Abstract
As the principal component of high-density lipoprotein (HDL), apolipoprotein (apo) A-I plays essential roles in lipid transport and metabolism. Because of its intrinsic conformational plasticity and flexibility, the molecular details of the tertiary structure of lipid-free apoA-I have not been fully elucidated. Previously, we demonstrated that the stability of the N-terminal helix bundle structure is modulated by proline substitution at the most hydrophobic region (residues around Y18) in the N-terminal domain. Here we examine the effect of proline substitution at S55 located in another relatively hydrophobic region compared to most of the helix bundle domain to elucidate the influences on the helix bundle structure and lipid interaction. Fluorescence measurements revealed that the S55P mutation had a modest effect on the stability of the bundle structure, indicating that residues around S55 are not pivotally involved in the helix bundle formation, in contrast to the insertion of proline at position 18. Although truncation of the C-terminal domain (Δ190–243) diminishes the lipid binding of apoA-I molecule, the mutation S55P in addition to the C-terminal truncation (S55P/Δ190–243) restored the lipid binding, suggesting that the S55P mutation causes a partial unfolding of the helix bundle to facilitate lipid binding. Furthermore, additional proline substitution at Y18 (Y18P/S55P/Δ190–243), which leads to a drastic unfolding of the helix bundle structure, yielded a greater lipid binding ability. Thus, proline substitutions in the N-terminal domain of apoA-I that destabilized the helix bundle promoted lipid solubilization. These results suggest that not only the hydrophobic C-terminal helical domain but also the stability of the N-terminal helix bundle in apoA-I are important modulators of the spontaneous solubilization of membrane lipids by apoA-I, a process that leads to the generation of nascent HDL particles.
Keywords: amphipathic α-helix, apolipoprotein A-I, helix bundle structure, lipid binding, proline substitution
1. Introduction
Elevated plasma concentrations of high-density lipoprotein (HDL) and apolipoprotein (apo) A-I, the major protein constituent of HDL, are inversely related to the risk of cardiovascular disease in humans [1,2]. The inherent anti-atherogenic functions of HDL and apoA-I are primarily attributed to their known involvement in the reverse cholesterol transport pathway [3]. In this pathway, HDL mediates efflux of cholesterol from peripheral cells by diffusion-mediated processes (for a review, see [4]) and lipid-free or lipid-poor apoA-I molecules remove cholesterol from peripheral cells via the ATP-binding cassette transporter A1 (ABCA1) (for a review, see [5]). The ability of apoA-I to solubilize membrane lipids plays a key role in cholesterol efflux from cells via ABCA1 and in nascent HDL particle assembly [6].
Human apoA-I is composed of 243 amino acids; residues 1–43 are encoded by exon 3 and residues 44–243 are encoded by exon 4 of the apoA-I gene. Analysis of the amino acid sequence reveals that residues 1–43 form a class G* amphipathic α-helix, while residues 44–243 contain a series of 22-mer and 11-mer class A and class Y amphipathic α-helices [7]. Several studies have elucidated that lipid-free apoA-I is folded into two structural domains, comprising an N-terminal part (residues around 1–189) forming a four-helix bundle and a discrete C-terminal part (residues around 190–243) [8–10]. Previously, we demonstrated that the extreme N-terminal region spanning residues 1–43 is important for maintaining the helix bundle structure [8,11]. On the other hand, the most C-terminal region spanning residues 223–243 is crucial for initiating lipid binding, which also involves the subsequent conformational opening of the N-terminal helix bundle [8,12].
A proline residue lacks an amide hydrogen atom essential for the formation of a hydrogen bond so that helix formation tends to be disrupted [13]. We reported previously that insertion of a proline residue into the most hydrophobic residue in the N-terminal domain (Y18P) disrupted the helix bundle structure [11]. Interestingly, although removal of the C-terminal domain (Δ190–243) significantly reduced the binding of apoA-I to lipids, the presence of the Y18P mutation offset the negative effects of this C-terminal truncation. Considering the two-step mechanism for apoA-I binding to lipid [8], it is conceivable that the α-helix around Y18 masks a potential lipid-binding region in the N-terminal domain by inhibiting the unfolding of the helix bundle.
Hydrogen-deuterium exchange (HX) experiments revealed that helical structures in the N-terminal domain are located in residues 7–44, 54–65, 70–78, 81–115, and 147–178 [14]. Hydropathy analysis shows that the region around residue 55 is relatively hydrophobic compared to most of the helix bundle domain [12]. In addition, a peptide corresponding to residues 44–65 binds to lipids well whereas peptides from the remainder of the N-terminal domain do not [15]. These observations led us to postulate that the segment around residues 54–65 is a potential lipid-binding region in the N-terminal domain, substituting for the lipid-binding function when the C-terminal domain is deleted. Y18 is located in the middle of the putative α-helical segment (residues 7–44), whereas S55 is located at the edge of the putative α-helical segment (residues 54–65), according to the HX results. In the present study, in comparison to the Y18P mutation, we examined the effects of proline insertion at position S55 on the stability and lipid-binding properties of apoA-I.
2. Materials and methods
2.1 Materials
Egg phosphatidylcholine (PC) and dimyristoylphosphatidylcholine (DMPC) were purchased from Sigma-Aldrich (St. Louis, MO) and NOF (Tokyo, Japan), respectively. Ultrapure guanidine hydrochloride (GdnHCl) was obtained from MP Biomedicals (Aurora, OH). 8-Anilino-1-naphthalenesulfonic acid (ANS) was purchased from Molecular Probes (Eugene, OR). [3H]-Cholesterol and [14C]-formaldehyde were purchased from PerkinElmer Life Sciences (Wellesley, MA). All other reagents were special grade.
2.2 Protein expression and purification
Mutations were introduced in the N-terminal (S55P), or both of the N- and C-terminal (S55P/Δ190–243, Y18P/S55P/Δ190–243) domains in human apoA-I. Wild-type (WT) human apoA-I and engineered mutants were expressed as thioredoxin fusion proteins in the E. coli strain BL21-DE3 and then cleaved and purified as described previously [8]. The apoA-I preparations were at least 95% pure as assessed by SDS-PAGE. In all experiments, apoA-I was freshly dialyzed from 6 M GdnHCl solution into Tris buffer (10 mM Tris, 150 mM NaCl, 1 mM EDTA, 0.02 % NaN3, pH 7.4) or phosphate buffer (10 mM sodium phosphate, pH 7.4) before use. Protein concentrations were determined either by the Lowry procedure using bovine serum albumin (Bio-Rad, Hercules, CA) as a standard [16,17] or by absorbance measurements at 280 nm.
2.3 Preparation of lipid vesicles
A film of egg PC or DMPC on the wall of a glass tube was dried under vacuum overnight and hydrated with buffer. Egg PC small unilamellar vesicles (SUV) with a diameter of approximately 25 nm were prepared as described [18]. Multilamellar vesicles of DMPC were prepared by vortexing vigorously. The PC concentration was determined using an enzymatic assay kit for choline from Wako Pure Chemicals (Osaka, Japan).
2.4 Circular dichroism (CD) spectroscopy
Far-UV CD spectra were obtained using an Aviv 62ADS spectropolarimeter. ApoA-I samples were dissolved at 50 μg/mL in phosphate buffer. The α-helix contents were calculated from the equation using molar ellipticities [θ] at 222 nm: % α-helix = [(−[θ]222 + 3000) / (36000 + 3000)] × 100. For lipid-binding experiments, proteins were incubated with SUV for 1 h prior to the measurement [18].
2.5 Fluorescence measurements
All fluorescence measurements were carried out at 25 °C using a Hitachi F-7000 or F-4500 spectrophotometer. For denaturation experiments, samples were preincubated with given concentrations of GdnHCl overnight at 4 °C. The emission spectra of tryptophan (Trp) were recorded from 300 to 420 nm at the excitation wavelength of 295 nm. ANS fluorescence spectra were recorded from 400 to 600 nm at the excitation wavelength of 395 nm in the absence or presence of 50 μg/mL protein and an excess amount of ANS (250 μM) [8].
2.6 Interactions of apoA-I with lipid
The binding of apoA-I labeled with [14C]-formaldehyde to eggPC SUV labeled with a trace amount of [3H]-cholesterol was assayed by gel-filtration chromatography as described [18]. The kinetics of solubilization of DMPC vesicles by the addition of apoA-I were measured by monitoring the time-dependent decrease in the turbidity at 24.0 °C [19]. Sample light scattering intensity was monitored at 325 nm on a Beckman Coulter DU-640 spectrophotometer. The 10 min time-courses were fitted to a mono-exponential decay equation. The 10 min decrease in absorbance was measured as a function of apoA-I concentration.
2.7 Isothermal titration calorimetry (ITC) measurements
Enthalpies of apoA-I binding to SUV were measured using a MicroCal MCS isothermal titration calorimeter at 25 °C as described [18]. Measurements were carried out by titrating 8 μL aliquots of apoA-I sample (0.8 mg/mL) into the cell (1.35 mL) containing an excess of SUV (15–20 mM) at constant time intervals of 540 sec. Enthalpies of apoA-I binding to SUV were corrected for heats of apoA-I dilution and dissociation at 25 °C; these values were determined by titrating apoA-I into buffer alone.
3. Results
3.1 Effect of S55P mutation on the secondary structure and stability of apoA-I
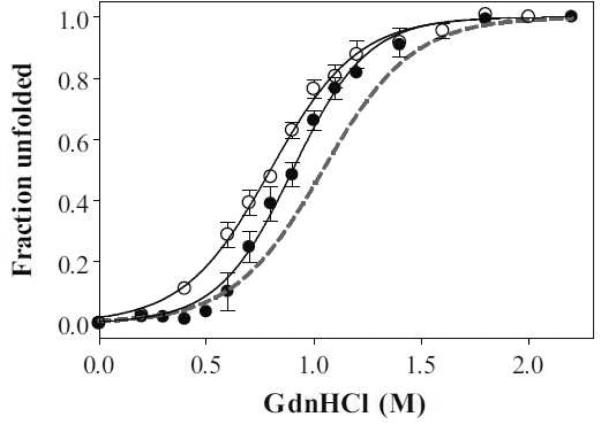
The secondary structures of apoA-I variants were analyzed by far-UV CD spectroscopy. α-Helix contents calculated from the molar ellipticity at 222 nm are summarized in Table 1. In the lipid-free state, the α-helical content of S55P and S55P/Δ190–243 apoA-I was comparable to that of WT and Δ190–243 apoA-I, respectively (Table 1). This suggests that the influence of a single proline substitution on the secondary structure in lipid-free state is not significant, consistent with the previous study [11]. Figure 1 shows the GdnHCl-induced denaturation curves of WT, S55P, and S55P/Δ190–243 apoA-I monitored by the change in Trp fluorescence intensity at 335 nm. The conformational stability, ΔGD°, the midpoint of denaturation, D1/2, and m values are listed in Table 1. All four Trp residues (positions 8, 50, 72, and 108) are located in the N-terminal half, which enabled us to estimate the stability of the N-terminal domain. It has been proposed that there are possible contributions to the stability of electrostatic interactions between the N- and C-terminal domains of apoA-I, as is seen in the apoE molecule [20,21]. However, deletion of the C-terminal domain (Δ190–243) had a negligible effect on the stability of apoA-I [8,22], because electrostatic interactions are shielded by the addition of GdnHCl. In Figure 1, the denaturation curves for S55P and S55P/Δ190–243 apoA-I slightly shifted toward lower GdnHCl concentration compared to WT apoA-I, indicating that the S55P mutation has a relatively minor influence on the stability of the N-terminal helix bundle, in contrast to our previous observation with the Y18P mutation [11].
Table 1.
α-Helix content and parameters of GdnHCl denaturation and ANS binding of apoA-I proline mutants
| α-Helixa |
GdnHCl denaturation |
|||||
|---|---|---|---|---|---|---|
| ApoA-I variants | Lipid-free | Lipid-bound | Δ GD ° | m | D1/2 | ANS fluorescenceb |
| % | % | kcal/mol | M | |||
| WTc | 44 ± 4 | 70 ± 4 | 3.1 ± 0.2 | 3.0 | 1.05 ± 0.13 | 1.0 |
| S55P | 47 ± 2 | 73 ± 5 | 2.9 ± 0.2 | 3.3 | 0.89 ± 0.14 | 1.2 |
| Δ190–243c | 51 ± 2 | 58 ± 2 | 3.4 ± 0.1 | 3.2 | 1.05 ± 0.05 | 0.6 |
| S55P/Δ190–243 | 55 ± 1 | 75 ± 1 | 2.3 ± 0.2 | 3.0 | 0.78 ± 0.09 | 0.9 |
| Y18P/S55P/Δ190–243 | 45 ± 5 | 61 ± 1 | 0.9 ± 0.1 | 2.5 | 0.37 ± 0.06 | 1.6 |
Figure 1.
GdnHCl-induced denaturation curves of WT (dashed line), S55P (•), and S55P/Δ190–243 (◯) apoA-I monitored by Trp fluorescence intensity. Proteins at a concentration of 50 μ/mL were preincubated with GdnHCl. The denaturation curves are representative of at least two separate experiments.
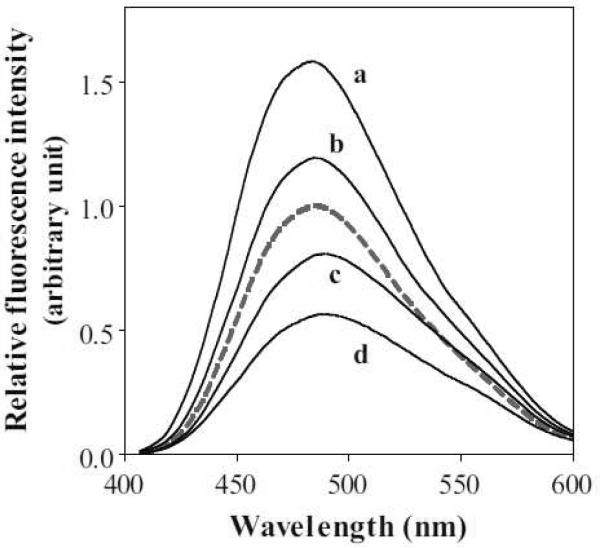
Furthermore, the extent of exposure of hydrophobic sites on a protein was determined by measuring ANS fluorescence spectra (Figure 2 and Table 1). This fluorescent probe, which is much less fluorescent in aqueous environment, binds to exposed hydrophobic surfaces of proteins and exhibits a significant fluorescence enhancement [23]. About 1.2- and 1.5-fold increases in ANS fluorescence were caused by the mutation of S55P relative to full-length and Δ190–243 apoA-I, respectively. These values are much smaller than those observed for Y18P mutations (about 1.7- and 2.5-fold increases, respectively) [11], but nonetheless still significant. These results suggest that the insertion of proline at S55 results in the reorganization of the helix bundle such that hydrophobic surface is exposed.
Figure 2.
Fluorescence spectra of ANS (250 μM) in the presence of 50 μg/mL of apoA-I variants. Fluorescence intensities are normalized to the maximum fluorescence observed with WT apoA-I (dashed line). (a) Y18P/S55P/Δ190–243, (b) S55P, (c) S55P/Δ190–243, and (d)Δ190–243.
3.2 Effect of S55P mutation on the interaction of apoA-I with lipid
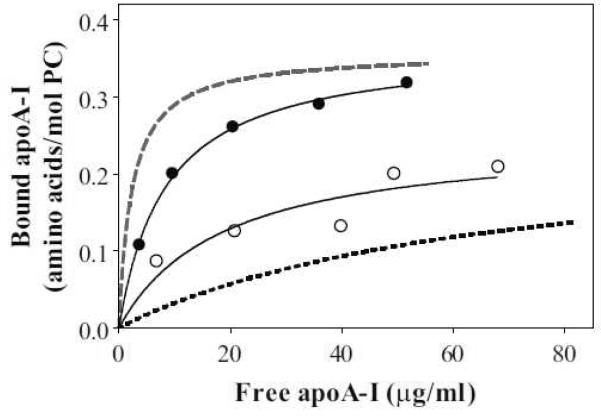
Binding isotherms of apoA-I variants to egg PC SUV were obtained by a gel-filtration method (Figure 3). The dissociation constants, Kd, which reflect the binding affinities, are listed in Table 2. Compared to WT, S55P apoA-I reduced the binding affinity without changing the binding capacity. On the other hand, S55P/Δ190–243 apoA-I bound to SUV with a higher affinity than Δ190–243 apoA-I, although both apoA-I variants entirely lack the C-terminal domain responsible for initiating lipid binding. However, the effect of proline insertion in the S55P mutation on lipid binding was much smaller than that in Y18P/Δ190–243 apoA-I, which showed high-affinity binding comparable to WT apoA-I [11].
Figure 3.
Binding isotherms of WT (dashed line), S55P (•), Δ190–243; (dotted line), and S55P/Δ190–243; (◯) apoA-I to egg PC SUV. Proteins at a concentration of 20–100 μg/ml were preincubated with SUV. The binding curves were obtained by nonlinear regression fitting to a one-site binding model.
Table 2.
Thermodynamic parameters of apoA-I binding to SUV
| Kd | ΔH | ΔG | ΔS | |
|---|---|---|---|---|
| μg/ml | kcal/mol | kcal/mol | cal/mol K | |
| WTa | 2.3 ± 1.1 | −92.6 ± 5.3 | −12.0 ± 0.3 | −270 ± 19 |
| S55P | 8.4 ± 0.5 | −86.7 ± 1.8 | −11.3 ± 0.1 | −253 ± 6 |
| Δ190–243a | 68.7 ± 36.8 | −39.2 ± 2.8 | −9.9 ± 0.5 | −98 ± 11 |
| S55P/Δ190–243 | 18.8 ± 11.4 | −81.7 ± 7.7 | −10.7 ± 0.4 | −238 ± 27 |
| Y18P/S55P/Δ190–243 | N.D. | −76.8 ± 5.2 | N.D. | N.D. |
Data are added from ref [11].
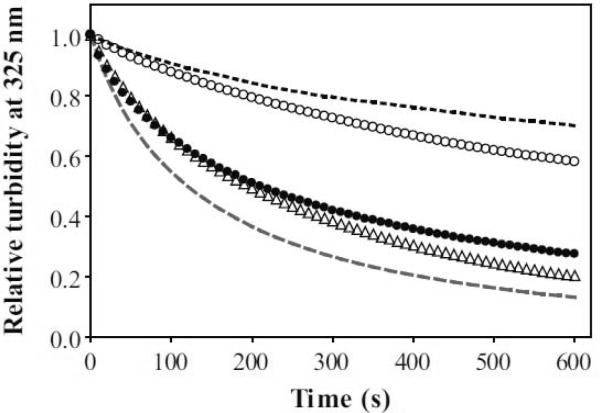
ApoA-I can solubilize unstable DMPC vesicles at the gel to liquid-crystalline phase transition temperature to form discoidal HDL-like particles [24–26], leading to a clarification of the vesicle turbidity. Figure 4 shows a time-dependent decrease in the turbidity monitored by the absorbance at 325 nm. The S55P mutation affected the clearance rate of full-length and the C-terminal deleted apoA-I in opposite ways; S55P in full-length apoA-I retarded the clearance rate, whereas S55P in the C-terminal deletion mutant slightly accelerated it. The effects of apoA-I concentration on the rates of DMPC solubilization are summarized in Figure 5. The Vmax, Km, and Vmax/Km values, which represent the solubilizing efficiency, are listed in Table 3. The Vmax/Km value for S55P apoA-I was significantly smaller than that for WT apoA-I, suggestive of possible involvement of the helical region around S55 in the solubilization process. However, S55P/Δ190–243 apoA-I showed a similar Vmax/Km value to Δ190–243 apoA-I, implying that partial unfolding of the helix bundle caused by the S55P mutation serves to enhance the solubilizing efficiency.
Figure 4.
Solubilization of DMPC vesicles by apoA-I variants at 24.0 °C. Representative time-courses of the decrease in absorbance at 325 nm: WT (dashed line), S55P (•), Δ190–243; (dotted line), S55P/Δ190–243; (◯), and Y18P/S55P/Δ190–243; (△). The relative absorbance values were fitted to a mono-exponential decay equation.
Figure 5.
Increase in fraction of turbidity cleared in 10 min with increasing concentration of protein. WT (dashed line), S55P (•), Δ190–243; (dotted line), S55P/Δ190–243; (◯), and Y18P/S55P/Δ190–243; (△). Curves were obtained by fitting to the Michaelis-Menten equation.
The effect of a single proline mutation on the interaction of apoA-I with SUV was also examined by far-UV CD and ITC measurements (Figure 6, Table 1 and 2). The α-helical content of apoA-I increases upon binding to lipids, generating large exothermic heats which is thought to be the driving force for the high-affinity binding of apoA-I [18,27]. In the full-length apoA-I, S55P mutation had no significant influence on the increase in α-helical content of apoA-I upon binding to SUV (Table 1). In contrast, S55P/Δ190–243 apoA-I exhibited a greater increase in α-helical content compared to Δ190–243 apoA-I (Table 1), accompanied by a greater exothermic heat (Table 2 and Figure 6). These results imply that the partial unfolding of the N-terminal helix bundle caused by the S55P mutation facilitates lipid binding of apoA-I lacking the C-terminal lipid-binding domain.
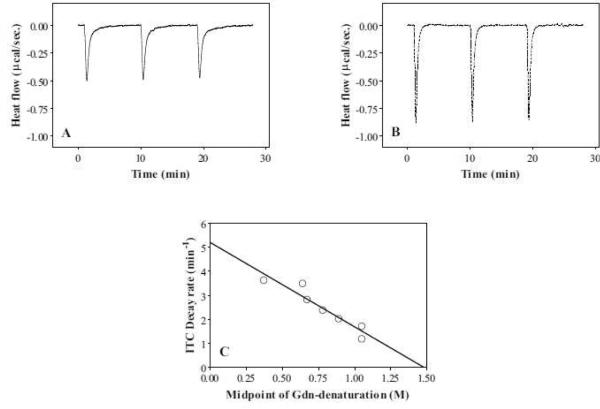
Figure 6.
Isothermal titration calorimetry of (A) S55P/Δ190–243; and (B) Y18P/S55P/Δ190–243; apoA-I injected into SUV. The heats generated upon binding of apoA-I to SUV was calculated by subtracting the dilution heat of the protein (Table 2). (C) Correlation between the midpoints of GdnHCl-induced denaturation of apoA-I variants and the decay rate constants of ITC curves. Corresponding data from Y18P and Y18P/Δ190–243; apoA-I are also plotted [11].
3.3 Effects of double proline mutation on the structure and lipid interaction of apoA-I
To further examine the relationship between the helix bundle stability and lipid binding efficiency of apoA-I, two prolines were inserted into the N-terminal helices to create Y18P/S55P/Δ190–243 apoA-I. Such double proline mutation profoundly destabilized the helix bundle structure as indicated by the GdnHCl denaturation (Table 1), and increased ANS binding (Figure 2), consistent with the previous observation that residues around Y18 in the apoA-I molecule play a critical role in the formation of the helix bundle structure [11].
As shown in Figure 4, Y18P/S55P/Δ190–243 significantly enhanced the clearance rate of DMPC vesicles compared to S55P/Δ190–243 apoA-I. Such faster clearance rates for Y18P/S55P/Δ190–243 versus S55P/Δ190–243 apoA-I were seen over a wide range of concentrations (Figure 5). The Vmax/Km value of Y18P/S55P/Δ190–243 apoA-I was much larger than that of S55P/Δ190–243 apoA-I and comparable to that of full-length apoA-I, demonstrating that the Y18P mutation enhances the ability of the N-terminal helix bundle to solubilize DMPC vesicles.
Far-UV CD and ITC measurements revealed that Y18P/S55P/Δ190–243 apoA-I retained the lipid-binding ability despite the double proline insertion and the lack of the C-terminal domain. We previously demonstrated that the rate of heat reduction in ITC curves reflects the conformational rearrangement of the N-terminal helix bundle structure in apoA-I [8]. As shown in Figure 6A and 6B, incorporation of Y18P affects the rate of heat reduction more than the S55P mutation does, as expected from the effect on the helical bundle stability (Table 1). The strong correlation between the midpoints of GdnHCl-induced denaturation and the decay rate constants of ITC curves for all apoA-I variants (Figure 6C) indicates that the more unstable helix bundles in the apoA-I variants unfold more quickly and interact with the lipids rapidly.
4. Discussion
4.1 Lipid-free structure of apoA-I
ApoA-I is a very flexible protein, which enables it to exert multiple functions and adopt a variety of conformations [28–30]. Recent HX experiments identified that residues 7–44, 54–65, 70–78, 81–115, and 147–178 form α-helices [14]; the helix content of approximately 50% is similar to that observed by CD measurements. The finding that the S55P mutation does not have much influence on apoA-I stability (Table 1) suggests that this residue is not pivotally involved in helix formation, in agreement with the helix locations proposed by HX results. In agreement with this idea, electron paramagnetic resonance spectroscopy of site-directed spin labels also revealed that V53 has a high degree of side chain motional freedom in the apoA-I molecule [31]. The occurrence of proline residue in the middle of helices in the apoA-I molecule is expected to make the helix bundle structure labile. Thus, no proline residues are observed in the middle of helices in the bundle structure of apoE molecule [32], and the helix bundle structure of human apoE is more stable than that of apoA-I [33]. Differences in the stability of the N-terminal domain are likely to be related to the functionalities of apoA-I and apoE; apoE needs to strictly regulate the conformation to modulate the low-density lipoprotein (LDL) receptor binding, whereas apoA-I needs to readily alter conformation to adapt to various sized and heterogeneous HDL particles [12,34].
4.2 Lipid binding of apoA-I
The finding that apoA-I variant containing the double Y18P/S55P mutation in the N-terminal helix bundle has significant lipid-binding ability appears to be inconsistent with the hypothesis that the segment around residues 54–65 substitutes for the lipid-binding function of the C-terminal domain when the latter is deleted [11]. Perhaps, disruption of the α-helical segment around S55 by a proline residue is not sufficient to impair lipid binding because proline substitution at one end of the peptide has little or no effect on lipid binding [35]. In support of this, the apoA-I (1–43/Y18P) peptide completely lacked lipid binding ability whereas the apoA-I (44–65/S55P) peptide still retained the ability to bind to lipid [36].
According to the two-step mechanism of binding of apoA-I to a lipid surface, lipid binding of the C-terminal domain, predominantly in the region spanning residues 220–241, allows the N-terminal domain to unfold and convert helix-helix interactions into helix-lipid interactions [12,37]. Thus, the conformational plasticity of the N-terminal helix bundle, which regulates the exposure of hydrophobic site, is thought to be important for apoA-I to complete lipid binding. In fact, the N-terminal domain of mouse apoA-I, which has less stable helix bundle structure than human apoA-I, can strongly interact with lipids [22]. A strong correlation between the unfolding stability of the N-terminal domain and the rate of heat reduction in binding observed in this study (Figure 6C) clearly demonstrates that the stability of the helix bundle structure is an important determinant for the N-terminal domain of apoA-I to bind to lipids.
The proline insertions into Y18 and S55 affect the helix bundle stability differently, resulting in the variations in the efficiency of DMPC solubilization. Since residue S55 is located at the edge of the helix [14], the effect of proline substitution on the helix bundle stability was relatively small (Table 1). The Vmax/Km value of S55P/Δ190–243 apoA-I for DMPC solubilization was similar to that of Δ190–243 apoA-I (Table 3). In contrast, Y18P/S55P/Δ190–243 apoA-I showed a much larger value. The Y18P mutation, which unfolds drastically and exposes hydrophobic sites, confers a stronger ability to solubilize DMPC vesicles. Thus, unfolding of the N-terminal helix bundle is an important determinant not only for its binding to lipids but also for solubilization of lipid membranes.
In addition to the helix bundle stability, the helix formation upon lipid binding also contributes to the strong lipid-binding affinity of apoA-I [18]. In fact, a good correlation between the binding enthalpy (Table 2) and the increase in the number of helical residues (calculated from Table 1) upon SUV binding of apoA-I variants used in the present study was observed (data not shown). However, although increase in helical structure upon binding to SUV and consequently the binding enthalpy for Y18P/S55P/Δ190–243 apoA-I was comparable to those for S55P/Δ190–243 apoA-I (Table 1 and 2), their abilities of DMPC clearance were significantly different (Figures 4 and 5). This difference is probably attributed to the effect of the unfolding stability as observed in the denaturation and ANS binding experiments (Table 1).
Lipid solubilization is central to ABCA1-mediated efflux of cellular lipids to apoA-I. In this process, which is sensitive to apoA-I structure [38], the step in which apoA-I acquires lipid is rate-limiting [6]. Mutations in the N-terminal and central part of apoA-I are likely to affect helix bundle stability, which might affect lipidation and HDL production rates. A naturally occurring apoA-I mutation associated with low HDL-cholesterol, apoA-I Milano (R173C) in the monomeric state has no effect on the ability of apoA-I to solubilize DMPC vesicles, leading to similar abilities to stimulate cellular lipid efflux [39,40]. In contrast, the apoA-I variant with displacement of proline (P165R) in the middle of the α-helical region spanning residues 147–178 has a significantly reduced capacity to solubilize DMPC [41], which may underlie the reduced HDL production rate. Thus, alterations in the stability of the N-terminal helix bundle structure of apoA-I caused by natural mutations appear to influence the production of nascent HDL particles because of modification of the lipid binding and solubilization properties of the protein.
Research highlights.
-
·
S55P mutation had a modest effect on the stability of the bundle structure of apoA-I
-
·
S55P/Δ190–243; apoA-I restored the lipid binding despite the C-terminal truncation
-
·
Destabilized N-terminal helix bundle promoted lipid solubilization
-
·
Alterations in the stability of apoA-I appear to influence the production of HDL
Acknowledgments
The authors thank Drs. Saburo Aimoto and Toru Kawakami (Institute for Protein Research, Osaka University, Japan) for their help with ITC measurements. This work was supported in part by NIH grant (HL22633) and Grant-in-Aid for Scientific Research (No. 22590046).
Abbreviations
- ABCA1
ATP-binding cassette transporter A1
- ANS
8-anilino-1-naphthalenesulfonic acid
- apo
apolipoprotein
- CD
circular dichroism
- DMPC
dimyristoylphosphatidylcholine
- GdnHCl
guanidine hydrochloride
- HDL
high-density lipoprotein
- HX
hydrogen-deuterium exchange
- ITC
isothermal titration calorimetry
- PC
phosphatidylcholine
- SUV
small unilamellar vesicle
- Trp
tryptophan
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lewis GF, Rader DJ. New insights into the regulation of HDL metabolism and reverse cholesterol transport. Circ. Res. 2005;96:1221–1232. doi: 10.1161/01.RES.0000170946.56981.5c. [DOI] [PubMed] [Google Scholar]
- [2].Navab M, Anantharamaiah GM, Reddy ST, Van Lenten BJ, Ansell BJ, Fogelman AM. Mechanisms of disease: proatherogenic HDL--an evolving field. Nat. Clin. Pract. Endocrinol. Metab. 2006;2:504–511. doi: 10.1038/ncpendmet0245. [DOI] [PubMed] [Google Scholar]
- [3].Lewis GF. Determinants of plasma HDL concentrations and reverse cholesterol transport. Curr. Opin. Cardiol. 2006;21:345–352. doi: 10.1097/01.hco.0000231405.76930.a0. [DOI] [PubMed] [Google Scholar]
- [4].Johnson WJ, Mahlberg FH, Rothblat GH, Phillips MC. Cholesterol transport between cells and high-density lipoproteins. Biochim. Biophys. Acta. 1991;1085:273–298. doi: 10.1016/0005-2760(91)90132-2. [DOI] [PubMed] [Google Scholar]
- [5].Yokoyama S. Assembly of high-density lipoprotein. Arterioscler. Thromb. Vasc. Biol. 2006;26:20–27. doi: 10.1161/01.ATV.0000195789.39418.e8. [DOI] [PubMed] [Google Scholar]
- [6].Vedhachalam C, Duong PT, Nickel M, Nguyen D, Dhanasekaran P, Saito H, Rothblat GH, Lund-Katz S, Phillips MC. Mechanism of ATP-binding cassette transporter A1-mediated cellular lipid efflux to apolipoprotein A-I and formation of high density lipoprotein particles. J. Biol. Chem. 2007;282:25123–25130. doi: 10.1074/jbc.M704590200. [DOI] [PubMed] [Google Scholar]
- [7].Segrest JP, Jones MK, De Loof H, Brouillette CG, Venkatachalapathi YV, Anantharamaiah GM. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 1992;33:141–166. [PubMed] [Google Scholar]
- [8].Saito H, Dhanasekaran P, Nguyen D, Holvoet P, Lund-Katz S, Phillips MC. Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J. Biol. Chem. 2003;278:23227–23232. doi: 10.1074/jbc.M303365200. [DOI] [PubMed] [Google Scholar]
- [9].Brouillette CG, Dong WJ, Yang ZW, Ray MJ, Protasevich, Cheung HC, Engler JA. Forster resonance energy transfer measurements are consistent with a helical bundle model for lipid-free apolipoprotein A-I. Biochemistry. 2005;44:16413–16425. doi: 10.1021/bi051018v. [DOI] [PubMed] [Google Scholar]
- [10].Silva RA, Hilliard GM, Fang J, Macha S, Davidson WS. A three-dimensional molecular model of lipid-free apolipoprotein A-I determined by cross-linking/mass spectrometry and sequence threading. Biochemistry. 2005;44:2759–2769. doi: 10.1021/bi047717+. [DOI] [PubMed] [Google Scholar]
- [11].Tanaka M, Dhanasekaran P, Nguyen D, Ohta S, Lund-Katz S, Phillips MC, Saito H. Contributions of the N- and C-terminal helical segments to the lipid-free structure and lipid interaction of apolipoprotein A-I. Biochemistry. 2006;45:10351–10358. doi: 10.1021/bi060726t. [DOI] [PubMed] [Google Scholar]
- [12].Saito H, Lund-Katz S, Phillips MC. Contributions of domain structure and lipid interaction to the functionality of exchangeable human apolipoproteins. Prog. Lipid Res. 2004;43:350–380. doi: 10.1016/j.plipres.2004.05.002. [DOI] [PubMed] [Google Scholar]
- [13].Schulman BA, Kim PS. Proline scanning mutagenesis of a molten globule reveals non-cooperative formation of a protein's overall topology. Nat. Struct. Biol. 1996;3:682–687. doi: 10.1038/nsb0896-682. [DOI] [PubMed] [Google Scholar]
- [14].Chetty PS, Mayne L, Lund-Katz S, Stranz D, Englander SW, Phillips MC. Helical structure and stability in human apolipoprotein A-I by hydrogen exchange and mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19005–19010. doi: 10.1073/pnas.0909708106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Palgunachari MN, Mishra VK, Lund-Katz S, Phillips MC, Adeyeye SO, Alluri S, Anantharamaiah GM, Segrest JP. Only the two end helixes of eight tandem amphipathic helical domains of human apo A-I have significant lipid affinity. Implications for HDL assembly. Arterioscler. Thromb. Vasc. Biol. 1996;16:328–338. doi: 10.1161/01.atv.16.2.328. [DOI] [PubMed] [Google Scholar]
- [16].Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- [17].Markwell MA, Haas SM, Bieber LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- [18].Saito H, Dhanasekaran P, Nguyen D, Deridder E, Holvoet P, Lund-Katz S, Phillips MC. Alpha-helix formation is required for high affinity binding of human apolipoprotein A-I to lipids. J. Biol. Chem. 2004;279:20974–20981. doi: 10.1074/jbc.M402043200. [DOI] [PubMed] [Google Scholar]
- [19].Segall ML, Dhanasekaran P, Baldwin F, Anantharamaiah GM, Weisgraber KH, Phillips MC, Lund-Katz S. Influence of apoE domain structure and polymorphism on the kinetics of phospholipid vesicle solubilization. J. Lipid Res. 2002;43:1688–1700. doi: 10.1194/jlr.m200157-jlr200. [DOI] [PubMed] [Google Scholar]
- [20].Dong LM, Weisgraber KH. Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J. Biol. Chem. 1996;271:19053–19057. doi: 10.1074/jbc.271.32.19053. [DOI] [PubMed] [Google Scholar]
- [21].Koyama M, Tanaka M, Dhanasekaran P, Lund-Katz S, Phillips MC, Saito H. Interaction between the N- and C-terminal domains modulates the stability and lipid binding of apolipoprotein A-I. Biochemistry. 2009;48:2529–2537. doi: 10.1021/bi802317v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tanaka M, Koyama M, Dhanasekaran P, Nguyen D, Nickel M, Lund-Katz S, Saito H, Phillips MC. Influence of tertiary structure domain properties on the functionality of apolipoprotein A-I. Biochemistry. 2008;47:2172–2180. doi: 10.1021/bi702332b. [DOI] [PubMed] [Google Scholar]
- [23].Stryer L. The interaction of a naphthalene dye with apomyoglobin and apohemoglobin. A fluorescent probe of non-polar binding sites. J. Mol. Biol. 1965;13:482–495. doi: 10.1016/s0022-2836(65)80111-5. [DOI] [PubMed] [Google Scholar]
- [24].Jonas A. Reconstitution of high-density lipoproteins. Methods Enzymol. 1986;128:553–582. doi: 10.1016/0076-6879(86)28092-1. [DOI] [PubMed] [Google Scholar]
- [25].Zhu K, Brubaker G, Smith JD. Large disk intermediate precedes formation of apolipoprotein A-I-dimyristoylphosphatidylcholine small disks. Biochemistry. 2007;46:6299–6307. doi: 10.1021/bi700079w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Massey JB, Pownall HJ. Role of oxysterol structure on the microdomain-induced microsolubilization of phospholipid membranes by apolipoprotein A-I. Biochemistry. 2005;44:14376–14384. doi: 10.1021/bi051169y. [DOI] [PubMed] [Google Scholar]
- [27].Arnulphi C, Jin L, Tricerri MA, Jonas A. Enthalpy-driven apolipoprotein A-I and lipid bilayer interaction indicating protein penetration upon lipid binding. Biochemistry. 2004;43:12258–12264. doi: 10.1021/bi036118k. [DOI] [PubMed] [Google Scholar]
- [28].Marcel YL, Kiss RS. Structure-function relationships of apolipoprotein A-I: a flexible protein with dynamic lipid associations. Curr. Opin. Lipidol. 2003;14:151–157. doi: 10.1097/00041433-200304000-00006. [DOI] [PubMed] [Google Scholar]
- [29].Brouillette CG, Anantharamaiah GM, Engler JA, Borhani DW. Structural models of human apolipoprotein A-I: a critical analysis and review. Biochim. Biophys. Acta. 2001;1531:4–46. doi: 10.1016/s1388-1981(01)00081-6. [DOI] [PubMed] [Google Scholar]
- [30].Lund-Katz S, Liu L, Thuahnai ST, Phillips MC. High density lipoprotein structure. Front. Biosci. 2003;8:d1044–1054. doi: 10.2741/1077. [DOI] [PubMed] [Google Scholar]
- [31].Lagerstedt JO, Budamagunta MS, Oda MN, Voss JC. Electron paramagnetic resonance spectroscopy of site-directed spin labels reveals the structural heterogeneity in the N-terminal domain of apoA-I in solution. J. Biol. Chem. 2007;282:9143–9149. doi: 10.1074/jbc.M608717200. [DOI] [PubMed] [Google Scholar]
- [32].Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- [33].Sakamoto T, Tanaka M, Vedhachalam C, Nickel M, Nguyen D, Dhanasekaran P, Phillips MC, Lund-Katz S, Saito H. Contributions of the carboxyl-terminal helical segment to the self-association and lipoprotein preferences of human apolipoprotein E3 and E4 isoforms. Biochemistry. 2008;47:2968–2977. doi: 10.1021/bi701923h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Narayanaswami V, Ryan RO. Molecular basis of exchangeable apolipoprotein function. Biochim. Biophys. Acta. 2000;1483:15–36. doi: 10.1016/s1388-1981(99)00176-6. [DOI] [PubMed] [Google Scholar]
- [35].Ponsin G, Hester L, Gotto AM, Jr., Pownall HJ, Sparrow JT. Lipid-peptide association and activation of lecithin:cholesterol acyltransferase. Effect of alpha-helicity. J. Biol. Chem. 1986;261:9202–9205. [PubMed] [Google Scholar]
- [36].Tanaka M, Tanaka T, Ohta S, Kawakami T, Konno H, Akaji K, Aimoto S, Saito H. Evaluation of lipid-binding properties of the N-terminal helical segments in human apolipoprotein A-I using fragment peptides. J. Pept. Sci. 2009;15:36–42. doi: 10.1002/psc.1092. [DOI] [PubMed] [Google Scholar]
- [37].Narayanaswami V, Kiss RS, Weers PM. The helix bundle: a reversible lipid binding motif. Comp. Biochem. Physiol. A. 2010;155:123–133. doi: 10.1016/j.cbpa.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Vedhachalam C, Liu L, Nickel M, Dhanasekaran P, Anantharamaiah GM, Lund-Katz S, Rothblat GH, Phillips MC. Influence of apoA-I structure on the ABCA1-mediated efflux of cellular lipids. J. Biol. Chem. 2004;279:49931–49939. doi: 10.1074/jbc.M406924200. [DOI] [PubMed] [Google Scholar]
- [39].Weibel GL, Alexander ET, Joshi MR, Rader DJ, Lund-Katz S, Phillips MC, Rothblat GH. Wild-type ApoA-I and the Milano variant have similar abilities to stimulate cellular lipid mobilization and efflux. Arterioscler. Thromb. Vasc. Biol. 2007;27:2022–2029. doi: 10.1161/ATVBAHA.107.148403. [DOI] [PubMed] [Google Scholar]
- [40].Alexander ET, Tanaka M, Kono M, Saito H, Rader DJ, Phillips MC. Structural and functional consequences of the Milano mutation (R173C) in human apolipoprotein A-I. J. Lipid Res. 2009;50:1409–1419. doi: 10.1194/jlr.M800578-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Daum U, Leren TP, Langer C, Chirazi A, Cullen P, Pritchard PH, Assmann G, von Eckardstein A. Multiple dysfunctions of two apolipoprotein A-I variants, apoA-I (R160L) Oslo and apoA-I (P165R), that are associated with hypoalphalipoproteinemia in heterozygous carriers. J. Lipid Res. 1999;40:486–494. [PubMed] [Google Scholar]