Abstract
The complete nucleotide sequences of three chimpanzee polyomavirus genetic variants were determined. Phylogenetic analysis indicated that the viruses form two different genotypes of ChPyV. Comparison with other primate polyomaviruses revealed a putative agnogene, and an unusually long VP1 open reading frame. The transcriptional control regions (TCR) of the viruses were extremely short (155 nucleotides), and highly conserved amongst the genotypes. Analysis of the TCR from different chimpanzee subspecies, and from a series of tissues from five individuals confirmed its genetic stability, and also indicates that double-infections with different genotypes can occur.
Findings
The number of primate polyomaviruses (PyV), including human polyomaviruses, has rapidly expanded in recent years. Six human viruses, KIPyV, WUPyV, Merkel cell polyomavirus (McPyV), Trichodysplasia Spinulosa-associated polyomavirus (TSPyV), HPyV6, and HPyV7 have been characterized in patients suffering from respiratory tract infections (KI and WU), Merkel cell carcinomas (MC), virus-associated trichodysplasia spinulosa (TSP), or were detected in the skin of healthy individuals (HPyV6, and HPyV7) [1-5]. Simultaneously, novel simian viruses have been discovered in healthy squirrel monkeys and orangutans [6,7], and in diarrheal stool from a chimpanzee [8]. From the chimpanzee polyomavirus (ChPyV) only the nucleotide sequence of the VP1 gene has been published [GenBank: AY691168]. We have investigated the genetic variation of ChPyV, and sequenced the genome of three chimpanzee polyomavirus variants. We also analyzed the genetic variation of ChPyV in relation to the host subspecies, and investigated ChPyV tissue tropism.
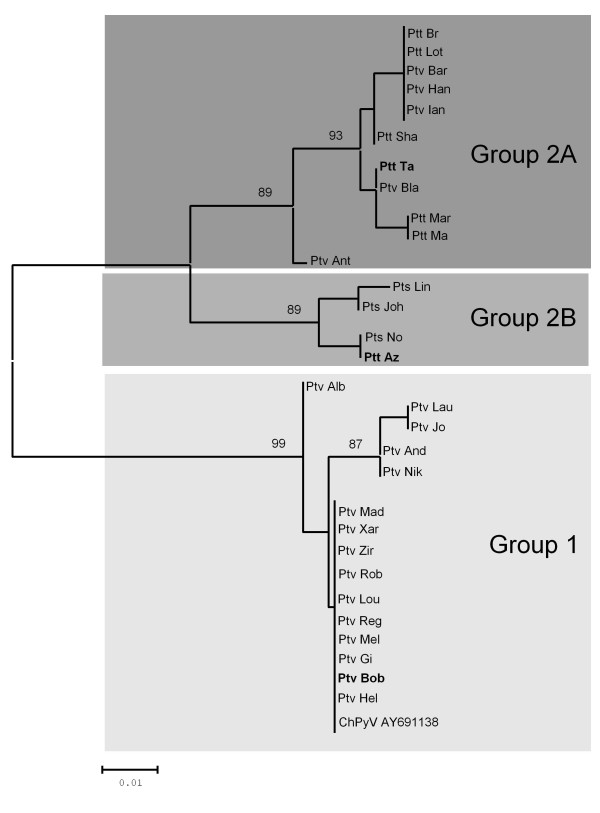
ChPyV VP1-specific PCR primers, based on the published VP1 sequence, were used to screen DNA isolated from blood samples collected from captive and wild-caught chimpanzees (QIAamp DNA Mini Kit, QIAGEN Benelux BV, Venlo, The Netherlands) (Table 1). Captive animals originated from former chimpanzee colonies kept at the BPRC (n = 66) and another primate facility in Europe (n = 24). Materials from wild-caught chimpanzees were obtained from animals housed in a rehabilitation centre in Africa (n = 22). The outer amplification reaction was performed in a 50 μl volume using 1 μg of DNA, 2 units Maxima™ Hot Start Taq DNA polymerase (Fermentas GMBH, St. Leon-Rot, Germany), 5 μl 10 × Hot Start PCR buffer, 1 pmol of each primer, 2 mM MgCl2, and 200 μM of each dNTP. Cycling conditions for both reactions were 95°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec. In a second amplification reaction, 2 μl of the PCR product of the outer PCR was used as template. Inner PCR conditions were identical to those for the outer PCR, except that 2.5 mM MgCl2 was used. The PCR fragments were gel-purified using the Zymoclean™ Gel DNA Recovery Kit (Zymo Research Corp, Orange, USA), and sequence analysis was performed directly on the purified amplicons (Baseclear BV, Leiden, The Netherlands). Thirty VP1 sequences were obtained and sequenced, and phylogenetic analysis revealed the presence of two genetic groups, one of which consisted of two smaller subclusters (genogroup 2A and 2B; Figure 1). We next investigated if there was a relationship between viral genotype and chimpanzee subspecies. The chimpanzee subspecies was determined by analysis of mitochondrial control region (D-loop) [9], and data showed that genogroup 1 solely consisted of viruses derived from individuals belonging to the Pan troglodytes verus subspecies, while group 2 was formed by viruses obtained from the three major subspecies Pt. verus, Pt. troglodytes, and Pt. schweinfurthii.
Table 1.
Primers used for PCR amplification of VP1 and TCR sequences
| Primer name | Sequence (5' > 3') |
|---|---|
| ChPyV VP1 assay | |
| ChPyV-Fout | GTTATTCATCATGCAGATGG |
| ChPyV-Rout | TCAGCTAATTTAGCTATATC |
| ChPyV-Fin | GAACACAGACATGACCTGTG |
| ChPyV-Rin | GTATAGCTGAAGCATATTTAG |
| ChPyV TCR assay | |
| TCRoutF | AAAGTTTTACATCATAGCAATCAGA |
| TCRoutR | AGAGGGCTTCAATAGTCAATCCAGA |
| TCRinF | GACCCTCTTGAAATTTTTGCCACAGT |
| TCRinR | TTAGTTCAGAAGCCATCACAATCATA |
Figure 1.
Phylogenetic tree constructed using partial VP1 gene sequences of chimpanzee polyomaviruses. Grey shading indicates genogroups described in the text, and isolates used for complete genome sequencing are in bold. The published ChPyV VP1 sequence (AY691138) is included in the tree. Sequence alignments were made by using MacVector version 10.6. Phylogenetic analysis was performed by the Neighbor-Joining method as implemented in MEGA version 4 [25]. Bootstrap values (as % of 1000 re-samplings) are indicated. Bar, 0.01 nucleotide replacements per site. First three letters of name indicate subspecies: Ptv, Pan troglodytes verus; Ptt, P.t. troglodytes; Pts, P.t. schweinfurthii. [EMBL: FR692245-FR692275].
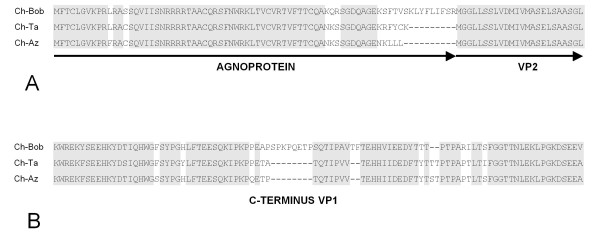
Full-length nucleotide sequences of representatives from each variant were determined using long-distance PCR [6,7]. Sequence comparison of the genomes confirmed that two variants ChPyV-Ta and -Az (genogroup 2A and 2B, respectively) were more similar to each other (96.6%), than to ChPyV-Bob, a genogroup 1 virus (92.6% and 92.7%, respectively). Sequences have been deposited under EMBL database accession numbers FR692334 to FR692336. Further analysis confirmed a typical polyomavirus genetic structure of each variant, with an early region encoding the small t- (t-Ag) and large T-antigens (T-Ag), and a late region encoding the VP1, VP2, and VP3 structural proteins. All three viruses accommodate a potential agnogene, encoding a protein of 64, 65, or 74 amino acids for ChPyV-Az, ChPyV-Ta and ChPyV-Bob, respectively. The first two agnogenes are located 5' to the VP2/VP3 open reading frame (orf), but curiously the agnogene of ChPyV-Bob is fused in-frame with the VP2/VP3 orf. An alignment of the agno-VP2 junction, illustrating the disparity between the viral genomes, is given in Figure 2A. The VP1 structural proteins encoded by the ChPyV genomes are considerably longer than VP1 from other polyomaviruses. The VP1 orf of ChPyV-Bob (nt. 1033-2526) encodes a protein of 498 amino acid residues (aa.) that has an additional 75 amino acids at its C-terminus compared to the longest VP1 described to date, that of the McPyV. Within the same C-terminus of ChPyV-Az and -Ta, an 8 aa. deletion (nt. 2356-2380) is found (Figure 2B). BLAST analysis of this region did not reveal any similarity with other known proteins. Search for specific polypeptide motifs or patterns (ExPASy proteomics server; http://www.expasy.ch/tools/) was also unsuccessful. The amino acid sequence similarity of the ChPyV structural proteins (represented by ChPyV-Ta) with known human and simian polyomaviruses is shown in table 2. Strikingly, within the early region the highest similarity is found with t-Ag and T-Ag from the human Merkel cell polyomavirus, while the late proteins, VP1- VP3, are most similar to the equivalent proteins of the polyomavirus from Sumatran orangutans.
Figure 2.
Alignments of chimpanzee polyomavirus proteins. A. Comparison of the agnoprotein-VP2 junction of ChPyV variants. The putative agnoproteins and the N-terminal 24 amino acid residues of VP2 are aligned. Areas with similarities and identities within the three agnoprotein-VP2 junctions are shaded grey. B. Alignment of the C-terminus of the chimpanzee polyomavirus VP1 proteins. Areas with similarities and identities within the three VP1 proteins are shaded grey.
Table 2.
Protein sequence similarity (%) between ChPyV-Az and known primate polyomaviruses.
| JCV | KIPyV | McPyV | TSPyV | HPyV6 | OraPyV-Sum | LPV | SquiPyV | |
|---|---|---|---|---|---|---|---|---|
| VP1 | 44,2 | 30,4 | 54,2 | 53,9 | 34,0 | 51,9 | 51,1 | 47,3 |
| VP2 | 31,8 | 21,5 | 58,0 | 43,2 | 24,5 | 44,0 | 35,7 | 33,3 |
| VP3 | 26,7 | 19,2 | 41,7 | 25,6 | 21,6 | 26,0 | 22,3 | 25,0 |
| t-Ag | 40,2 | 39,3 | 47,0 | 40,2 | 42,4 | 49,8 | 45,8 | 40,0 |
| T-Ag | 49,7 | 53,3 | 50,5 | 58,8 | 54,6 | 59,5 | 57,5 | 54,5 |
Proteins with highest percentage similarity with ChPyV proteins are given in bold italic.
The transcriptional control region (TCR) of polyomaviruses controls gene expression and viral replication. This region, located between the start of the t-Ag orf, and the start of the putative agnoprotein orf, is only 155 bp long for all three ChPyV variants, and is the shortest TCR of all PyV presently known. It is practically conserved between the viral variants; the TCR of ChPyV-Bob differs only at nucleotide 128 with the other TCRs. Consequently, the architecture of the TCR is simple (Figure 3). A 22-bp palindromic sequence is located at nt. 96-117, and contains two tandemlypositioned T-ag binding sites. An additional binding site is found at nt. 68-72, and is directed towards the early region. In contrast to other polyomavirus TCRs no repeated sequences are distinguishable. This feature makes the ChPyV the most basic TCR yet characterized, exceeding the proto-archetypal SV40 TCR in simplicity [10-12]. The SV40 TCR is a highly variable region that is mainly due to extensive rearrangements of enhancer elements caused by propagation of the virus in cell culture [13,14]. Evidence also indicates that rearrangements play a role in viral pathogenesis [15,16], and, recently it was found that in kidney transplant recipients a re-arranged TCR conferred BKV with a higher replicating capacity [17]. From a group of 23 animals, consisting of 16 Pt. verus, 6 Pt. troglodytes, and one Pt. schweinfurthii, the TCR region was amplified in a nested PCR assay (Table 1). PCR mixes were identical to the VP1 assay, except that 2 mM MgCl2 was used. Amplification conditions were: an enzyme activation step of 4 min at 96°C, followed by 40 amplification cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 45 sec. Sequence analysis revealed minimal TCR variation was observed [EMBL: FR692222-FR692244]. In 13 animals, an adenine instead of a guanine was seen at nucleotide 128, which is located within the AT-rich region. Of interest, all animals that had the adenine at this position belonged to the Pt. verus subspecies.
Figure 3.
Architecture of the ChPyV transcriptional control region. The > indicate the direction T-Ag binding site. The variable nucleotide 128 is boxed.
We also investigated the presence of ChPyV in different tissues taken at autopsy from five Pt. verus chimpanzees. The animals, varying from 7 to 43 years old, died of various causes. Histopathological examination did not reveal any lesions related to polyomavirus infection, like interstitial lymphoplasmacytic nephritis with occasional epithelial intranuclear inclusion bodies, proliferative interstitial pneumonia with intranuclear inclusions within type II pneumocytes, areas of demyelination of subcortical white matter, and/or intranuclear inclusions within astrocytes and oligodendrocytes (typical for progressive multifocal leucoencephalopathy; PML). The findings, as well as the cause of dead and the age are summarized in Table 3. All tissue samples were screened with the VP1 and TCR assays. An overview of the tissues analyzed from each individual, and PCR results is given in Table S1 (Additional file 1). Although the type and number of tissues analyzed from each animal varied considerable, it was evident that virus tissue distribution in Regina was most widespread. Regina was a 42-year-old female who was euthanized because of deteriorating body condition. The virus was easily detectable in 31 of 35 tissues tested, including the skin, and was undetectable in only a few tissues (parotid, muscle, aorta and sciatic nerve). Notably, the same tissues from the other chimpanzees were also negative, with the exception of the sciatic nerve sample from Antoine, which was positive in the TCR test, but not in the VP1 assay. Recent data from human polyomaviruses point to the skin as a target organ for PyV persistence and replication [2,4,5]. Interestingly, the skin was positive in all samples (n = 4) that were analyzed from our chimpanzees. In Melanie and Gina, both chimpanzees with a low number of tissues infected (4 of 32, and 4 of 33, respectively), the skin belonged to the few PCR-positive tissues. This suggests a similar skin tropism for ChPyV as for the human viruses. In addition, in 4 of 5 spleen samples the virus was easily detectable by PCR, while a more generally accepted target organ like the kidney, scored only 1 out of 5 DNA samples positive.
Table 3.
Animals examined in this study
| Animal | Age | Cause of dead | Histopathological findings |
|---|---|---|---|
| Regina | 42 y | Euthanised because of deteriorating body condition | Mostly age related lesions: focal endocarditis; chronic interstitial nephritis; myodegeneration; mild lymphoid depletion in spleen No polyomavirus-associated lesions |
| Gina | 43 y | Drowned | Bronchopneumonia; No polyomavirus-associated lesions |
| Melanie | 12 y | Severely emaciated; died during anesthesia | Subacute pneumonia; hemosiderosis (spleen, liver) |
| Antoine | 7 y | Died during anesthesia | No polyomavirus-associated lesions |
| Bob | 14 y | Euthanised after episode of severe hematuria | Immune-mediated hemolytic anemia; No polyomavirus-associated lesions |
The TCR of all 32 positive tissue samples was sequenced [EMBL: FR692190-FR692221]. Variation was minimal and similar to the abovementioned results in animals from different origin. In 25 TCR sequences (1 from Bob, 2 from Gina and Melanie, and 20 from Regina) an adenosine was identified at nucleotide 128, while in all 6 sequences from Antoine, and in 1 out of 2 TCRs obtained from Bob a guanine was found at this site. This strongly suggests that Bob was double infected with two viral variants, although a point mutation, occurred during viral replication, cannot be completely ruled out.
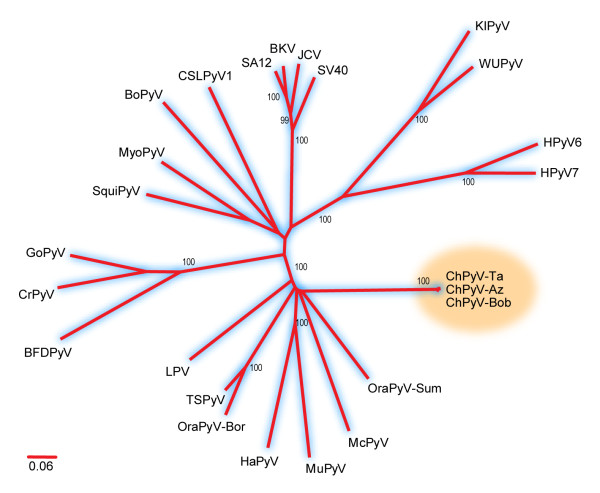
In this study we have molecularly characterized three variants of the chimpanzee polyomavirus, and took a glimpse at some biological and evolutionary properties of this virus. Phylogenetic analysis of the concatenated VP1 and T-Ag protein sequences from avian and mammalian polyomaviruses show that the chimpanzee viruses form a distinct group of viruses, distantly related to the human McPyV and TSPyV, the orangutan polyomaviruses and LPV from African green monkeys. Interestingly, both rodent viruses (MuPyV and HaPyV) also fall within this large cluster (Figure 4) The chimpanzee polyomavirus genomes have some unique features, as they encode for unusually long VP1 structural proteins, and, in contrast, possess an exceptionally short TCR. The exact significance of these finding needs to be substantiated, and goes beyond the scope of this paper. Most interesting is the short and conserved TCR of the chimpanzee virus. Because the polyomavirus TCR regulates viral replication and pathogenesis, and its sequence variation in other PyV is likely the cause or consequence of these processes [15-17], it is an intriguing question how ChPyV with such a 'basic' and apparently genetically constant TCR regulates these processes. Our findings add to the increasing awareness that the Polyomaviridae are a genetically diverse family of viruses. In a recent study, van der Meijden et al. distinguished seven PyV clades, and pointed towards a complex evolutionary history [5]. The number of PyV has increased in the last few years; viruses have been detected in Californian sea lions (CSLPyV1) [18], bats (MyoPyV) [19], birds [20,21], in addition to the novel simian viruses. We have detected new polyomaviruses in apes (gorillas and bonoboos), Old World monkeys, like hamadryas baboon and mandrill, and in capuchin monkeys and spider monkeys, both New World monkeys (unpublished data; EMBL: FR692182-FR692189). With the help of improved diagnostic techniques and the use of metagenomic approaches [22-24] it can be expected that more polyomaviruses will be detected in the near future.
Figure 4.
Phylogenetic analysis of concatenated VP1 and Large T proteins from avian and mammalian polyomaviruses. Sequence alignments were made by using MacVector version 10.6. The GapStreeze program (Los Alamos HIV Sequence Database; http://www.hiv.lanl.gov/content/sequence/GAPSTREEZE/gap.html) was used to remove columns with a gap tolerance of 0%. Phylogenetic analysis was performed by the neighbor-joining method using the JTT matrix model as implemented in MEGA version 4 [25]. Bootstrap values (as % of 1000 re-samplings) are indicated. Bar, 0.06 amino acid residue replacements per site. The GenBank accession numbers of the viruses used are: NC_001515 (MuPyV), NC_001663 (HaPyV), HM355825 (McPyV), M30540 (LPV), NC_001442 (BoPyV), NC_009951 (SquiPyV), NC_011310 (MyoPyV), NC_007922 (CrPyV), NC_004800 (GoPyV), AB453166 (BFDPyV), NC_001669 (SV40), NC_001699 (JCV), AY614708 (SA12), NC_001538 (BKV), EF127906 (KIPyV), EF444549 (WUPyV), FN356900 (OraPyV-Bor), FN356901 (OraPyV-Sum), GU989205 (TSPyV), NC_014407 (HPyV6), NC_014407 (HPyV7), NC_013796 (CSLPyV1). Chimpanzee polyomaviruses are highlighted.
Abbreviations
PyV: polyomavirus; TCR: transcriptional control region.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
ID and ZF contributed in obtaining PCR data and sequencing. HN provided chimpanzee blood samples. MJG performed long PCR and genome sequencing. IK was responsible for histopathological analysis and provided tissue samples. WB was helpful in interpreting the data. EJV was responsible for the planning of the study, data analysis, and drafted the manuscript. All authors have read and approved the final manuscript
Supplementary Material
Table S1. PCR analysis of chimpanzee tissues.
Contributor Information
Ilona Deuzing, Email: ilona_deuzing@hotmail.com.
Zahra Fagrouch, Email: fagrouch@bprc.nl.
Marlous J Groenewoud, Email: M.J.Groenewoud-11@umcutrecht.nl.
Henk Niphuis, Email: niphuis@bprc.nl.
Ivanela Kondova, Email: kondova@bprc.nl.
Willy Bogers, Email: bogers@bprc.nl.
Ernst J Verschoor, Email: verschoor@bprc.nl.
Acknowledgements
This study was supported by the European Community Research Infrastructures Program grant RII3-CT-2006-026155 'European Primate Network: Specialized Infrastructures and Procedures for Biological and Biomedical Research (EUPRIM-NET)'.
References
- Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MAA, Dalianis T, Ramqvist T, Andersson B. Identification of a third human polyomavirus. J Virol. 2007;81:4130–4136. doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathogens. 2007;3:e64. doi: 10.1371/journal.ppat.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog. 2010;6:e1001024. doi: 10.1371/journal.ppat.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenewoud MJ, Fagrouch Z, van Gessel S, Niphuis H, Bulavaite A, Warren KS, Heeney JL, Verschoor EJ. Characterization of novel polyomaviruses from Bornean and Sumatran orang-utans. J Gen Virol. 2010;91:653–658. doi: 10.1099/vir.0.017673-0. [DOI] [PubMed] [Google Scholar]
- Verschoor EJ, Groenewoud MJ, Fagrouch Z, Kewalapat A, van Gessel S, Kik MJ, Heeney JL. Molecular characterization of the first polyomavirus from a New World primate: squirrel monkey polyomavirus. J Gen Virol. 2008;89:130–137. doi: 10.1099/vir.0.83287-0. [DOI] [PubMed] [Google Scholar]
- Johne R, Enderlein D, Nieper H, Muller H. Novel polyomavirus detected in the feces of a chimpanzee by nested broad-spectrum PCR. J Virol. 2005;79:3883–3887. doi: 10.1128/JVI.79.6.3883-3887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot NG, Garcia CA, Verschoor EJ, Doxiadis GG, Marsh SG, Otting N, Bontrop RE. Reduced MIC gene repertoire variation in West African chimpanzees as compared to humans. Mol Biol Evol. 2005;22:1375–1385. doi: 10.1093/molbev/msi127. [DOI] [PubMed] [Google Scholar]
- Lednicky JA, Butel JS. Consideration of PCR methods for the detection of SV40 in tissue and DNA specimens. Dev Biol Stand. 1998;94:155–164. [PubMed] [Google Scholar]
- Lednicky JA, Butel JS. Simian virus 40 regulatory region structural diversity and the association of viral archetypal regulatory regions with human brain tumors. Semin Cancer Biol. 2001;11:39–47. doi: 10.1006/scbi.2000.0345. [DOI] [PubMed] [Google Scholar]
- White MK, Safak M, Khalili K. Regulation of gene expression in primate polyomaviruses. J Virol. 2009;83:10846–10856. doi: 10.1128/JVI.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lednicky JA, Butel JS. Tissue culture adaptation of natural isolates of simian virus 40: changes occur in viral regulatory region but not in carboxy-terminal domain of large T-antigen. J Gen Virol. 1997;78(Pt 7):1697–1705. doi: 10.1099/0022-1317-78-7-1697. [DOI] [PubMed] [Google Scholar]
- O'Neill FJ, Greenlee JE, Carney H. The archetype enhancer of simian virus 40 DNA is duplicated during virus growth in human cells and rhesus monkey kidney cells but not in green monkey kidney cells. Virology. 2003;310:173–182. doi: 10.1016/S0042-6822(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Gosert R, Rinaldo CH, Funk GA, Egli A, Ramos E, Drachenberg CB, Hirsch HH. Polyomavirus BK with rearranged noncoding control region emerge in vivo in renal transplant patients and increase viral replication and cytopathology. J Exp Med. 2008;205:841–852. doi: 10.1084/jem.20072097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogo Y, Zhong S, Shibuya A, Kitamura T, Homma Y. Transcriptional control region rearrangements associated with the evolution of JC polyomavirus. Virology. 2008;380:118–123. doi: 10.1016/j.virol.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Olsen GH, Hirsch HH, Rinaldo CH. Functional analysis of polyomavirus BK non-coding control region quasispecies from kidney transplant recipients. J Med Virol. 2009;81:1959–1967. doi: 10.1002/jmv.21605. [DOI] [PubMed] [Google Scholar]
- Wellehan JF Jr, Yu F, Venn-Watson SK, Jensen ED, Smith CR, Farmerie WG, Nollens HH. Characterization of San Miguel sea lion virus populations using pyrosequencing-based methods. Infect Genet Evol. 2010;10:254–260. doi: 10.1016/j.meegid.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra V, Dumonceaux T, Dubois J, Willis C, Nadin-Davis S, Severini A, Wandeler A, Lindsay R, Artsob H. Detection of polyoma and corona viruses in bats of Canada. J Gen Virol. 2009;90:2015–2022. doi: 10.1099/vir.0.010694-0. [DOI] [PubMed] [Google Scholar]
- Arroube AS, Halami MY, Johne R, Dorrestein GM. Mortality due to polyomavirus infection in two nightjars (Caprimulgus europaeus) J Avian Med Surg. 2009;23:136–140. doi: 10.1647/2008-007.1. [DOI] [PubMed] [Google Scholar]
- Halami MY, Dorrestein GM, Couteel P, Heckel G, Muller H, Johne R. Whole genome characterization of a novel polyomavirus detected in fatally diseased canary birds. J Gen Virol. 2010;91:3016–3022. doi: 10.1099/vir.0.023549-0. [DOI] [PubMed] [Google Scholar]
- Ambrose HE, Clewley JP. Virus discovery by sequence-independent genome amplification. Rev Med Virol. 2006;16:365–383. doi: 10.1002/rmv.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Victoria JG, Wang C, Jones M, Fellers GM, Kunz TH, Delwart E. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol. 2010;84:6955–6965. doi: 10.1128/JVI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. PCR analysis of chimpanzee tissues.