Abstract
Fc receptor-like A (FCRLA) is an unusual member of the extended Fc receptor family. FCRLA has homology to receptors for the Fc portion of Ig (FCR) and to other FCRL proteins. However, unlike these other family representatives, which are typically transmembrane receptors with extracellular ligand-binding domains, FCRLA has no predicted transmembrane domain or N-linked glycosylation sites and is an intracellular protein. We show by confocal microscopy and biochemical assays that FCRLA is a soluble resident endoplasmic reticulum (ER) protein, but it does not possess the amino acid sequence KDEL as an ER retention motif in its C-terminus. Using a series of deletion mutants, we found that its ER retention is most likely mediated by the amino terminal partial Ig-like domain. We have identified ER-localized Ig as the FCRLA ligand. FCRLA is unique among the large family of Fc receptors, in that it is capable of associating with multiple Ig isotypes, IgM, IgG and IgA. Among hemopoietic cells, FCRLA expression is restricted to the B lineage and is most abundant in germinal center B lymphocytes. The studies reported here demonstrate that FCRLA is more broadly expressed among human B lineage cells than originally reported; it is found at significant levels in resting blood B cells and at varying levels in all B-cell subsets in tonsil.
Keywords: B lymphocyte, ER retention, Fc receptor, membrane and secretory Ig, microsomes
Introduction
The presence of receptors on phagocytic cells for the Fc portion of IgG was demonstrated >50 years ago (1). With the recent cloning of a human and mouse receptor for the Fc portion of Ig (FcR) for IgM (2, 3), cell surface Fc receptors for all the Ig isotypes except IgD have now been molecularly identified (4, 5). In addition to these ‘classical’ Ig-binding Fc receptors, a new family of FcR-related genes, now called Fc receptor-like (FCRL), has more recently been identified, primarily by mining of genomic databases (6–8). The founding members of this relative large family, designated FCRL1–6 in humans, are transmembrane proteins expressed on the cell surface of lymphoid cells and several of them have been shown to have activating and inhibitory signaling function (9–11). However, unlike the classical FcR, these FCRL have not been found to bind Ig (12) and their ligands have remained a mystery until the very recent identification of MHC class II as a ligand for FCRL6 (13).
Two additional members of the FCRL family, now designated Fc receptor-like A (FCRLA) and Fc receptor-like B (FCRLB) (7), have unique features that warrant their inclusion in a separate subfamily. Most notably, FCRLA and FCRLB lack conventional transmembrane regions and are not expressed on the cell surface but instead localize intracellularly (14–19). Structurally, both molecules are Ig gene superfamily members but FCRLB has three Ig domains whereas FCRLA has only two; instead, the first domain of FCRLA is a novel domain type. In addition to their Ig-like domains, both FCRLA and FCRLB have an unusual bipartite C-terminus consisting of an N-terminal mucin-like region, rich in proline, serine and threonine residues and a C-terminal region predicted to form an α-helix (see Fig. 9a for a model of the FCRLA molecule). FCRLB has two potential N-linked glycosylation sites but FCRLA has none. Both FCRLA and FCRLB are preferentially expressed in B lineage cells, although there have been reports of their expression in melanocytes and melanoma cells (14, 20). Until now, the ligands for these two intracellular orphan receptors have been unknown. In one study, FCRLA was artificially expressed on the surface of transfected 293 cells where it was unable to bind IgM, IgG, IgA or IgE (18). However, this negative finding is difficult to interpret since FCRLA was removed from its physiological intracellular location for this assay. Here, we show that FCRLA is a resident endoplasmic reticulum (ER) protein and define its ligand-binding specificity in this native intracellular context. Unique among the entire large family of FcR and FCRL molecules, FCRLA can bind multiple Ig isotypes, IgM, IgG and IgA.
Fig. 9.
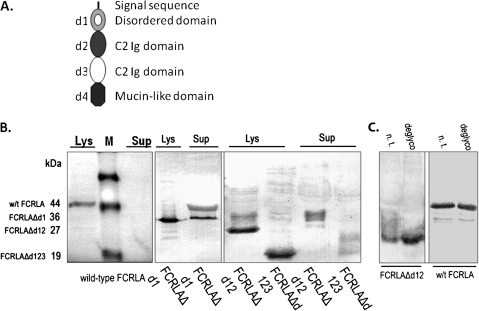
The N-terminal domain mediates FCRLA intracellular retention. (A) Model of FCRLA showing its domain structure. (B and C) 293T cells were transfected with cDNA expression vectors encoding the indicated FCRLA deletion mutants. (B) Two days after transfection, lysate of cells (Lys) and supernatants of conditioned media (Sup) were subjected to immunoblotting with FCRLA antibody. (C) Deglycosylation of cell lysates of wild-type FCRLA and the FCRLAΔd12 deletion mutant. Not treated (n.t.) and deglycosylated (deglyco) cell lysates.
Materials and methods
Cells and reagents
Human lymphoid cell lines and HeLa cells were cultured in RPMI 1640 or DMEM supplemented media with 10% heat-inactivated fetal bovine serum (FBS; Gibco-BRL Life Technologies), 50 μm β-mercaptoethanol (Millipore), 2 mM l-glutamine and 100 Uml−1 antibiotic-antimycotic (Gibco-BRL Life Technologies) at 37°C in a 5% CO2 atmosphere. For metabolic labeling experiments, cells were labeled with [35S]-TransLabel (MP Biomedicals) as described previously (21).
Immunoprecipitation and western blots
Antibodies used for immunoprecipitation were anti-human FCRLA mAbs M101 or M116 (22) or polyclonal goat antibodies specific for human Ig: μ, γ or α heavy chains (HCs) or κ or λ light chains (LCs) (Southern Biotech, Birmingham, AL, USA). For metabolic labeling immunoprecipitation experiments, antibodies were used in conjunction with protein A-coupled sepharose. For the immunoprecipitation–western blot analysis, antibodies were covalently attached to an activated affinity chromatography media, Affi-Gel 10 (BioRad, Hercules, CA, USA). This method was used since the Ig produced by some of the cell lines bound protein A directly due to VH3 usage by the HC (23–25). Cells were washed twice with PBS and lysed at 4°C in lysis buffer (50 mM Tris–HCl, 0.15 M NaCl, 5 mM EDTA, 0.5% NP40 and protease inhibitor cocktail) for 30 min followed by centrifugation at 14 680×g for additional 30 min at 4°C. Total cell lysates were immunoprecipitated overnight under constant gentle agitation. After incubation, samples were centrifuged and the pellets were washed with ice-cold wash buffer 3× and heated to 100°C for 5 min in Laemmli SDS sample buffer. The proteins obtained were separated by SDS–PAGE under reducing conditions and transferred to polyvinylidene fluoride membranes. Blots were blocked with 5% skim milk in PBS for 1 h at room temperature and then incubated with either HRP-conjugated goat anti-human IgM (1:500, Southern Biotech) unlabeled mouse monoclonal or rabbit anti-human FCRLA (15) overnight at 4°C. Membranes were washed 3× with 5% milk in PBS and incubated with HRP-labeled goat anti-mouse IgG or goat anti-rabbit IgG (1:1000) for 2 h at room temperature. Before developing, the blots were washed again 3× with 5% milk in PBS. All membranes were visualized using Pierce SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL, USA) and exposed to film.
For the analysis of transfected 293T and BJAB, the cells were lysed for 5 min in a loading SDS buffer at 100°C. For western blotting, the samples were resolved on 10 or 11% SDS–polyacrylamide gel under reducing conditions and transferred to a Hybond-C nitrocellulose membrane (GE Healthcare Bio-Sciences Corp, Piscataway, NJ, USA). The membrane was blocked overnight at 4°C in 0.1 M Na2CO3 containing 0.5% gelatin and 1% casein. The membrane was then incubated with rabbit anti-FCRLA Ig diluted 1:500 in freshly prepared blocking solution supplemented with 0.1% Triton X-100 for 1 h at 37°C. Following incubation with primary antibodies, the membrane was washed several times with 0.1 M Na2CO3 containing 0.1% Triton X-100 and incubated with peroxidase-conjugated goat anti-rabbit antibodies. Enzyme activity was visualized by staining with 3,3′-diaminobenzidine tetrahydrochloride in a 0.1 M Tris–HCl, pH 7.4, buffer containing 0.05 M imidazole.
Immunofluorescent staining, flow cytometry and confocal microscopy
For immunofluorescent staining and flow cytometry, cells were fixed with 1% PFA, washed and then permeabilized with 0.1% saponin prior to intracellular staining. The M101 FCRLA mAb was conjugated with Alexa 488 using an Alexa Fluor® 488 protein labeling kit (Molecular Probes Invitrogen, Eugene, OR, USA). In some cases, cells were stained for cell surface markers prior to permeabilization. The following commercially available antibodies were used: PE-labeled goat antibodies to human IgM and an IgD mAb (Southern Biotech) and PE-labeled CD3, CD19 and CD38 antibodies (BD PharMingen, San Diego, CA, USA). Stained cells were washed and re-suspended in cold PBS 0.5% BSA before analysis on a FACSCalibur (BD Bioscience). Sorting of normal blood B and T cells was performed on a MoFlo instrument (DAKO Cytomation, Fort Collins, CO, USA) after cell surface staining for CD3 and CD19. The purity of the sorted cells was routinely >98%. For confocal microscopy, FCRLA-transfected HeLa cells were seeded onto coverslips. Cells were washed 3× with PBS, fixed with methanol/acetone 1:1 and blocked with 5% BSA (Calbiochem) in PBS. Alexa 488-conjugated monoclonal anti-human FCRLA, PE-conjugated anti-ER (calreticulin) and Golgi intermediate compartment (giantin) antibodies (a kind gift of Dr Elizabeth Sztul, University of Alabama at Birmingham) were used. Cells were examined using a confocal laser scanning microscope (Leica SP2; Leica, Bannockburn, IL, USA). Cells (293T) were grown on coverslips and transiently transfected with pCI-neo-FCRLA, using Unifectin M-56 reagent. Cells were harvested 48 h after the transfection, washed several times and fixed for 20 min with ice-cold acetone–methanol (1:1) and then air-dried and washed with PBS 3×. Cells were then incubated with FCRLA-specific rabbit antibody and either anti-58K to label Golgi (Abcam, Cambridge, UK) or anti-calnexin (BD TransductionLab) to label the ER, for 1 h at room temperature, washed twice with PBS and 1% FBS and labeled with secondary anti-mouse FITC conjugate (BD Biosciences) for 1 h at RT. Stained and washed cells were mounted onto cover slips. Confocal microscopic analysis was performed using a LSM 510 (Zeiss, Jena, Germany).
RT–PCR
For detection of FCRLA transcripts in sorted T and B cells and the Ramos cell line, cells were deposited into TRIzol reagent (Life Technologies, Grand Island, NY, USA) for RNA isolation. Total cellular RNA was primed with random hexamers and oligo(dT) primers and reverse transcribed with SuperScript II (Life Technologies) into single-stranded cDNA. Two primer pairs located in exons 2 and 3 and 4 and 5 were used to amplify FCRLA transcripts (35 cycles at an annealing temperature of 52°C). HFCRXD12U: 5′-TTGGACTGTGATAGCCACAAC-3′, HFCRXD12L: 5′-GCCAGTTTTGAGACGCTGCA-3′, HFCRXD34U: 5′-CTATTCAGCAGTAGCCTTTGT-3′, HFCRXD34L: 5′-GAACTGTTTCCAGCGCCAATT 3′.
Mutant construction and transfection
Cloning of wild-type FCRLA was described earlier (15). To obtain deletion mutants hFCRLAΔ1d, hFCRLAΔ1dΔ2d and hFCRLAΔ1dΔ2dΔ3d, the coding sequence of the full length hFCRLA was amplified by polymerase chain reaction with upstream oligo1 (5′-cacggcccagccggccGACTGGCTGATCCTCCAAG-3′), oligo2 (5′-cacggcccagccggccGAACTGTTTCCAGCGCCAAT-3′) or oligo3 (5′-cacggcccagccggccGGTGCTTCCAGCTCTGCTG-3′) respectively, together with reverse oligo (5′-cacctcgagCATGGATGAACTGTTTACTTC-3′). PCR products were digested by SfiI–XhoI and cloned in pAP-Tag5 (GenHunter, Nashville, TN, USA).
Transient transfections of wild-type FCRLA and deletion mutants were performed using Unifectin M-56 (Unifect, Moscow, Russia) according to the manufacturer's protocol. After transfection, cells were cultured in the medium 48–72 h prior to Immunofluorescence and western blotting.
Proteinase K digestion, sodium carbonate extraction and solubilization of ER proteins
Microsomes were produced as described elsewhere (26, 27). The cells were washed once with PBS and 10 μg ml−1 leupeptin, 5 μg ml−1 pepstatin, 2 μg ml−1 aprotinin, re-suspended with of 25 mM HEPES–KOH, 10 mM KOAc, 1.5 mM Mg(OAc)2, 250 mM sorbitol, pH7.2, plus protease inhibitors and homogenized by 10 strokes in a tightly fitting glass Dounce homogenizer. The post-nuclear supernatant fraction was obtained by centrifugation at 500×g for 5 min in an Eppendorf centrifuge. The supernatant was centrifuged at 16 000 × g for 5 min to pellet the microsomes. The ER microsomes were then re-suspended in the appropriate buffer for later experiments.
Extraction of ER proteins with sodium carbonate was done according to a published protocol (28). The microsomes were re-suspended in 0.1 M sodium carbonate (pH 11.5) and homogenized by five strokes in a 2 ml Dounce homogenizer. After 30 min incubation at 4°C, the homogenates were centrifuged at 400 000×g at 4°C in the TLA-100.2 rotor of a Beckman tabletop ultracentrifuge. The pellets were re-suspended in 0.5 ml of 100 mM sodium bicarbonate buffer, pH 11.5, and pellet and supernatant fractions were immediately mixed with an equal volume of SDS–PAGE sample buffer.
For protease treatment, proteinase K (100 μg ml−1), with or without Triton X-100, was added to microsomes re-suspended in PBS. The samples were then incubated on ice for 1 h before the addition of sample buffer.
Protein deglycosylation
Immunoprecipitated proteins were washed and re-suspended in 0.1 M sodium acetate buffer, pH 5.5, and treated with endoglycosidase H (Endo H) as described previously (29). O- and N-deglycosylation of proteins was performed according to the Prozyme Glyco Kit denaturing protocol (Prozyme, Hayward, CA, USA). Briefly, 30 μl conditioned media or cell lysate was mixed with 10 μl of 5× incubation buffer and 2.5 μl of denaturing solution. A total of 2.5 μl of detergent solution was added after heating at 100°C and then cooled to room temperature. A total of 1 μg of each glycosidase including O-glycanase was added and incubated at 37°C for 3 h in the presence of protease inhibitors (Pierce Biotechnology, Rockford, IL, USA). Deglycosylated proteins were visualized by western blotting as described above.
Results
Reanalysis of FCRLA expression in normal human B cells
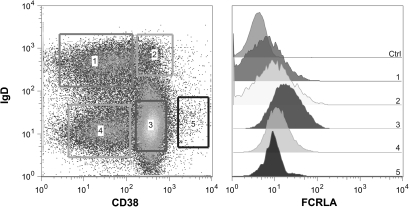
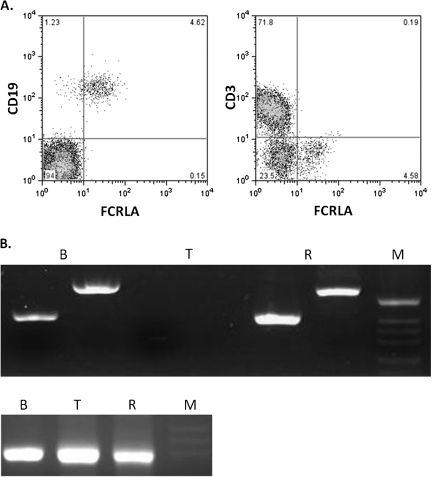
Initial descriptions of human FCRLA established that it is not a cell surface receptor but instead is located intracellularly (15, 16, 18). FCRLA was shown to be highly expressed in germinal center (GC) B cells; however, our subsequent analysis of tissue sections indicated that it is also found in marginal zone B cells and more weakly in mantle zone B cells (30). We have developed FCRLA-specific mAbs (22) and directly conjugated the M101 sub-clone with Alexa 488, allowing us to analyze FCRLA expression more quantitatively by flow cytometry. After demonstrating the efficacy of this reagent on permeabilized FCRLA transfectants and Ramos B cells (not shown), we examined FCRLA expression by CD19+ tonsillar B cells that were also stained for cell surface IgD and CD38 to discriminate the naive, pre-GC, GC, memory and plasma cell sub-populations (31) (Fig. 1). The highest level of FCRLA expression was seen among GC and pre-GC cells but, consistent with our previous RT–PCR analysis (16), the naive and memory B cells also expressed FCRLA albeit at lower levels. Also, consistent with our previous studies, plasma cells only express low levels of FCRLA (30). We next examined FCRLA expression by peripheral blood B cells, which have previously been reported to be FCRLA negative unless activated (18). Surprisingly, we found readily detectable, although relatively dim staining in most of the CD19-positive B cells (Fig. 2A, panel A). The CD3-positive T cells did not express FCRLA, as expected (panel B). In order to confirm this finding by an independent method, peripheral blood B and T cells were sort-purified and then analyzed, in parallel with the Ramos B cell line, by RT–PCR using two different primer pairs to amplify FCRLA transcripts. Primary B cells and Ramos cells gave strong signals, whereas the T cells were negative (Fig. 2B). Thus, FCRLA expression in human B cells is more ubiquitous than originally reported.
Fig. 1.
Analysis of intracellular FCRLA expression by flow cytometry. Single-cell suspensions from tonsils were prepared and stained for cell surface CD19, IgD, and CD38. The cells were then fixed with 1% PFA, washed and permeabilized with 0.1% saponin prior to intracellular staining with Alexa 488-conjugated M101 FCRLA mAb. The staining gates for the CD19+ tonsil B-cell sub-populations are depicted on the left side and FCRLA expression by cells in each subpopulation is shown on the right. 1. naive B cells, 2. Pre-GC cells, 3. GC cells, 4. Memory cells and 5. Plasma cells. The profiles shown are representative of five separate tonsil preparations.
Fig. 2.
Resting peripheral blood B cells express FCRLA. (A) PBMC were stained for cell surface CD19 (left panel) or CD3 (right panel) and then fixed, permeabilized and stained for intracellular FCRLA. Most B cells express FCRLA but T cells are negative. (B) (Top panel) RT–PCR was performed on FACS sorted PBMC B and T cells and the Ramos B cell line. Two primer pairs, one located in exons 2 and 3 (first lane in each group) and the other in exons 4 and 5 (second lane), amplified FCRLA from B cells (B) and Ramos cells (R) but not from the T cells (T). The bottom panel illustrates amplification of β-actin from the same samples. M, size markers.
FCRLA associates with intracellular Ig
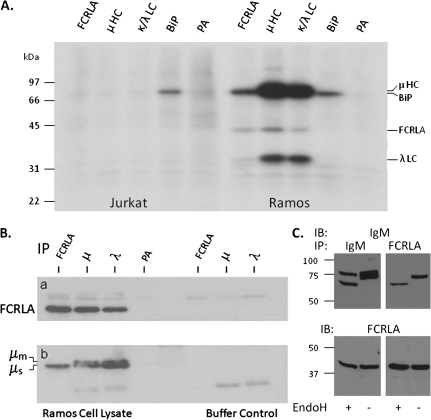
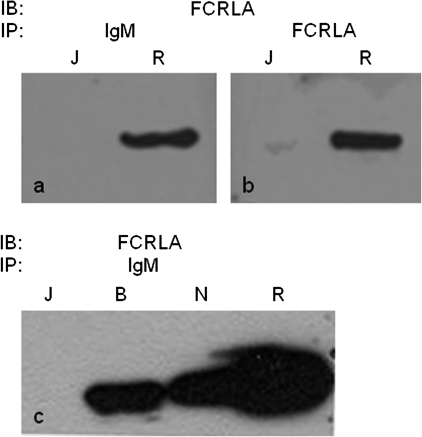
Despite its phylogenetic relationship and molecular similarity to members of the FcR family, thus far, FCRLA has not been found to bind Ig and its ligand(s) have remained undefined. To identify proteins associated with FCRLA in its native intracellular environment, we performed metabolic labeling and immunoprecipitation studies (Fig. 3A). The Jurkat T cell line was used as a negative control (left side) and, as expected since these cells are FCRLA and Ig negative, only the ubiquitously expressed protein HC-binding protein (BiP) was immunoprecipitated. Two proteins were found to co-immunoprecipitate with FCRLA in Ramos B cells, and these migrate identically with μ heavy chain (μ HC) and the ∼32 kDa λ light chain (λ LC) (right side) (32). In the reciprocal experiment, both anti-μ and anti-LC co-precipitated a protein with the same Mr as FCRLA. The finding that LC antibodies co-immunoprecipitate FCRLA suggests that some FCRLA molecules are associated with partially or fully assembled (μλ) IgM or that FCRLA also interacts directly with LC. In order to confirm the identity of the bands that co-immunoprecipitate with FCRLA and IgM in Ramos, we performed immunoprecipitation and western blotting (Fig. 3B). Ramos cell lysates (left side) or lysing buffer (right side) were immunoprecipitated with FCRLA, μ HC or λ LC antibodies and, after electrophoresis and transfer, membranes were probed with polyclonal anti-FCRLA (a) or anti-μ (b). As can be seen on the left side of panel a, in keeping with the metabolic labeling experiments, FCRLA is co-immunoprecipitated by both anti-μ and anti-λ. The finding that FCRLA is co-immunoprecipitated with anti-λ again suggests that either FCRLA binds to light chains or some of the FCRLA is associated with μ chains that are at least partially assembled. Ramos is known to synthesize both the membrane (μm) and the secretory (μs) forms of μHC (33), and the anti-μ blot (panel b) shows, as expected, that anti-μ and anti-λ immunoprecipitate both μm and μs. However, in striking contrast, FCRLA antibodies exclusively immunoprecipitate the μs isoform (lower band). The assignment of these bands as μm and μs is based on differences in their SDS–PAGE mobility and is consistent with previous studies (33–36).
Fig. 3.
FCRLA binds intracellular IgM. (A) Jurkat T cells and Ramos B cells were metabolically labeled overnight, lysed with NP-40, cleared with protein A-sepharose beads (PA) and then immunoprecipitated with antibodies to FCRLA (M116), μHC, κ and λ LC and BiP. The samples were analyzed by SDS–PAGE under reducing conditions. The migration of molecular weight markers is indicated on the left side of the gel. (B and C) FCRLA preferentially binds to μs in Ramos B cells. (B) Ramos NP-40 cell lysates (left) or buffer only controls (right) were precleared with protein A-sepharose beads (PA) and then immunoprecipitated with the monoclonal FCRLA and polyclonal anti-Ig antibodies indicated along the top of the figure. The blots were probed with polyclonal FCRLA (a) or μHC (b) antibodies. The right side of this figure contains the precipitating antibodies but no cell lysate, a necessary negative control as the second antibodies used to develop the blots cross react with the immunoprecipitating antibodies. (C) Ramos cell lysates were immunoprecipitated with polyclonal anti-μ (left panels) and monoclonal FCRLA antibodies (right panels). Immunoprecipitates were treated with EndoH (+) or were mock treated (−). The blots were probed (IB) with anti-μ (upper panel) and then stripped and probed with anti-FCRLA (lower panel).
In order to more clearly resolve the closely migrating μm and μs molecules, proteins were immunoprecipitated with either anti-μ or anti-FCRLA and then treated with (Fig. 3c, lanes 1 and 3) or without (Fig. 3c, lanes 2 and 4) endoglycosidase H (EndoH). After electrophoresis and transfer, the membranes were first blotted with anti-μ (Fig. 3C, top panels). The anti-μ precipitated material migrated as a doublet in mock-treated samples (lane 2), and when a parallel sample was treated with EndoH, the mobility of the upper band was unchanged (lane 1) demonstrating that in Ramos cells under steady state conditions, the μm has exited the ER and been processed in the Golgi. However, EndoH treatment shifted the entire lower band into a single faster migrating species, arguing that this band is only comprised of μs, which is retained in the ER in B cells due to the absence of J chain expression. When anti-FCRLA immunoprecipitates were similarly analyzed (lanes 3 and 4), again, only the lower ER-localized μs band (lane 3) was found to be associated with FCRLA. After stripping, the blots were reprobed for FCRLA and the results confirmed the predicted absence of any N-linked sugars on FCRLA (Fig 3C, bottom panels).
We next wished to confirm that intracellular IgM binding by FCRLA is a general phenomenon and not an idiosyncrasy of Ramos cells. Indeed, when cell lysates from BJAB and NALM-6 were immunoprecipitated with anti-μ, FCRLA was co-immunoprecipitated (Fig. 4, panel c). This was also the case for BL-41 cells, another IgM-producing Burkitt's lymphoma cell line (data not shown).
Fig. 4.
FCRLA associates with IgM in multiple cell lines. Cell lysates were immunoprecipitated (IP) with IgM (a and c) or FCRLA (b) mAbs. The membranes were probed (IB) with anti-FCRLA. R, Ramos; J, Jurkat T cell line; B, BJAB and N, NALM 6.
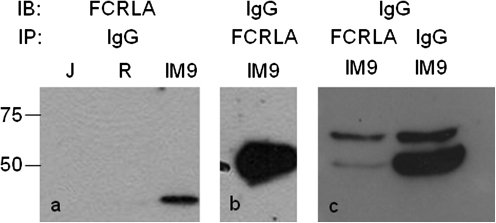
The classical transmembrane FcRs that bind Ig via their extracellular Ig domains have a restricted isotype specificity, thus, it was important to define this parameter for FCRLA. The isotype specificity of the FCRLA–Ig association was first examined using the IgG-producing EBV transformed cell line IM-9 (37). FCRLA was found to co-immunoprecipitate with IgG when either anti-IgG or anti-FCRLA was used as the precipitating antibody (Fig. 5). IM-9 synthesizes both the membrane and the secretory forms of γ heavy chain but primarily the latter. However, unlike the case for IgM in the Ramos cell line, FCRLA showed no unambiguous preferential association with γs.
Fig. 5.
FCRLA associates with IgG in the IM-9 cell line. Cell lysates were immunoprecipitated (IP) with FCRLA mAb or polyclonal anti-IgG antibodies and the membranes were probed (IB) with polyclonal anti-FCRLA or anti-IgG as indicated. J, Jurkat T cell line; R, Ramos and IM9, an EBV immortalized IgGκ producing B lymphoblastoid cell line.
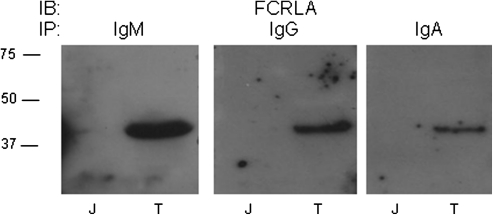
Our analysis of human cell lines demonstrated that FCRLA can associate with intracellular IgM and IgG. We next examined whether this association also occurs in primary B lineage cells and additionally expanded our isotype analysis to include IgA. For these experiments, control Jurkat T cells or single-cell suspensions prepared from tonsil were detergent lysed and immunoprecipitated with antibodies to IgM, IgG or IgA. After electrophoresis and transfer, membranes were probed with the anti-FCRLA antibody and in each case, FCRLA was co-immunoprecipitated (Fig. 6). Thus, FCRLA can associate with the major Ig isotypes, IgM, IgG and IgA, in normal human B lineage cells.
Fig. 6.
FCRLA associates with IgM, IgG and IgA in tonsil B cells. Cell lysates prepared from single-cell suspensions of human tonsil were immunoprecipitated (IP) with anti-IgM, IgG or IgA antibodies. The membranes were probed (IB) with anti-FCRLA. T, tonsil and J, Jurkat T cell line.
FCRLA is a lumenally disposed resident ER protein
Knowing that FCRLA binds intracellular Ig, we next sought clues to its function by determining its subcellular localization using microscopic and biochemical approaches. FCRLA contains a single hydrophobic segment, which is positioned at the N-terminus and is predicted to be a cleavable signal peptide by the SignalP analysis program (http://www.cbs.dtu.dk/services/SignalP/) (38), indicating that FCRLA is targeted to the ER. The molecule may be a resident ER protein or could also populate other compartments along the secretory pathway, including the Golgi and/or exosome/lysosome compartments.
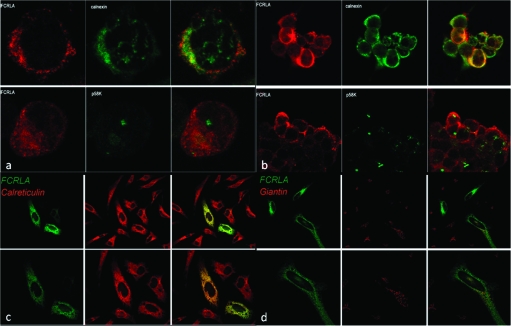
To examine this issue, we first performed double immunofluorescent staining with the anti-FCRLA antibody and reagents specific for the resident ER proteins calreticulin and calnexin, the intermediate compartment protein p58 and the Golgi complex protein giantin. This analysis was performed with both endogenous FCRLA in BJAB cells (Fig 7A) and transfected 293T (Fig 7B) and HeLa (Fig 7C and D) cells. Using laser scanning confocal microscopy, both endogenous and transfected FCRLA were found to co-localize with calreticulin and calnexin in these B and non-B cell lines, but it did not appear to associate with other markers of the secretory pathway. These results are also in agreement with those obtained by immunoelectron microscopy on BJAB cells (data not shown). Coupled with its association with EndoH-sensitive forms of Ig (Fig. 3C), these data indicate that FCRLA is a resident ER protein that is not transported further along the secretory pathway.
Fig. 7.
Intracellular localization of FCRLA as determined by confocal microscopy. The localization of FCRLA in BJAB (a) and transfected 293T cells (b) was determined by staining with anti-FCRLA antibody (red) and anti-calnexin or anti-p58k antibodies (green). The localization of FCRLA in transfected HeLa cells was determined by staining with anti-FCRLA Alexa 488-conjugated M101 FCRLA mAb (green) and anti-calreticulin (c) or anti-giantin (d) antibodies (red). Images of sections of the stained cells captured by a laser scanning confocal microscope are shown. Yellow color shows co-localization of FCRLA with the ER markers.
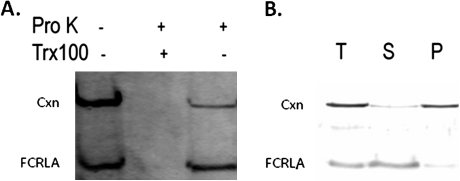
We next sought to determine the orientation of FCRLA with respect to the ER lumen. Microsomes prepared from BJAB cells were treated with proteinase K, either in the presence or in the absence of Triton X-100, and proteins protected from protease digestion were detected by western blot analysis. Lumenally oriented proteins were expected to be protected from proteinase K because this protease cannot cross the microsomal membrane. As a control, Triton X-100 was added to disrupt the membrane and allow access of the protease to intralumenal proteins. As shown in Fig. 8 (A), FCRLA was not digested unless the microsomal membrane was lysed by Triton X-100. Although calnexin is a transmembrane protein that is largely within the ER lumen, the signal for calnexin was dramatically reduced in these experiments because the epitope recognized by the anti-calnexin antibody is cytosolically oriented. These results indicate that FCRLA is localized within the lumen of the ER.
Fig. 8.
Orientation and topology of FCRLA. BJAB cells were homogenized and a microsomal extract was prepared as described in the Methods. (a) FCRLA is localized within the ER. The resulting microsomes were incubated with (+) or without (−) proteinase K (ProK) or 1% Triton X-100 (Trx100) lysis buffer for 1 h at 4°C as indicated and subsequently subjected to anti-FCRLA and anti-calnexin western blot analysis. (b) FCRLA is a lumenal ER protein. Microsomes were subjected to carbonate extraction at pH 11.5 followed by ultracentrifugation. Samples of total homogenate (T), supernatant (S) and pellet (P) were resolved on SDS–polyacrylamide gels, transferred to nitrocellulose membranes and probed with antibodies specific for FCRLA and calnexin.
To ascertain whether FCRLA in the ER is integrated into the ER membrane, microsomes were subjected to a carbonate/pH 11 extraction procedure. This method retains the general integrity of the ER membranes but opens them into sheets, allowing release of soluble proteins but not integral membrane proteins. Therefore, after centrifugation, membrane proteins are found in the pellet and soluble or membrane-associated molecules accumulate in the supernatant (28). Supernatants and pellets from BJAB microsomes were resolved by SDS–PAGE and then blotted with FCRLA and calnexin antibodies (Fig. 8B). Calnexin pelleted with the membranes as expected for an integral membrane protein. By contrast, FCRLA was quantitatively re-distributed to the supernatant fraction, strongly suggesting that the FCRLA signal peptide is cleaved upon translocation into the ER and does not serve as a membrane anchor in this organelle. This interpretation is further supported by our finding that the molecular mass of FCRLA produced from the full-length cDNA in a bacterial expression system, where there is no signal peptide cleavage, is ∼2 KDa larger than the recombinant protein produced in eukaryotic cells (data not shown).
In spite of its retention in the ER, FCRLA contains no known ER retention signals (39, 40). To examine which domain of FCRLA mediates its retention, we generated and characterized a series of FCRLA mutants lacking the first partial Ig domain (Δd1), the first and second domains (Δd1d2) or the first three N-terminal domains (Δd1d2d3) (Fig. 9a). Transfection of 293T cells with these deletion mutants revealed that all three were efficiently secreted (Fig. 9b), arguing that domain 1 possessed the ER localization signal. In contrast to wild-type intracellular FCRLA, the mutants appeared to be glycosylated as they migrated as broad bands in SDS–PAGE. Since FCRLA lacks canonical N-glycosylation sites and is resistant to digestion with EndoH (Fig. 3C, bottom), this increase in molecular mass, in particular its smeared appearance, is most consistent with O-glycosylation of its C-terminal mucin-like domain. Indeed, treatment of transfectant lysates with a cocktail of O-glycosidases resulted in reduction of the molecular mass of the deletion mutants but did not affect migration of wild-type FCRLA (Fig. 9c). This result is entirely consistent with our finding that wild-type FCRLA is retained in the ER and does not reach the Golgi complex, where O-glycosylation occurs (41). It appears that the arrest of FCRLA in the ER is determined by thus far unknown signal(s) residing in the N-terminal domain of the protein.
Discussion
These studies demonstrate that FCRLA is more broadly expressed among human B lineage cells than was originally suspected. We also show that FCRLA is a soluble resident ER protein, retained there by unknown mechanisms that involve the amino terminal partial Ig-like domain of the protein. In the ER, FCRLA can associate with multiple Ig isotypes, IgM, IgG and IgA.
The finding that FCRLA is expressed at multiple stages of B-cell development has several implications as we attempt to understand the function of this unusual protein. The concept that B cells only express FCRLA when activated is clearly no longer tenable; the protein is expressed at varying levels at many stages of B-cell differentiation. This relatively more ubiquitous expression pattern is, in most respects, consistent with our very recently described findings with murine Fcrla (A. V. Taranin, manuscript submitted). We observed that mouse FCRLA is expressed at varying levels in most B cells, although the high-level expression observed in human GC B cells was not seen in mice. There could be several explanations for the failure to detect FCRLA expression in resting human blood B cells in a previous study (18). These investigators used an indirect immunofluorescence assay, which may have diminished sensitivity. Alternatively, but probably less likely, the FCRLA epitope(s) recognized by their mAb may be obscured in resting B cells. Nonetheless, it is clear from their studies and ours here that FCRLA protein levels can be up-regulated following B-cell activation, either in vitro (18) or in vivo in the GC (Fig. 1).
The mechanism(s) by which FCRLA is retained in the ER are unknown. The KDEL amino acid sequence motif found on the C-terminus of some soluble resident ER proteins is perhaps the most well-characterized ER retention signal (42, 43). However, the C-terminal amino acid sequences of human (ATAE) and mouse (VADK) FCRLA do not correspond to this motif; moreover, our deletion mutagenesis studies implicate the N-terminal domain 1, which has several unusual features. From a phylogenetic perspective, D1 may derive from an ancestral FcR-like protein Ig domain. It is somewhat similar to the subtype D1 domain of the receptor for the Fc portion of IgG (FcγR) I, II and III in overlapping regions and the corresponding domain of FCRLB is an authentic Ig domain. However, D1 is only 47 amino acids long compared with the ∼100 amino acids of a typical Ig domain. Moreover, it lacks a properly positioned second cysteine residue that would typically form a disulfide bond to stabilize the Ig domain fold. In most species, FCRLA instead has two closely spaced cysteine residues separated by only 10 amino acids near the D1 amino terminus. Human FCRLA contains an additional cysteine located between these two conserved residues. Secondary structure predictions suggest that D1 is a disordered domain, which is largely unfolded. Thus, ER chaperones, most likely BiP since the D1 is not glycosylated, would bind and retain the protein in the ER. Alternatively, the multiple cysteine residues in D1 could be involved in its ER retention either via thiol-mediated retention mechanisms (44) or by disulfide bond formation with other ER-resident proteins. One thing that clear from our studies is that the ER retention of FCRLA does not require participation of B cell-specific proteins since it also occurs in kidney and epithelia cell lines (Fig. 7). In the confocal microscopy studies of the subcellular localization of the endogenous protein in BJAB cells, there was incomplete co-localization of FCRLA with the ER marker calnexin (Fig. 7A). We suspect that this is an artifact introduced by the cell preparation procedure; the BJAB cells grow in suspension, unlike the 293T and HeLa cells, which are adherent and therefore had to be spun down onto microscope slides. Alternatively, FCRLA may reside in a subregion of the ER where there is incomplete overlap with calnexin. Although it remains a possibility that when FCRLA is complexed with Ig it exits the ER, we consider this unlikely since we saw no microscopic evidence of FCRLA in the Golgi. Moreover, the full-length protein is not modified by O-glycans, a process that occurs in the Golgi, whereas the FCRLA deletion mutants that enter the secretory pathway are O-glycosylated. Another possibility is that FCRLA is located in endosomes/lysosomes. However, we are not aware of any Golgi-independent pathway to these compartments, and moreover, our immunoelectron microscopic analysis failed to detect FCRLA in endosomes/lysosomes.
Early studies failed to demonstrate Ig binding to FCRLA when the molecule was ectopically expressed as a transmembrane domain-containing fusion protein on the surface of transfected 293 cells (18). There could be several explanations for this negative result. The environment of the ER in vivo and that of PBS/FBS in vitro are clearly different, and FCRLA may only be competent for Ig binding in its native ER setting. Alternatively, FCRLA may not bind fully assembled and glycosylated antibodies, in keeping with its ER location and our failure to observe its association with the mature form of μm. In any case, our demonstration that FCRLA can bind multiple isotypes of Ig makes it unique among the large family of Fc receptors. The poly Ig receptor can bind polymeric IgM and IgA because they contain J chain to facilitate their polymerization (45, 46). The Fc α/μ receptor can also bind these two Ig isotypes by unknown structural motif(s) (47–49). However, FcR binding to three dissimilar isotypes is unprecedented, although admittedly, we have not formally demonstrated that this binding is to the Fc region of these Igs. FCRLA is termed ‘FCRL’ because it has significant protein sequence homology (32% sequence identity) with the high affinity IgG FcR, FcγRI (CD64) and because the FCRLA gene is located within the FcR gene cluster on human chromosome 1q21–23 along with the genes encoding the FcγRs, FCER1A, FCRL1-6 and FCRLB. Indeed, the binding of FCRLA to conserved motifs in V regions would more easily explain its ability to bind to multiple isotypes, although not its apparent preferential association with μs.
Another feature that must be taken into account when considering the possible function of FCRLA, as for example an Ig-binding chaperone, is that it is only expressed at low levels in plasma cells, where multiple quality control checkpoints, e.g. BiP (50, 51), are present to prevent secretion of defective Ig molecules (52). Perhaps, FCRLA has some unique function in non-plasma B cells, although it is unclear why a B-cell-restricted ER chaperone would be specifically required since BiP is also expressed in cells at these earlier stages of differentiation.
The preferential association of FCRLA with μs in Ramos cells is intriguing and quite unexpected since the μm and μs proteins are identical except at the very carboxy terminus. However, these results must be interpreted with caution since the immunoprecipitation/westerns can only measure associations in the steady state. For example, if most of the membrane form of IgM in Ramos has already left the ER, as our EndoH experiments would suggest (Fig. 3C), then the apparent lack of association with mIgM would be due to the fact that FCRLA and secretory IgM are in different compartments. This however begs the question of whether there is a role for FCRLA in the selective ER retention of the secretory isoform of IgM. A further complication is that there was no unambiguous preferential association with the secretory isoform of IgG. (For technical reasons, we have not yet been able to examine whether FCRLA shows preferential association with αm or αs.) Certain features of the quality control of Ig assembly may be relevant in a consideration of which Ig isoform(s) associate with FCRLA. For example, the secreted form of μ HC has an additional quality control step that requires J chain addition before the pentameric IgM molecule is released from the ER (53). By contrast, the membrane isoforms of all Ig isotypes are non-covalently associated with the Igα/Igβ heterodimer to form the B-cell receptor and this association, which occurs in the ER, normally occurs before further transit along the secretory pathway (54). If the levels of either J chain or Igα/Igβ are limiting at a particular B-cell differentiation stage, the secretory form of IgM and IgA or the membrane forms of all Ig isotypes, respectively, will have difficulty passing their respective final quality control checkpoints and may then become ligands for FCRLA and ER retention.
Resolution of these issues will require short-term pulse/chase metabolic labeling studies combined with non-reducing gels to examine the potential association of FCRLA with assembly intermediates or, ideally, the development of heterologous cellular systems where membrane and secretory forms of HC, alone or in combination with or without LC can be transduced together with FCRLA and their intermolecular associations examined in vivo. Such a system would also be invaluable for mutagenesis studies to define regions of the molecules involved in these associations.
Note added in proof
While this manuscript was under final review, Wilson et al. published a paper demonstrating that FCRLA interacts with IgM and IgG in human B cell lines.
Wilson, T.J., Gilfillan, S. and Colonna, M. 2010. Fc receptor-like A associates with intracellular IgG and IgM but is dispensable for antigen-specific immune responses. J Immunol. 185:2960.
Funding
RFBR grants (08-04-00522, 10-04-01398); RAS Program ‘Molecular and cellular biology’ (22.16) to A.V.T.; CLL Global Research Foundation (R.S.D.); National Institutes of Health grants (AI069365 to P.D.B., GM054068 to L.M.H., CA131656 to R.S.D.).
Acknowledgments
In Birmingham, we thank our UAB colleagues Drs John Kearney and Andre Vale for helpful suggestions and reagents, Dr Elizabeth Sztul for the anti-calnexin and giantin antibodies, Drs William Britt, Magdalena Krzyzaniak and Glen Bantug for assistance with confocal microscopy and Mr Haitao Li and Ms Dreama White for technical assistance. In Novosibirsk, we thank Dr Nikolai Rubtsov for assistance with confocal microscopy and Dr Tatyana Duzhak for mass spectrometry.
References
- 1.Berken A, Benacerraf B. Properties of antibodies cytophilic for macrophages. J. Exp. Med. 1966;123:119. doi: 10.1084/jem.123.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kubagawa H, Oka S, Kubagawa Y, et al. Identity of the elusive IgM Fc receptor (FcmuR) in humans. J. Exp. Med. 2009;206:2779. doi: 10.1084/jem.20091107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shima H, Takatsu H, Fukuda S, et al. Identification of TOSO/FAIM3 as an Fc receptor for IgM. Int. Immunol. 2010;22:149. doi: 10.1093/intimm/dxp121. [DOI] [PubMed] [Google Scholar]
- 4.Daeron M. Fc receptor biology. Annu. Rev. Immunol. 1997;15:203. doi: 10.1146/annurev.immunol.15.1.203. [DOI] [PubMed] [Google Scholar]
- 5.Nimmerjahn F, Ravetch JV. Analyzing antibody-Fc-receptor interactions. Methods Mol. Biol. 2008;415:151. doi: 10.1007/978-1-59745-570-1_9. [DOI] [PubMed] [Google Scholar]
- 6.Davis RS, Dennis G, Jr., Kubagawa H, Cooper MD. Fc receptor homologs (FcRH1-5) extend the Fc receptor family. Curr. Top. Microbiol. Immunol. 2002;266:85. doi: 10.1007/978-3-662-04700-2_7. [DOI] [PubMed] [Google Scholar]
- 7.Maltais LJ, Lovering RC, Taranin AV, et al. New nomenclature for Fc receptor-like molecules. Nat. Immunol. 2006;7:431. doi: 10.1038/ni0506-431. [DOI] [PubMed] [Google Scholar]
- 8.Miller I, Hatzivassiliou G, Cattoretti G, Mendelsohn C, Dalla-Favera R. IRTAs: a new family of immunoglobulinlike receptors differentially expressed in B cells. Blood. 2002;99:2662. doi: 10.1182/blood.v99.8.2662. [DOI] [PubMed] [Google Scholar]
- 9.Ehrhardt GR, Davis RS, Hsu JT, Leu CM, Ehrhardt A, Cooper MD. The inhibitory potential of Fc receptor homolog 4 on memory B cells. Proc. Natl Acad. Sci. USA. 2003;100:13489. doi: 10.1073/pnas.1935944100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leu CM, Davis RS, Gartland LA, Fine WD, Cooper MD. FcRH1: an activation coreceptor on human B cells. Blood. 2005;105:1121. doi: 10.1182/blood-2004-06-2344. [DOI] [PubMed] [Google Scholar]
- 11.Haga CL, Ehrhardt GR, Boohaker RJ, Davis RS, Cooper MD. Fc receptor-like 5 inhibits B cell activation via SHP-1 tyrosine phosphatase recruitment. Proc. Natl Acad. Sci. USA. 2007;104:9770. doi: 10.1073/pnas.0703354104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schreeder DM, Pan J, Li FJ, Vivier E, Davis RS. FCRL6 distinguishes mature cytotoxic lymphocytes and is upregulated in patients with B-cell chronic lymphocytic leukemia. Eur. J. Immunol. 2008;38:3159. doi: 10.1002/eji.200838516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schreeder DM, Cannon JP, Wu J, Li R, Shakhmatov MA, Davis RS. Cutting edge: FcR-like 6 is an MHC class II receptor. J. Immunol. 2010;185:23. doi: 10.4049/jimmunol.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chikaev NA, Bykova EA, Najakshin AM, et al. Cloning and characterization of the human FCRL2 gene. Genomics. 2005;85:264. doi: 10.1016/j.ygeno.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 15.Mechetina LV, Najakshin AM, Volkova OY, et al. FCRL, a novel member of the leukocyte Fc receptor family possesses unique structural features. Eur. J. Immunol. 2002;32:87. doi: 10.1002/1521-4141(200201)32:1<87::AID-IMMU87>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 16.Davis RS, Li H, Chen CC, Wang Y-H, Cooper MD, Burrows PD. Definition of an Fc receptor related gene (FcRX) expressed in human and mouse B cells. Int. Immunol. 2002;14:1075. doi: 10.1093/intimm/dxf074. [DOI] [PubMed] [Google Scholar]
- 17.Masuda K, Davis RS, Maruyama T, et al. FcRY, an Fc receptor related gene differentially expressed during B lymphocyte development and activation. Gene. 2005;363:32. doi: 10.1016/j.gene.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 18.Facchetti F, Cella M, Festa S, Fremont DH, Colonna M. An unusual Fc receptor-related protein expressed in human centroblasts. Proc. Natl Acad. Sci. USA. 2002;99:3776. doi: 10.1073/pnas.022042699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson TJ, Colonna M. A new Fc receptor homolog, FREB2, found in germinal center B cells. Genes Immun. 2005;6:341. doi: 10.1038/sj.gene.6364185. [DOI] [PubMed] [Google Scholar]
- 20.Inozume T, Matsuzaki Y, Kurihara S, et al. Novel melanoma antigen, FCRL/FREB, identified by cDNA profile comparison using DNA chip are immunogenic in multiple melanoma patients. Int. J. Cancer. 2005;114:283. doi: 10.1002/ijc.20735. [DOI] [PubMed] [Google Scholar]
- 21.Lee YK, Brewer JW, Hellman R, Hendershot LM. BiP and immunoglobulin light chain cooperate to control the folding of heavy chain and ensure the fidelity of immunoglobulin assembly. Mol. Biol. Cell. 1999;10:2209. doi: 10.1091/mbc.10.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volkova OY, Reshetnikova ES, Mechetina LV, et al. Generation and characterization of monoclonal antibodies specific for human FCRLA. Hybridoma (Larchmt) 2007;26:78. doi: 10.1089/hyb.2006.043. [DOI] [PubMed] [Google Scholar]
- 23.Sasso EH, Silverman GJ, Mannik M. Human IgM molecules that bind staphylococcal protein A contain VHIII H chains. J. Immunol. 1989;142:2778. [PubMed] [Google Scholar]
- 24.Sasso EH, Silverman GJ, Mannik M. Human IgA and IgG F(ab')2 that bind to staphylococcal protein A belong to the VHIII subgroup. J. Immunol. 1991;147:1877. [PubMed] [Google Scholar]
- 25.Ibrahim S, Seppala I, Makela O. V-region-mediated binding of human Ig by protein A. J. Immunol. 1993;151:3597. [PubMed] [Google Scholar]
- 26.Yu M, Haslam RH, Haslam DB. HEDJ, an Hsp40 co-chaperone localized to the endoplasmic reticulum of human cells. J. Biol. Chem. 2000;275:24984. doi: 10.1074/jbc.M000739200. [DOI] [PubMed] [Google Scholar]
- 27.Ryan NM. Subcellular fractionation of animal tissues. In: Cutler P, editor. Protein Purification Protocols. Totowa, NJ: Humana Press Inc.; 2003. p. 47. [Google Scholar]
- 28.Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 1982;93:97. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung KT, Shen Y, Hendershot LM. BAP, a mammalian BiP-associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J. Biol. Chem. 2002;277:47557. doi: 10.1074/jbc.M208377200. [DOI] [PubMed] [Google Scholar]
- 30.Masir N, Jones M, Pozzobon M, et al. Expression pattern of FCRL (FREB, FcRX) in normal and neoplastic human B cells. Br. J. Haematol. 2004;127:335. doi: 10.1111/j.1365-2141.2004.05193.x. [DOI] [PubMed] [Google Scholar]
- 31.Pascual V, Liu YJ, Magalski A, de Bouteiller O, Banchereau J, Capra JD. Analysis of somatic mutation in five B cell subsets of human tonsil. J. Exp. Med. 1994;180:329. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerr WG, Cooper MD, Feng L, Burrows PD, Hendershot LM. Mu heavy chains can associate with a pseudo-light chain complex (ψL) in human pre-B cell lines. Int. Immunol. 1989;1:355. doi: 10.1093/intimm/1.4.355. [DOI] [PubMed] [Google Scholar]
- 33.Kerr WG, Hendershot LM, Burrows PD. Regulation of IgM and IgD expression in human B-lineage cells. J. Immunol. 1991;146:3314. [PubMed] [Google Scholar]
- 34.McCune JM, Fu SM, Kunkel HG, Blobel G. Biogenesis of membrane-bound and secreted immunoglobulins: two primary translation products of the human delta chain, differentially N-glycosylated to four discrete forms in vivo and in vitro. Proc. Natl Acad. Sci. USA. 1981;78:5127. doi: 10.1073/pnas.78.8.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hendershot L, Levitt D. Differential regulation of membrane and secretory μ chain synthesis in human B cell lines. Regulation of membrane μ or secreted μ. J. Exp. Med. 1982;156:1622. doi: 10.1084/jem.156.6.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stockdale AM, Dul JL, Wiest DL, Digel M, Argon Y. The expression of membrane and secreted immunoglobulin during the in vitro differentiation of the murine B cell lymphoma CH12. J. Immunol. 1987;139:3527. [PubMed] [Google Scholar]
- 37.Drexler HG, Dirks WG, Matsuo Y, MacLeod RA. False leukemia-lymphoma cell lines: an update on over 500 cell lines. Leukemia. 2003;17:416. doi: 10.1038/sj.leu.2402799. [DOI] [PubMed] [Google Scholar]
- 38.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: signalP 3.0. J. Mol. Biol. 2004;340:783. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 39.Ellgaard L, Molinari M, Helenius A. Setting the standards: quality control in the secretory pathway. Science. 1999;286:1882. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- 40.Wrzeszczynski KO, Rost B. Annotating proteins from endoplasmic reticulum and Golgi apparatus in eukaryotic proteomes. Cell. Mol. Life Sci. 2004;61:1341. doi: 10.1007/s00018-004-4005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rottger S, White J, Wandall HH, et al. Localization of three human polypeptide GalNAc-transferases in HeLa cells suggests initiation of O-linked glycosylation throughout the Golgi apparatus. J. Cell Sci. 1998;111:45. doi: 10.1242/jcs.111.1.45. [DOI] [PubMed] [Google Scholar]
- 42.Pelham HR. The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem. Sci. 1990;15:483. doi: 10.1016/0968-0004(90)90303-s. [DOI] [PubMed] [Google Scholar]
- 43.Munro S, Pelham HR. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 44.Anelli T, Alessio M, Bachi A, et al. Thiol-mediated protein retention in the endoplasmic reticulum: the role of ERp44. EMBO J. 2003;22:5015. doi: 10.1093/emboj/cdg491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hendrickson BA, Conner DA, Ladd DJ, et al. Altered hepatic transport of immunoglobulin A in mice lacking the J chain. J. Exp. Med. 1995;182:1905. doi: 10.1084/jem.182.6.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johansen FE, Braathen R, Brandtzaeg P. The J chain is essential for polymeric Ig receptor-mediated epithelial transport of IgA. J. Immunol. 2001;167:5185. doi: 10.4049/jimmunol.167.9.5185. [DOI] [PubMed] [Google Scholar]
- 47.Shibuya A, Sakamoto N, Shimizu Y, et al. Fcα/μ receptor mediates endocytosis of IgM-coated microbes. Nat. Immunol. 2000;1:441. doi: 10.1038/80886. [DOI] [PubMed] [Google Scholar]
- 48.Kinet JP, Launay P. Fc α/μR: single member or first born in the family? Nat. Immunol. 2000;1:371. doi: 10.1038/80805. [DOI] [PubMed] [Google Scholar]
- 49.Kikuno K, Kang DW, Tahara K, et al. Unusual biochemical features and follicular dendritic cell expression of human Fcalpha/mu receptor. Eur. J. Immunol. 2007;37:3540. doi: 10.1002/eji.200737655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haas IG, Wabl M. Immunoglobulin heavy chain binding protein. Nature. 1983;306:387. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- 51.Bole DG, Hendershot LM, Kearney JF. Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J. Cell Biol. 1986;102:1558. doi: 10.1083/jcb.102.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feige MJ, Hendershot LM, Buchner J. How antibodies fold. Trends Biochem. Sci. 2010;35:189. doi: 10.1016/j.tibs.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brewer JW, Randall TD, Parkhouse RM, Corley RB. Mechanism and subcellular localization of secretory IgM polymer assembly. J. Biol. Chem. 1994;269:17338. [PubMed] [Google Scholar]
- 54.Melnick J, Argon Y. Molecular chaperones and the biosynthesis of antigen receptors. Immunol. Today. 1995;16:243. doi: 10.1016/0167-5699(95)80167-7. [DOI] [PubMed] [Google Scholar]