/deoxyhypusine modification and eIF5A in eukaryotic and mammalian cell proliferation has been established from gene disruption studies of eIF5A and DHS in the yeast, Saccharomyces cerevisiae, and from studies involving the inhibition of polyamine or hypusine biosynthesis [20-25].
Although eIF5A is essential for eukaryotic cell growth and survival, the precise cellular function of this putative initiation factor is not fully understood. eIF5A stimulates methionyl-puromycin synthesis, a model assay for the first peptide bond formation, in a hypusine-dependent manner [26, 27]. Recent evidence for the association of eIF5A with actively translating ribosomes [28, 29] and the increase in the polysome/monosome ratio in certain eIF5A mutant yeast strains suggest a role for eIF5A in the elongation step of translation [30, 31].
eIF5A is an abundant protein with a long half life (>>24h) and the hypusine modification is an irreversible process. Thus it is difficult to manipulate the cellular level of eIF5A by the use of inhibitors in the pathways of polyamine biosynthesis or hypusine modification. Therefore, we explored other potential mechanisms of regulation of eIF5A activity. Lys47 and Lys68 of eIF5A have been reported as targets for acetylation [32, 33]. Comparison of the three different human eIF5A mutants involving Lys47, K47A, K47D and K47R provided an interesting insight into the role of Lys47 [4]. The fact that eIF5A activity was partially impaired by Ala substitution of Lys47 and totally by Asp substitution, but not by Arg substitution, suggests that the basic charge of Lys47 is important for its activity and that eIF5A activity is negatively regulated by acetylation in cells.
In searching for the cellular acetyltransferase that is involved in eIF5A acetylation, we tested four histone acetyltransferase recombinant enzymes, p300, CBP, PCAF and GCN5, and also the polyamine acetyltransferase SSAT1 and its sequence-related enzyme SSAT2 (also termed as TLAT, thialysine acetyltransferase) [34] in vitro. We observed acetylation of the non-hypusinated eIF5A precursor, eIF5A(Lys), by p300 and CBP. Interestingly, unlike eIF5A(Lys), the hypusine-containing eIF5A was effectively acetylated by SSAT1 (but not by any of the other enzymes tested) and the site of acetylation was determined to be the terminal amino group of the hypusine side chain. Acetylation of the hypusine residue by SSAT1 inactivates eIF5A activity in the methionyl-puromycin synthesis assay in vitro and suggests a potential regulation of eIF5A activity by reversible acetylation/deacetylation at this site.
EXPERIMENTAL
Materials
[1,8-3H]Spermidine.HCl (15-25 Ci/mmol) and [acetyl-3H]acetyl-Coenzyme A (AcCoA)(3.6 Ci/mmol) were purchased from PerkinElmer/NEN. Recombinant histone acetyltransferase (HAT) enzymes, p300, CBP, PCAF and hGCN5, were purchased from Biomol Research Labs, Inc. Lipofectamine 2000, precast Tris-glycine and NuPAGE (Bis-Tris) gels, and electrophoresis buffers were from Invitrogen and ECL Plus Western Blotting Detection system from GE Healthcare. A monoclonal antibody against recombinant human eIF5A (aa58-154) was purchased from BD Biosciences, AcLys antibody from Santa Cruz. The anti-FLAG antibody (mouse monoclonal) and polyamines, putrescine, spermidine and spermine were from Sigma. Deoxyhypusine and hypusine were chemically synthesized as described previously [35]. Recombinant SSAT1 and SSAT2 enzymes were purified as described previously [34]. Radiolabeled eIF5A([3H]Dhp) was produced in an in vitro DHS reaction [36]. Recombinant eIF5A proteins produced in E. coli, eIF5A(Lys), eIF5A(Dhp), and eIF5A(Hpu) were purified from E. coli cells using a polycistronic vector pST39 encoding human eIF5A-1 alone, two proteins (human eIF5A-1 and DHS) and all three proteins (human eIF5A-1, DHS and DOHH), respectively (JH Park et al., unpublished results). These recombinant eIF5A proteins were used for the majority of experiments in Fig 1–Fig 4. Mammalian eIF5A (hypusine form) was purified from human red blood cells [37], Chinese hamster ovary cells [27] and rabbit reticulocyte lysates [38] as described. BENSpm was kindly provided by Dr. Patrick M Woster (Wayne State University).
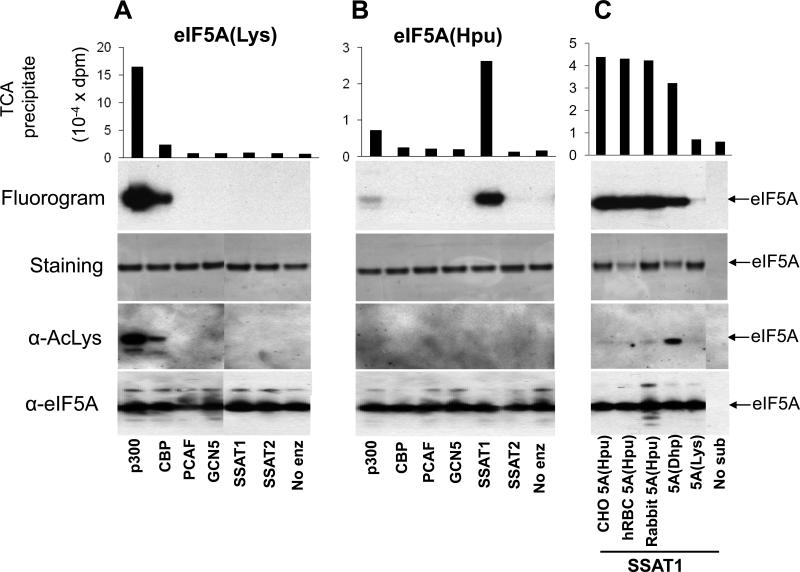
Figure 1. Specificity of acetylation of eIF5A(Lys) and eIF5A(Hpu) by different acetyltransferases in vitro.
The acetyltransferase assays were carried out as described in “Experimental”, using [3H]AcCoA, 0.05 μg of each enzyme as indicated, with 2 μg each of (A) recombinant eIF5A(Lys) or (B) recombinant eIF5A(Hpu). (C) eIF5A (hypusine form) purified from Chinese hamster ovary (CHO) cells, human red blood cells (hRBC), or rabbit reticulocyte, eIF5A(Dhp) prepared by an in vitro DHS reaction, or eIF5A(Lys) were used as substrates for SSAT1 under the same condition as in A and B, except that 0.1 μg of enzyme was used. After 1 h incubation, radioactivity incorporated into eIF5A protein was measured in the TCA-precipitated proteins and also by fluorography after SDS-PAGE. Aliquots of reaction mixtures were used for western blot analyses with AcLys antibody or eIF5A antibody. The experiments were carried out two times with similar results and representative data are shown. Abbreviation: 5A, eIF5A
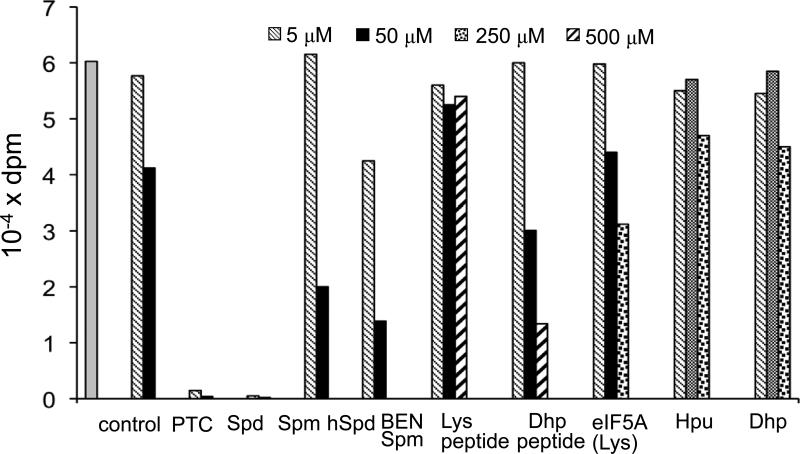
Figure 4. Effects of polyamines, hypusine, deoxyhypusine, eIF5A synthetic peptides and proteins on the acetylation of eIF5A(Hpu) by SSAT1.
The reaction mixture contained recombinant eIF5A(Hpu) protein (3 μM), 0.1 μg of SSAT1, 5 μM of [3H]AcCoA and indicated concentration of inhibitors (5, 50, 250 or 500 μM). After 1 h at 30 °C, the reaction was stopped by addition of 10 % TCA and the radioactivity incorporated into protein was measured. Representative data is shown from two independent experiments whose results were similar. Arreviations: Ptc, putrescine; Spd, spermidine; Spm, spermine; hSpd, sym-homospermidine; Dhp, deoxyhypusine; Hpu, hypusine
Methods
Transfection and pull down assays
The HeLa cells were cultured in Dulbecco's modified Eagle's medium(DMEM) supplemented with 10% heat-inactivated fetal bovine serum. For DNA transfection, HeLa cells were trypsinized, plated in 100mm dishes, and grown to 70-80% confluence. The cells were transiently transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendation with the indicated expression vector (2μg) and treated with or without the polyamine analog, BENSpm. At 48h after transfection, cells were harvested and lysed in a buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1% NP-40, 5 mM EDTA, and cocktail of protease inhibitor). After the removal of cell debris by centrifugation at 13,000 g for 15 minutes, equal amounts of proteins (20-50 μg) were separated by SDS-PAGE and transferred to nitrocellulose membranes. Blotted membranes were blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.1% Tween 20 (TBST) and then incubated with the primary antibodies overnight at 4 °C. After washing with TBST buffer three times, blotted membrabes were incubated with the anti-mouse secondary antibodies for 1hr at room temperature and were visualized by chemiluminescence. For immunoprecipitation, cell lysate containing 2 mg total protein was preincubated with protein A/G beads for 30 minutes to remove non-specifically binding proteins. The lysate was then incubated with anti-FLAG or anti-eIF5A1 antibody and protein A/G beads overnight. After washing precipitates three times, the precipitates were dissolved in SDS-sample buffer for SDS-PAGE and western blotting.
In vitro HAT, SSAT1 and SSAT2 acetyltransferase reactions
A typical reaction mixture in 50 μl contained 50 mM Tris-HCl pH 8.0, 1 mM DTT, 10 % glycerol, 0.1 mM EDTA, [3H]AcCoA (1 μCi, 3.6 Ci/mmol, 280 pmol, 5.6 μM), with indicated amounts of enzyme and eIF5A substrate protein. The reaction mixture was incubated at 30 °C for 1h or the indicated times. The reaction was stopped by addition of 10 % TCA. 500 μg of carrier BSA was added and the samples kept on ice for 30 min. The TCA-precipitated proteins were centrifuged in a microfuge (15,000 g) at 4 °C for 5 min and the precipitates were washed three times with 10 % TCA to remove free [3H]AcCoA. Aliquots of the precipitates were used for detection of radiolabeled protein by SDS/PAGE and fluorography, for western blotting, and for counting of radioactivity to measure the amount of [3H]acetyl moiety (of [3H]AcCoA) incorporated into proteins.
Chemical synthesis of acetylhypusine
Acetylhypusine [(4-acetamido-2-hydroxybutyl)lysine] was prepared from hypusine using a selective acetylating reagent. To (2S,9R)-hypusine(free-base)(2) 316 mg (1.36mmol) in 5 ml of ethanol at 5°C was added 365mg (1.49mmol,1.1 eq) of 2-trifluoroethyl-N,N-diacetylaniline [39] in one portion with stirring. After continued stirring for 2 h at room temperature the solvent was evaporated and the residue was chromatographed on silica gel with a mixture of methylene chloride: methanol: conc. ammonium hydroxide (2:2:1). The residue, after removal of solvent, was dissolved in 1 ml of water and the pH was adjusted to 5.2 with dilute HCl. Removal of solvent and addition of ethanol afforded a crystalline product as the mono HCl salt which was recrystallized from water/ethanol. Yield 212mg (45%); mp. 235-236 °C decomp.; 1H-NMR (500 MHz, D2O) δ 3.93 (t, J = 9.3 Hz, 1 H), 3.71 (t, J = 6.0 Hz, 1 H), 3.27 (t, J = 6.9 Hz, 1 H), 3.14 (dd, J = 12.9, 2.4 Hz, 1 H), 3.07 (t, J = 7.7 Hz, 2 H), 2.97 (dd, J = 12.8, 10.4 Hz, 1 H), 1.96 (s, 3 H), 1.89-1.84 (m, 2 H), 1.76-1.70 (comp, 3 H), 1.66-1.58 (m, 1 H), 1.53-1.35 (comp, 2 H); 13C-NMR (125 MHz, D2O) δ 174.18, 173.78, 64.16. 53.99, 51.69, 46.67, 35.13, 32.86, 29.42, 24.53, 21.34, 21.02.;HRMS (CI) m\z (M+1) calculated for C12H25N3O4: 276.1923. found 276.1931; Analysis calculated for C12H25N3O4. HCl.0.2 H2O: C45.7, H 8.44, N 13.32. Found C 45.84, H 8.52, N 13.20; [α]D + 4.25 (C= 1.0, H2O)
Structural verification of this product as 11-acetamido-2- amino-7aza-9-hydroxyundecanoic acid was attained through identification of lysine as the sole dinitrophenylated amino acid produced upon dinitrophenylation of its oxidation products, under conditions described previously [40]. CH3CONH-CH2-CH2-CHOH-CH2-NH-CH2-CH2-CH2-CH2-CHNH2-COOH + HIO4 + KMnO4 + FDNB= CH2CH2CH2CH2CHCOOH- NHDNP
Chemical synthesis of eIF5A peptides, IVEMSTSKTGK50HGHAK and IVEMSTSKTGDhp50HGHAK
The two 16-mer peptides were prepared by a solid-phase procedure as previously described for the 9-mer eIF5A peptides [41] and their purity and identity were validated by thin layer chromatography and by amino acid analysis after acid hydrolysis.
Identification of acetyllysine, acetylhypusine, and acetyldeoxyhypusine, hypusine and deoxyhypusine by ion exchange chromatographic separation
For identification of radiolabeled amino acid component of the HAT, SSAT1 and SSAT2 reaction products, TCA-precipitated proteins were exhaustively digested with pronase as follows: TCA precipitated proteins (washed free of [3H]AcCoA with repeated TCA wash) were washed with acetone to remove TCA, and the protein pellet was resuspended in 0.1 ml of 0,1M Tris.HCl pH 8.0 buffer. To this, 10 μl of pronase (50 mg/ml in the same buffer) and thimerosal (0.1 mg/ml) were added and the mixture was incubated for 24 h at 37 C. The pronase addition was repeated three times and after 72 h of digestion, 30 μl of 50 % TCA was added. After removal of TCA precipitates, the radioactive component in the TCA supernatant was analyzed by ion exchange chromatography on a Dionex D-400 analyzer using a column (0.4 × 8 cm) of DC-6A resin (Dionex) as described [42] and the buffer system (10 min, 66 mM sodium citrate pH 4.31 and 10 min, 13.2 mM sodium citrate buffer pH 5.55 containing 1.5 M NaCl, 20 min, 66 mM sodium citrate pH 4.31). 1 min fractions were collected for counting of the radioactivity or the amino acids were detected by post-column fluorometric detection after reaction with ortho-phthaldehyde.
Purification of eIF5A from bovine testis
We purified bovine testis eIF5A (hypusine form) using the radiolabed, eIF5A([3H]Dhp) as a tracer, since the deoxyhypusine form and the hypusine form have virtually the same pIs and ion exchange chromatographic properties. Bovine testis (1kg) was chopped and ground in a blender in 1.0 liter of ice-cold Buffer A (Tris-acetate, pH 6.7, 0.1 mM EDTA, 1 mM DTT, 1 mM benzamidine, 1 mM PMSF). The lysate was centrifuged at 15,000 g for 40 min. The supernatant was passed through glass wool to remove debris. A radioactive tracer protein, eIF5A([3H]Dhp) (106 dpm, 62 pmol, 1 μg) was added to the clarified lysates and the mixture was applied to a 120 ml size column of DEAE sepharose. Throughout the purification steps, the fractions containing eIF5A were identified by SDS-PAGE and by the counting of radioactivity. After washing the column with 200 ml of Buffer A, the proteins were eluted with a salt gradient (100 ml each step) of 0.1, 0.2, 0.3, 0.4 and 0.5 M KCl in Buffer A, and 25 ml fractions were collected. A 100 μl aliquot of each fraction was counted and the radioactive fractions were pooled. This pool was treated with 40-80 % ammonium sulfate. The proteins from ammonium sulfate precipitation (~2 g) after dialysis in Buffer A were applied to a Q-sepharose column (70 ml size). After washing the column with 200 ml of buffer A, the proteins were eluted with a step-wise gradient (50 ml each step) of 0.1, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4 and 0.5 M KCl in Buffer A and 25 ml fractions were collected. The radioactivity in each fraction was counted and 2 fractions with the highest radioactivity were pooled and dialyzed against Buffer B (50 mM Tris acetate pH 7.0, 1 mM DTT, 0.1 mM EDTA) and applied to a SP-sepharose column (20 ml). The sample was applied, and column was washed with 100 ml of Buffer B. The bound proteins were eluted with a KCl gradient of 0.1 M (25 ml), 0.2 M (25 ml), 0.3 M (50 ml), 0.4 M (50 ml), and 0.5 M (50 ml). 5 ml fractions were collected. Two fractions with the highest radioactivity were pooled and ammonium sulfate added to 1.5 M for phenyl sepharose chromatography (10 ml size). The column was equilibrated in Buffer B containing 1.5 M ammonium sulfate. The pooled proteins were applied and the column was washed with 20 ml of the initial buffer (1.5 M ammonium sulfate). The bound proteins were eluted with decreasing concentration of ammonium sulfate, steps (10 ml each) of 1.35, 1.2, 1.05, 0.9, 0.75, 0.6, 0.45, 0.3 and 0.15 M and 5 ml fractions were collected. The three fractions containing eIF5A protein and the radioactivity were pooled and concentrated and equilibrated in 50 mM Tris.HCl, pH 7.5, 0.1 M KCl, 1 mM DTT.
Acetylation of bovine testis eIF5A with SSAT1 and separation of acetylated eIF5A
Acetylated bovine testis eIF5A was produced by an in vitro SSAT1 reaction and separated from the non-acetylated eIF5A and SSAT1 enzymes as follows. The large scale SSAT1 reaction mixture contained in 0.3 ml, 50 mM Tris-Cl pH 8.0, 1 mM DTT, 10% glycerol, 0.1mM EDTA, 1 mg (60 nmol) of eIF5A and 3×105 dpm (0.3 μg) of radioactive tracer protein, eIF5A([3H]Dhp), 200 nmol of unlabeled AcCoA, and 40 μg of SSAT1. The reaction mixture was incubated at 30 °C for 2 h. A small aliquot (10 μl) of the reaction mixture was mixed with [3H]Ac CoA (8 μl) and 2 μl of 5x buffer to monitor the progress of acetylation at 0, 2 and 4 h. At 2 and 4 h, extra unlabeled AcCoA (100 nmol) and enzyme (40 μg) were added and a majority of eIF5A was acetylated at 6 h. As a control for non-acetylated eIF5A, 0.1 mg of eIF5A was incubated in a parallel reaction mixture (30 μl, but without SSAT1) for the same period at 30° C. The SSAT1 reaction mixture was applied to a column of SP-sepharose (8 ml size) equilibrated with Tris buffer C (50 mM Tris-HCl, pH 7.4, 1 mM DTT) and the column was washed with 10 ml of Buffer C. The acetylated and non-acetylated eIF5A proteins were separated upon elution with a KCl gradient (5 ml each step) of 0.1, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4 and 0.5 M in Buffer C. The acetylated and non-acetylated eIF5A was concentrated and equilibrated in 50 mM Tris.HCl, pH 7.5, 0.1 M KCl, 1 mM DTT prior to the activity assay.
In vitro methionyl-puromycin synthesis assay for eIF5A activity
The reaction mixture (in 25 μl) contained 20 mM Tris-HCl pH7.4, 0.1 M KCl, 1 mM DTT, 3 mM MgCl2, 0.8 mM GTP, 1.5 mM puromycin, 0.15 A260 unit of 60S, 0.07 A260 unit of 40 S, 33 μM AUG, 0.375 μg of eIF1A, 0.75 μg of eIF2, 3.3 μg of eIF3, 0.48 μg of eIF5B and 8 pmol of Met-tRNA (120,000 dpm) and the indicated amounts of bovine testis eIF5A and its acetylated counterpart. The mixture was incubated at 37 °C for 20 min. To each tube, 0.4 ml of 0.2 M Na2HPO4 buffer, pH 8.0, was added. 1 ml of ethyl acetate was added and the tubes shaken for 10 min. The tubes were centrifuged in a microfuge for 10 min. The upper layer (~0.9 ml) was carefully removed and the radioactivity was counted.
RESULTS
Acetylation of eIF5A by histone acetyltransferases and spermidine/spermine acetyltransferase
Several lines of evidence indicating the acetylation of eIF5A at specific lysine residues, Lys47 [4] and Lys68 [43] have been reported. In an effort to identify the cellular acetyltransferase that may be responsible for the acetylation of eIF5A, we tested four HAT enzymes, p300, CBP, PCAF, hGCN5 as well as SSAT1 and a related enzyme, SSAT2, using recombinant eIF5A proteins produced in E. coli, eIF5A(Lys) and eIF5A(Hpu) in vitro (Fig. 1 A and B). All four of the HAT enzymes were effective in the acetylation of histone III as a substrate (data not shown). However, a highly selective pattern of acetylation was observed with recombinant eIF5A(Lys) or eIF5A(Hpu) as substrate. The eIF5A(Lys), was effectively acetylated by p300 and moderately by CBP, as judged by incorporation of radioactivity from [3H]AcCoA into protein and also by western blotting with an AcLys antibody (Fig.1 A).
A totally distinct acetylation pattern was observed for recombinant eIF5A(Hpu) (Fig. 1 B). Only very faint labeling of eIF5A(Hpu) was observed with p300, but strong labeling was observed with SSAT1. No other enzyme resulted in eIF5A(Hpu) acetylation. Curiously, eIF5A(Hpu) acetylated by SSAT1 was not detected by the AcLys antibody, suggesting that acetylation did not occur at any of its Lys residues. This observation, together with the fact that only the hypusine-containing protein, but not the precursor, was acetylated by SSAT1, indicates that acetylation occurs at the hypusine residue. We also tested natural mammalian eIF5A as substrates for SSAT1. In mammalian cells and tissues, natural eIF5A exists predominantly as the fully modified hypusine-containing form[25, 27]. Mammalian eIF5A proteins (hypusine form) purified from three different sources, Chinese hamster ovary (CHO) cells , human red blood cells (hRBC) or from rabbit reticulocytes, all acted as effective substrates for SSAT1 (Fig. 1 C) as the recombinant eIF5A(Hpu) produced in E. coli (Fig 1B). Deoxyhypusine-containing eIF5A, eIF5A(Dhp), was also acetylated by SSAT1, although it was not as good a substrate as the hypusine-containing protein. Interestingly, unlike the acetylated eIF5A(Hpu), the acetylated eIF5A(Dhp) was recognized by the AcLys antibody (Fig. 1 C), probably due to the acetylated butyl amino side chain present in both eIF5A(Lys) and eIF5A(Dhp) .
Acetylation of eIF5A(Hpu) or eIF5A(Dhp) by SSAT1 occurs selectively at the hypusine/deoxyhypusine residue
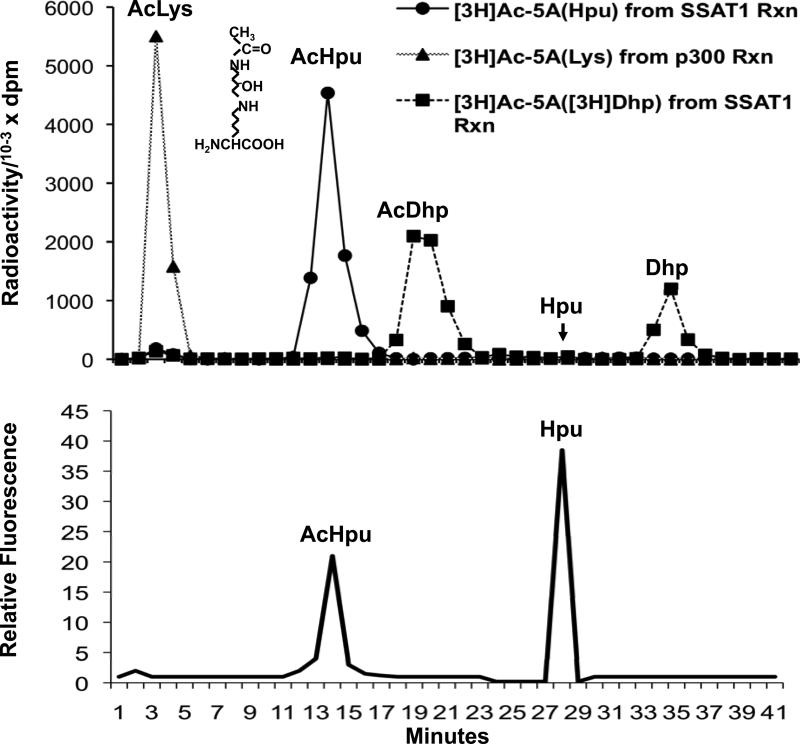
In order to confirm the sites of acetylation, acetylated eIF5A proteins, as shown in Fig 1, were exhaustively digested using pronase and the digests were separated by ion-exchange chromatography (Fig. 2 upper panel). The eIF5A(Lys) acetylated by p300 contained only acetyl lysine as the labeled component, consistent with the western blot results with AcLys antibody. In contrast, the major radioactive component in the eIF5A(Hpu) acetylated by SSAT1 was identified as acetylhypusine with the acetyl moiety on the terminal amino group of the hypusine side chain (see structure in Fig. 2). The radioactivity eluted at the exact position of a synthetic acetylhypusine standard, whose chemical structure was validated by NMR (bottom panel, Fig. 2). Furthermore, it was destroyed by acid hydrolysis, as the acetyl moiety was cleaved from acetylhypusine, as expected. Only a small amount of acetyl lysine (<3 % of the total) was found in the digest of acetylated eIF5A(Hpu) and it is not clear whether it is derived from eIF5A or from SSAT1 enzyme autoacetylation. When radiolabeled eIF5A(Dhp), with radioactivity in the terminal aminobutyl side chain of deoxyhypusine, was used as a substrate for the SSAT1 reaction with [3H]AcCoA, two radioactive components were detected after exhaustive proteolytic digestion, deoxyhypusine and an early peak at the expected position of acetyldeoxyhypusine. This radiolabeled component of acetylated eIF5A(Dhp) elutes at a time 4-5 minutes later than that for acetylhypusine, as expected, just as deoxyhypusine elutes 4-5 min later than hypusine. This may be because the absence of the hydroxyl group in deoxyhypusine, or its acetylated form, permits stronger hydrophobic interaction with the resin. The radiolabeled material in the early peak (acetyl deoxyhypusine) was completely converted to deoxyhypusine after acid hydrolysis, confirming its identity as acetyl deoxyhypusine.
Figure 2. Ion exchange chromatographic separation of acetyllysine, acetylhypusine, acetyldeoxyhypusine, hypusine and deoxyhypusine.
(A) The radiolabeled, acetylated eIF5A proteins produced from SSAT1 reactions with recombinant eIF5A(Hpu) or with eIF5A([3H]Dhp) and that from a p300-catalyzed reaction with eIF5A(Lys) (the same or similar reaction as shown in Fig 1) were exhaustively digested with pronase and the radioactive amino acids in the digests were separated by ion exchange chromatography. (B) Acetyl hypusine was identified by its elution time, which was identical to that of the chemically synthesized acetyl hypusine standard determined by fluorometric detection. The position of each amino acid is indicated and the chemical structure of acetyl hypusine is shown. Abbreviations: AcLys, acetyllysine; AcHpu, acetylhypusine; AcDhp, acetyldeoxyhypusine; Hpu, hypusine; Dhp, deoxyhypusine
Specificity of the SSAT1 reaction: inhibition of eIF5A acetylation by polyamines, hypusine, deoxyhypusine, eIF5A(Lys) and eIF5A-derived peptides
The acetylation reaction by the SSAT1 increased with time and was dependent on the substrate concentration (Fig. 3). The Km for eIF5A(Hpu) was estimated to be approximately 60 μM (data not shown). Thus, eIF5A(Hpu) is a fairly good substrate for SSAT1. The long side chain of the hypusine or deoxyhypusine residue resembles the polyamine spermidine and this may be the molecular basis of its acetylation by SSAT1. In order to test this hypothesis and to compare its substrate efficiency with those of polyamines, we tested polyamines, the analog BENSpm, free hypusine and deoxyhypusine, eIF5A(Lys) and eIF5A peptides (16 mer) as inhibitors (Fig. 4). SSAT1 was reported to be specific for the aminopropyl moiety; it does not effectively acetylate the aminobutyl moiety of the polyamines, putrescine, spermidine and sym-homospermidine [44]. Consistent with this finding, putrescine and sym-homospermidine were poor inhibitors of eIF5A(Hpu) acetylation causing little or no inhibition at 5 μM. In contrast, spermidine and spermine (at 5 and 50 μM) strongly inhibited eIF5A(Hpu) acetylation (substrate at 3 μM) , indicating that they are better substrates for the SSAT1 than the eIF5A(Hpu) (Fig. 4). BENSpm exhibited a moderate inhibition.
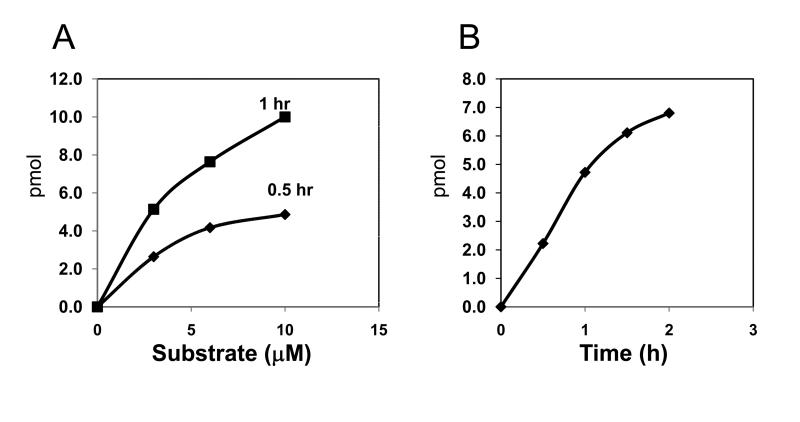
Figure 3. The dependence of the SSAT1 reaction on the concentration of substrate and time.
The reaction was conducted as outlined under “Experimental” (A) with varying amounts of the substrate at 0.1 μg of SSAT1 and (B) with varying incubation time at 3 μM of substrate and 0.1 μg of enzyme. The product formed is indicated in pmol (1 pmol equals 7200 dpm). Representative data from two similar sets of experiments are shown.
Hypusine or deoxyhypusine, as the free amino acid, did not inhibit acetylation of eIF5A(Hpu) (no or little inhibition at 5 -50 uM, 10-20 % inhibition at 250 μM). This finding indicates that eIF5A(Hpu) acetylation by SSAT1 is not merely by virtue of the similarity of the long side chain of these amino acids to the polyamines and that unlike the protein eIF5A(Hpu), these free amino acids are not good substrates for SSAT1. Then we tested two synthetic peptides (16 mers) modeled on the sequence of amino acids surrounding the hypusine residue 50, (IVEMSTSKTGK50HGHAK and IVEMSTSKTGDhp50HGHAK) one with Lys 50 and the other with deoxyhypusine 50. The Lys peptide did not inhibit eIF5A acetylation by SSAT1 (even at 0.5 mM), whereas the deoxyhypusine-containing counterpart caused moderate inhibition (50% and 80 % at 50 and 500 μM, respectively). Thus the deoxyhypusine or hypusine residue of eIF5A seems to be critical for its binding to SSAT1, even though they are not substrates as free amino acids. The intact eIF5A(Lys) protein was better than the 16 mer Lys peptide, but not as inhibitory as the deoxyhypusine peptide. These findings suggest that both the extended amino acid sequences (macromolecular structure) of eIF5A and the target residues (hypusine and deoxyhypusine) are important for the binding and modification of eIF5A(Hpu) and eIF5A(Dhp) by SSAT1.
Molecular interaction between eIF5A and SSAT1
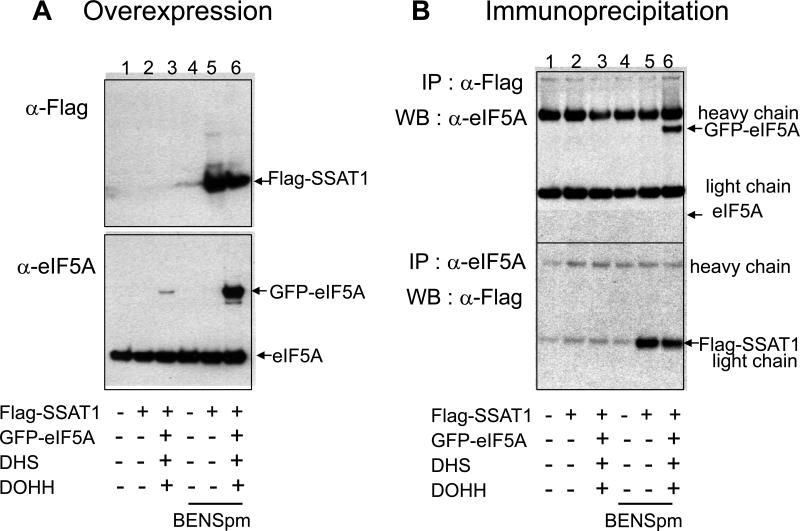
The observation that eIF5A(Hpu) and eIF5A(Dhp) are good substrates for SSAT1 whereas free hypusine, deoxyhypusine and short peptide derivatives are not, suggests that SSAT1 recognizes the eIF5A macromolecule as a substrate, not just the hypusine or deoxyhypusine residue. Therefore, we examined the interaction of eIF5A and SSAT1 by pull-down assays after co-transfection of GFP-eIF5A and FLAG-SSAT1 in HeLa cells (Fig. 5). As was reported earlier [45], GFP-eIF5A was cotransfected with the hypusine modification enzymes, DHS and DOHH, to produce the hypusine-modified form. Expression level of FLAG-SSAT1 was very low in cells transfected with either the SSAT1 vector alone (Fig. 5A, top panel, lane 2) or in those cotransfected with the vectors encoding eIF5A, DHS and DOHH (Fig. 5A, top panel, lane 3). Expression of GFP-eIF5A was remarkably low in cells cotransfected with SSAT1 (Fig. 5A bottom panel, lane 3) due to a novel activity of SSAT1 in suppression of exogenous gene expression [46]. Since SSAT1 has a very short half life and is stabilized by a polyamine analog BENSpm [1], we treated cells with this compound after transfection. Indeed, the expression level of SSAT1 was restored in the presence of BENSpm (Fig 5A, top panel, lanes 5 and 6) and so was the expression of GFP-eIF5A (Fig 5A, bottom panel lane 6). Upon immuno-precipitation of the cell lysate with the FLAG antibody, GFP-eIF5A (hypusine form) was pulled down with FLAG-SSAT1 (lane 6 in Fig. 5 B). Reciprocally, FLAG-SSAT1 was immuno-precipitated along with eIF5A when the lysate was immuno-precipitated with eIF5A antibody (Fig. 5B). Taken together, these results suggest a specific physical interaction between eIF5A protein and SSAT1.
Figure 5. Physical interaction between eIF5A and SSAT1 in cells.
HeLa cells were cotransfected with GFP-eIF5A, DHS, DOHH and FLAG-SSAAT1 vectors as indicated and treated with or without 10 μM BENSpm, which was added at the time of medium change after transfection. (A) The overexpression of Flag-SSAT1 and GFP-eIF5A was detected by western blotting of cell lysates. (B) The cell lysates were immuno-precipitated with either with anti-FLAG antibody (top panel) or with eIF5A antibody (bottom panel) and immuno-precipitates were analyzed by western blotting with antibodies indicated. The experiments were repeated once with consistent results.
The effects of hypusine acetylation on eIF5A activity in vitro
Considering the stringent requirement for the hypusine for eIF5A activity, acetylation of the terminal amino-group of the hypusine side chain is expected to affect molecular interactions involving the hypusine residue and therefore the biological activity of eIF5A. We have prepared acetylated eIF5A, by an in vitro SSAT1 reaction with eIF5A (hypusine form) isolated from bovine testis. The acetylated eIF5A protein, with a more acidic pI, was separated from its non-acetylated counterpart upon ion exchange chromatography on SP-sepharose column (Fig. 6 A). The activity of the acetylated eIF5A was compared with that of non-acetylated bovine eIF5A incubated in a parallel reaction mixture without SSAT1, using the methiony-puromycin synthesis assay (Fig. 6B). The activity of acetylated eIF5A (Fig. 6 B) was lost, indicating that acetylation of the hypusine side chain inactivates eIF5A. The activity of recombinant eIF5A(Hpu) produced in E. coli was also lost upon hypusine acetylation by SSAT1(data not shown).
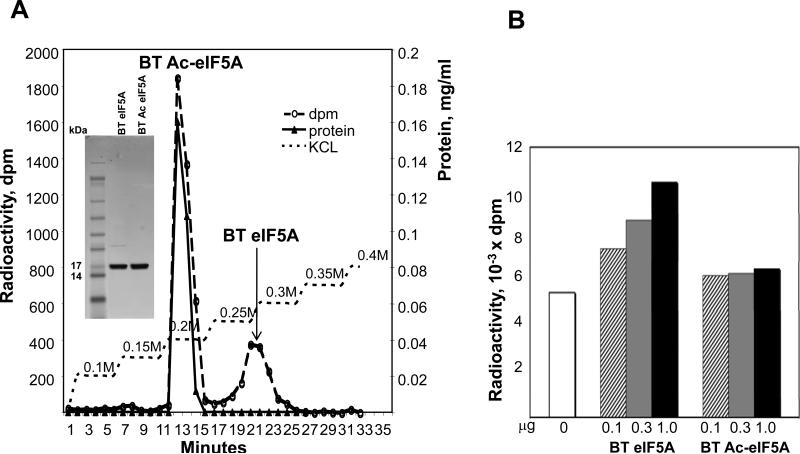
Figure 6. Purification of acetylated bovine testis eIF5A by ion exchange chromatography and comparison of activities of acetylated vs non-acetylaated eIF5A.
(A) The bovine testis eIF5A (hypusine form) was mixed with radioactive tracer, eIF5A([3H]Dhp) and was incubated with SSAT1 as described under Experimental. The acetylated eIF5A (F13-15) was separated by ion exchange chromatography on SP-sepharose resin. Bovine testis eIF5A was completely acetylated, whereas the radioactive tracer, eIF5A([3H]Dhp) was not, probably because the hypusine-containing eIF5A is a better substrate for SSAT1 than the deoxyhypusine-containing form. The purity of nonacetylated and acetylated eIF5A on SDS gel is shown in the inset.
(B) The methionyl-puromycin synthesis assay was conducted using acetylated eIF5A prepared in (A) and non-acetylated counterpart (from a parallel acetylation reaction mixture but without SSAT1), as described under “Experimental” in duplicate with 0.1, 0.3 and 1.0 μg of bovine testis eIF5A and its acetylated counterpart. Average values are of duplicates.
Abbreviation: BT, bovine testis; BT-AceIF5A, acetylated bovine testis eIF5A
DISCUSSION
In this paper we present definitive biochemical evidence for acetylation of eIF5A at its hypusine residue by SSAT1. eIF5A is the first reported target substrate protein of SSAT1 to be completely inactivated by acetylation. SSAT1 selectively targets the terminal amino group of the deoxyhypusine/hypusine side chain of eIF5A, which is located at the tip of an exposed hypusine loop in the N-terminal domain. The specificity may be due, in large part, to the similarity of the hypusine side chain to the polyamine spermidine. However, the acetylation of eIF5A by SSAT1 probably involves a specific macromolecular interaction between the enzyme and substrate, since free hypusine or deoxyhypusine does not appear to act as a substrate for SSAT1. Even the 16-mer deoxyhypusine peptide exerted only modest inhibition of eIF5A hypusine acetylation. Acetylation of the hypusine/deoxyhypusine residue abolished the activity of eIF5A in methionyl-puromycin synthesis in vitro. The acetylation reduces the strong basic charge of the hypusine loop and may disturb the binding of eIF5A to the ribosome and to other acidic binding partners.
The hypusine modification is essential for eIF5A activity and thereby defines a key function of polyamines in cell proliferation [47]. In cells depleted of natural polyamines by inhibitors of their biosynthetic enzymes or by induction of their catabolic enzyme, SSAT1, only spermidine or those closely related analogs that serve as a substrate for deoxyhypusine/hypusine modification can support growth of mammalian cells in the long term [7, 20]. Polyamine biosynthetic enzymes, such as ornithine decarboxylase and adenosylmethionine decarboxylase, as well as the polyamine catabolic enzyme, SSAT1, have been targeted for anti-tumor therapy and chemoprevention[48]. Furthermore, the hypusine biosynthetic enzymes, deoxyhypusine synthase and deoxyhypusine hydroxylase, have been also explored as targets for intervention in aberrant cell proliferation. A drawback in targeting the hypusine modification is that it is not possible to deplete eIF5A rapidly, since eIF5A is abundant and has a long half life. Furthermore, there is no pathway known to convert hypusine-containing eIF5A back to an inactive precursor and the activity of cellular eIF5A cannot be regulated by reversing the hypusine modification.
In this regard, reversible modification of eIF5A, such as acetylation or phosphorylation, may be invoked for a rapid regulation of eIF5A activity without a change in its protein level. Although phosphorylation of yeast eIF5A at a serine (the second residue from the N-terminus) was detected, it probably is not required for eIF5A activity or its regulation, since the phosphorylated eIF5A is fully active [49] and serine to Ala substitution mutant supports yeast growth [50]. Acetylation of eIF5A at two specific lysine residues, K47 [33] and K68 [43] has been reported. K68D and K68A mutant forms support yeast growth (data not shown), suggesting that acetylation at this site does not affect eIF5A activity. In contrast, the loss of activity of the K47D mutant supports the notion that eIF5A activity is negatively regulated by acetylation at Lys47 [4]. Indeed, our previous data demonstrated that eIF5A(Lys) is the major acetylated protein in cells detected by an AcLys antibody [45]. Furthermore, eIF5A was identified as the main target substrate of the NAD-dependent deacetylase SIRT2 in S. cerevisiae [51]. Thus, eIF5A may be potentially regulated by two different acetylation mechanisms, one at Lys47 by a cellular HAT enzyme and the other at the hypusine/deoxyhypusine residue by SSAT1, depending on special physiological conditions.
SSAT2 (TLAT) was initially considered to be a candidate enzyme to acetylate eIF5A, because it is quite inefficient toward polyamines but can efficiently acetylate thialysine, 5-hydroxy-l-lysine, S-(aminoethyl)-homocysteine, and O-(aminoethyl)-l-serine, and has a modest activity on Lys itself. However, no acetylation of eIF5A(Hpu) or eIF5A(Lys) was observed with SSAT2. SSAT1 has been reported to exert specificity toward the aminopropyl moiety of spermidine, spermine or other low molecular weight amine substrates. The aminobutyl moiety in putrescine or sym-homospermidine was a poor substrate for the rat liver enzyme [44]. The weak inhibition of eIF5A(Hpu) acetylation by putrescine and sym-homospermidine, compared to the strong inhibition by spermidine and spermine, is consistent with the aminopropyl specificity. Interestingly, such structural specificity does not seem to apply to protein substrates, considering the acetylation of its own Lys26, and of the eIF5A deoxyhypusine or hypusine residue. SSAT1 was also reported to metabolize a drug, amantadine hydrochloride, used in the profilaxis and treatment of influenza A virus infection, by acetylating an achiral polycyclic aliphatic primary amine [52].
SSAT1 may have cellular functions in addition to polyamine metabolism. SSAT1 has been proposed to be involved in the integrin-mediated cell migration by its binding to the α9β1 integrin which functions in conjunction with the potassium channel Kir4.2 modulated by local polyamine concentrations[11]. Both SSAT1 and SSAT2 (TLAT) were implicated in the regulation of HIF-1α, supposedly by promoting its ubiquitination and degradation by the 26S proteosome. Although SSAT1 was reported to interact with these proteins, HIF-1α, integrin β9, SLC3A2, and the SSAT1 activity appeared to be critical for HIF-1α degradation [13] or integrin β9-mediated cell migration [11], there is no evidence for acetylation of these binding proteins by SSAT1. Furthermore, cotransfection with SSAT1 suppresses exogenous expression of other reporter proteins [46]. Although this effect was also dependent on SSAT1 enzyme activity, the loss of exogenous gene expression could not be simply attributed to a decrease in the overall cellular polyamine pools [46]. Thus, it is unclear whether it was mediated by polyamine acetylation (and consequent depletion of local polyamines) and/or by acetylation of other unknown protein substrates. The autoacetylation of its own K26 residue and the hypusine/deoxyhypusine residue of eIF5A by SSAT1 and the spacious active site of the SSAT1 crystal structure leave open the possibility of other SSAT1 target substrate proteins [14].
Whereas hypusine modification is tied to spermidine and its biosynthesis, it is intriguing to note that a potential eIF5A inactivation mechanism is linked to the polyamine catabolic enzyme SSAT1. SSAT1 induction leads to growth inhibition by depletion of cellular polyamines, which in turn leads to decrease in hypusine-containing eIF5A. Under normal cellular conditions, the SSAT1-mediated hypusine acetylation is probably not significant, considering that the SSAT1 enzyme level is low and that the eIF5A concentration is much lower than those of free cellular polyamines. However, upon super-induction of SSAT1 in cells, hypusine acetylation by SSAT1 may lead to inactivation of eIF5A (further reducing the level of biologically active eIF5A), while new synthesis of hypusine is blocked due to depletion of spermidine. Whether such regulation of the eIF5A level and activity contributes to growth inhibition in the case of SSAT1 super-induction and polyamine depletion warrants further investigation.
ACKNOWLEDGMENT
We thank Drs. Edith C. Wolff (NIDCR, NIH) for critical reading of the manuscript and helpful suggestions.
FUNDING
The research was supported in part by the Intramural Research Program of National Institute of Dental and Craniofacial Research (NIDCR), National Institute of Drug Abuse (NIDA) and National Institute on Alcohol Abuse and Alcoholism (NIAA), NIH, NIH extramural grant [grant number R01 GM092927 (to C.S F.)] and by the Human Frontier Science Program fellowship [fellowship number LT00575/2007-L (to M. S.)]i.
Abbreviations
- eIF5A
eukaryotic translation initiation factor 5A
- eIF5A(Lys)
eIF5A precursor
- eIF5A(Dhp)
eIF5A intermediate containing deoxyhypusine
- eIF5A(Hpu)
eIF5A containing hypusine
- DHS
deoxyhypusine synthase
- DOHH
deoxyhypusine hydroxylase
- SSAT1
spermidine/spermine-N1-acetyltransferase-1
- SSAT2
spermidine/spermine-N1-acetyltransferase-2
- N1-AcSpd
N1-Acetylspermidine
- HAT
histone acetyltransferase
- CBP
CREB binding protein
- PCAF
P300/CBP-associated factor
- BENSpm
N1, N11-bis(ethyl)norspermine
- AcCoA
acetyl coenzyme A
- TCA
trichloroacetic acid
- DTT
dithiothreitol
- GFP
green fluorescent protein
Footnotes
The term eIF5A usually refers to the native, naturally occurring eukaryotic protein that contains hypusine. Where it is necessary to indicate the specific form, e.g, for recombinant proteins produced in E. coli, the terms, eIF5A(Lys), eIF5A(Dhp) or eIF5A(Hpu) are used. In certain context, eIF5A may also refer to the eIF5A polypeptide irrespective of its modification status.
REFERENCES
- 1.Pegg AE. Spermidine/spermine-N(1)-acetyltransferase: a key metabolic regulator. Am J Physiol Endocrinol Metab. 2008;294:E995–1010. doi: 10.1152/ajpendo.90217.2008. [DOI] [PubMed] [Google Scholar]
- 2.Wallace HM, Fraser AV, Hughes A. A perspective of polyamine metabolism. Biochem J. 2003;376:1–14. doi: 10.1042/BJ20031327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pirinen E, Kuulasmaa T, Pietila M, Heikkinen S, Tusa M, Itkonen P, Boman S, Skommer J, Virkamaki A, Hohtola E, Kettunen M, Fatrai S, Kansanen E, Koota S, Niiranen K, Parkkinen J, Levonen AL, Yla-Herttuala S, Hiltunen JK, Alhonen L, Smith U, Janne J, Laakso M. Enhanced polyamine catabolism alters homeostatic control of white adipose tissue mass, energy expenditure, and glucose metabolism. Mol Cell Biol. 2007;27:4953–4967. doi: 10.1128/MCB.02034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cano VS, Jeon GA, Johansson HE, Henderson CA, Park JH, Valentini SR, Hershey JW, Park MH. Mutational analyses of human eIF5A-1--identification of amino acid residues critical for eIF5A activity and hypusine modification. Febs J. 2008;275:44–58. doi: 10.1111/j.1742-4658.2007.06172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kramer DL, Diegelman P, Jell J, Vujcic S, Merali S, Porter CW. Polyamine acetylation modulates polyamine metabolic flux, a prelude to broader metabolic consequences. J Biol Chem. 2008;283:4241–4251. doi: 10.1074/jbc.M706806200. [DOI] [PubMed] [Google Scholar]
- 6.Jell J, Merali S, Hensen ML, Mazurchuk R, Spernyak JA, Diegelman P, Kisiel ND, Barrero C, Deeb KK, Alhonen L, Patel MS, Porter CW. Genetically altered expression of spermidine/spermine N1-acetyltransferase affects fat metabolism in mice via acetyl-CoA. J Biol Chem. 2007;282:8404–8413. doi: 10.1074/jbc.M610265200. [DOI] [PubMed] [Google Scholar]
- 7.Hyvonen MT, Merentie M, Uimari A, Keinanen TA, Janne J, Alhonen L. Mechanisms of polyamine catabolism-induced acute pancreatitis. Biochem Soc Trans. 2007;35:326–330. doi: 10.1042/BST0350326. [DOI] [PubMed] [Google Scholar]
- 8.Pegg AE, Feith DJ, Fong LY, Coleman CS, O'Brien TG, Shantz LM. Transgenic mouse models for studies of the role of polyamines in normal, hypertrophic and neoplastic growth. Biochem Soc Trans. 2003;31:356–360. doi: 10.1042/bst0310356. [DOI] [PubMed] [Google Scholar]
- 9.Babbar N, Murray-Stewart T, Casero RA., Jr. Inflammation and polyamine catabolism: the good, the bad and the ugly. Biochem Soc Trans. 2007;35:300–304. doi: 10.1042/BST0350300. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, Young BA, Coleman CS, Pegg AE, Sheppard D. Spermidine/spermine N1-acetyltransferase specifically binds to the integrin alpha9 subunit cytoplasmic domain and enhances cell migration. J Cell Biol. 2004;167:161–170. doi: 10.1083/jcb.200312166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.deHart GW, Jin T, McCloskey DE, Pegg AE, Sheppard D. The alpha9beta1 integrin enhances cell migration by polyamine-mediated modulation of an inward-rectifier potassium channel. Proc Natl Acad Sci U S A. 2008;105:7188–7193. doi: 10.1073/pnas.0708044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baek JH, Liu YV, McDonald KR, Wesley JB, Hubbi ME, Byun H, Semenza GL. Spermidine/spermine-N1-acetyltransferase 2 is an essential component of the ubiquitin ligase complex that regulates hypoxia-inducible factor 1alpha. J Biol Chem. 2007;282:23572–23580. doi: 10.1074/jbc.M703504200. [DOI] [PubMed] [Google Scholar]
- 13.Baek JH, Liu YV, McDonald KR, Wesley JB, Zhang H, Semenza GL. Spermidine/spermine N(1)-acetyltransferase-1 binds to hypoxia-inducible factor-1alpha (HIF-1alpha) and RACK1 and promotes ubiquitination and degradation of HIF-1alpha. J Biol Chem. 2007;282:33358–33366. doi: 10.1074/jbc.M705627200. [DOI] [PubMed] [Google Scholar]
- 14.Bewley MC, Graziano V, Jiang J, Matz E, Studier FW, Pegg AE, Coleman CS, Flanagan JM. Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target. Proc Natl Acad Sci U S A. 2006;103:2063–2068. doi: 10.1073/pnas.0511008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park MH. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A). J Biochem. 2006;139:161–169. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolff EC, Kang KR, Kim YS, Park MH. Posttranslational synthesis of hypusine: evolutionary progression and specificity of the hypusine modification. Amino Acids. 2007;33:341–350. doi: 10.1007/s00726-007-0525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen KY, Liu AY. Biochemistry and function of hypusine formation on eukaryotic initiation factor 5A. Biol Signals. 1997;6:105–109. doi: 10.1159/000109115. [DOI] [PubMed] [Google Scholar]
- 18.Joe YA, Wolff EC, Park MH. Cloning and expression of human deoxyhypusine synthase cDNA. Structure- function studies with the recombinant enzyme and mutant proteins. J Biol Chem. 1995;270:22386–22392. doi: 10.1074/jbc.270.38.22386. [DOI] [PubMed] [Google Scholar]
- 19.Park JH, Aravind L, Wolff EC, Kaevel J, Kim YS, Park MH. Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: a HEAT-repeat-containing metalloenzyme. Proc Natl Acad Sci U S A. 2006;103:51–56. doi: 10.1073/pnas.0509348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byers TL, Lakanen JR, Coward JK, Pegg AE. The role of hypusine depletion in cytostasis induced by S-adenosyl-L- methionine decarboxylase inhibition: new evidence provided by 1- methylspermidine and 1,12-dimethylspermine. Biochem J. 1994;303:363–368. doi: 10.1042/bj3030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chattopadhyay MK, Park MH, Tabor H. Hypusine modification for growth is the major function of spermidine in Saccharomyces cerevisiae polyamine auxotrophs grown in limiting spermidine. Proc Natl Acad Sci U S A. 2008;105:6554–6559. doi: 10.1073/pnas.0710970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerner EW, Mamont PS, Bernhardt A, Siat M. Post-translational modification of the protein-synthesis initiation factor eIF-4D by spermidine in rat hepatoma cells. Biochem J. 1986;239:379–386. doi: 10.1042/bj2390379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasaki K, Abid MR, Miyazaki M. Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae. FEBS Lett. 1996;384:151–154. doi: 10.1016/0014-5793(96)00310-9. [DOI] [PubMed] [Google Scholar]
- 24.Schnier J, Schwelberger HG, Smit-McBride Z, Kang HA, Hershey JW. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishimura K, Murozumi K, Shirahata A, Park MH, Kashiwagi K, Igarashi K. Independent roles of eIF5A and polyamines in cell proliferation. Biochem J. 2005;385:779–785. doi: 10.1042/BJ20041477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smit-McBride Z, Schnier J, Kaufman RJ, Hershey JW. Protein synthesis initiation factor eIF-4D. Functional comparison of native and unhypusinated forms of the protein. J Biol Chem. 1989;264:18527–18530. [PubMed] [Google Scholar]
- 27.Park MH. The essential role of hypusine in eukaryotic translation initiation factor 4D (eIF-4D). Purification of eIF-4D and its precursors and comparison of their activities. J Biol Chem. 1989;264:18531–18535. [PubMed] [Google Scholar]
- 28.Jao DL, Chen KY. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J Cell Biochem. 2006;97:583–598. doi: 10.1002/jcb.20658. [DOI] [PubMed] [Google Scholar]
- 29.Zanelli CF, Maragno AL, Gregio AP, Komili S, Pandolfi JR, Mestriner CA, Lustri WR, Valentini SR. eIF5A binds to translational machinery components and affects translation in yeast. Biochem Biophys Res Commun. 2006;348:1358–1366. doi: 10.1016/j.bbrc.2006.07.195. [DOI] [PubMed] [Google Scholar]
- 30.Gregio APB, Cano VPS, Avaca JS, Valentini SR, Zanelli CF. eIF5A has a function in the elongation step of translation in yeast. BBRC. 2009;380:785–790. doi: 10.1016/j.bbrc.2009.01.148. [DOI] [PubMed] [Google Scholar]
- 31.Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park MH. The identification of an eukaryotic initiation factor 4D precursor in spermidine-depleted Chinese hamster ovary cells. J Biol Chem. 1988;263:7447–7449. [PubMed] [Google Scholar]
- 33.Klier H, Csonga R, Joao HC, Eckerskorn C, Auer M, Lottspeich F, Eder J. Isolation and structural characterization of different isoforms of the hypusine-containing protein eIF-5A from HeLa cells. Biochemistry. 1995;34:14693–14702. doi: 10.1021/bi00045a010. [DOI] [PubMed] [Google Scholar]
- 34.Coleman CS, Stanley BA, Jones AD, Pegg AE. Spermidine/spermine-N1-acetyltransferase-2 (SSAT2) acetylates thialysine and is not involved in polyamine metabolism. Biochem J. 2004;384:139–148. doi: 10.1042/BJ20040790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park MH, Cooper HL, Folk JE. The biosynthesis of protein-bound hypusine (N epsilon -(4-amino-2- hydroxybutyl)lysine). Lysine as the amino acid precursor and the intermediate role of deoxyhypusine (N epsilon -(4-aminobutyl)lysine). J Biol Chem. 1982;257:7217–7222. [PubMed] [Google Scholar]
- 36.Park MH, Wolff EC, Smit-McBride Z, Hershey JW, Folk JE. Comparison of the activities of variant forms of eIF-4D. The requirement for hypusine or deoxyhypusine. J Biol Chem. 1991;266:7988–7994. [PubMed] [Google Scholar]
- 37.Park MH, Liu TY, Neece SH, Swiggard WJ. Eukaryotic initiation factor 4D. Purification from human red blood cells and the sequence of amino acids around its single hypusine residue. J Biol Chem. 1986;261:14515–14519. [PubMed] [Google Scholar]
- 38.Benne R, Brown-Luedi ML, Hershey JW. Purification and characterization of protein synthesis initiation factors eIF-1, eIF-4C, eIF-4D, and eIF-5 from rabbit reticulocytes. J Biol Chem. 1978;253:3070–3077. [PubMed] [Google Scholar]
- 39.Murakami Y, Kondo K, Miki K, Akiyama Y, Watanabe T, Yokuyama Y. The ortho-Substituted N,N-Diacetylaniline as a Selective Acetylating Reagent. Tetrahedron Letters. 1997;38:3751–3754. [Google Scholar]
- 40.Park MH, Cooper HL, Folk JE. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc Natl Acad Sci U S A. 1981;78:2869–2873. doi: 10.1073/pnas.78.5.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abbruzzese A, Park MH, Beninati S, Folk JE. Inhibition of deoxyhypusine hydroxylase by polyamines and by a deoxyhypusine peptide. Biochim Biophys Acta. 1989;997:248–255. doi: 10.1016/0167-4838(89)90195-7. [DOI] [PubMed] [Google Scholar]
- 42.Folk JE, Park MH, Chung SI, Schrode J, Lester EP, Cooper HL. Polyamines as physiological substrates for transglutaminases. J Biol Chem. 1980;255:3695–3700. [PubMed] [Google Scholar]
- 43.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 44.Della Ragione F, Pegg AE. Studies of the specificity and kinetics of rat liver spermidine/spermine N1-acetyltransferase. Biochem J. 1983;213:701–706. doi: 10.1042/bj2130701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee SB, Park JH, Kaevel J, Sramkova M, Weigert R, Park MH. The effect of hypusine modification on the intracellular localization of eIF5A. Biochem Biophys Res Commun. 2009;383:497–502. doi: 10.1016/j.bbrc.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee SB, Park JH, Woster PM, Casero RA, Jr., Park MH. Suppression of exogenous gene expression by spermidine/spermine-N1-acetyltransferase 1 (SSAT1) cotransfection. J Biol Chem. doi: 10.1074/jbc.M109.092007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pegg AE. Mammalian polyamine metabolism and function. IUBMB Life. 2009;61:880–894. doi: 10.1002/iub.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerner EW. Cancer chemoprevention locks onto a new polyamine metabolic target. Cancer Prev Res (Phila Pa) 3:125–127. doi: 10.1158/1940-6207.CAPR-09-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang HA, Schwelberger HG, Hershey JW. Translation initiation factor eIF-5A, the hypusine-containing protein, is phosphorylated on serine in Saccharomyces cerevisiae. J Biol Chem. 1993;268:14750–14756. [PubMed] [Google Scholar]
- 50.Klier H, Wohl T, Eckerskorn C, Magdolen V, Lottspeich F. Determination and mutational analysis of the phosphorylation site in the hypusine-containing protein Hyp2p. FEBS Lett. 1993;334:360–364. doi: 10.1016/0014-5793(93)80712-4. [DOI] [PubMed] [Google Scholar]
- 51.Shirai A, Matsuyama A, Yashiroda Y, Hashimoto A, Kawamura Y, Arai R, Komatsu Y, Horinouchi S, Yoshida M. Global analysis of gel mobility of proteins and its use in target identification. J Biol Chem. 2008;283:10745–10752. doi: 10.1074/jbc.M709211200. [DOI] [PubMed] [Google Scholar]
- 52.Bras AP, Janne J, Porter CW, Sitar DS. Spermidine/spermine n(1)-acetyltransferase catalyzes amantadine acetylation. Drug Metab Dispos. 2001;29:676–680. [PubMed] [Google Scholar]