Abstract
Background
Catabolite repression control (CRC) is an important global control system in Pseudomonas that fine tunes metabolism in order optimise growth and metabolism in a range of different environments. The mechanism of CRC in Pseudomonas spp. centres on the binding of a protein, Crc, to an A-rich motif on the 5' end of an mRNA resulting in translational down-regulation of target genes. Despite the identification of several Crc targets in Pseudomonas spp. the Crc regulon has remained largely unexplored.
Results
In order to predict direct targets of Crc, we used a bioinformatics approach based on detection of A-rich motifs near the initiation of translation of all protein-encoding genes in twelve fully sequenced Pseudomonas genomes. As expected, our data predict that genes related to the utilisation of less preferred nutrients, such as some carbohydrates, nitrogen sources and aromatic carbon compounds are targets of Crc. A general trend in this analysis is that the regulation of transporters is conserved across species whereas regulation of specific enzymatic steps or transcriptional activators are often conserved only within a species. Interestingly, some nucleoid associated proteins (NAPs) such as HU and IHF are predicted to be regulated by Crc. This finding indicates a possible role of Crc in indirect control over a subset of genes that depend on the DNA bending properties of NAPs for expression or repression. Finally, some virulence traits such as alginate and rhamnolipid production also appear to be regulated by Crc, which links nutritional status cues with the regulation of virulence traits.
Conclusions
Catabolite repression control regulates a broad spectrum of genes in Pseudomonas. Some targets are genus-wide and are typically related to central metabolism, whereas other targets are species-specific, or even unique to particular strains. Further study of these novel targets will enhance our understanding of how Pseudomonas bacteria integrate nutritional status cues with the regulation of traits that are of ecological, industrial and clinical importance.
Background
The genus Pseudomonas is an important group of microorganisms that occupy a wide variety of habitats including soil [1], the rhizosphere [2], food [3] and mammalian hosts [4]. Some species are important plant or human pathogens, whereas others are involved in processes such as bioremediation [5], biocontrol [6-8], nutrient cycling [9] or biotechnological processes [10]. A key aspect of the lifestyle of Pseudomonads is their ability to adapt, grow and compete in a wide variety of habitats. Thus, Pseudomonads require great flexibility in controlling their diverse array of metabolic pathways and, like most microorganisms, have global regulatory systems that ensure that the best nutrient source is utilised and almost depleted before less favoured nutrient sources are exploited [11-13].
Pseudomonads favour the utilisation of organic acids, particularly tricarboxylic acid (TCA) cycle intermediates, and amino acids over various other carbon sources such as carbohydrates or hydrocarbons [14]. This is in contrast to the majority of well-studied Enterobacteriaceae and Firmicutes, which favour glucose and use a system known as carbon catabolite repression (CCR) or catabolite repression control (CRC) to regulate carbon utilisation. The mechanism of CCR in Enterobacteriaceae and Firmicutes centres on a protein phosphorylation cascade and also involves transcriptional regulation mediated through cyclic AMP (cAMP) binding to the cAMP receptor protein (Crp) (for review see [11,12]). Although Pseudomonads possess a Crp homolog, Vfr, this protein is not involved in carbon source regulation, at least in P. aeruginosa PAO1 [15]. In fact, the CRC mechanism used by Pseudomonads to regulate carbon source utilisation is fundamentally different to CCR of Enterobacteriaceae and Firmicutes.
A central mediator of CRC is the Crc protein, which acts as a post-transcriptional regulator of target genes [16]. The post-transcriptional action of Crc relies on the binding of Crc to an unpaired A-rich motif in the 5'-end of a target mRNA causing inhibition of the initiation of translation [17,18]. It is still not fully understood how Crc activity is regulated in different Pseudomonas species, nor whether a common unified regulatory system is employed. In P. aeruginosa, activity is regulated by small RNA, CrcZ, which has five A-rich motifs, that binds to the Crc protein and sequesters it [17]. Levels of the CrcZ sRNA, in turn, are regulated by a two-component system (CbrA/CbrB) and by RpoN. Interestingly, CbrAB and NtrBC form a network to control the C/N balance in both P. aeruginosa and P. fluorescens [19-21]. Furthermore, the presence of a readily available nitrogen source enhances the magnitude of CRC [22], two observations that are suggestive of a link between regulatory systems controlling C and N utilisation. Although the crcZ gene is present in other Pseudomonads, its role in regulating CRC outside of P. aeruginosa has not yet been demonstrated. Indeed, in P. putida, crc mRNA and Crc protein levels are higher under conditions where CRC is active, a phenomenon not observed in P. aeruginosa, suggesting that an alternative system of regulating CRC may be used in this species [23,24].
Much of what is known about CRC comes from work on mutants lacking the Crc protein in P. aeruginosa and P. putida. Initially, the key work in identifying the CRC system came from the isolation and characterisation of a P. aeruginosa crc mutant [25]. In this mutant, the succinate-mediated catabolite repression control (CRC) of glucose and mannitol transport and Entner-Doudoroff pathway enzymes was alleviated, thereby establishing the importance of Crc. More recently, the role of Crc has been examined on a global scale in P. putida [26] and P. aeruginosa [27] by carrying out transcriptome and proteome analyses of crc mutants. No less than 134 targets in P. putida and 65 targets in P. aeruginosa were differentially altered in expression in rich media as a result of a crc mutation. This indicates that crc is an important global regulator that superimposes an additional layer of regulation over many metabolic pathways that are otherwise regulated locally by specific regulatory elements that control only one or a few genes. The global analyses of the P. putida and P. aeruginosa crc mutants indicates that CRC is responsible for the hierarchical assimilation of amino acids from rich media, with pathways required for assimilation of valine, isoleucine, leucine, tyrosine, phenylalanine, threonine, glycine and serine inhibited by Crc [26,27]. Additionally, the P. aeruginosa crc mutation was shown to alter the expression of targets with roles in anaerobic respiration, antibiotic resistance and virulence [27]. Recent work on a crc mutant of P. putida DOT-T1E established that Crc is not involved in the induction of pathways for nutrient utilisation since the mutant grows on the same range of carbon and nitrogen sources as the wild type strain [28]. This is in contrast to the E. coli CCR system where the cAMP-CRP complex is responsible for the induction of genes for utilisation of less favoured carbon sources such as lactose [29].
The role of CRC in regulating linear and aromatic hydrocarbon utilisation pathways in P. putida has received a lot of attention because of the potential implications of CRC on bioremediation processes. The utilisation of alkanes and a wide range of aromatic compounds including benzene and toluene are subject to CRC in P. putida [16,30-34]. Indeed Crc mediated post-transcriptional control of the pheA and pheB toluene degradation genes [31], the benR activator of benzene degradation [33], the alkS activator of alkane degradation [16], the xylR activator of the TOL genes and xylB (benzyl alcohol dehydrogenase) [34] and the bkdR activator of branched-chain keto acid dehydrogenase [35] has been demonstrated. The Crc protein is known to bind in vitro to mRNAs of the PU, PM, PS1, PR1, promoters and mRNAs of the xylMABN (and weakly to xylL and xylQ) genes of the pWW0 toluene degradation plasmid in P. putida indicating that these targets may also be regulated by directly by Crc [34]. Besides the role that CRC plays in the bioremediation activities of P. putida, little else is known about the control that CRC imposes on the ecological functions of Pseudomonads other than for virulence-associated functions in Pseudomonas aeruginosa. A crc mutant of P. aeruginosa PA14 was defective in biofilm formation and type IV pilus-mediated twitching motility [36]. and a crc mutant of P. aeruginosa PAO1 displayed increased susceptibility to some antibiotics as well as defects in type III secretion, motility and expression of quorum sensing-regulated virulence factors [27]. Given the range of ecological functions that Pseudomonas may perform there is great scope for Crc to be a significant regulator beyond the realm of primary metabolism. For instance, glucose metabolism is subject to CRC and gluconate is a product of glucose metabolism. Gluconate itself is linked to phosphate solubilisation [9] and biocontrol [37] and there is a link between the ability to produce gluconate and the levels of antimicrobial compounds produced such as 2,4-diacetylphloroglucinol and pyoluteorin [38]. Additionally, recent evidence indicates that there is a link between primary metabolism and secondary metabolism controlled by the GacS/Rsm system [39]. This suggests that there is great potential for CRC to interact with other regulatory networks, at least indirectly, and it is therefore a high priority to better understand the Crc regulon.
Based on the size of the Crc product and the proposed mechanism of action, it is thought that Crc binding must occur within -70 to +16 bp relative to the origin of translation [18]. There remain, however, very few known direct targets of Crc: only benR [33], alkS [18], xylR and xylB [34] mRNAs from P. putida and amiE mRNA [17] (product of the amidase gene amiE), in P. aeruginosa have been demonstrated to bind Crc. To extend the number of direct targets known, we carried out a bioinformatic analysis using genome information from sequenced Pseudomonas strains. By identifying the specific targets in pathways that are known to be regulated by CRC, it will be possible to determine precisely how different Pseudomonads control nutrient uptake and utilisation. Furthermore, the analysis is expected to identify new pathways and processes, not previously known to be CRC-regulated. A better understanding of how Pseudomonas species use CRC will enhance knowledge of the ecology of these bacteria and will facilitate efforts to exploit the metabolic capacity of these bacteria in industrial and environmental microbiology.
Results and discussion
Crc binding site detection: estimation of the numbers of Crc regulated genes in Pseudomonas
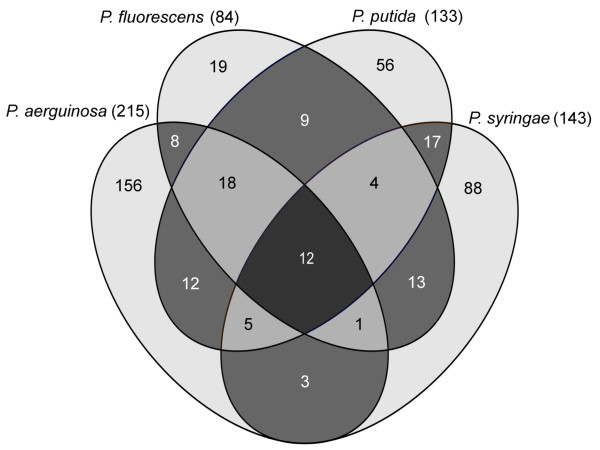
Following the recent discovery that the Crc protein binds to an A-rich motif in the -70 to +16 region of target mRNAs [17,18] a search was performed to catalogue the occurrences of this motif in the forward orientation in the upstream regions of all protein-encoding genes in Pseudomonas strains present in the RSAT database [40] at the time of analysis. These were P. aeruginosa (PAO1, PA14, PA7 and LESB58), P. fluorescens (Pf0-1, Pf-5 and SBW25), P. putida (KT2440, F1 and W619) and P. syringae (B728a and DC3000). It was reasoned that if a gene is under direct Crc control, the binding site should be present in that gene in all representatives of a particular species. Accordingly, only genes with the A-rich motif (AAnAAnAA) in the upstream region of intraspecies orthologs for all strains of a given species were considered as candidates (Additional file 1). In total, 421 candidate genes were identified, with an estimated false discovery rate of 27% (see materials and methods). P. aeruginosa has the highest number (215) of Crc candidates, P. syringae and P. putida had 143 and 133, respectively while P. fluorescens has the lowest number (84) (Figure 1). This difference in the number of possible CRC-regulated genes is likely to be a consequence of the taxonomic organisation within the genus, in particular the diversity of P. fluorescens species. A consequence of this diversity is that the core genome of P. fluorescens is significantly smaller than that of P. aeruginosa and so the pool of orthologous genes that are potentially regulated by Crc is lower [41-45]. Twelve Crc candidates are common to all four Pseudomonas species while a further 28 Crc candidates are present in three out of the four species examined (Figure 1). Taken together, these 40 Crc candidates represent the predicted core Crc regulon of Pseudomonas (Table 1). Many of these Crc candidates are annotated as having roles in nutrient transport and metabolism, fitting with the idea of CRC as a means of controlling hierarchical assimilation of nutrients from the environment. Most putative Crc targets are not part of the core regulon and are confined to a single or two species. These include the three Crc target genes (alkS, benR of P. putida and amiE of P. aeruginosa) that have been experimentally shown to bind Crc in the 5' region of the mRNA [17,18,33]. No orthologues of benR or amiE were detected outside of P. putida or P. aeruginosa species, respectively, and so these are species-specific targets. The absence of alkS in our dataset is due to its location on a mobile element (the P. putida OCT plasmid) that is only present in some strains of P. putida. In summation, the Pseudomonas regulatory network controlled by Crc ranges from genes that are regulated at a genus-wide level, down to genes that may only be regulated in certain strains within a particular species.
Figure 1.
Interspecific variations of the Crc regulon. Venn diagram showing a four way comparison of Crc candidates in P. aeruginosa, P. fluorescens, P. putida and P. syringae. Numbers in parentheses correspond to total number of Crc candidates in each species.
Table 1.
Predicted core Crc regulon of Pseudomonas.
| Gene name | Function | PAO1 | PA14 | PA7 | LESB58 | Pf0-1 | Pf-5 | SBW25 | KT2440 | F1 | W619 | B728a | DC3000 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino acid transport and metabolism | ||||||||||||||
| putP | Sodium/proline symporter | 0783 | 54150 | 4736 | 45601 | 0453 | 0496 | 0452 | 4946 | 4818 | 0522 | NM | NM | |
| Probable amino acid permease | 0789 | 54040 | 4729 | 45531 | 4561 | 4906 | 1103 | 1059 | 1100 | 1089 | NO | NO | ||
| phhA | Phenylalanine-4-hydroxylase | 0872 | 52990 | 4644 | 44441 | 1499 | 1611 | 4458 | 4490 | 1424 | 3779 | NM | NM | Y |
| gltI | Probable binding protein component of ABC transporter | 1342 | 46910 | 4043 | 38391 | 4535 | 4871 | 1139 | 1071 | 1112 | 1100 | NM | NM | |
| Probable sodium/alanine symporter | 2252 | 35460 | 2989 | 30521 | NM | NM | NM | 0496 | 0530 | 0551 | 5052 | 5500 | ||
| Probable glutamine synthetase | 2040 | 38140 | 3247 | 32821 | 5408 | 5930 | 5849 | 5184 | 5091 | 0279 | 4868 | 5310 | ||
| Carbohydrate transport and metabolism | ||||||||||||||
| mltE | Probable binding protein component of ABC maltose/mannitol transporter | 2338 | 34420 | 2925 | 29651 | 2640 | 3070 | 2745 | NO | NO | 2041 | 2440 | 2707 | |
| oprB | Glucose/carbohydrate outer membrane porin precursor | 3186 | 23030 | 1943 | 18821 | 4366 | 4613 | 4842 | 1019 | 1057 | 4205 | 1117 | 1296 | Y |
| Probable binding protein component of ABC sugar transporter | 3190 | 22980 | 1939 | 18781 | 4370 | 4617 | 4846 | 1015 | 1053 | 4209 | 1113 | 1292 | Y | |
| lldP | L-lactate permease | 4770 | 63080 | 5490 | 51551 | 0753 | 0817 | 5278 | 4735 | 4601 | 0696 | NO | NO | |
| fruA | PTS fructose IIC component | NM | NM | NM | NM | 0795 | 0861 | 0806 | 0795 | 0818 | 4398 | 0823 | 0956 | Y |
| glpF | Glycerol uptake facilitator protein | NM | NM | NM | NM | 4531 | 4867 | 1143 | 1076 | 1117 | 1105 | 3904 | 4167 | |
| Energy production and conservation | ||||||||||||||
| coxB | Cytochrome c oxidase, subunit II | 0105 | 01290 | 0180 | 01061 | 0079 | 0061 | 0058 | 0103 | 0119 | 0122 | NO | NO | |
| pntB | Pyridine nucleotide transhydrogenase, beta subunit | 0196 | 02470 | 0277 | 01971 | 0112 | 0113 | 0111 | 0155 | 0173 | 5072 | NM | NO | |
| acsA | Acetyl-coenzyme A synthetase | 0887 | 52800 | 4627 | 44291 | 4293 | 4522 | 4766 | 4487 | 1428 | 3776 | 3572 | 1825 | |
| Putative glycerate 2-kinase | 1499 | 45030 | 3833 | 39131 | 1595 | 1698 | 1800 | 4300 | 1569 | 3625 | NO | NO | ||
| Probable D-beta-hydroxybutyrate permease | 2004 | 38580 | 3285 | 33191 | 3078 | 3575 | 2629 | 3074 | 2650 | 2717 | NO | NO | ||
| Putative acetate transporter | 3235 | 22340 | 1890 | 18321 | 1607 | 1711 | 1813 | 1742 | 3977 | 1292 | 3757 | 1623 | ||
| cstA | Probable carbon starvation protein | 4606 | 60950 | 5246 | 49911 | 4883 | 5352 | 5333 | 4641 | 4503 | 0798 | 4273 | 4638 | |
| phaC1 | Poly(3-hydroxyalkanoic acid) synthase 1 | 5056 | 66820 | 5794 | 54461 | 0394 | 0434 | 0396 | 5003 | 4877 | 0461 | NO | NO | |
| phaF | polyhydroxyalkanoate synthesis protein PhaF | 5060 | 66875 | 5799 | 54501 | NM | NM | NM | 5007 | 4881 | 0457 | 0391 | 5147 | |
| Alginate metabolism | ||||||||||||||
| algP | Alginate regulatory protein | 5253 | 69370 | 5998 | 56471 | NM | NM | NM | 0194 | 0215 | 0263 | 0054 | 0136 | |
| Lipid metabolism | ||||||||||||||
| fadD2 | Long-chain-fatty-acid--CoA ligase | 3300 | 21340 | 1824 | 17661 | 4354 | 4599 | 4830 | 4550 | 1339 | 3845 | 3836 | 4098 | |
| estA | Esterase | 5112 | 67510 | 5845 | 55021 | NM | NM | NM | 0418 | 0452 | 4784 | 4606 | 0569 | |
| Polyamine metabolism | ||||||||||||||
| aphA | Acetylpolyamine aminohydrolase | 1409 | 46230 | 3930 | 40041 | 5631 | 6145 | 6061 | 5340 | 5249 | 0133 | NO | NO | |
| Xenobiotic degradation and transport | ||||||||||||||
| pcaK | Benzoate transport | NM | NM | NM | NM | 1266 | 1316 | 1362 | 1376 | 4347 | 1016 | 2124 | 2340 | |
| DNA replication, recombination and repair | ||||||||||||||
| recA | RecA protein | 3617 | 17530 | 1523 | 14181 | 1175 | 1231 | 1189 | 1629 | 4088 | 4030 | 1378 | 4033 | |
| hupA | HU family DNA-binding protein | 5348 | 70600 | 6125 | 57431 | 5600 | 6102 | 6032 | 5313 | 5222 | 0160 | NM | NM | Y |
| nucleotide transport and metabolism | ||||||||||||||
| Probable transporter | 1519 | 44800 | 3815 | 38911 | 1711 | 4364 | 4355 | 4284 | 1584 | 3610 | NO | NO | ||
| xdhA | Xanthine dehydrogenase | 1524 | 44710 | 3809 | 38041 | 1797 | 1889 | 4592 | 4278 | 1590 | 3604 | NM | NM | |
| Translation | ||||||||||||||
| rplR | 50S ribosomal protein L18 | 4247 | 09010 | 0853 | 06811 | 5063 | 5566 | 5511 | 0470 | 0503 | 4733 | 4532 | 0642 | |
| tufB | Elongation factor Tu | 4277 | 08830 | 0835 | 06631 | 5081 | 5584 | 5529 | 0452 | 0485 | 4751 | 4550 | 0624 | |
| Unknown function | ||||||||||||||
| Hypothetical protein | 0754 | 54540 | 4766 | 45891 | NM | NM | NM | 1418 | 4303 | 1058 | 3966 | 4232 | Y | |
| Probable transporter | 1507 | 44950 | 3824 | 39041 | 1701 | 4371 | 4364 | 4290 | 1578 | 3616 | NO | NO | ||
| Probable major facilitator superfamily (MFS) transporter | 3709 | 16410 | 1427 | 12731 | 3359 | 2486 | 2159 | 0057 | 0073 | 0076 | NO | NO | ||
| Hypothetical protein | 3923 | 13130 | 1184 | 10541 | 4708 | 5116 | 0923 | 0765 | 0792 | 4424 | 0991 | 1149 | ||
| Probable ATP-binding component of ABC transporter | 4461 | 57930 | 5034 | 48401 | 0858 | 0916 | 0883 | 0953 | 0992 | 4262 | 4146 | 4452 | ||
| Hypothetical protein | 4570 | 60480 | 5210 | 49531 | 4863 | 5332 | 5174 | 0685 | 0716 | 4500 | NO | NO | ||
| Hypothetical protein | 5052 | 66760 | 5789 | 54421 | 0398 | 0438 | 0400 | 5090 | 4963 | 0375 | NM | NM | ||
| Phosphotransferase domain-containing protein | NM | NM | NM | NM | 1487 | 1597 | 4880 | 4500 | 1412 | 3789 | 3586 | 1810 |
Loci with an A-rich motif in the upstream region in all strains tested for at least three species are shown. The numbers under strain names indicate the locus id, according to Genbank annotation, of the locus with the A-rich motif in the upstream region. NO (no ortholog) indicates that no orthologous locus was found. NM (no motif) indicates that the orthologous locus did not have the A-rich motif in the upstream region. Y indicates that the locus is has increased transcript and/or protein levels in a crc mutant of P. putida KT2442 (a spontaneous rifampicin resistant mutant of KT2440) [26].
Comparison of predicted Crc-regulated candidates to experimental datasets
Other studies have made use of crc mutant strains derived from P. putida KT2440 and P. aeruginosa PAO1 to experimentally identify possible targets of the CRC system. To assess the level of concordance between the bioinformatic and experimental approaches, the datasets were compared (Table 2 and Table 3). Recently, transcriptome and proteome comparisons of transcript and protein abundances of P. putida KT2442, a spontaneous rifampicin-resistant mutant of strain KT2440, and an isogenic crc mutant have been performed in rich media [26]. The transcriptome and proteome data sets identified 134 genes that were differentially altered in expression either at transcriptional or translational level in the crc mutant. We compared this list of 134 genes to the lists of genes identified in our bioinformatic analysis, with the results presented in table 2. The initial comparison was to the 133 candidate genes that were bioinformatically predicted to be the core Crc regulon of P. putida and then to ensure that possible positive matches were not overlooked, we extended the comparison to the longer list of 294 candidates identified in P. putida strain KT2440 (only targets present in all three P. putida strains were shown in additional file 1). 18 common targets between the predicted P. putida Crc regulon and the transcriptome/proteome data were identified, and another 5 possible targets are seen when the comparison is with the full KT2440 list of candidates.
Table 2.
Comparison of predicted Crc regulon of P. putida with transcriptome and proteome data.
| Gene name | putidaa | KT2440b | Function | mRNA | Protein |
|---|---|---|---|---|---|
| NO | PP_0267 | outer membrane ferric siderophore receptor | nd | 1.6 | |
| fruR | NM | PP_0792 | FruR transcriptional regulator | nd | 2.3 |
| fruA | PP_0795 | PP_0795 | PTS fructose IIC component | 2.1 | nd |
| gap-1 | PP_1009 | PP_1009 | glyceraldehyde-3-phosphate dehydrogenase, type I | 2.7 | 3.3 |
| PP_1015 | PP_1015 | probable binding protein component of ABC sugar transporter | 2.3 | 4.9 | |
| oprB-1 | PP_1019 | PP_1019 | Glucose/carbohydrate outer membrane porin OprB precursor | 3.5 | 2.9 |
| PP_1059 | PP_1059 | probable amino acid permease | 6.4 | nd | |
| aatJ | PP_1071 | PP_1071 | probable binding protein component of ABC transporter | 3.3 | 7.7 |
| NM | PP_1400 | dicarboxylate MFS transporter | 2.5 | nd | |
| tctC | PP_1418 | PP_1418 | hypothetical protein | 1.6 | 3.4 |
| cspA-1 | PP_1522 | PP_1522 | cold shock protein CspA | 1.9 | 3.5 |
| ansA | PP_2453 | PP_2453 | L-asparaginase, type II | 2.4 | 3.1 |
| PP_3123 | PP_3123 | 3-oxoacid CoA-transferase subunit B | 9.1 | 4.5 | |
| NO | PP_3434 | hypothetical protein | 6.7 | nd | |
| NM | PP_3530 | conserved hypothetical protein | 2.0 | nd | |
| PP_3593 | PP_3593 | amino acid ABC transporter, periplasmic amino acid-binding protein | nd | 6.3 | |
| bkdA-1 | PP_4401 | PP_4401 | 3-methyl-2-oxobutanoate dehydrogenase | 3.2 | 1.6 |
| phhA | PP_4490 | PP_4490 | phenylalanine-4-hydroxylase | 2.8 | 1.9 |
| PP_4495 | PP_4495 | aromatic amino acid transport protein AroP2 | 2.6 | nd | |
| hmgA | PP_4621 | PP_4621 | homogentisate 1,2-dioxygenase | 5.0 | 7.8 |
| PP_4636 | PP_4636 | acetyl-CoA acetyltransferase | 3.6 | 2.3 | |
| hupA | PP_5313 | PP_5313 | probable DNA-binding protein | 3.8 | nd |
| accC-2 | PP_5347 | PP_5347 | acetyl-CoA carboxylase subunit A | 2.4 | nd |
Genes differentially regulated, based on transcriptome and proteome data, in rich media in a crc mutant of P. putida KT2442 [26] are cross referenced with (a) predicted Crc targets from three P. putida strains (KT2440, F1 and W619) and (b) with predicted Crc targets from P. putida KT2440 alone. Values of mRNA and protein indicate the relative levels of transcripts and protein in transcriptome and proteome analyses respectively [26]. NO (no ortholog) indicates that no orthologous loci were detected in either or both of P. putida F1 and W619. NM (no motif) indicates that no A-rich motif was detected in the upstream region of the orthologous loci in P. putida F1 and W619.
Table 3.
Comparison of predicted Crc regulon of P. aeruginosa with proteome data.
| Gene name | PAO1 | Function | protein |
|---|---|---|---|
| PA0534 | conserved hypothetical protein | 2.03 | |
| hpd | PA0865 | 4-hydroxyphenylpyruvate dioxygenase | 4.71 |
| oprD | PA0958 | Basic amino acid, basic peptide and imipenem outer membrane porin OprD precursor | 1.75 |
| PA1069 | hypothetical protein | 4.28 | |
| PA2553a | probable acyl-CoA thiolase | 1.59 | |
| PA2555 | probable AMP-binding enzyme | 1.54 | |
| PA2776 | conserved hypothetical protein | 1.71 | |
| PA3187b | probable ATP-binding component of ABC transporter | 10.28 | |
| edd | PA3194 | phosphogluconate dehydratase | 2.17 |
| PA4500 | probable binding protein component of ABC transporter | 3.48 | |
| PA4502c | probable binding protein component of ABC transporter | 3.35 | |
| PA4506c | probable ATP-binding component of ABC dipeptide transporter | 8.43 | |
| dadA | PA5304 | D-amino acid dehydrogenase, small subunit | 2.36 |
Genes differentially regulated, based on proteome data, in rich media in a crc mutant of P. aeruginosa PAO1 [27] are cross referenced with predicted targets from all P. aeruginosa strains considered in this study. Values of protein indicate relative levels of protein in the crc mutant relative to levels in the wild type strain. Some genes are proximal to, and possibly in operons with, bioinformatically predicted Crc targets: (a) PA2553 is proximal to PA2555, (b) PA3187 is proximal to PA3186 and (c) PA4502 and PA4506 are proximal to PA4501.
A proteomic analysis comparing the wild type strain P. aeruginosa PAO1 to an isogenic crc mutant in LB broth was also recently performed [27]. Under these conditions, 46 proteins were present at higher levels in the crc mutant compared to the wild type strain, suggesting that these targets are negatively regulated by the CRC system. Comparing those 46 experimentally-identified targets with the 215 predicted Crc targets identified in our bioinformatic study, it is seen that 13 of the 46 targets overlap (Table 3). Of these, 9 common targets have a predicted Crc binding site in the gene itself and a further 4 targets are in operons downstream of predicted Crc targets (Table 3). When the comparison is expanded to include all 279 candidates identified in PAO1 no new matches were found. The authors of that study identified putative Crc-binding sites in the 5' region of 23 of the 46 genes, and suggested that these may be subject to direct Crc mediated regulation [27]. The criteria applied for identifying putative Crc-binding sites was less strict than our study (with respect to consensus and distance from AUG codon), which explains the difference between the 13 binding sites we propose and the 23 postulated by these authors.
The fact that 18/23 overlaps are in the core P. putida regulon (and a further 2 are only excluded because orthologues are absent) and that no new overlaps with experimental data are introduced when the predicted Crc-regulon of P. aeruginosa is considered reinforces the validity of using the presence of the motif in orthologous genes within a species as a selection criterion in the global bioinformatic screen. Although the overlap between the experimental and bioinformatic datasets appears low for P. putida - 18(23)/267 genes - this should not be entirely unexpected. Genes predicted by the bioinformatics but not identified experimentally could simply be because they were below experimental detection limits, or more likely because the growth conditions used favoured some classes of genes. Of course, some hits may represent false positives, and our analysis predicted that there are rates of 18% and 26% false positive hits for P. aeruginosa and P. putida respectively. These are also possible explanations for differences between our data set and the PAO1 proteome data despite the higher level of overlap between our data with PAO1 (13/46) than between our data with KT2440. It is interesting that all three studies identify amino acid metabolism as an important component of the Crc-regulon. This reflects Crc metabolic adaptations in a nutrient rich environment (which was the experimental condition) where various amino acids are the major carbon sources. Performing the transcriptome/proteome experiments under different growth conditions, would be likely to yield a different set of genes. Conversely, there were also targets identified in the experimental studies that did not feature in the bioinformatic analysis. The most likely explanation for this is that these are indirect rather than direct targets of Crc as they lack the predicted Crc binding site. It is also possible, however, that the strict criteria used in the bioinformatic analysis excluded some genuine targets, or that Crc has alternative or additional binding sites, perhaps used only under certain conditions. From comparing all the data, we can already see that this was probably the case with the bkdA1 gene, which was identified as a target experimentally in both P. putida and P. aeruginosa, but bioinformatically only in P. putida (Table 2). The proposed Crc binding site in P. aeruginosa is AACAAGAGAAACAA [27], which differs in some positions to the consensus AAnAAnAA used in the bioinformatic analysis. Ultimately, protein-mRNA binding studies will be needed to resolve all these Crc-binding possibilities.
Crc regulates carbohydrate and amino acid utilisation
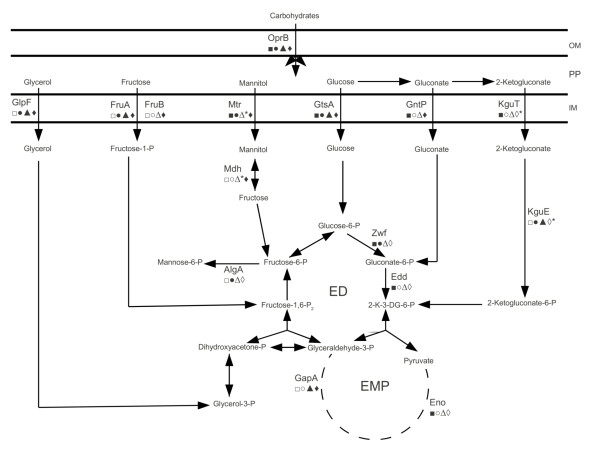
In order to find a common pattern of Crc regulation in Pseudomonas spp., we examined the function associated with the Crc candidates. In Pseudomonads, intermediates of the TCA cycle such as succinate or citrate cause catabolic repression of pathways involved in metabolism of carbohydrates, amino acids and other carbon sources [14,46]. Therefore, it is not surprising to find predicted Crc targets involved in such pathways. Indeed, our analysis highlights six interspecies Crc candidates involved in carbohydrate metabolism (Table 1). Since these candidates are all related to transport, it is tempting to speculate that Crc is responsible for direct down-regulation of transporters of carbohydrate utilisation, which would result in the indirect down-regulation of the relevant catabolic enzymes due to the lack of inducing molecules in the cytoplasm. Closer inspection of the intra-species Crc candidates, however, shows that some genes linked to carbohydrate metabolism could also be directly regulated by Crc (Additional file 1). For example, in P. aeruginosa and P. fluorescens species, the gene, zwf, encoding glucose-6-phosphate dehydrogenase has a Crc motif, whereas in P. putida and P. syringae species, the gene, gap-1, encoding glyceraldehyde-3-phosphate dehydrogenase has a Crc motif. When viewed in an integrated way, it is seen that there are two distinct patterns to the regulation of genes in this class (Figure 2). When present, sugar transporters are generally subject to CRC control, whereas the regulation of downstream sugar metabolism is species-specific with respect to genes encoding catabolic enzymes. Interestingly, the same trend is observed for amino acid metabolism where most of the interspecies Crc candidates are involved in transport (Table 1), whereas intraspecies candidates are involved in metabolism (Additional file 1).
Figure 2.
Predicted Crc regulon of carbohydrate metabolism in Pseudomonas. Selected genes involved in carbohydrate transport and metabolism are shown along with their status vis a vis (predicted) Crc regulation. Genes from P. aeruginosa (squares), P. fluorescens (circles), P. putida (triangles) and P. syringae (diamonds) are shown, with filled/unfilled symbols indicating that the target in that species is/is not predicted to be regulated by Crc. An asterisk (*) after a symbol indicates where an orthologous locus is absent in the relevant species. OM - outer membrane; PP - periplasm; IM - inner membrane; ED - Entner-Doudoroff pathway; EMP - Embden-Meyerhoff pathway; 2-K-3-DG-6-P - 2-keto-3-deoxygluconate-6-phosphate. OprB - carbohydrate porin B; GlpF - glycerol transporter; FruAB - fructose phosphotransferase system; Mtr - mannitol transporter subunit; GtsA - glucose transporter subunit; GntP - gluconate transporter; KguT - 2-ketogluconate transporter; Mdh - mannitol dehydrogenase; AlgA - mannose-6-P isomerase; Zwf - glucose-6-P dehydrogenase; Edd - gluconate-6-P dehydratase; KguE - xylose isomerase; GapA - glyceraldehyde-3-P dehydrogenase; Eno - phosphopyruvate hydratase. Some steps of the Embden-Meyerhoff pathway are abbreviated with a dashed line for clarity.
It is notable that another gene, cstA, with a predicted role in carbon starvation stress alleviation was also implicated as a Crc candidate. The CstA protein is involved in peptide transport that would assist the cell in escaping carbon starvation [47]. In Escherichia coli, induction of the cstA gene depends on cAMP and Crp [48] indicating that this locus is subject to CCR in E. coli. It is intriguing that a locus is implicated as being subject to catabolite repression in both Escherichia and Pseudomonas, despite differences in preferred nutrient sources and mechanisms of catabolite repression.
Crc regulates transcriptional activators that are induced during stationary phase
Crc also seems to regulate proteins involved in transcriptional regulation, as previously described [33]. Indeed the gene, hupA, encoding a bacterial histone like protein (HU-like protein), possesses a Crc motif in the P. aeruginosa, P. putida and P. fluorescens species. HU proteins are ubiquitous DNA binding factors that are involved in the structural maintenance of the bacterial chromosome and other events that require DNA binding [49]. In contrast to the structurally related integration host factor (IHF), HU proteins bind DNA in a sequence-independent manner. Generally, Pseudomonas possesses five HU/IHF copies per genome [50]. Two of these ORFs encode the two subunits of the IHF (integration host factor) protein (ihfA and ihfB), whereas hupA (or hupP), hupB and hupN encode HU-like proteins. Although the precise role of hupA is not known, HU-like proteins are required for transcription from the σ54-dependent Ps promoter of the toluene degradation pathway in P. putida [51], which is known to be subject to control by the CRC system. Identification of the Crc motif would be consistent with the idea that Crc impacts indirectly on the transcription level of a subset of genes through translational regulation of the regulatory genes hupA or ihfB. This may also explain some of the indirect targets of Crc identified in the transcriptome/proteome analysis discussed earlier [26]. The expression of hupA, hupB and hupN has been monitored during P. putida KT2440 growth [52]. Interestingly, whereas hupB and hupN transcript abundances are maximal in exponential phase, hupA expression seems to be activated during stationary phase. Remarkably, another Crc candidate of P. aeruginosa and P. syringae, ihfB, has increased expression during transition of cells from exponential growth to stationary phase [53]. This observation is not an isolated phenomenon as other predicted Crc targets, for example cstA [47,48] and polyhydroxyalkanoate biosynthesis (phaC1 or phaZ) [54], are also induced at the onset of stationary phase. CRC is depressed during stationary phase [24] so these observations on expression are consistent with a role for Crc in repressing expression of target genes during active growth.
Crc regulates virulence-related traits
It was shown previously that a crc mutant of P. aeruginosa PA14 was defective for biofilm formation and type IV pilus-mediated twitching motility [36] and a crc mutant of P. aeruginosa PAO1 is compromised in type III secretion, motility, expression of quorum sensing-regulated virulence factors and was less virulent in a Dictyostelium discoideum model [27]. Therefore, we searched for bioinformatic evidence that Crc integrates nutritional status cues with the regulation of virulence-related traits. We postulate that Crc might regulate some steps in alginate biosynthesis in Pseudomonas. Alginate production is linked to the conversion of microcolonies from a non-mucoid to a mucoid phenotype. In P. aeruginosa this phenotype marks the transition to a more persistent state during pulmonary infection, characterised by antibiotic resistance and accelerated pulmonary decline [55]. The regulation of alginate production in Pseudomonas is highly complex and involves the interaction of many regulatory systems [56]. In this study, the transcriptional activator AlgP, involved in the transcription of a key alginate biosynthetic gene, algD [57] encoding GDP-mannose 6-dehydrogenase, is predicted, to be directly regulated by Crc in P. aeruginosa, P. putida and P. syringae species. In this case, the interspecific Crc regulation blocks the synthesis of a transcriptional regulator which leads to indirect regulation of the biosynthetic pathway, reminiscent of the cases of alkS and benR in P. putida [18]. Nevertheless, at the species level, Crc is also predicted to regulate some enzymes directly. In P. aeruginosa, Crc also is predicted to bind to alg8 and algF transcripts which encode a subunit of alginate polymerase [58,59] and an alginate acetylation protein [60] respectively. The synthesis of the alginate precursor, mannose-6-phosphate, encoded by algA, is predicted to be under the control of Crc in P. fluorescens only (Figure 2). The additional levels of regulation of alginate in P. aeruginosa, could reflect the importance of this exopolysaccharide for persistence in specialised ecological niches, including inside the host.
Another interesting Crc target is estA encoding an autotransporter protein with esterase activity [61] that is indispensable for rhamnolipid production [62]. Rhamnolipids are surface-active molecules that play a role in biofilm fluidity [63] and are toxic against a variety of microorganisms [64]. Preliminary experiments confirm that rhamnolipid production is a Crc-regulated trait in P. aeruginosa (data not shown). Moreover, inactivation of the estA gene in P. aeruginosa also influenced other virulence-related functions like swimming, twitching and swarming in a rhamnolipid-independent fashion [62]. Rhamnolipids have numerous features in common with polyhydroxyalkanoic acid (PHAs), a metabolic storage material involved in bacterial stress-resistance and biofilm formation [65]. Firstly they are both synthesised in response to the presence of excess carbon where other nutrients, such as nitrogen or phosphorus, are growth limiting [54,64,66]. Secondly, both molecules are composed of 3-hydroxydecanoic acids connected by ester bonds. Interestingly, phaC1 [67] and phaF [68] encoding a PHA polymerase and PHA transcriptional regulator respectively are also predicted to be Crc regulated in P. aeruginosa, P. putida and P. syringae species. Notwithstanding the role of PHA in attachment of P. aeruginosa to surfaces [65], the implication of Crc in PHA production may also be interesting from an industrial point of view since it is hoped that PHA accumulating bacteria may be exploited in bioplastic production [69]. Under high carbon:nitrogen ratios, PHA and rhamnolipids are produced and represent carbon sinks to accommodate an inability to metabolise an excess of carbon over nitrogen. One possible function of the CRC system is to integrate C/N metabolism by regulating the production of carbon sink compounds such as PHA and rhamnolipid. This could be mediated by the CbrAB/NtrBC links outlined earlier.
Conclusions
CRC is an important global control network employed by Pseudomonas to optimise growth with available nutrients in a variety of environments. This analysis aimed to predict the set of targets that are directly regulated by the Crc protein in four species of Pseudomonas. As expected, genes involved in the metabolism of less favoured nutrients were identified. An interesting feature, however, was that the regulation of transporters is a conserved feature of Crc regulation in Pseudomonas spp. while the regulation of particular enzymatic steps and transcriptional activators is generally present in a more species-dependent manner. This suggests that different Pseudomonas species have fine-tuned CRC to reflect the ecology of that particular species. In addition to anticipated effects on sugar metabolism, there are indications from the data that Crc may play a role in maintaining the carbon/nitrogen balance in Pseudomonas and this is worthy of further study. It was postulated that identifying Crc targets might enhance knowledge of some applied aspects of Pseudomonas and one example of this was the prediction that Crc regulates steps in polyhydroxyalkanoate (PHA) synthesis in P. putida, as this is of interest for the production of biodegradable bioplastics. In the case of P. aeruginosa, the analysis revealed that alginate production and other traits linked to virulence may be under CRC control. It was especially intriguing to discover that Crc may play a role in regulation of globally important DNA binding proteins such as HU and IHF and thus regulate, indirectly, many pathways that depend on the DNA bending properties of these proteins for transcription or repression. These novel aspects of Crc regulation therefore deserve further investigation given the potential that it may enhance our understanding of the integration of nutritional status cues with the regulation of important activities of the Pseudomonas.
Methods
Positions -70 to +16 relative to the origin of translation of all protein encoding genes of available Pseudomonas spp. were downloaded from the regulatory sequence analysis tool (RSAT) [40] using the retrieve sequence function. Genes containing an A-rich (AAnAAnAA) motif in the -70 to +16 region were identified using a script in Perl. Translated protein encoding sequences were downloaded from the Pseudomonas genome database [70] and used to create local blast databases with formatdb [71]. Orthologous genes were identified as best hits using blastp analysis (blastall v2.2.22) [71,72] against local databases. Cut-offs of 50% identity over at least 80% of the sequence length and an expected value (e-value) of 1e-10 were applied. Orthology was confirmed by reciprocating the blastp analysis. Since the A-rich motif is short and degenerate it is expected that occurrences of the A-rich motif that are unrelated to Crc binding will be detected in this analysis, giving rise to false positive hits. In order to estimate the rate of false positive hits in our analysis we searched for the A-rich motif in the reverse orientation of the upstream regions of orthologous loci [73]. Since the A-rich motif in the reverse orientation is unrelated to Crc binding it is reasoned that this estimates the rate of occurrence of the A-rich motif in the sequence fragments tested. Predictably it was found that the use of more strains per species resulted in lower estimated rates of false positives (P. aeruginosa - 4 strains, 18% estimated false positives; P. fluorescens - 3 strains, 32% estimated false positives; P. putida - 3 strains, 26% estimated false positives; P. syringae - 2 strains, 41% estimated false positives). Thus, it is estimated, based on the weighted mean false discovery rate, that approximately 73% of the Crc candidates in additional file 1 are genuine targets for Crc binding. Functional information about the translated protein sequences was obtained from the sequence headers and by performing Blast2GO analysis [74].
Authors' contributions
PB, JPM and FOG conceived the study. PB performed the bioinformatic analyses, PB and MB interpreted the data and JPM and FOG oversaw the study. PB and MB prepared figures, tables and additional files presenting the data and PB, MB, JPM and FOG drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Crc candidates identified in every Pseudomonas spp. List of every locus bearing a Crc motif in P. aeruginosa, P. fluorescens, P. putida and P. syringae species. The numbers under strain names on the left indicate the locus id, according to Genbank annotation, of the locus with the A-rich motif in the upstream region. The numbers under the strain names on the right indicate the position of the A-rich motif relative to the origin of translation.
Contributor Information
Patrick Browne, Email: p.browne@umail.ucc.ie.
Matthieu Barret, Email: m.barret@ucc.ie.
Fergal O'Gara, Email: f.ogara@ucc.ie.
John P Morrissey, Email: j.morrissey@ucc.ie.
Acknowledgements
This research was supported in part by grants awarded by the Science Foundation of Ireland (grants 04/BR/B0597, 07/IN.1/B948, 08/RFP/GEN1295, 08/RFP/GEN1319 and 09/RFP/BMT2350), the Department of Agriculture, Fisheries and Food (RSF grants 06-321 and 06-377; FIRM grants 06RDC459, 06RDC506 and 08RDC629), the European Commission (grant FP6#O36314 and Marie Currie TOK:TRAMWAYS), Irish Research Council for Science Engineering and Technology (grant 05/EDIV/FP107/INTERPAM), the Marine Institute (Beaufort award C&CRA2007/082), the Health Research Board (grants RP/2006/271 and RP/2007/290). P.B. is supported by a STRIVE Doctoral Scholarship from the Environmental Protection Agency, Ireland and the Department of Environment, Heritage and Local Government provided by the Irish Government under the National Development Plan 2007-2013 (EPA-2006-S-21). We thank Pat Higgins for ongoing techncial support and members of our groups for useful discussions.
References
- Tarnawski S, Hamelin J, Locatelli L, Aragno M, Fromin N. Examination of Gould's modified S1 (mS1) selective medium and Angle's non-selective medium for describing the diversity of Pseudomonas spp. in soil and root environments. FEMS Microbiol Ecol. 2003;45:97–104. doi: 10.1016/S0168-6496(03)00130-2. [DOI] [PubMed] [Google Scholar]
- Browne P, Rice O, Miller SH, Burke J, Dowling DN, Morrissey JP, O'Gara F. Superior inorganic phosphate solubilization is linked to phylogeny within the Pseudomonas fluoresence complex. Appl Soil Ecol. 2009;43:131–138. doi: 10.1016/j.apsoil.2009.06.010. [DOI] [Google Scholar]
- Rajmohan S, Dodd C, Waites W. Enzymes from isolates of Pseudomonas fluorescens involved in food spoilage. J Appl Micro. 2002;93:205–213. doi: 10.1046/j.1365-2672.2002.01674.x. [DOI] [PubMed] [Google Scholar]
- Mulcahy H, O'Callaghan J, O'Grady EP, Maciá MD, Borrell N, Gómez C, Casey PG, Hill C, Gahan CGM, Oliver A, O'Gara F. Pseudomonas aeruginosa RsmA plays an important role during murine infection by influencing colonization, virulence, persistence and pulmonary inflammation. Infect Immun. 2008;76:632–638. doi: 10.1128/IAI.01132-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haritash A, Kaushik C. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): A review. J Hazard Mater. 2009;169:1–15. doi: 10.1016/j.jhazmat.2009.03.137. [DOI] [PubMed] [Google Scholar]
- Walsh UF, Morrissey JP, O'Gara F. Pseudomonas for biocontrol of phytopathogens: from functional genomics to commercial exploitation. Curr Opin Biotechnol. 2001;12:289–295. doi: 10.1016/S0958-1669(00)00212-3. [DOI] [PubMed] [Google Scholar]
- Cronin D, Moënne-Loccoz Y, Fenton A, Dunne C, Dowling DN, O'Gara F. Role of 2,4-diacetylphloroglucinol in the interactions of the biocontrol pseudomonad strain F113 with the potato cyst nematode Globodera rostochiensis. Appl Environ Microbiol. 1997;63:1357–1361. doi: 10.1128/aem.63.4.1357-1361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas D, Défago G. Biological control of soil-borne pathogens by fluorescent pseduomonads. Nat Rev Microbiol. 2005;3:307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- Miller SH, Browne P, Prigent-Combaret C, Combes-Meynet E, Morrissey JP, O'Gara F. Biochemical and genomic comparison of inorganic phosphate solubilization in Pseudomonas species. Environ Microbiol Rep. 2010;2:403–411. doi: 10.1111/j.1758-2229.2009.00105.x. [DOI] [PubMed] [Google Scholar]
- Villacieros M, Whelan C, Mackova M, Molgaard J, Sánchez-Contreras M, Lloret J, Aguirre de Cárcer DA, Oruezábal RI, Bolaños L, Macek T, Karlson U, Dowling DN, Martín M, Rivilla R. Polychlorinated biphenyl rhizoremediation by Pseudomonas fluorescens F113 derivatives, using a Sinorhizobium meliloti system to drive bph gene expression. Appl Environ Microbiol. 2005;71:2687–2694. doi: 10.1128/AEM.71.5.2687-2694.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol. 2008;11:87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- Rojo F. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol Rev. 2010;34:658–684. doi: 10.1111/j.1574-6976.2010.00218.x. [DOI] [PubMed] [Google Scholar]
- Collier D, Hager P, Phibbs P Jr. Catabolite repression control in the Pseudomonads. Res Microbiol. 1996;147:551–561. doi: 10.1016/0923-2508(96)84011-3. [DOI] [PubMed] [Google Scholar]
- Suh SJ, Runyen-Janecky LJ, Maleniak TC, Zielinski-Monzy NA, Phibbs P Jr, West SEH. Effect of vfr mutation on global gene expression and catabolite repression control of Pseudomonas aeruginosa. Microbiology. 2002;148:1561–1569. doi: 10.1099/00221287-148-5-1561. [DOI] [PubMed] [Google Scholar]
- Moreno R, Ruiz-Manzano A, Yuste L, Rojo F. The Pseudomonas putida Crc global regulator is an RNA binding protein that inhibits translation of the AlkS transcriptional regulator. Mol Micro. 2007;64:665–657. doi: 10.1111/j.1365-2958.2007.05685.x. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Abdou L, Hass D. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. PNAS. 2009;106:21866–21871. doi: 10.1073/pnas.0910308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno R, Marzi S, Romby P, Rojo F. The Crc global regulator binds to an unpaired A-rich motif at the Pseudomonas putida alkS mRNA coding sequence and inhibits translation initiation. Nucl Acids Res. 2009;37:7678–7690. doi: 10.1093/nar/gkp825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishijyo T, Haas D, Itoh Y. The CbrA-CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol Microbiol. 2001;40:917–931. doi: 10.1046/j.1365-2958.2001.02435.x. [DOI] [PubMed] [Google Scholar]
- Li W, Lu CD. Regulation of carbon and nitrogen utilization by CbrAB and NtrBC two-component systems in Pseudomonas aeruginosa. J Bacteriol. 2007;189:5413–5420. doi: 10.1128/JB.00432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XX, Rainey PB. Dual involvement of CbrAB and NtrBC in the regulation of histidine utilization in Pseudomonas fluorescens SBW25. Genetics. 2008;178:185–195. doi: 10.1534/genetics.107.081984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts J, Clarke P. The effect of nitrogen limitation on catabolite repression of amidase, histidase and urocanase in Pseudomonas aeruginosa. J Gen Microbiol. 1976;93:377–387. doi: 10.1099/00221287-93-2-377. [DOI] [PubMed] [Google Scholar]
- Aranda-Olmedo I, Ramos JL, Marqués S. Integration of signals through Crc and PtsN in catabolite repression of Pseudomonas putida TOL Plasmid pWW0. Appl Environ Microbiol. 2005;71:4191–4198. doi: 10.1128/AEM.71.8.4191-4198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Manzano A, Yuste L, Rojo F. Levels an activity of the Pseudomonas putida global regulatory protein Crc vary according to growth conditions. J Bacteriol. 2005;187:3678–3686. doi: 10.1128/JB.187.11.3678-3686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff J, MacGregor C, Eisenberg R, Phibbs P Jr. Isolation and characterization of catabolite repression control mutants of Pseudomonas aeruginosa PAO. J Bacteriol. 1991;173:4700–4706. doi: 10.1128/jb.173.15.4700-4706.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno R, Martínez-Gomariz M, Yuste L, Gil C, Rojo F. The Pseudomonas putida Crc global regulator controls the hierarchical assimilation of amino acids in a complete medium: Evidence from proteomic and genomic analyses. Proteomics. 2009;9:2910–2928. doi: 10.1002/pmic.200800918. [DOI] [PubMed] [Google Scholar]
- Linares J, Moreno R, Fajardo A, Martínez-Solano L, Escalante R, Rojo F, Martínez J. The global regulator Crc modulates metabolism, susceptibility to antibiotics and virulence in Pseudomonas aeruginosa. Environ Microbiol. 2010. [DOI] [PubMed]
- Daniels C, Godoy P, Duque E, Molina-Henares MA, de la Torre J, del Arco JM, Herrera C, Segura A, Guazzaroni ME, Ferrer M, Ramos JL. Global regulation of food supply by Pseudomonas putida DOT-T1E. J Bacteriol. 2010;192:2169–2181. doi: 10.1128/JB.01129-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Sample A, Runyen-Janecky L. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor portein family. J Bacteriol. 1994;176:7532–7542. doi: 10.1128/jb.176.24.7532-7542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste L, Rojo F. Role of the crc gene in catabolic repression of the Pseudomonas putida GPo1 alkane degradation pathway. J Bacteriol. 2001;183:6197–6206. doi: 10.1128/JB.183.21.6197-6206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putrinš M, Tover A, Tegova R, Saks Ü, Kivisaar M. Study of factors which negatively affect expression of the phenol degardation operon pheBA in Pseudomonas putida. Microbiology. 2007;153:1860–1871. doi: 10.1099/mic.0.2006/003681-0. [DOI] [PubMed] [Google Scholar]
- Morales G, Linares JF, Beloso A, Albar JP, Martínez JL, Rojo F. The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J Bacteriol. 2004;186:1337–1344. doi: 10.1128/JB.186.5.1337-1344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno R, Rojo F. The target for the Pseudomonas putida Crc global regulator in the benzoate degradation pathway is the BenR transcriptional regulator. J Bacteriol. 2008;190:1539–1545. doi: 10.1128/JB.01604-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno R, Fonseca P, Rojo F. The Crc global regulator inhibits the Pseudomonas putida pWW0 toluene/xylene assimilation pathway by repressing the translation of regulatory and structural genes. J Biol Chem. 2010;285:24412–24419. doi: 10.1074/jbc.M110.126615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester K, Madhusudhan K, Sokatch J. Catabolite repression control by Crc in 2xYT medium is mediated by posttranscriptional regulation of bkdR expression in Pseudomonas putida. J Bacteriol. 2000;182:1150–1153. doi: 10.1128/JB.182.4.1150-1153.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole G, Gibbs K, Hager P, Phibbs P Jr, Kolter R. The global carbon metabolism regulator Crc is a component of a singnal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J Bacteriol. 2000;182:425–431. doi: 10.1128/JB.182.2.425-431.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R, Macleod J, Foley W, Nayudu M. Gluconic acid: An antifungal agent produced by Pseudomonas species in biological control of take-all. Phytochemistry. 2006;67:595–604. doi: 10.1016/j.phytochem.2005.12.011. [DOI] [PubMed] [Google Scholar]
- de Werra P, Péchy-Tarr M, Keel C, Maurhofer M. Role of gluconic acid production in the regulation of biocontrol traits of Pseudomonas fluorescens CHA0. Appl Environ Microbiol. 2009;75:4162–4174. doi: 10.1128/AEM.00295-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Kiefer P, Reimmann C, Keel C, Rolli J, Vorholt JA, Haas D. Small RNA-dependent expression of secondary metabolism is controlled by Krebs cycle function in Pseudomonas fluorescens. J Biol Chem. 2009;284:34976–34985. doi: 10.1074/jbc.M109.052571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M, Sand O, Turatsinze JV, Janky R, Defrance M, Vervisch E, Broheé S, van Helden J. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119–W127. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silby MW, Cerdeño-Tárraga AM, Vernikos GS, Giddens SR, Jackson RW, Preston GM, Zhang XX, Moon CD, Gehrig SM, Godfrey SAC, Knight CG, Malone JG, Robinson Z, Spiers AJ, Harris S, Challis GL, Yaxley AM, Harris D, Seeger K, Murphy L, Rutter S, Squares R, Quail MA, Saunders E, Mavromatis K, Brettin TS, Bentley SD, Hothersall J, Stephens E, Thomas CM, Parkhill J, Levy SB, Rainey PB, Thomson NR. Genomic and genetic analysis of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol. 2009;10:R51. doi: 10.1186/gb-2009-10-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathee K, Narasimhan G, Valdes C, Qiu X, Matewish JM, Koehrsen M, Rokas A, Yandava CN, Engels R, Zeng E, Olavarietta R, Doud M, Smith RS, Montgomery P, White JR, Godfrey PA, Kodira C, Birren B, Galagan JE, Lory S. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci USA. 2008;105:3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan JA, Morrissey JP, Coppoolse ER, Stiekema WJ, O'Gara F, Boyd EF. Evolutionary history of the phl gene cluster in the plant-associated bacterium Pseudomonas fluorescens. Appl Environ Microbiol. 2009;75:2122–2131. doi: 10.1128/AEM.02052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy PH, Tetu SG, Larouche A, Elbourne L, Tremblay S, Ren Q, Dodson R, Harkins D, Shay R, Watkins K, Mahamoud Y, Paulsen IT. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA14. PLoS One. 2010;5:e8842. doi: 10.1371/journal.pone.0008842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Guttman D. Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. App Env Microbiol. 2004;70:1999–2012. doi: 10.1128/AEM.70.4.1999-2012.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo F, Dinamarca A. In: Pseudomonas: virulence and gene regulation. Ramos JL, editor. Vol. 2. Kluwer Academic/Plenum Publishers; 2004. Catabolite repression and physiological control; pp. 365–387. [Google Scholar]
- Schultz JE, Matin A. Molecular and functional characterization of a carbon starvation gene of Escherichia coli. J Mol Biol. 1991;218:129–140. doi: 10.1016/0022-2836(91)90879-B. [DOI] [PubMed] [Google Scholar]
- Schultz JE, Latter GI, Matin A. Differential regulation by cyclic AMP of starvation protein synthesis in Escherichia coli. J Bacteriol. 1988;170:3903–3909. doi: 10.1128/jb.170.9.3903-3909.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam TA, Ishihama A. Twelve species of nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affininty. J Biol Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- Cases I, de Lorenzo V. The genomes of Pseudomonas encode a third HU protein. Micriobiology Comment. 2002;148:1243–1245. doi: 10.1099/00221287-148-5-1243. [DOI] [PubMed] [Google Scholar]
- Pérez-Martín J, de Lorenzo V. The σ54-dependent promoter Ps of the TOL plasmid of Pseudomonas putida requires HU for transcriptional activation in vivo by xylR. J Bacteriol. 1995;177:3758–3763. doi: 10.1128/jb.177.13.3758-3763.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste L, Hervás AB, Canosa I, Tobes R, Nogales J, Pérez-Pérez MM, Santero E, Díaz E, Ramos JL, de Lorenzo V, Rojo F. Growth phase-dependent expression of the Pseudomonas putida KT2440 transcriptional machinery analysed with a genome-wide DNA microarray. Environ Microbiol. 2006;8:165–177. doi: 10.1111/j.1462-2920.2005.00890.x. [DOI] [PubMed] [Google Scholar]
- Valls M, Buckle M, de Lorenzo V. In vivo UV laser footprinting of the Pseudomonas putida σ54 promoter reveals that integration host factor couples transcriptional activity to growth phase. J Biol Chem. 2002;277:2169–2175. doi: 10.1074/jbc.M108162200. [DOI] [PubMed] [Google Scholar]
- Ward PG, de Roo G, O'Connor KE. Accumulation of polyhydroxyalkanoate from sytrene and phenylacetic acid by Pseudomonas putida CA-3. Appl Environ Microbiol. 2005;71:2046–2052. doi: 10.1128/AEM.71.4.2046-2052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey DM, Wozniak DJ. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol. 2005;56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- Muhammadi Ahmed N. Genetics of bacterial alginate: alginate genes distribution, organization and biosynthesis in bacteria. Curr Genomics. 2007;8:191–202. doi: 10.2174/138920207780833810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konyecsni WM, Deretic V. DNA sequence and expression of algP and algQ, components of the multigene system transcriptionally regulating mucoidy in Pseudomonas aeruginosa: algP contains multiple direct repeats. J Bacteriol. 1990;172:2511–2520. doi: 10.1128/jb.172.5.2511-2520.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remminghorst U, Rehm BHA. In vitro alginate polymerization and the functional role of Alg8 in alginate production by Pseudomonas aeruginosa. Appl Environ Microbiol. 2006;72:298–305. doi: 10.1128/AEM.72.1.298-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oglesby LL, Sumita J, Ohman DE. Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology. 2008;154:1605–1615. doi: 10.1099/mic.0.2007/015305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin MJ, Ohman DE. Identification of algF in the alginate biosynthetic gene cluster of Pseudomonas aeruginosa which is requried for alginate acetylation. J Bacteriol. 1993;175:5057–5065. doi: 10.1128/jb.175.16.5057-5065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm S, Tommassen J, Jaeger K. A novel lipolytic enzyme located in the outer membrane of Pseudomonas aeruginosa. J Bacteriol. 1999;181:6977–6986. doi: 10.1128/jb.181.22.6977-6986.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm S, Gdynia A, Tielen P, Rosenau F, Jaeger K. The autotransporter esterase EstA of Pseudomonas aeruginosa is required for rhamnolipid production, cell motility, and biofilm formation. J Bacteriol. 2007;189:6695–6703. doi: 10.1128/JB.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey ME, Caizza NC, O'Toole GA. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol. 2003;185:1027–1036. doi: 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberón-Chávez G, Lépine F, Déziel E. Production of rhamnolipids by Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2005;68:718–725. doi: 10.1007/s00253-005-0150-3. [DOI] [PubMed] [Google Scholar]
- Pham TH, Webb JS, Rehm BHA. The role of polyhydroxyalkanoate biosynthesis by Pseudomonas aeruginosa in rhamnolipid and alginate production as well as stress tolerance and biofilm formation. Microbiology. 2004;150:3405–3413. doi: 10.1099/mic.0.27357-0. [DOI] [PubMed] [Google Scholar]
- de Smet MJ, Eggink G, Witholt B, Kingma J, Wyngerg H. Characterization of intracellular inclusions formed by Pseudomonas oleovorans during growth on Octane. J Bacteriol. 1983;154:870–878. doi: 10.1128/jb.154.2.870-878.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary ND, O'Connor KE, Ward P, Goff M, Dobson ADW. Genetic characterization of accumulation of polyhydroxyalkanoate from styrene in Pseudomonas putida CA-3. Appl Environ Microbiol. 2005;71:4380–4387. doi: 10.1128/AEM.71.8.4380-4387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto MA, Bühler B, Jung K, Witholt B, Kessler B. PhaF, a polyhydroxyalkanoate-granule-associated protein of Pseudomonas oleovorans GPo1 involved in the regulatory expression system for pha genes. J Bacteriol. 1999;181:858–868. doi: 10.1128/jb.181.3.858-868.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim SJ, Snell KD, Hogan SA, Stubbe J, Rha C, Sinskey A. PHA synthase activity controls the molecular weight and polydispersity of polyhydroxybutyrate in vivo. Nat Biotechnol. 1997;15:63–67. doi: 10.1038/nbt0197-63. [DOI] [PubMed] [Google Scholar]
- Winsor G, Van Rossum GL, Lo R, Khaira B, Whiteside MD, Hancock REW, Brinkman FSL. Pseudomonas Genome Database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucl Acids Res. 2009;37:D483–D488. doi: 10.1093/nar/gkn861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421–429. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Cases I, Ussery DW, de Lorenzo V. The σ54 regulon (stimulon) of Pseudomonas putida. Environ Microbiol. 2003;5:1281–1293. doi: 10.1111/j.1462-2920.2003.00528.x. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, Miguel García-Gómez J, Terol J, Talón M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crc candidates identified in every Pseudomonas spp. List of every locus bearing a Crc motif in P. aeruginosa, P. fluorescens, P. putida and P. syringae species. The numbers under strain names on the left indicate the locus id, according to Genbank annotation, of the locus with the A-rich motif in the upstream region. The numbers under the strain names on the right indicate the position of the A-rich motif relative to the origin of translation.