Abstract
The C-X-C chemokine receptor type 4 (CXCR4)/stromal cell derived factor-1 (SDF-1 or CXCL12) interaction and the resulting cell signaling cascade play a key role in metastasis and inflammation. Based on the previously published CXCR4 antagonist 5 (WZ811), a series of novel non-peptidic anti-CXCR4 small molecules have been designed and synthesized to improve potency. Following a structure-activity profile around 5, more advanced compounds in the N, N'-(1, 4-phenylenebis(methylene)) dipyrimidin-2-amines series were discovered and shown to possess higher CXCR4 binding potential and specificity than 5. Compound 26 (508MCl) is the leading compound, and exhibits subnanomolar potency in three in vitro assays including competitive binding, Matrigel invasion, and Gαi cyclic adenosine monophosphate (cAMP) modulation signaling. Furthermore, compound 26 displays promising effects by interfering with CXCR4 function in three mouse models: paw inflammation, Matrigel plug angiogenesis, and uveal melanoma micrometastasis. These data demonstrate that dipyrimidine amines are unique CXCR4 antagonists with high potency and specificity.
Keywords: CXCR4 antagonist, Metastasis, Angiogenesis, Inflammation, Lung fibrosis
Introduction
Chemokines are a superfamily of small cytokines that induce cytoskeleton rearrangements and directional migration of several cell types through their interaction with G-protein-coupled receptors.1 These secreted proteins act in a coordinated fashion with cell-surface proteins, including integrins, to direct the specific homing of various subsets of hematopoietic cells to specific anatomical sites.2,3 One member of the chemokine family, stromal cell derived factor-1 (SDF-1; also named CXCL12), is a chemokine that interacts specifically with C-X-C chemokine receptor type 4 (CXCR4) which was previously identified as a major co-receptor for the entry of T cell line-tropic HIV.4–7 The CXCL12/CXCR4 interaction was later shown to direct cellular recruitment and trafficking, and it offers a new target for indications where such processes play a key role. This interaction also plays a central role in the locomotion and homing of metastatic cells. Therefore, interruption of this interaction may provide a means of intervening in the metastatic process.
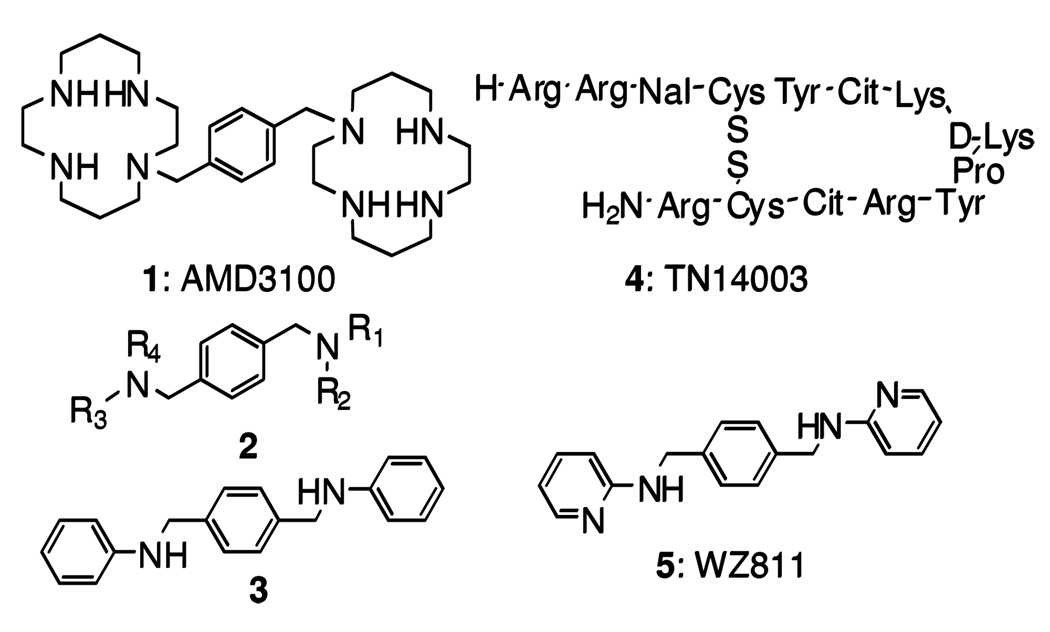
Currently, cyclams and bicyclams represent the most thoroughly studied class of non-peptide anti-CXCR4 molecules. However, this class of inhibitors is not ideal for long-term treatment due to cyclam’s metal ion chelating property, which may result in cardiotoxicities. 8 Two years ago, we reported the identification of a novel class of small molecule CXCR4 antagonists.9 In view of aspects of the molecular mechanism of the bicyclam CXCR4 antagonist 1 (AMD31008), we designed a template with the general structure 2 and identified the first lead compound 3 by means of an affinity binding assay against the potent, peptidic CXCR4 antagonist 4 (TN1400310) (Figure 1). An extensive structure-activity profile around 3 indicated the central aromatic ring to be critical for high CXCR4 affinity, and a one-carbon separation between the central aromatic phenyl ring and the nitrogen of the acyclic linker is essential for high potency. These SAR features finally led to identification of compound 5 (WZ8119) as a potential CXCR4 antagonist that shows excellent potency in an affinity binding assay and in vitro function assays. Unfortunately, further preclinical studies suggested that compound 5 failed to exhibit any in vivo efficacy, due to poor bioavailability (data not shown). We explored further reiteration of the compounds.
Figure 1.
Structures of nonpeptidic CXCR4 antagonist 1, potential CXCR4 antagonist template 2, potential CXCR4 antagonist lead compound 3, peptidic CXCR4 antagonist 4, and potential CXCR4 antagonist 5.
Based on the working hypothesis that the poor pharmacokinetic profile of 5 might be the result of rapid oxidative metabolism, various electron deficient moieties have been introduced to the terminal aromatic ring of 5. The synthetic pathways employed to prepare the final compounds are depicted in Schemes 1–3. For the primary screening, a competitive binding assay utilizing the potent, peptidic CXCR4 antagonist 4 was employed. Previously, we described the rationale for using this assay as our primary assay. 9, 11 In addition, two functional assays measuring cAMP modulation and Matrigel invasion were performed to determine the rank order of anti-CXCR4 efficacy of the newly designed and synthesized compounds.12–14 Furthermore, the in vivo effects of the selected compounds were tested in two mouse models; paw edema for inflammation and matrigel plug for angiogenesis. Finally, the leading compound 26(508MCl) was tested in mouse lung fibrosis and uveal melanoma micrometastasis models.
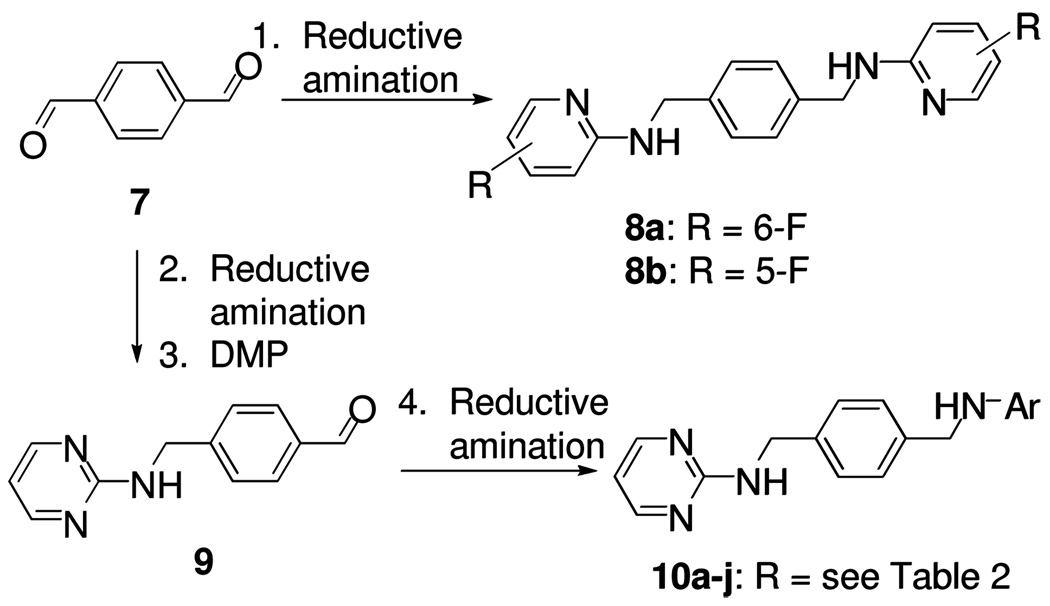
Scheme 1a.
a reagents and conditions: 1. 2-amino-fluoropyridines, NaBH(OAc)3, HOAc, ClCH2CH2Cl, 61–64%; 2. 2-amino-pyrimidine, NaBH(OAc)3, HOAc, ClCH2CH2Cl, 82%; 3. DMP, CH2Cl2, 94%; 4. ArNH2, NaBH(OAc)3, HOAc, ClCH2CH2Cl, 65–69%.
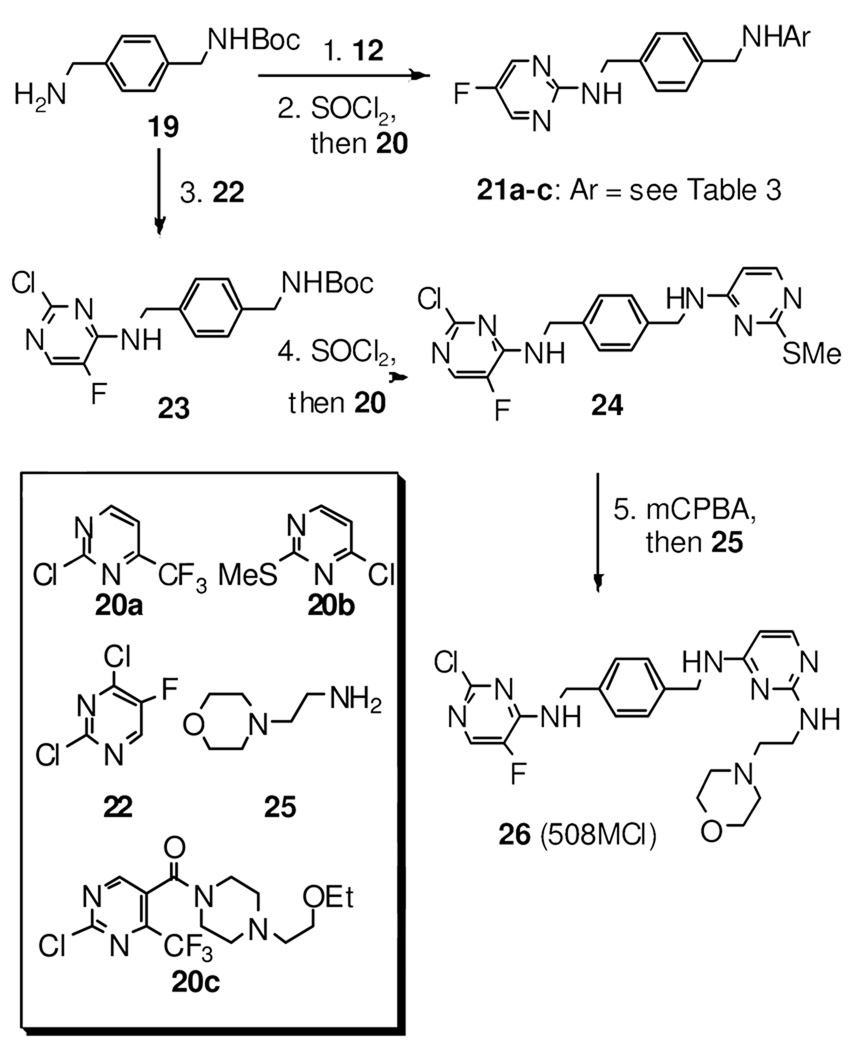
Scheme 3a.
a reagents and conditions: 1. 12, DIPEA, DMF, 96%; 2. SOCl2, MeOH, then 20a–c, DIPEA, DMF,63–72% (2 steps); 3. 22, DIPEA, DMF, 94%; 4. SOCl2, MeOH, then 20b, DIPEA, DMF, 65% (2 steps); 5. mCPBA, CH2Cl2, then 25, dioxane, 25% (2 steps).
Results and Discussion
Chemistry
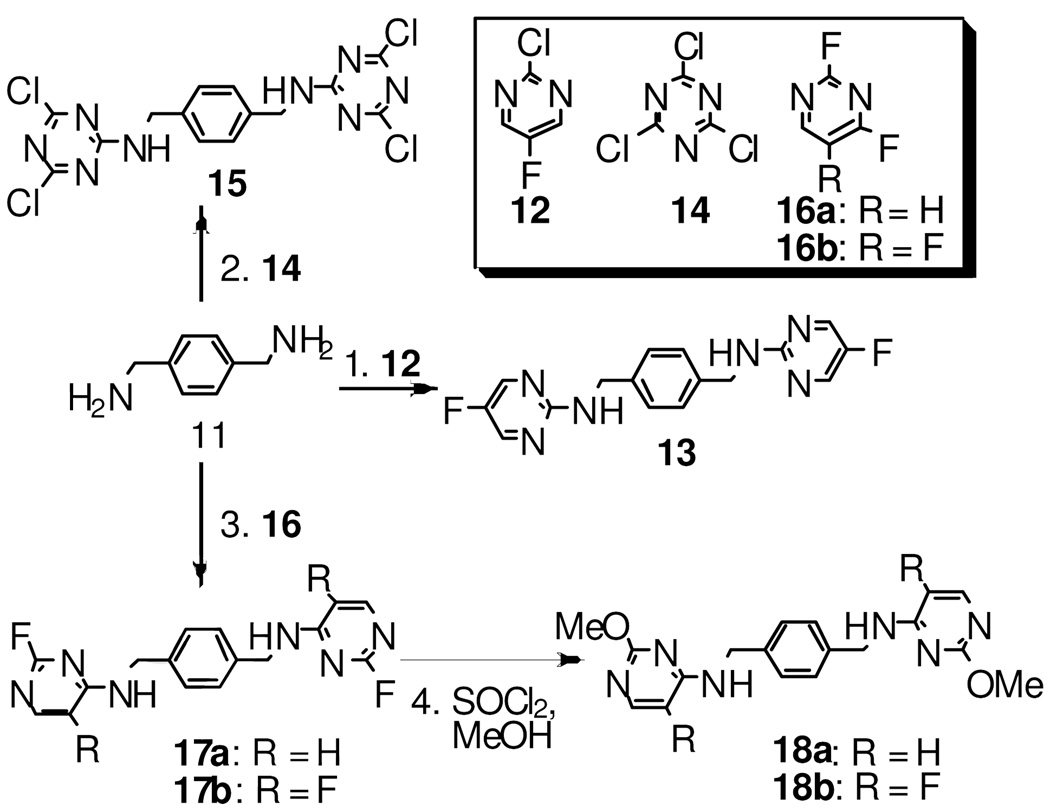
The synthesis of the target compounds are illustrated in Schemes 1–3. Compound 8a and 8b were prepared from the one-pot reductive amination of aldehyde 7 and fluoropyridin-2-amines in the presence of the reducing reagent NaBH(OAc)3.15 To prepare compounds 10a–j, aldehyde 9 was first generated by a two-step sequence from aldehyde 7, followed by reductive amination to give the desired compounds 10a–j (Scheme 1). As shown in Scheme 2, compound 13 was synthesized by treating amine 11 with 2-chloro-5-fluoropyrimidine 12 in a good yield. Following a similar strategy, compounds 15, 17a, and 17b were prepared efficiently. Subsequently, treating compound 17a/b with SOCl2 in methanol delivered the methoxyl-substituted analog 18a/b (Scheme 2). The synthesis of compounds 21a–c starts from mono-Boc-protected amine 19. Amine 19 was first treated with 2-chloro-5-fluoropyrimidine 12, and the Boc was subsequently removed in the presence of SOCl2, followed by an addition-elimination sequence to furnish the desired target compounds 21a–c (Scheme 3). The similar synthetic strategy also efficiently delivers compounds 23 and 24. Subsequent mCPBA mediated oxidation converted 24 into a mixture of 2-(methylsulfinyl)pyrimidine and 2-(methylsulfonyl)pyrimidine derivatives which were treated with 2-morpholinoethanamine 25 without further purification to afford compound 26 in 25% yield over two steps (Scheme 3).
Scheme 2a.
a reagents and conditions: 1. 12, Cs2CO3, DMF, 75%; 2. 14, NaHCO3, THF, 94%; 3.fluoropyrimidines (16a–b), DIPEA, DMF, 65–70%; 4. SOCl2, MeOH, quant.
Primary screening
Based on the behavior of 5, we know that the central 1,4-bis-(aminomethyl)benzene group is critical for CXCR4 binding affinity. Consequently, the distal pyridinyl ring was modified in several ways. For primary compound screening, the previously reported assay was utilized.9 MDA-MB-231 cells were preincubated with compounds at concentrations of 1, 10, 100, and 1000 nM, following incubation with biotinylated 4 and streptavidin-conjugated rhodamine to determine the binding efficiency of the newly synthesized chemical entities to the CXCL12 binding domain of CXCR4. The effective concentration (EC) is defined as the concentration at which the compound blocks more than 50% of 4 binding on CXCR4. Thus, the EC values of compounds meeting this criterion were determined. The Matrigel invasion assay, as the secondary functional assay, was performed for those compounds with an EC value lower than 100 nM to test whether they could block the CXCR4/CXCL12- mediated chemotaxis and invasion as utilized previously. 9 The results of competitive binding and Matrigel invasion are summarized in Table 1. It should be noted that more electron deficient functional groups were introduced to compound 5 to maintain the symmetric chemical structure (Table 1). Pyrimidinyl compound 13 was identified as a potent CXCR4 antagonist with high CXCR4 binding affinity and efficient blocking of Matrigel invasion (> 75%) at 10 nM. With the discovery of the pyrimidinyl group as a potent pharmacophore for CXCR4 antagonists, a series of unsymmetrical compounds were designed and prepared with a pyrimidinyl ring at one terminus of the scaffold and a pyridinyl ring on the other with different functional groups. All these substances exhibited excellent antagonist activity (>60%, Table 2) at 100 nM and >40% at 10 nM against CXCR4/CXCL12-mediated Matrigel invasion. Furthermore, we designed and synthesized dual pyrimidinyl compounds with different functional groups such as methoxy and morpholinyl to adjust their hydrophilicity. We assumed that increased hydrophilicity could increase the compounds’ binding affinity to CXCR4 (Table 3). Most of the corresponding compounds show exceptional binding affinity to CXCR4 with EC values at 1 nM except 18a with an EC value at 10 nM. While these compounds also scored well in the Matrigel invasion assay with >65% inhibition at 100 nM, 21a, 21b, 21c, 17a, 18b, and 26 are especially effective at blocking invasion between 81% and 100%. At a concentration as low as 10 nM, 21a, 21b, 17a, 18b, and 26 inhibit invasion >60%; 26 blocking 84% invasion at 10 nM. Interestingly, all dipyrimidines demonstrated high potency without being significantly influenced by variable substitution.
Table 1.
Effects of symmetrical compounds as blockers of CXCR4 binding and matrigel invasion.a
 | |||||
|---|---|---|---|---|---|
| Entry | Compd | Ar | TN-binding Blocking EC (nM) |
Invasion Inhibition | |
| 100nM | 10nM | ||||
| 1 | 3 | 10 | 64% | 54% | |
| 2 | 6 |  |
100 | 58% | 32% |
| 3 | 5 | 10 | 90% | 72% | |
| 4 | 8a |  |
100 | 85% | 55% |
| 5 | 8b | 100 | 68% | 45% | |
| 6 | 13 | 1 | 75% | 75% | |
| 7 | 15 |  |
10 | 82% | 61% |
In this paper, EC (effective concentration) is defined as the concentration at which the compound blocks >50% of 4 binding to CXCR4.
Table 2.
Effects of asymmetrical compounds as blockers of CXCR4 binding and matrigel invasion.a
 | |||||
|---|---|---|---|---|---|
| Entry | Compd | Ar | TN-binding Blocking EC(nM) |
Invasion Inhibition | |
| 100nM | 10nM | ||||
| 1 | 10a | 10 | 100% | 55% | |
| 2 | 10b |  |
100 | 62% | 41% |
| 3 | 10c | 100 | 64% | 58% | |
| 4 | 10d | 1 | 78% | 90% | |
| 5 | 10e |  |
1 | 80% | 53% |
| 6 | 10f | 10 | 100% | 82% | |
| 7 | 10g |  |
1 | 100% | 100% |
| 8 | 10h |  |
100 | 73% | 61% |
| 9 | 10i |  |
10 | 78% | 41% |
| 10 | 10j | 1 | 82% | 57% | |
In this paper, EC (effective concentration) is defined as the concentration at which the compound blocks > 50% of 4 binding to CXCR4.
Table 3.
Effects of bis-pyrimidines as blockers of CXCR4 binding and matrigel invasion. a
 | ||||||
|---|---|---|---|---|---|---|
| Entry | Compd | R | R’ | TN-binding Blocking EC (nM) |
Invasion Inhibition | |
| 100nM | 10nM | |||||
| 1 | 21a |  |
 |
1 | 93% | 73% |
| 2 | 21b |  |
 |
1 | 92%, | 62% |
| 3 | 21c |  |
 |
1 | 95% | 34% |
| 4 | 17a |  |
 |
1 | 81% | 65% |
| 5 | 18a |  |
 |
10 | 65% | 24% |
| 6 | 18b |  |
 |
1 | 100% | 77% |
| 7 | 26 |  |
 |
1 | 100% | 84% |
In this paper, EC (effective concentration) is defined as the concentration at which the compound blocks > 50% of 4 binding to CXCR4.
cAMP assay
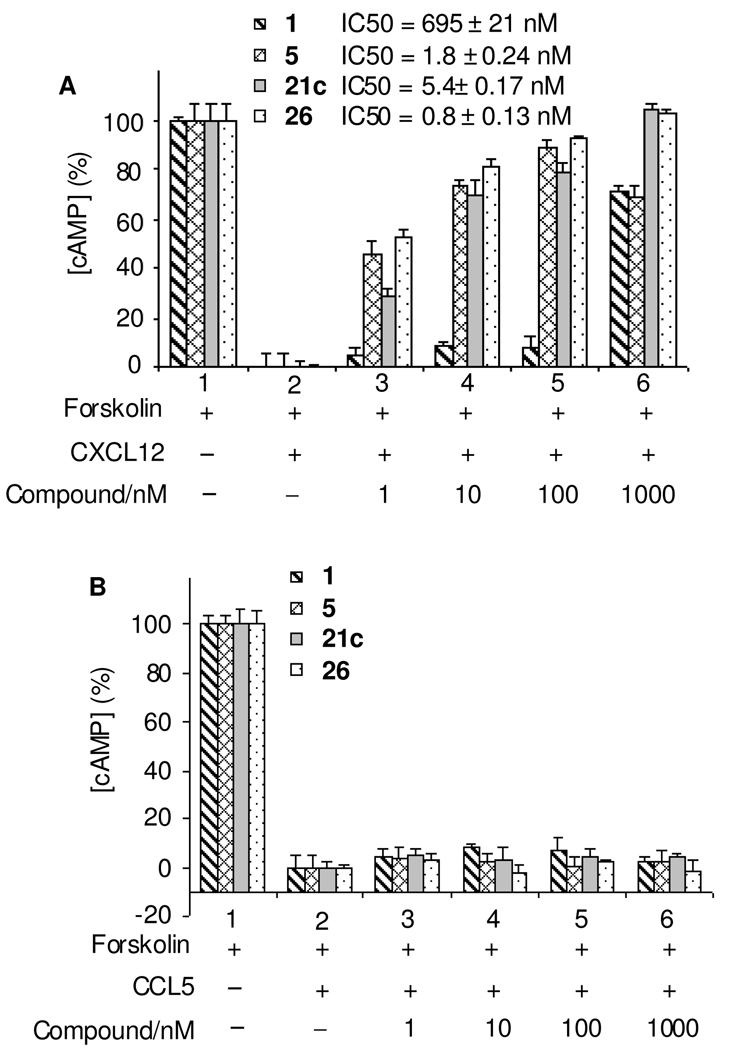
As a GPCR, CXCR4 binds CXCL12 and activates G-protein mediated signaling through the Gαi pathway that reduces cAMP levels within cells. Therefore, we utilized the cAMP assay as the secondary functional assay as we reported previously.9 The selection criteria for the compounds subjected to the cAMP assay was an EC ≤ 10 nM binding affinity and >50% inhibition of invasion at 10 nM. After pre-treatment for 15 min at room temperature with 1 and selected compounds, the effect of CXCL12 on cAMP reduction was blocked significantly in a dose-dependent manner. Compounds 21c with an IC50 at 5.4 ± 0.17 nM and 26 with an IC50 at 0.8 ± 0.13 nM counteracted CXCL12 function effectively at nanomolar concentrations, while 1 required nearly 1000 nM to significantly block CXCL12 function with an IC50 at 695 ± 21 nM. These compounds are almost 1000-fold more efficient in blocking the Gαi pathway than 1 (Figure 2A). Compounds 21c and 26 are comparable to or even better than the previously reported 5 with an IC50 at 1.8 ± 0.24 nM.9 The specificity of 21c and 26 to CXCR4 was tested using U87CD4 cell lines from the AIDS Consortium overexpressing the CCR3 or CCR5 chemokine receptors. These cells were stimulated by 150 ng/mL CCL5 instead of CXCL12. Figure 2B shows the cAMP assay results of compounds 1, 5, 21c, 26 on U87CD4CCR5 cells, all showing no significant effect on blocking CCL5 stimulating CCR5. The compounds were unable to counteract the effect of CCL5 on CCR3 as well (data not shown).
Figure 2.
Comparison of inhibition of CXCR4/CXCL12-mediated cAMP modulation by anti-CXCR4 compounds. (A) CXCR4 positive cell line U87CD4CXCR4 was treated with chemokine CXCL12; (B) CCR5 positive cell line U87CD4CCR5 was treated with CCL5. With pre-treatment (15 min at room temperature) of compounds, 1, 5, 21c, or 26 at various concentrations, the effect of 150 ng/ml of CXCL12 or CCL5 on cAMP reduction in the presence of 5 µM forskolin was measured by using the TR-FRET based LANCE assay kit. While our anti-CXCR4 compounds are effective in counteracting CXCL12 function at as low as 10 nM, 1 requires almost 1000 nM to significantly block CXCL12 function, which is 1000 times less efficient than our compounds in blocking the Gαi pathway. Compound 5 is our previously reported analog; compounds 21c and 26 are comparable or better than compound 5 in blocking CXCL12 function. The tested compounds have no effect on CCR5 cells.
Carrageenan-induced paw edema model
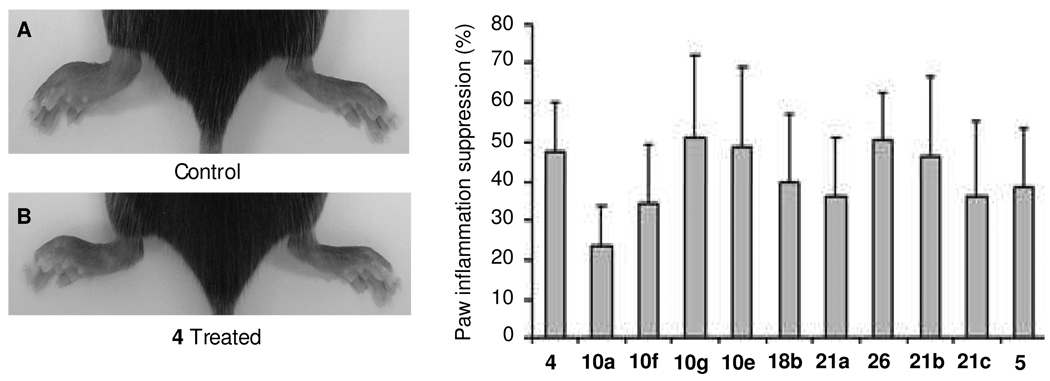
Carrageenan-induced mouse paw edema is a widely used test to assess anti-inflammatory activity in vivo. It is well known that CXCR4 plays a key role in the recruitment of inflammatory cells to sites of inflammation. An apparent edema response was seen 24 h after the λ-carrageenan injection (compared to the contralateral paw that was injected with saline). Based on the rank order of in vitro assay results, ten of the best compounds were investigated. Figure 3 shows the inflammation inhibition percentage by compound. Substances 10g, 10e, and 26 have more than 50% inhibitory effect on inflammation, which is superior to 5 and comparable to 4, while the rest of the compounds revealed variable inhibition effects ranging from 20% to 50%. These data confirm that selected anti-CXCR4 drugs can inhibit inflammation as anticipated.
Figure 3.
Suppression effect of anti-CXCR4 compounds on carrageenan-induced mouse paw inflammation. Acute paw inflammation was induced by subcutaneous injection of 50 µL of λ-carrageenan in one hind paw. The mice in the treatment group were all administered CXCR4 antagonists at 10 mg/kg i.p., while 4 was administered at 300 µg/kg and 30 min following carrageenan challenge and daily thereafter. Control animals received corresponding i.p. injections of vehicle. Panel (A) shows the control mouse with left paw induced inflammation by carrageenan; panel (B) shows the 4 treated mouse with left paw induced inflammation by carrageenan with about 50% suppression. The bar graph shows that all tested compounds are effective at suppressing paw inflammation at different degrees. Compounds 10g, 10e, 26, and 21b showed effects comparable to 4.
In vivo tumor angiogenesis assay (Matrigel plugs)
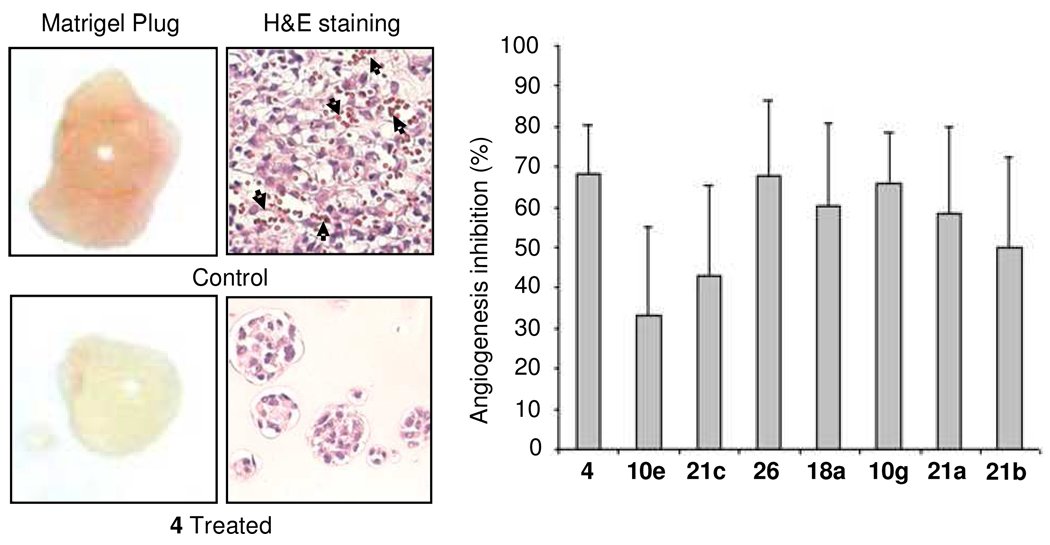
To determine the effect of the CXCR4/CXCL12 interaction on angiogenesis in vivo, an assay using Matrigel plugs to measure anti-angiogenic activity of related compounds was performed in nude mice as previously reported.16 A mixture of 2 × 105 MDA-MB-231 cells in 0.5 mL of growth factor-reduced Matrigel was implanted at two subcutaneous sites with the selected compound mixed-in at 1 µM. The rationale for initial mixing of the compounds into the Matrigel is that there is no route for the compounds to reach the cells within the Matrigel. From the following day, mice were treated with 10 mg/kg daily subcutaneous injections of selected compounds between the two plugs on the back of the mice. Ten days after the Matrigel implant, the mice were sacrificed, and the Matrigel plugs were excised, photographed, weighed and processed to measure hemoglobin content by using Drabkin’s solution.17 When MDA-MB-231 cells successfully promote neovasculature formation within Matrigel plug, these neovasculatures allow tumor cells to proliferate much better than those without neovasculatures.16 Therefore, the control group with better angiogenesis in the Matrigel plug showed more tumor cells than the treated group (Figure 4). The column graph in Figure 4 summarizes the percentage of anti-angiogenic efficacy based on hemoglobin content in ten Matrigel plugs per compound. All the selected compounds show a reasonable anti-angiogenic effect with >30% inhibition. Compound 26 furnishes the best inhibition efficacy of about 70%, which is comparable to the peptidic CXCR4 antagonist 4. This result is consistent with all the in vitro and in vivo test results for compound 26, which exhibits the best anti-CXCR4 activities in the full panel of in vitro assays.
Figure 4.
Inhibitory effect of anti-CXCR4 compounds on Matrigel plug angiogenesis assay in vivo. The mice in the CXCR4 antagonist-treated group received daily subcutaneous injections of the selected compounds (two plugs per mouse) at 10 mg/kg, and 4 at 300 µg/kg. Ten days after matrigel injection, the animals were sacrificed, and the Matrigel plugs were excised, photographed and sliced for H&E staining and processed for hemoglobin assay. Analog 4-treated Matrigel plugs revealed no significant angiogenesis, while the control group exhibited significantly more blood vessels clearly shown by H&E staining (the black arrows in the picture). The column graph shows the tested compounds can inhibit angiogenesis from 30% to 68%, while compound 26 delivers almost the same efficacy as 4 at about 70% angiogenesis inhibition.
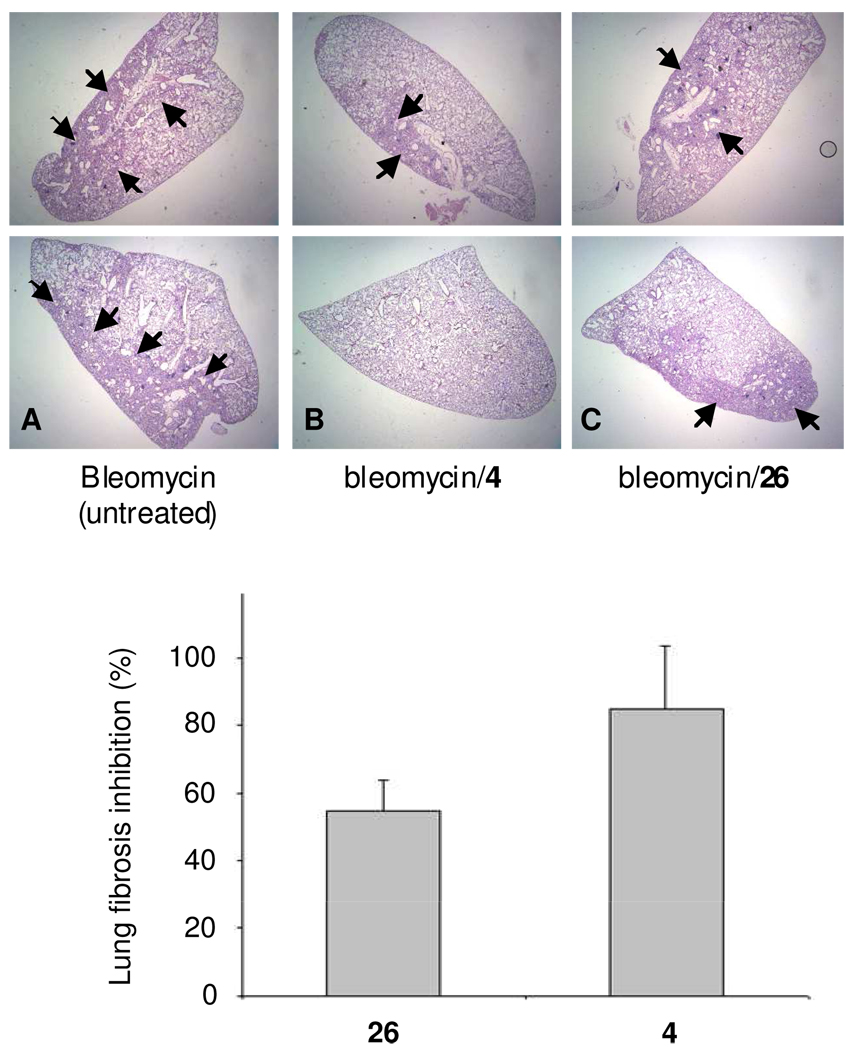
Bleomycin induced lung fibrosis model
The bleomycin induced pulmonary fibrosis model in rodents has been widely used for evaluation of potential therapies. Treatment by 4 in mice with bleomycin-induced lung injury has been reported to significantly attenuate lung fibrosis.18 We used the same method to evaluate the therapeutic effect of compound 26 on bleomycin induced lung fibrosis, because this compound is the most promising compound in all previous assays and in comparison to 4. Lungs harvested 20 days after bleomycin treatment were analyzed histologically by H&E-staining. Figure 5 shows representative photomicrographs of H&E-stained sections. As reported in the literature, bleomycin caused marked alterations in lung architecture with increased interstitial wall thickness and mononuclear cell infiltrates.19 Mice receiving bleomycin and treated separately with 4 and compound 26 led to a decrease in interstitial and alveolar structural distortion (Figure 5 B and C) compared to the lung tissue of untreated mice (5A). Compound 26 blocked lung fibrosis by 55% compared to 4 at 85% inhibition.
Figure 5.
Inhibition effect of compound 26 compared to 4 on lung fibrosis induced by Bleomycin. Representative H&E-stained histopathologic sections of untreated (A), 4 was administered at 300 µg/kg, i.p. (B), and 26 administered at 10 mg/kg, i.p. (C) lung tissues on Day 20 after bleomycin treatment. Lung fibrosis is shown by small black arrows in the images. Column graph shows 4 and 26-treated groups experience a significant decrease in lung fibrosis.
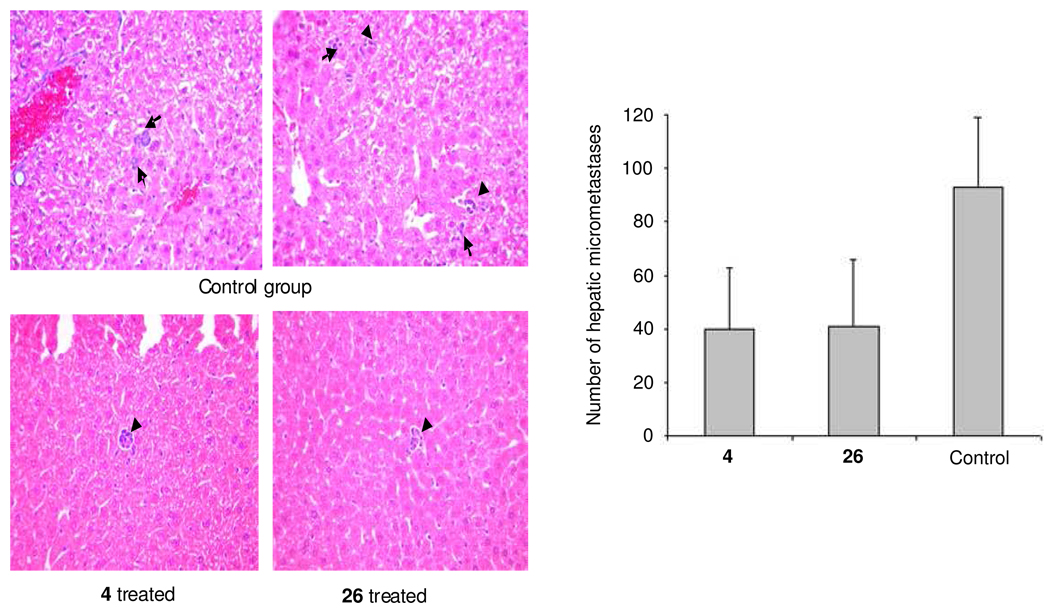
Uveal melanoma micrometastasis mouse model
Blocking CXCR4 has been shown to block metastasis of various cancers. Here, we tested its anti-metastatic efficacy in a uveal melanoma micrometastasis nude mouse model.20 Melanoma OMM2.3 cells overexpressing HGF/TGF-β/CXCR4/MMP2 were inoculated into the posterior chamber of the right eye. After 3 days, the treatment was started (and continued until sacrifice), and after one week, the eyes that developed tumor were enucleated. After 4 weeks, the mice were sacrificed and hepatic tissues were collected, fixed in 10% formalin and paraffin-embedded. The samples were then sectioned and H&E stained (Figure 6). Six sections through the center of the liver were microscopically examined for the presence of micrometastses (<100 µm diameter), and the average number of micrometastases per section was determined. This has been shown to be a reliable and reproducible method for detecting hepatic micrometastases in this animal model.21,22 The micrometastases are very small cohorts of cells (a few to several) and the frequency of the micrometastatic colonies is not high. Due to the small size of the micrometastatic colonies, images were taken at 40× magnification (Figure 6). With the high magnification images, we could not find any field of view that shows a clear difference between treated and untreated livers (it is uncommon to find more than one colony per field of view). The column graph in Figure 6 shows the average number of the hepatic micrometastases throughout the liver counted under the microscope. The number of mice per group was 10. The total number of hepatic micrometastases in the treatment group, which was administered compound 26 or 4 at day-4 after uveal melanoma inoculation, was around 40. This was significantly less than the control group at around 90 (p<0.01). Compound 26 decreased hepatic micrometastases by about 50% in a mouse model carrying human uveal melanoma with an efficacy similar to 4.
Figure 6.
Inhibition effect of 26 compared to 4 in a uveal melanoma micrometastasis animal model. Hepatic tissues were collected and fixed in 10% formalin, processed, and H&E stained, and the number of hepatic micrometastasis was counted under a microscope. Micrometastatic clones are shown by small black arrows in the images. The murine model with ocular melanoma metastatic to the liver was treated with 4 at 300 µg/kg i.p. or compound 26 at 10 mg/kg, i.p. once per day starting at 4th day after uveal melanoma inoculation. The animals presented significantly fewer micrometastases than those in the control (PBS-treated) group. Compound 26 decreased the numbers of hepatic micrometastases in a mouse model of human uveal melanoma with efficacy similar to 4, both are about 50% inhibition.
Conclusions
We and others have shown that the CXCR4 antagonist T140 analogs, including 4, bind to the CXCL12 binding site on CXCR4, block the CXCR4/CXCL12 interaction,23,11 and intervene in the progression of cancer metastasis.11,24–26 Encouraged by these promising results observed with peptidic agents, we sought to identify a novel series of potent, small molecule antagonists that might prove to be practical and safe as anti-metastatic agents. Currently, the metal-chelating cyclams and bicyclams represent the most studied class of non-peptide CXCR4 inhibitors.12 One of these, 1, has advanced as far as Phase II clinical evaluation as an HIV entry inhibitor.13,14,27,28 However, it was withdrawn from that indication in response to adverse effects including cardiotoxicity.8 Currently, 1 has been FDA-approved for stem cell mobilization that requires only one-time administration. We have developed a series of small molecule CXCR4 antagonists that are devoid of metal chelating properties in an effort to develop a safer drug for long term therapy. The present study reveals that N, N'-(1,4-phenylenebis(methylene))dipyrimidin-2-amines show potent CXCR4 antagonism and efficiency in a number of relevant in vitro and in vivo studies. Significantly, these compounds inhibit CXCL12-mediated chemotaxis and a series of pathway signaling events including cAMP action and angiogenesis. The results suggest that this class of compounds potently inhibits the signaling cascade induced by CXCR4/CXCL12 and the associated invasion and homing capabilities.
Most chemokine receptors are shown to signal through the Gαi pathway, which results in an inhibition of adenylyl cyclase and a corresponding decrease in cAMP in the cells.29 Another pathway that is believed to be activated is the Gq pathway, which causes an increase in cytosolic Ca2+ levels. In the last two decades, many reports have demonstrated that the calcium flux assay cannot measure all G-protein coupled receptor function.30 However, industrial and pharmaceutical companies still utilize the calcium flux assay as a gold standard to measure chemokine receptor function. Our CXCR4 compounds 26 and 21a are effective in interrupting CXCL12/CXCR4-mediated cAMP modulation. However, they do not block Ca2+ flux induced by CXCR4/CXCL12 interaction through Gq pathway (data not shown). Furthermore, our anti-CXCR4 compounds do not block 125I-CXCL12 from binding to CXCR4, but CXCL12 can block these same compounds from binding to CXCR4.31 This may be due to the fact that CXCL12, a 9 KDa peptide, has multiple interaction sites on CXCR4,32 whereas our small CXCR4 compounds only block a single site related to homing and chemotaxis. Therefore, CXCL12 is still able to bind to CXCR4 when small molecular weight CXCR4 compounds are preincubated with cells before adding CXCL12. Intriguingly, the present class of anti-CXCR4 compounds can intervene in the Gαi signaling pathway (cAMP modulation), but not the Gq pathway (Ca2+ flux). These findings may present an opportunity for developing a safe drug that partially interrupts CXCL12/CXCR4, especially because the interplay between CXCL12 and CXCR4 is critical to normal physiology. For example, CXCR4 modulates contractility in adult cardiac myocytes by mediating Ca2+ flux.33 Therefore, anti-CXCR4 compounds that do not interrupt Ca2+ flux may well be safer than 1, which exhibits myocardial toxicity in addition to metal-chelating properties. In addition, the CXCR4/CXCL12 interaction is involved in the homing and retention of hematopoietic progenitor cells in the bone marrow, while CXCL12 also acts as a major chemoattractant for stem cells and some differentiated cells in the pathological contexts of inflammation and tissue regeneration or repair.34–37 Therefore, the partial CXCR4 inhibitors reported here, which block chemotaxis and homing of the CXCR4-positive cells to the distant organ sites enriched with CXCL12 in their stroma, may present an alternative, safe option as chemopreventive drugs for cancer metastasis and tumor angiogenesis. Furthermore, we have demonstrated that 26 suppresses paw inflammation by 56%, inhibits angiogenesis by 70%, arrests lung fibrosis by 55% and blocks uveal melanoma OMM2.3 (HGF/TGF-β/CXCR4/MMP2) micrometastases by 50%, despite the fact that compound 26 has a fast blood clearance in mice (t1/2 ~ 15mins with intravenous injection). These data further support the role of CXCR4 in chemotaxis, motility, and invasion, and reinforce its value as a target for therapeutic intervention in disease states where these phenomena play key roles, such as cancer metastasis and inflammation.
Although the compound database represented by Tables 1–3 is sparse and the activity range is narrow, an intriguing SAR element is evident. It arises by classifying the blockade of 4 binding into two categories: high (1–10 nM) and low (100 nM). In this context, when one or both of the terminal aromatic rings are nitrogen deficient (i.e. either benzenoid or pyridinoid), then ortho substitution by a ring nitrogen, fluorine or N-alkyl can lead to less effective competition with 4. On the other hand, when both terminal rings are dipyrimidines, activity is high and substitution effects are marginal for the latter as shown, among others, by lead 26 (Table 3). Of course, the ring substitution pattern is decisive for phenomena associated with membrane passage and biodistribution. The improved antagonistic effects of double pyrimidine substitution cannot be rationalized on a molecular level, since the geometry of the specific binding site is presently unknown. However, a model of the binding of 1 to CXCR4 suggests a possible role for the dipyrimidine structure.23
In conclusion, a novel class of small molecules, N, N'-(1,4-phenylenebis(methylene)) dipyrimidin-2-amines, especially compound 26, have been identified by rational design and analysis of emerging structural and pharmacologic data as putative inhibitors of CXCR4/CXCL12 functions. The compelling features of 26 include effectiveness for (i) potent inhibition of CXCR4/CXCL12 functions in vitro (IC50 ~ 1 nM); (ii) anti-angiogenesis in vivo; (iii) suppression of inflammation in vivo; (iv) inhibition of lung fibrosis in vivo; and (v) blockade of uveal melanoma micrometastasis in vivo. In brief, compound 26 is an excellent anti-CXCR4 agent based on our in vitro and in vivo tests. However, the shortcomings of this compound are its fast blood clearance (t1/2 ~ 15 minutes with intravenous injection) and the lack of oral bioavailability. Thus, further improvement is needed to develop an orally available therapeutic drug. The physical properties of compound 26 make it suitable for nuclear imaging. Therefore, we are in a process of developing it as a Positron Emission Tomography (PET) tracer to specifically image CXCR4-positive cells.
Experimental section
Initial screening of anti-CXCR4 small molecules based on a binding affinity assay and cell invasion assay
Binding affinity and cell invasion assays are basic assay tools that apply to the initial screening. MDA-MB-231 cells cultured in an 8-well slide chamber were preincubated with the testing compounds at 1, 10, 100, and 1000 nM. Then the cells were fixed with 4% formaldehyde and incubated with 50 nM biotinylated 4, and followed by Rhodamine staining. Matrigel invasion chambers from BD Biocoat Cellware (San Jose, CA) were used for invasion assays. MDA-MB-231 cells were cultured on a layer of Matrigel in the upper chamber with testing compounds at 10 or 100 nM while 200 ng/mL CXCL12 was added in the lower chamber as a chemoattractant. Detailed procedures for the binding and invasion assays have been described in previous publications.11,38,16
cAMP assay to measure Gαi function
Perkin-Elmer’s LANCE cAMP assay kit (Cat # AD0262), based on time-resolved fluorescence resonance energy transfer (TR-FRET), was used to determine whether our compounds could block cAMP modulation induced by the CXCR4/CXCL12 interaction. U87CD4CXCR4 cells (AIDS Consortium), U87CD4CCR3 cells (AIDS Consortium), or U87CD4CCR5 cells (AIDS Consortium) were used. The detailed procedures of the cAMP assay have been described in previous publications.9
Paw inflammation suppression test
Acute inflammation was induced by subcutaneous injection of 50 µL of λ-carrageenan (1% w/v in saline) into one of the hind paws of female C57BL/6J mice (Jackson Laboratories); the other hind paw was used as a non-inflammation control. In the treatment group, CXCR4 antagonists were administered i.p. at 10 mg/kg, while 4 was at 300 µg/kg, 30 min following carrageenan challenge and continued daily.18,39 The rationale for using 300 µg/kg for compound 4 was that we found this concentration to be the minimum concentration to achieve the maximum efficacy of this compound in a breast cancer metastasis animal model.11 All CXCR4 antagonists were dissolved in 10% DMSO and 90% of 45% (2-Hydroxypropyl)-beta-cyclodextrin (CD) in PBS. Control animals received corresponding i.p. injections of vehicle. The animals were sacrificed 74 h after induction of inflammation and 2h after the last injection of CXCR4 antagonists. The final paws were photographed and measured for thickness from the "palm" to the back of the paw by a caliper. These were compared to the volume of carrageenan untreated contralateral paw to obtain the edema volume; the volume of the contralateral paw was subtracted from the volume of the carrageenan injected paw to obtain the edema volume. The inflammation suppression percentage was calculated by comparing the drug treated group to the control group.
In vivo angiogenesis assay (Matrigel plug)
2×105 MDA-MB-231 cells were mixed with the compound in 0.5 mL of growth factor-reduced matrigel (BD Biosciences, San Jose, CA) at 1 µM concentration and implanted subcutaneously into the flanks of nude mice (two plugs per mouse, 6 mice per group). The mice in the CXCR4 antagonist-treated group received daily subcutaneous injections of the selected drugs in the middle of the two plugs (two plugs per mouse) at 10 mg/kg, while 4 was administered at 300 µg/kg. Ten days after matrigel injection, the animals were sacrificed, and the Matrigel plugs were excised. The excised plugs were photographed and processed for hemoglobin assay. The samples were homogenized in 100 µl of deionized water and cleared by centrifugation at 10,000 rpm for 10 min. 20 µL of the supernatant was mixed with 100 µL of Drabkin's solution (Sigma, St. Louis, MO) to measure hemoglobin content. The mix was incubated at 52 °C for 10 min and continuously at room temperature for 20 min. The absorbance was read at 540 nm in an ELISA plate reader.
Bleomycin induced lung fibrosis mouse model
Mice were anesthetized by isofluorane inhalation; the trachea was exposed using sterile techniques and 4 U/kg bleomycin (Sigma) in 100 µL PBS or PBS vehicle was injected into the tracheal lumen. After inoculation, the incision was closed and the animals were allowed to recover. Each group has 10 mice that received 300 µg/kg of 4, 10 mg/kg of compound 26 or saline intraperitoneally 1 day before bleomycin treatment and daily for 20 days. All mice tolerated the antagonist well. Lungs harvested 20 days after bleomycin treatment were analyzed histologically by H&E-staining and imaged under microscope.
Uveal melanoma micrometastasis mouse model
The female nude mice were divided into 3 groups: two treatment groups and one control group, with 10 mice in each group. On day 0, each mouse was inoculated with 1×106 OMM2.3 cells expressing HGF/TGF-β/CXCR4/MMP2 into the posterior chamber of right eye. On day 3, mice were treated with 10 mg/kg compound 26 or 300 µg/kg 4 in 0.1 mL volume of 45% CD daily by I.P. injection, whereas the control mice were I.P. injected with 0.1 mL 45% CD only. On day 7, eyes with tumor were enucleated. The growth of tumor was checked by histological methods. On day 28, hepatic tissues were collected and fixed in 10% formalin, processed, H&E stained, and the number of hepatic micrometastases was counted under microscope. Six sections through the center of the liver were microscopically examined (Olympus BX41, Tokyo, Japan) for the presence of micrometastses (<100 µm diameter) and the average number of micrometastases per section was determined.
Chemistry: General
Proton and carbon NMR spectra were recorded on INOVA-400 (400 MHz) or INOVA-600 (600 MHz) Spectrometers. The spectra obtained in deuteriochloroform (CDCl3) or dimethyl sulfoxide-d6 (DMSO-d6) were referenced to the residual solvent peak. Mass spectra were recorded on a JEOL spectrometer at Emory University Mass Spectrometry Center. Elemental analyses were performed by Atlantic Mircolab, Inc. Norcross, GA. Flash column chromatography was carried out with Scientific Absorbent Incorporated Silica Gel 60. Analytical thin layer chromatography (TLC) was performed on precoated glass backed plates from Scientific Adsorbents Incorporated (Silica Gel 60 F254; 0.25 mm thickness). Plates were visualized using ultraviolet, iodine vapors or phosphomolybdic acid (PMA). Preparation of compounds 3, 5 and 6 has been documented in our previous report.9 All final compounds were > 98% pure, which was confirmed by a Beckman HPLC system using Nova Pak C18 4 µm 3.9 × 150 mm column (Waters) and methanol/wanter (65:35) with 0.1% triethylamine as an eluent. Elemental analyses were performed by Atlantic Mircolab, Inc. Norcross, GA and were within ± 0.4% of the theoretical values.
N,N'-(1,4-phenylenebis(methylene))bis(6-fluoropyridin-2-amine) (8a)
A mixture of terephthaldicarboxaldehyde (268 mg, 2.0 mmol) and 6-fluoro-2-aminopyridin (0.47 g, 4.2 mmol) in 1, 2-dichloethane (20 mL) was treated with triacetoxyborohydride (1.27 g, 6.0 mmol) and HOAc (0.24 mL, 4.0 mmol). After stirring at room temperature under an argon atmosphere until the disappearance of the starting materials, the reaction was quenched by adding aqueous NaOH (10 mL, 1.0 N). The resulting mixture was extracted with ethyl acetate (3 × 15 mL), and the combined organic layers were washed with brine, dried over anhydrous MgSO4 and concentrated in vacuo. Purification by flash column chromatography (silica gel, hexane/ethyl acetate, 3/1 with 0.5% NH4OH) gave the title compound 8a (0.42 g, 64 %,) as a white solid: mp 182–185 °C (dec); 1H NMR (400 MHz, DMSO-d6) δ 7.48 (dd, J = 16.8, 7.9 Hz, 2H), 7.43 (t, J = 6.0, 6.0 Hz, 2H), 7.26 (s, 4H), 6.35 (dd, J = 8.1, 2.4 Hz, 2H), 6.08 (dd, J = 7.6, 2.2 Hz, 2H), 4.37 (d, J = 6.0 Hz, 4H); 13C NMR (100 MHz, DMSO-d6) δ 162.79, 158.23, 141.43, 138.30, 127.26, 104.49, 93.56, 43.96; HRMS Calcd for C18H17F2N4 327.14213 [M+H]+, found 327.14156. Anal. (C18H16F2N4) C, H, N.
N,N'-(1,4-phenylenebis(methylene))bis(5-fluoropyridin-2-amine) (8b)
Starting with terephthaldicarboxaldehyde (0.27 g, 2.0 mmol) and 5-fluoropyridin-2-amine (0.45 g, 4.0 mmol) by treating with triacetoxyborohydride (1.27 g, 6.0 mmol), the same procedure to compound 8a furnished the title product 8b (0.40 g, 61%) as a white solid: mp 163–165 °C (dec); 1H NMR (600 MHz, CDCl3) δ 7.90 (d, J = 3.0 Hz, 2H), 7.33 (dt, J = 8.8, 8.8, 3.1 Hz, 2H), 7.24 (s, 4H), 7.01 (t, J = 5.9, 5.9 Hz, 2H), 6.51 (dd, J = 9.2, 3.6 Hz, 2H), 4.38 (d, J = 6.0 Hz, 4H); 13C NMR (150 MHz, CDCl3) δ 155.69, 152.47, 138.70, 133.42, 125.03, 108.78, 44.40; HRMS Calcd for C18H17F2N4 327.14213 [M+H]+, found 327.14117; Anal. (C18H16F2N4) C, H, N.
4-((pyrimidin-2-ylamino)methyl)benzaldehyde (9)
In a 500 mL one-necked round-bottomed flask equipped with a stirrer, terephthaldicarboxaldehyde (5.36 g, 40 mmol), 2-amino-pyrimidine (3.91 g, 41 mmol), and acetic acid (4.7 mL, 80 mmol) were mixed in 1,2-dichloroethane (250 mL). After being stirred at room temperature until 2-amino-pyrimidine completely dissolved (approximately 10 minutes), 4Å molecular sieves (20 g) were added to the mixture. After being stirred for 10 min, the resulting mixture was treated with sodium triacetoxyborohydride (25.43 g, 120 mmol). After being stirred for 24 hours at room temperature under an argon atmosphere, the reaction was quenched by adding aqueous NaOH (150 mL, 1.0 N). The resulting mixture was extracted with ethyl acetate (3 × 200 mL), and the combined organic layers were washed with brine, dried over anhydrous MgSO4 and concentrated in vacuo to give the alcohol intermediate S1 which was purified by column chromatography (ethyl acetate) to give alcohol S1 as a white solid (7.32 g, 86%): mp 120–121 °C (dec); 1H NMR (400 MHz, CDCl3,) δ 8.30 (d, J = 4.8 Hz, 2H),7.35 (s, 4H), 6.57 (t, J = 4.8 Hz, 1H), 5.42 (bs, 1H), 4.69 (d, J = 5.6 Hz, 2H), 4.64 (d, J = 5.6 Hz, 2H), 1.83 (t, J = 5.6 Hz, 1H) ; 13C NMR (100 MHz, CDCl3) δ 162.27, 157.93, 140.71, 138.75, 126.74, 126.36, 110.14, 62.73, 43.65; HRMS Calcd for C12H14N3O 216.11369 [M+H]+, found 216.11284. To a solution of the alcohol S1 (2.22 g, 10.3 mmol) in CH2Cl2 (50 mL) was added Dess-Martin periodinane (5.24 g, 12.3 mmol) at 0 °C. After being stirred for 10 min, the reaction mixture was allowed to warm to room temperature and was stirred for another 1h. The reaction mixture was diluted with diethyl ether (100 mL) and quenched by saturated aqueous NaHCO3 (50 mL) and Na2SO3 (50 mL). The organic phase was separated, and the aqueous phase was further extracted with diethyl ether (3 × 20 mL). The combined organics were dried over MgSO4, filtered and concentrated under reduced pressure. The residue was purified through flash column chromatography (Ethyl acetate) to furnish aldehyde 9 (2.07 g, 94%) as a white solid: mp 114–115 °C (dec); 1H NMR (400 MHz, CDCl3) δ 10.00 (s, 1H), 8.29 (d, J = 4.8 Hz, 2H), 7.85 (d, J = 8.2 Hz, 2H), 7.52 (d, J = 8.1 Hz, 2H), 6.60 (t, J = 4.8, 4.8 Hz, 1H), 5.82 (bs, 1H), 4.75 (d, J = 6.3 Hz, 2H); 13C NMR (100 MHz, CDCl3) δ 192.13, 162.30, 158.35, 146.69, 135.69, 130.31, 127.86, 111.52, 45.25; HRMS Calcd for C12H12N3O 214.09804 [M+H]+, found 214.09718.
N-(4-((phenylamino)methyl)benzyl)pyrimidin-2-amine (10a). General procedure A
A mixture of aldehyde 9 (0.32 g, 1.5 mmol) and aniline (0.15 g, 1.6 mmol) in 1,2-dichloroethane (15 mL) was treated with sodium triacetoxyborohydride (0.48 mg, 2.25 mmol). After being stirred at room temperature overnight, the reaction mixture was quenched by adding aqueous NaOH (10 mL, 1.0 N), extracted with ethylacetate (2 × 30 mL). The combined organic phases were washed by brine, dried over anhydrous MgSO4, and filtered and concentrated under reduced pressure. The residue was purified by column chromatography (Hexanes/ethyl acetate, 1/1) to afford product 10a (0.41 mg, 94%) as a white solid: mp 131–132 °C (dec); 1H NMR (400 MHz, CDCl3) δ 8.27 (d, J = 4.7 Hz, 2H), 7.34 (s, 4H), 7.21-7.15 (m, 2H), 6.72 (tt, J = 7.4, 7.4, 1.0, 1.0 Hz, 1H), 6.65-6.0 (m, 2H), 6.54 (t, J = 4.8, 4.8 Hz, 1H), 5.59 (s, 1H), 4.63 (d, J = 5.9 Hz, 2H), 4.32 (s, 2H), 4.04 (s, 1H); 13C NMR (100 MHz, CDCl3) δ 162.48, 160.40, 158.33, 148.27, 138.66, 138.29, 129.47, 127.96, 117.77, 113.02, 111.12, 48.19, 45.31; HRMS Calcd for C18H19N4 291.16097, [M+H]+ found 291.1033; Anal. (C18H18N4) C, H, N.
N-(4-((2-fluorophenylamino)methyl)benzyl)pyrimidin-2-amine (10b)
Starting from aldehyde 9 (53.4 mg, 0.25 mmol) and 2-fluoroaniline (29.2 mg, 0.26 mmol), general procedure A gave 10b (44.2 mg, 57%) as a white solid: mp 126–128 °C (dec); 1H NMR (400 MHz, CDCl3) δ 8.30 (d, J = 4.8 Hz, 2H), 7.35 (s, 4H), 7.01-6.94 (m, 2H), 6.69-6.61 (m, 2H), 6.57 (t, J = 4.8, 4.8 Hz, 1H), 5.52 (bs, 1H), 4.65 (d, J = 6.0 Hz, 2H), 4.36 (s, 2H), 4.32 (bs, 1H); 13C NMR (100 MHz, CDCl3) δ 162.47, 158.26, 151.68 (d, J = 237.5 Hz), 138.58 (d, J = 32.6 Hz), 136.75, 136.64, 128.02, 127.81, 124.76, 116.98, 114.55, 112.44, 110.98, 47.70, 45.29; HRMS Calcd for C18H18FN4 309.15155 [M+H]+, found 309.15050; Anal. (C18H17FN4) C, H, N.
N-(4-((pyridin-2-ylamino)methyl)benzyl)pyrimidin-2-amine (10c)
Starting from aldehyde 9 (0.43 g, 2.0 mmol) and 2-aminopyridine (0.23 g, 2.4 mmol) by treatment with sodium triacetoxyborohydride (0.64 g, 3.0 mmol) and HOAc (0.13 mL, 2.2 mmol), general procedure A gave white solid, which was washed by methanol to afford 10c (0.35 g, 61%) as a white solid: mp 172–174 °C (dec); 1H NMR (400 MHz, CDCl3) δ 8.30 (d, J = 4.8 Hz, 2H), 8.11 (d, J = 4.8 Hz, 1H), 7.43-7.39 (m, 1H), 7.34 (s, 4H), 6.62-6.58 (m, 1H), 6.57 (t, J = 4.8, 4.8 Hz, 1H), 6.37 (d, J = 8.4 Hz, 1H), 5.41 (bs, 1H), 4.88 (bs, 1H), 4.64 (d, J = 6.0 Hz, 2H), 4.50 (d, J = 6.0 Hz, 2H); 13C NMR (150 MHz, CDCl3) δ 162.48, 158.65, 158.33, 148.05, 138.36, 138.29, 137.90, 127.96, 127.86, 113.37, 111.14, 107.11, 46.24, 45.30; HRMS Calcd for C17H18N5 292.15622 [M+H]+, found 292.15518; Anal. (C17H17N5) C, H, N.
N-(4-((5-fluoropyridin-2-ylamino)methyl)benzyl)pyrimidin-2-amine (10d)
Starting from aldehyde 9 (0.22 mg, 1.0 mmol) and 5-fluoro-2-aminopyridin (0.12 g, 1.0 mmol) by treatment with sodium triacetoxyborohydride (0.32 g, 1.5 mmol) and HOAc (0.06 mL, 1.0 mmol), general procedure A gave 10d (0.28 mg, 90%) as a white solid: mp 167–169 °C (dec); 1H NMR (400 MHz, CDCl3) δ 8.29 (d, J = 4.8 Hz, 2H), 7.96 (d, J = 2.8 Hz, 1H), 7.33 (s, 4H), 7.21-7.16 (m, 1H), 6.57 (t, J = 4.8, 4.8 Hz, 1H), 6.32 (dd, J = 9.2, 3.2 Hz, 1H), 5.49 (bs, 1H), 4.85 (bs, 1H), 4.63 (d, J = 6.0 Hz, 2H), 4.46 (d, J = 6.0 Hz, 2H); 13C NMR (150 MHz, CDCl3) δ 162.43, 158.35, 155.44, 153.72, 138.38, 138.26, 134.80, 127.96, 127.88, 125.61, 111.19, 107.40, 46.73, 45.30; HRMS Calcd for C17H17FN5 310.14680 [M+H]+, found 310.14588; Anal. (C17H16FN5) C, H, N.
N-(4-((6-fluoropyridin-2-ylamino)methyl)benzyl)pyrimidin-2-amine (10e)
Starting from aldehyde 9 (0.43 g, 2.0 mmol) and 6-fluoro-2-aminopyridine (0.25 g, 2.4 mmol) by treatment with sodium triacetoxyborohydride (0.64 g, 3.0 mmol) and HOAc (0.12 mL, 2.2 mmol), general procedure A gave 10e (0.46 g, 73%) as a white solid: mp 170–171 °C (dec); 1H NMR (400 MHz, CDCl3) δ 8.24 (d, J = 4.4 Hz, 2H), 7.66 (t, J = 6.2 Hz, 1H), 7.51-7.40 (m, 2H), 7.24 (s, 4H), 6.55 (t, J = 4.8 Hz, 1H), 6.34 (dd, J = 8.0, 2.4 Hz, 1H), 6.08 (dd, J = 8.0, 2.8 Hz, 1H), 4.45 (d, J = 6.4 Hz, 2H), 4.35 (d, J = 6.0 Hz, 2H); 13C NMR (150 MHz, CDCl3) δ 162.77, 162.26, 158.22, 157.96, 141.42, 138.88, 137.99, 127.14, 126.99, 110.17, 104.46, 93.52, 43.96, 43.62; HRMS Calcd for C17H18FN5 310.14680 [M+H]+, found 310.14954; Anal. (C17H16FN5) C, H, N.
N-(4-((5-chloropyridin-2-ylamino)methyl)benzyl)pyrimidin-2-amine (10f)
Starting from aldehyde 9 (0.22 g, 1.0 mmol) and 5-chloro-2-aminopyridine (0.14 g, 1.1 mmol) by treatment with sodium triacetoxyborohydride (0.32 g, 1.5 mmol) and HOAc (0.06mL, 1.1 mmol), general procedure A gave 10f (0.20 g, 60%) as a white solid: mp 163–165 °C (dec); 1H NMR (400 MHz, DMSO-d6) δ 8.24 (d, J = 4.8 Hz, 2H), 7.93 (d, J = 2.8 Hz, 1H), 7.66 (t, J = 6.4 Hz, 1H), 7.41 (dd, J = 8.8, 2.4 Hz, 1H), 7.25 (d, J = 6.0 Hz, 1H), 7.23 (s, 4H), 6.55 (t, J = 4.8, 4.8 Hz, 1H), 6.51 (d, J = 8.8 Hz, 1H), 4.44 (d, J = 6.4 Hz, 2H), 4.39 (d, J = 6.4 Hz, 2H); 13C NMR (150 MHz, DMSO-d6) δ 162.27, 158.00, 157.26, 145.46, 138.81, 138.29, 136.48, 127.12, 126.97, 117.43, 110.18, 109.61, 44.08, 43.63; HRMS Calcd for C17H17ClN5 326.11725 [M+H]+, found 326.11638; Anal. (C17H16ClN5) C, H, N.
N-(4-((6-chloropyridin-2-ylamino)methyl)benzyl)pyrimidin-2-amine (10g)
Starting from aldehyde 9 (0.21 g, 1.0 mmol) and 6-chloro-2-aminopyridine (0.14 g, 1.1 mmol) by treatment with sodium triacetoxyborohydride (0.32 g, 1.5 mmol) and HOAc (0.064 mL, 1.1 mmol), general procedure A gave 10g (0.22 g, 67%) as a white solid: mp 148–150 °C (dec); 1H NMR (400 MHz, DMSO-d6) δ 8.24 (d, J = 4.8 Hz, 2H), 7.67 (t, J = 6.0 Hz, 1H), 7.43 (t, J = 6.0 Hz, 1H), 7.37 (t, J = 7.6 Hz, 1H), 7.25 (s, 4H), 6.55 (t, J = 4.8 Hz, 1H), 6.49 (d, J = 7.2 Hz, 1H), 6.42 (d, J = 8.0 Hz, 1H), 4.46 (d, J = 6.4 Hz, 2H), 4.37 (d, J = 6.0 Hz, 2H); 13C NMR (100 MHz, DMSO-d6) δ 162.26, 158.89, 157.97, 148.39, 139.59, 138.93, 137.89, 127.24, 126.99, 110.20, 110.17, 106.42, 43.99, 43.62; HRMS Calcd for C17H17ClN5 326.11725 [M+H]+, found 326.11640; Anal. (C17H16ClN5) C, H, N.
N-(4-((2-(pyrrolidin-1-yl)phenylamino)methyl)benzyl)pyrimidin-2-amine (10h)
A solution of boc-protected 2-(pyrrolidin-1-yl)aniline (0.26 g, 1.0 mmol) in CH2Cl2 (5 mL) was treated with HCl solution in dioxane (5 mL, 4M in dioxane). After being stirred for 6 h at room temperature, the solvent was removed under reduced pressure to afford a yellow solid (0.27 g). The yellow solid prepared above was dissolved in 1, 2-dichloroethane (20 mL), to which was added aldehyde 9 (0.20 g, 0.9 mmol) and HOAc (0.06 mL, 1.0 mmol). The resulting light brown solution was treated with triacetoxyborohydride (0.32 mg, 1.5 mmol), and general procedure A gave the title product 10h (0.31 g, 85%, 2 steps) as a pale white solid: mp 124–126 °C (dec); 1H NMR (400 MHz, CDCl3) δ 8.27 (d, J = 4.8 Hz, 2H), 7.33–7.39 (m, 4H), 7.05 (dd, J =7.6, 1.6 Hz, 1H), 6.95 (td, J =7.6, 1.6 Hz, 1H), 6.70 (td, J =7.6, 1.6 Hz, 1H), 6.59 (dd, J =7.6, 1.6 Hz, 1H), 6.55 (t, J = 4.8 Hz, 1H), 5.68 (bs, 1H), 4.91 (bs, 1H), 4.65 (d, J = 6.0 Hz, 2H), 4.36 (d, J = 5.2 Hz, 2H), 3.04–3.07 (m, 4H), 1.88–1.95 (m, 4H); 13C NMR (100 MHz,CDCl3) δ 162.48, 158.22, 143.51, 139.23, 137.97, 137.42, 127.93, 127.69, 124.22, 118.56, 117.10, 110.86, 110.47, 51.42, 48.22, 45.34, 24.23; HRMS Calcd for C22H26N5 360.21882 [M+H]+, found 360.21805; Anal. (C22H25N5) C, H, N.
N-(4-((3-(pyrrolidin-1-yl)phenylamino)methyl)benzyl)pyrimidin-2-amine (10i)
The boc-protected 3-(pyrrolidin-1-yl)aniline (0.29mg, 1.1 mmol) was converted to 10i (0.19 g, 58%, 2 steps) according to the procedure described above for 10h as a pale white solid: mp 136–138 °C (dec.); 1H NMR (400 MHz, CDCl3) δ 8.19 (bs, 2H), 7.38-7.33 (m, 4H), 7.06 (t, J =8.0 Hz, 1H), 6.50 (t, J = 4.8, 4.8 Hz, 1H), 6.28 (t, J = 5.6, 5.6 Hz, 1H), 6.03 (dd, J = 6.0, 2.0 Hz, 2H), 5.88 (t, J = 2.0, 2.0 Hz, 1H), 4.64 (d, J = 5.6, 5.6 Hz, 2H), 4.34 (s, 2H), 4.01 (bs, 1H), 3.30-3.24 (m, 4H), 2.01-1.95 (m, 4H); 13C NMR (100 MHz,CDCl3) δ 162.41, 158.14, 149.36, 149.15, 139.10, 138.04, 129.97, 127.90, 110.77, 102.24, 101.17, 96.34, 48.23, 47.65, 45.29, 25.54; HRMS Calcd for C22H26N5 360.21882 [M+H]+, found 360.21805; Anal. (C22H25N5) C, H, N.
N-(4-((4-(pyrrolidin-1-yl)phenylamino)methyl)benzyl)pyrimidin-2-amine (10j)
The boc-protected 4-(pyrrolidin-1-yl)aniline (0.26 g, 1.0 mmol) was converted to 10j (0.30 g, 92%, 2 steps) according to the procedure described above for 10h as a pale white solid: mp 149–153 °C (dec); 1H NMR (400 MHz, CDCl3) δ 8.30 (d, J = 4.8 Hz, 2H), 7.31–7.37 (m, 4H), 6.65 (bs, 2H), 6.57 (t, J = 4.8 Hz, 1H), 6.55 (bs, 2H), 5.41 (bs, 1H), 4.63 (d, J = 5.6 Hz, 2H), 4.27 (bs, 2H), 3.63 (bs, 1H), 3.20 (bs, 4H), 1.95–1.99 (m, 4H); 13C NMR (100 MHz, CDCl3) δ 162.48, 158.26, 142.07, 139.21, 138.00, 128.04, 127.87, 115.19, 113.31, 110.97, 49.63, 48.51, 25.44 HRMS Calcd for C22H26N5 360.21882 [M+H]+, found 360.21829; Anal. (C22H25N5) C, H, N.
N,N'-(1,4-phenylenebis(methylene))bis(5-fluoropyrimidin-2-amine) (13)
A mixture of 2-chloro-5-fluoropyrimidine (0.92 g, 6.9 mmol), 1,4-phenylenedimethanamine (0.45 g, 3.3 mmol), and cesium carbonate (2.58 g, 7.9 mmol) in DMF (25 mL) was stirred at 100 °C overnight. After removing the solvent under reduced pressure, the yellow residue was washed with H2O and hot ethanol to give the title product 13 (0.65 g, 60%) as pale yellow solid: mp 220–226 °C (dec.); 1H NMR (400 MHz, DMSO-d6) δ 8.33 (s, 4H), 7.78 (t, J = 6.0 Hz, 2H), 7.21 (s, 4H), 4.41 (d, J = 6.0 Hz, 4H); 13C NMR (100 MHz,DMSO-d6) δ 159.47, 151.60, 145.49, 138.44, 126.86, 44.24; HRMS Calcd for C16H15F2N6 329.13263 [M+H]+, found 329.13192; Anal. (C16H14F2N6) C, H, N.
N,N'-(1,4-phenylenebis(methylene))bis(4,6-dichloro-1,3,5-triazin-2-amine) (15)
To a solution of cyanuric chloride (1.94 g, 10.5 mmol) in THF (70 mL) was added 1,4-phenylenedimethanamine (0.681 g, 5.0 mmol) at 0 °C in portionwise, followed by an addition of NaHCO3 (1.05 g, 12.5 mmol). After being stirred overnight at 0 °C, the reaction mixture was warmed to room temperature and stirred for additional 2h. The solvent was removed under reduced pressure, and the white solid residue was sequentially washed with water and ethanol to afford pure title compound 15 (2.04 g, 94%) as a white solid: mp > 400 °C (dec); 1H NMR(400 MHz, DMSO-d6) δ 9.61 (t, J = 6.1, 6.1 Hz, 2H), 7.27 (s, 4H), 4.50 (d, J = 6.2 Hz, 4H); 13C NMR (150 MHz, DMSO-d6) δ 169.51, 168.63, 165.46, 136.51, 127.45, 43.71. HRMS Calcd for C14H11Cl4N8 430.98608 [M+H]+, found 430.96623; Anal. (C14H10Cl4N8) C, H, N.
N,N'-(1,4-phenylenebis(methylene))bis(2-fluoropyrimidin-4-amine) (17a)
To a solution of 1,4-phenylenedimethanamine (1.36 g, 10 mmol) in DMF (50 mL) was added 2,4-difluoropyrimidine (2.56 g, 22 mmol) and N,N-diisopropylethylamine (6.10 mL, 35 mmol) at room temperature. The resulting mixture was heated to 60 °C and stirred until the starting material disappeared from TLC plate. The reaction mixture was cooled down to ambient temperature, poured into ice water (20 mL) and extracted with ethyl acetate (3 × 5 mL). The combined organic layers were washed with brine, dried over anhydrous MgSO4 and concentrated in vacuo. The crude was purified by flash column chromatography (silica gel, MeOH/CH2Cl2, 0 → 25%) to give the title compound 17a (2.49 g, 76%), as a white solid: mp 267–268 °C (dec); 1H NMR (600 MHz, DMSO-d6) δ 7.93 (d, J = 4.2 Hz, 2H), 7.28 (s, 4H), 6.38(bs, 1H), 6.34(bs, 1H), 6.47 (t, J = 4.8Hz, 2H), 4.47(d, J = 6 Hz, 4H); 13C NMR (150 MHz, DMSO-d6) δ 165.22, 161.98, 156.20, 137.43, 127.60, 104.25, 43.17. HRMS Calcd for C16H15F2N6 329.13263 [M+H]+, found 329.13230; Anal. (C16H14F2N6) C, H, N.
N,N'-(1,4-phenylenebis(methylene))bis(2,5-difluoropyrimidin-4-amine) (17b)
Starting with 1,4-phenylenedimethanamine (1.36 g, 10 mmol) and 2,4,5-trifluoropyrimidine (2.95 g, 22 mmol), the same procedure to compound 17a furnished the title product 17b (2.77 g, 76%) as a white solid: 1H NMR (600 MHz, DMSO-d6) δ 8.06 (d, J = 3 Hz, 2H), 7.27 (s, 4H), 6.82(bs, 2H), 4.52 (d, J = 6 Hz 4H); 13C NMR (150 MHz,DMSO-d6) δ 157.89, 156.52, 145.00, 143.38, 137.17, 127.39, 43.05; HRMS Calcd for C16H13F4N6 365.11378 [M+H]+, found 365.11325.
N,N'-(1,4-phenylenebis(methylene))bis(2-methoxypyrimidin-4-amine) (18a)
A solution of 17a (1.31 g, 4 mmol) in methanol (20 mL) was treated with thionyl chloride (2.91 mL, 40 mmol) at room temperature. The resulting mixture was stirred, at which time all starting material was consumed. The solvent was removed under reduced pressure. The residue was dissolved in ethyl acetate (20 mL) and washed with aqueous NaOH (10 mL, 1N). The organic layer was separated, dried over anhydrous MgSO4 and concentrated in vacuo. The crude was purified by flash column chromatography (silica gel, MeOH/CH2Cl2, 0→25%) to give the title compound 18a (1.35 g, 96%) as a white solid: mp 187–188 °C (dec); 1H NMR (600 MHz, DMSO-d6) δ 7.89 (t, J = 6Hz, 2H), 7.83 (bs, 2H), 7.26 (s, 4H), 6.16 (bs, 2H), 4.45 (bs, 4H), 3.74 (s, 6H); 13C NMR (150 MHz,DMSO-d6) δ 165.06, 163.81, 155.29, 138.12, 127.46, 100.32, 53.53, 42.92; HRMS Calcd for C18H21N6O2 353.17260 [M+H]+, found 353.17221; Anal. (C18H20N6O2) C, H, N.
N,N'-(1,4-phenylenebis(methylene))bis(5-fluoro-2-methoxypyrimidin-4-amine) (18b)
Starting with compound 17b (1.68 g, 4.6 mmol), the same procedure to compound 18a furnished the title product 18b (1.66 g, 93%) as a white solid: mp 158–160 °C (dec); 1H NMR (600 MHz, DMSO-d6) δ 8.24 (t, J = 6 Hz, 2H), 7.92 (d, J = 3.6 Hz, 2H), 7.26 (s, 4H), 4.50 (d, J = 6 Hz, 4H), 3.72 (s, 6H); 13C NMR (150 MHz,DMSO-d6) δ 160.49, 153.21, 143.34, 139.00, 137.82, 127.31, 54.23, 42.78; HRMS Calcd for C18H19F2N6O2 389.15376 [M+H]+, found 389.15330; Anal. (C18H18F2N6O2) C, H, N.
N-(4-((5-fluoropyrimidin-2-ylamino)methyl)benzyl)-4-(trifluoromethyl)pyrimidin-2-amine (21a)
To a solution of mono boc-protected 1,4-phenylenedimethanamine 19 (11.80 g, 50 mmol) in DMF (50 mL) was added 2-chloro-5-fluoropyrimidine (7.29 g, 55 mmol) and N,N-diisopropylethylamine (30.5 mL, 175 mmol) at room temperature. The resulting mixture was heated to 60 °C and stirred until the starting material disappeared from TLC plate. The reaction mixture was cooled down to ambient temperature, poured into ice water (200 mL) and extracted with ethyl acetate (3 × 50 mL). The combined organic layers were washed with brine, dried over anhydrous MgSO4 and concentrated in vacuo. The crude was purified by flash column chromatography (silica gel, MeOH/CH2Cl2, 0→20%) to give the intermediate 19a (4.25 g, 23 %) as a white solid: 1H NMR (600 MHz, CDCl3) δ 8.15 (s, 2H), 7.30(d, J = 6 Hz, 2H), 7.25 (d, J = 7.8 Hz, 2H), 5.57 (bs, 1H), 4.87 (bs, 1H), 4.57(d, J = 5.4 Hz, 2H), 4.30 (d, J = 5.4 Hz, 2H), 1.46 (s, 9H); 13C NMR (600 MHz, CDCl3) δ 159.43, 156.08, 153.23, 151.59, 145.76 (d, J = 85.2 Hz), 138.27, 127.98, 127.90, 79.73, 45.89, 44.56, 28.61; HRMS Calcd for C17H21FN4O2 333.16485 [M+H]+, found 333.17232.
To a solution of the intermediate 19a (1.66 g, 5.0 mmol) in methanol (15 mL) was added thionyl chloride (0.73 mL, 10.0 mmol) dropwise at 0 °C. The resulting mixture was warmed to ambient temperature and stirred until complete consumption of the starting material from TLC plate. The reaction mixture was diluted with diethyl ether (150 mL). The off-white solid was collected, washed with diethyl ether, dried under vacuum and used in the next step without further purification. The crude solid obtained as above was dissolved in DMF (15 mL), to which was added 2-chloro-4-(trifluoromethyl)pyrimidine (1.03 g, 5.5 mmol) and N,N-diisopropylethylamine (4.4 mL, 25 mmol) at room temperature. The resulting mixture was heated to 90 °C and stirred until the starting material disappeared from TLC plate. The reaction mixture was cooled down to ambient temperature, poured into ice water (60 mL) and extracted with ethyl acetate (3 × 15 mL). The combined organic layers were washed with brine, dried over anhydrous MgSO4 and concentrated in vacuo. The crude was purified by flash column chromatography (silica gel, MeOH/CH2Cl2, 0→20%) to give the intermediate 21a (1.27 g, 67%) as a white solid: mp 164–165 °C (dec); 1H NMR (600 MHz, DMSO-d6) δ 8.57 (d, J = 4.2 Hz, 1H), 8.42(bs, 1H), 8.32 (s, 2H), 7.76 (t, J = 6 Hz,1H), 7.23 (m, 4H), 6.96 (d, J = 5.4 Hz, 1H) 4.49 (bs,1H), 4.44 (bs, 1H), 4.41 (d, J = 6 Hz, 2H); 13C NMR (150 MHz, DMSO-d6) δ 162.21, 161.47, 159.48, 152.43, 150.81, 145.58, 138.68, 137.78, 127.42, 126.92, 121.58, 104.87, 44.24, 43.75; HRMS Calcd for C17H15F4N6 379.12943 [M+H]+, found 379.12905; Anal. (C17H14F4N6) C, H, N.
5-fluoro-N-(4-((2-(methylthio)pyrimidin-4-ylamino)methyl)benzyl)pyrimidin-2-amine (21b)
Starting with compound 19a (1.66 g, 5.0 mmol), the same procedure to compound 21a furnished the title product 21b (1.19 g, 67%) as a white solid: mp 162–163 °C (dec); 1H NMR (600 MHz, DMSO-d6) δ 8.16 (s, 2H), 7.99 (d, J = 3 Hz, 1H), 7.30 (m, 4H), 6.01 (m, 1H), 5.55(bs, 1H), 5.24(bs, 1H), 4.59 (m, 2H), 4.54 (bs, 2H), 2.49 (s, 3H); 13C NMR (150 MHz, DMSO-d6) δ 171.90, 159.44, 155.70, 153.29, 151.64, 145.86, 145.71, 138.74, 137.23, 128.04, 100.00, 45.83, 45.16, 14.16. HRMS Calcd for C17H18FN6S 357.12977 [M+H]+, found 357.12931; Anal. (C17H17FN6S) C, H, N.
(4-(2-ethoxyethyl)piperazin-1-yl)(2-(4-((5-fluoropyrimidin-2-ylamino)methyl)benzylamino)-4-(trifluoromethyl)pyrimidin-5-yl)methanone (21c)
Starting with compound 19a (1.66 g, 5.0 mmol), the same procedure to compound 21c furnished the title product 21c (1.77 g, 63%) as a white solid: m.p. 158–159 °C; 1H NMR (600 MHz, CDCl3) δ 8.18 (s, 2H), 7.32 (s, 4H), 7.27 (s, 1H), 5.94 (bs, 1H), 5.48 (bs, 1H), 4.65 (s, 2H), 4.60 (d, J = 6 Hz, 2H), 3.80 (d, J = 22.2 Hz, 2H), 3.56 (t, J = 6 Hz, 2H), 3.50 (dd, J = 7.2 Hz, 2H), 3.33 (s, 2H), 2.63 (t, J = 5.4 Hz, 2H ), 2.58 (s, 2H), 2.46 (bs, 2H), 1.20 (t, J = 7.2 Hz, 3H); 13C NMR (150 MHz, DMSO-d6) δ 163.50, 161.39, 159.49, 159.11, 152.44, 150.83, 145.64, 138.79, 137.42, 127.51, 126.98, 115.88, 67.69, 65.45, 57.02, 52.91, 52.51, 46.85, 44.24, 43.82, 41.42; HRMS Calcd for C26H29F4N8O2 563.25061 [M+H]+, found 563.24907; Anal. (C26H30F4N8O2) C, H, N.
tert-butyl 4-((2-chloro-5-fluoropyrimidin-4-ylamino)methyl)benzylcarbamate (23)
To a mixture of mono boc-protected 1,4-phenylenedimethanamine 19 (20.0 g, 80 mmol) N,N’-diisopropylethylamine (52 mL, 300 mmol) in DMF (200 mL) was slowly added 2,4-dichloro-5-fluoropyrimidine (14.9 g, 89 mmol). The resulting mixture was heated to 60 °C and stirred for 1 hour. TLC indicated complete consumption of the starting material. The reaction was cooled to ambient temperature, poured into ice water (800 mL), extracted with ethyl acetate (3 × 150 mL). The combined organic layers were washed with brine, dried over anhydrous MgSO4 and concentrated in vacuo. The crude was purified by flash column chromatography (silica gel, MeOH/CH2Cl2, 0→20%) to give the title compound 23 (29.1 g, 94 %) as a white solid: mp 127 °C (dec); 1H NMR (600 MHz, CDCl3) δ 7.89 (d, J = 3Hz, 1H), 7.31-7.27 (m, 4H), 5.51 (s, 1H), 4.49 (s, 1H), 4.66 (d, J = 5.4Hz, 2H), 4.31 (d, J = 6Hz, 2H), 1.45(s, 9H); 13C NMR (150 MHz, DMSO-d6) δ 156.10, 153.59, 146.25, 144.55, 140.09, 140.02, 139.20, 136.26, 128.59, 128.13, 79.84, 44.71, 44.47, 28.60; HRMS Calcd for C17H21ClFN4O2 367.13371 [M+H]+, found 367.13325.
2-chloro-5-fluoro-N-(4-((2-(methylthio)pyrimidin-4-ylamino)methyl)benzyl)pyrimidin-4-amine (24)
Thionyl chloride (11.62 mL, 160 mmol) was added dropwise to a solution of compound 19 (29.1 g, 80 mmol) in methanol (300 mL) at 0 °C. The resulting mixture was warmed to ambient temperature and stirred until complete consumption of the starting material was observed from TLC plate. The reaction mixture was diluted with 700 mL of ether. The off-white solid was collected, washed with diethyl ether, dried under vacuum and used in next step without further purification.
The crude solid obtained as above was dissolved in DMF (200 mL), to which was added 4-chloro-2-thiomethylpyrimidine (14.1 g, 88 mmol) and N,N-diisopropylethylamine (59.2 mL, 340 mmol) at room teperature. The resulting mixture was heated to 90 °C and stirred until the starting material disappeared from TLC plate. The reaction mixture was cooled down to ambient temperature, poured into ice water (1.0 L) and extracted with ethyl acetate (3 × 200 mL). The combined organic layers were washed with brine, dried over anhydrous MgSO4 and concentrated in vacuo. The crude was purified by flash column chromatography (silica gel, MeOH/CH2Cl2, 0→20%) to give the desired product 24 (20.1 g, 66%, 2 steps) as a white solid: 1H NMR (600 MHz, DMSO-d6) δ 8.73 (t, J = 5.4Hz, 1H), 8.09 (d, J = 3.6Hz, 1H), 7.95 (t, J = 6Hz, 1H), 7.86 (bs, 1H), 7.27 (s, 4H), 6.22 (s, 1H), 4.53 (d, J = 60 Hz, 2H), 4.49 (bs, 2H), 2.36 (s, 3H); 13C NMR (150 MHz, DMSO-d6) δ 170.12, 160.67, 153.96, 153.43, 146.06, 144.37, 139.78, 138.30, 136.98, 127.41, 127.39, 101.92, 43.00, 39.92, 13.28; HRMS Calcd for C17H17ClFN6S 391.09080 [M+H]+, found 391.09050.
Preparation of compound 26
To a mixture of compound 24 (5.0 g, 12.8 mmol) in methylene chloride (50 mL) was added 3-chloroperoxybenzoic acid (3.3 g, 19.2 mmol) at a rate to maintain the temperature lower 0 °C. The resulting mixture was stirred at that temperature until no more starting material was detected from TLC. The reaction then was quenched by the addition of saturated aqueous NaHCO3. After separation, the aqueous layer was extracted with additional methylene chloride (2 × 20 mL). The combined organic layers were dried over MgSO4 and concentrated under reduced pressure to afford a mixture of sulfoxide and sulfone as a yellow solid, which was used without additional purification.
The crude mixture obtained as above was dissolved in 1,4-dioxane (50 mL), to which N,N’-diisopropylethylamine (6.8 mL, 38.4 mmol) and 4-(2-aminoethyl)morpholine (2.0 g, 15.4 mmol) was added slowly. The resulting mixture was heated to 95 °C and stirred for 18 h. After cooling to ambient temperature, the reaction mixture was partitioned using ethyl acetate (100 mL) and saturated aqueous NaHCO3 (100 mL). The aqueous layer was extracted with additional ethyl acetate (2 × 20 mL). The combined organic layers were further washed with brine, dried over MgSO4, filtered and concentrated under reduced pressure. The crude was purified by flash column chromatography (silica gel, MeOH/CH2Cl2, 0 → 40%) to give the desired product 26 (1.5 g, 25%, 2 steps) as an off-white solid: mp 68 °C (dec); 1H NMR (300 MHz, CDCl3) δ 7.92(d, J = 2.7 Hz, 1H), 7.85 (d, J = 6.0 Hz, 1H), 7.33 (s, 4H), 5.70 (d, J = 5.4 Hz, 1H), 5.51 (bs, 1H), 5.33 (bs, 1H), 4.97 (bs, 1H), 4.69 (d, J =5.7 Hz, 2H), 4.54 (d, J = 5.4 Hz, 2H), 3.70 (t, J = 4.5 Hz, 4H), 3.45 (dd, J = 5.7, 5.7 Hz, 2H), 2.57-2.45 (m, 6H); 13C NMR (150 MHz, DMSO-d6) δ 162.23, 161.98, 153.42, 146.061, 144.360, 139.92, 139.78, 136.70, 127.31, 127.19, 66.19, 57.64, 53.34, 42.99, 40.04, 37.59; HRMS Calcd for C22H27ClFN8O 473.19804 [M+H]+, found 473.20026; Anal. (C22H26ClFN8O) C, H, N.
Supplementary Material
Acknowlegements
This study was supported by Distinguished Cancer Scientist Development Fund of Georgia Cancer Coalition and NCI R01 CA 109366 (Shim, H.), NIH R01 CA126447 (Grossniklaus, H. E.). We are grateful to Ms. Jessica Paulishen for careful reading of the manuscript and helpful remarks.
Abbreviations
- GPCRs
G protein-coupled receptors
- CXCR4
C-X-C chemokine receptor type 4
- CXCL12
C-X-C chemokine ligand 12
- CCR3
C-C chemokine receptor type 3
- CCR5
C-C chemokine receptor type 5
- SDF-1
stromal-derived factor-1
- cAMP
cyclic adenosine monophosphate
- AIDS
acquired immune deficiency syndrome
- HIV
human immunodeficiency virus
- EC
effective concentrations
- TR-FRET
time resolved-fluorescence resonance energy transfer
- TLC
thin layer chromatography
- H&E
hematoxylin and eosin
- CD
(2-hydroxypropyl)-β-cyclodextrin
- FDA
Food and Drug Administration
- PBS
phosphate buffered saline
Footnotes
Supporting Information Available: Elemental analysis of all final compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Butcher EC, Williams M, Youngman K, Rott L, Briskin M. Lymphocyte trafficking and regional immunity. Adv Immunol. 1999;72:209–253. doi: 10.1016/s0065-2776(08)60022-x. [DOI] [PubMed] [Google Scholar]
- 2.Forster R, Schubel A, Breitfeld D, Kremmer E, Renner-Muller I, Wolf E, Lipp M. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 3.Peled A, Petit I, Kollet O, Magid M, Ponomaryov T, Byk T, Nagler A, Ben-Hur H, Many A, Shultz L, Lider O, Alon R, Zipori D, Lapidot T. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999;283:845–848. doi: 10.1126/science.283.5403.845. [DOI] [PubMed] [Google Scholar]
- 4.Davis CB, Dikic I, Unutmaz D, Hill CM, Arthos J, Siani MA, Thompson DA, Schlessinger J, Littman DR. Signal transduction due to HIV-1 envelope interactions with chemokine receptors CXCR4 or CCR5. J Exp Med. 1997;186:1793–1798. doi: 10.1084/jem.186.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaitseva M, Blauvelt A, Lee S, Lapham CK, Klaus-Kovtun V, Mostowski H, Manischewitz J, Golding H. Expression and function of CCR5 and CXCR4 on human Langerhans cells and macrophages: implications for HIV primary infection. Nat Med. 1997;3:1369–1375. doi: 10.1038/nm1297-1369. [DOI] [PubMed] [Google Scholar]
- 6.Sanchez X, Cousins-Hodges B, Aguilar T, Gosselink P, Lu Z, Navarro J. Activation of HIV-1 coreceptor (CXCR4) mediates myelosuppression. J Biol Chem. 1997;272:27529–27531. doi: 10.1074/jbc.272.44.27529. [DOI] [PubMed] [Google Scholar]
- 7.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 8.De Clercq E. The bicyclam AMD3100 story. Nat Rev Drug Discov. 2003;2:581–587. doi: 10.1038/nrd1134. [DOI] [PubMed] [Google Scholar]
- 9.Zhan W, Liang Z, Zhu A, Kurtkaya S, Shim H, Snyder JP, Liotta DC. Discovery of small molecule CXCR4 antagonists. J Med Chem. 2007;50:5655–5664. doi: 10.1021/jm070679i. [DOI] [PubMed] [Google Scholar]
- 10.Fujii N, Tamamura H. Peptide-lead CXCR4 antagonists with high anti-HIV activity. Curr Opin Investig Drugs. 2001;2:1198–1202. [PubMed] [Google Scholar]
- 11.Liang Z, Wu T, Lou H, Yu X, Taichman RS, Lau SK, Nie S, Umbreit J, Shim H. Inhibition of breast cancer metastasis by selective synthetic polypeptide against CXCR4. Cancer Res. 2004;64:4302–4308. doi: 10.1158/0008-5472.CAN-03-3958. [DOI] [PubMed] [Google Scholar]
- 12.Onuffer JJ, Horuk R. Chemokines, chemokine receptors and small-molecule antagonists: recent developments. Trends Pharmacol Sci. 2002;23:459–467. doi: 10.1016/s0165-6147(02)02064-3. [DOI] [PubMed] [Google Scholar]
- 13.Donzella GA, Schols D, Lin SW, Este JA, Nagashima KA, Maddon PJ, Allaway GP, Sakmar TP, Henson G, De Clercq E, Moore JP. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 14.Fujii N, Nakashima H, Tamamura H. The therapeutic potential of CXCR4 antagonists in the treatment of HIV. Expert Opin Investig Drugs. 2003;12:185–195. doi: 10.1517/13543784.12.2.185. [DOI] [PubMed] [Google Scholar]
- 15.Abdel-Magid AF, Carson KG, Harris BD, Maryanoff CA, Shah RD. Reductive Amination of Aldehydes and Ketones with Sodium Triacetoxyborohydride. Studies on Direct and Indirect Reductive Amination Procedures(1) J Org Chem. 1996;61:3849–3862. doi: 10.1021/jo960057x. [DOI] [PubMed] [Google Scholar]
- 16.Liang Z, Brooks J, Willard M, Liang K, Yoon Y, Kang S, Shim H. CXCR4/CXCL12 axis promotes VEGF-mediated tumor angiogenesis through Akt signaling pathway. Biochem Biophys Res Commun. 2007;359:716–722. doi: 10.1016/j.bbrc.2007.05.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chavance M, Escolano S, Romon M, Basdevant A, de Lauzon-Guillain B, Charles MA. Latent variables and structural equation models for longitudinal relationships: an illustration in nutritional epidemiology. BMC Med Res Methodol. 2010;10:37. doi: 10.1186/1471-2288-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J, Mora A, Shim H, Stecenko A, Brigham KL, Rojas M. Role of the SDF-1/CXCR4 axis in the pathogenesis of lung injury and fibrosis. Am J Respir Cell Mol Biol. 2007;37:291–299. doi: 10.1165/rcmb.2006-0187OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimoto N, Jin H, Liu T, Chensue SW, Phan SH. Bone marrow-derived progenitor cells in pulmonary fibrosis. J Clin Invest. 2004;113:243–252. doi: 10.1172/JCI18847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H, Grossniklaus HE. Combined immunologic and anti-angiogenic therapy reduces hepatic micrometastases in a murine ocular melanoma model. Curr Eye Res. 2006;31:557–562. doi: 10.1080/02713680600718962. [DOI] [PubMed] [Google Scholar]
- 21.Yang J, Zhang B, Lin Y, Yang Y, Liu X, Lu F. Breast cancer metastasis suppressor 1 inhibits SDF-1alpha-induced migration of non-small cell lung cancer by decreasing CXCR4 expression. Cancer Lett. 2008;269:46–56. doi: 10.1016/j.canlet.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 22.Dithmar S, Rusciano D, Grossniklaus HE. A new technique for implantation of tissue culture melanoma cells in a murine model of metastatic ocular melanoma. Melanoma Res. 2000;10:2–8. [PubMed] [Google Scholar]
- 23.Trent JO, Wang ZX, Murray JL, Shao W, Tamamura H, Fujii N, Peiper SC. Lipid bilayer simulations of CXCR4 with inverse agonists and weak partial agonists. J Biol Chem. 2003;278:47136–47144. doi: 10.1074/jbc.M307850200. [DOI] [PubMed] [Google Scholar]
- 24.Mori T, Doi R, Koizumi M, Toyoda E, Ito D, Kami K, Masui T, Fujimoto K, Tamamura H, Hiramatsu K, Fujii N, Imamura M. CXCR4 antagonist inhibits stromal cell-derived factor 1-induced migration and invasion of human pancreatic cancer. Mol Cancer Ther. 2004;3:29–37. [PubMed] [Google Scholar]
- 25.Oonuma T, Morimatsu M, Nakagawa T, Uyama R, Sasaki N, Nakaichi M, Tamamura H, Fujii N, Hashimoto S, Yamamura H, Syuto B. Role of CXCR4 and SDF-1 in mammary tumor metastasis in the cat. J Vet Med Sci. 2003;65:1069–1073. doi: 10.1292/jvms.65.1069. [DOI] [PubMed] [Google Scholar]
- 26.Tamamura H, Hori A, Kanzaki N, Hiramatsu K, Mizumoto M, Nakashima H, Yamamoto N, Fujii N. T140 analogs as CXCR4 antagonists identified as anti-metastatic agents in the treatment of breast cancer. FEBS Lett. 2003;550:79–83. doi: 10.1016/s0014-5793(03)00824-x. [DOI] [PubMed] [Google Scholar]
- 27.Hatse S, Princen K, Bridger G, De Clercq E, Schols D. Chemokine receptor inhibition by AMD3100 is strictly confined to CXCR4. FEBS Lett. 2002;527:255–262. doi: 10.1016/s0014-5793(02)03143-5. [DOI] [PubMed] [Google Scholar]
- 28.Schols D, Este JA, Henson G, De Clercq E. Bicyclams, a class of potent anti-HIV agents, are targeted at the HIV coreceptor fusin/CXCR-4. Antiviral Res. 1997;35:147–156. doi: 10.1016/s0166-3542(97)00025-9. [DOI] [PubMed] [Google Scholar]
- 29.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 30.Richard CL, Blay J. CXCR4 in Cancer and Its Regulation by PPARgamma. PPAR Res. 2008;2008:769413. doi: 10.1155/2008/769413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu A, Yoon Y, Voll R, Williams L, Liang Z, Camp V, Liotta D, Goodman M, Shim H. SNM 57th Annual Meeting; Salt Lake City, Utah. 2010. [Google Scholar]
- 32.Gupta SK, Pillarisetti K, Thomas RA, Aiyar N. Pharmacological evidence for complex and multiple site interaction of CXCR4 with SDF-1alpha: implications for development of selective CXCR4 antagonists. Immunol Lett. 2001;78:29–34. doi: 10.1016/s0165-2478(01)00228-0. [DOI] [PubMed] [Google Scholar]
- 33.Pyo RT, Sui J, Dhume A, Palomeque J, Blaxall BC, Diaz G, Tunstead J, Logothetis DE, Hajjar RJ, Schecter AD. CXCR4 modulates contractility in adult cardiac myocytes. J Mol Cell Cardiol. 2006;41:834–844. doi: 10.1016/j.yjmcc.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao C, Li Y. SDF-1 plays a key role in the repairing and remodeling process on rat allo-orthotopic abdominal aorta grafts. Transplant Proc. 2007;39:268–272. doi: 10.1016/j.transproceed.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 35.Imitola J, Raddassi K, Park KI, Mueller FJ, Nieto M, Teng YD, Frenkel D, Li J, Sidman RL, Walsh CA, Snyder EY, Khoury SJ. Directed migration of neural stem cells to sites of CNS injury by the stromal cell-derived factor 1alpha/CXC chemokine receptor 4 pathway. Proc Natl Acad Sci U S A. 2004;101:18117–18122. doi: 10.1073/pnas.0408258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kajiyama H, Shibata K, Ino K, Nawa A, Mizutani S, Kikkawa F. Possible involvement of SDF-1alpha/CXCR4-DPPIV axis in TGF-beta1-induced enhancement of migratory potential in human peritoneal mesothelial cells. Cell Tissue Res. 2007;330:221–229. doi: 10.1007/s00441-007-0455-x. [DOI] [PubMed] [Google Scholar]
- 37.Moyer RA, Wendt MK, Johanesen PA, Turner JR, Dwinell MB. Rho activation regulates CXCL12 chemokine stimulated actin rearrangement and restitution in model intestinal epithelia. Lab Invest. 2007;87:807–817. doi: 10.1038/labinvest.3700595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang Z, Yoon Y, Votaw J, Goodman MM, Williams L, Shim H. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res. 2005;65:967–971. [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon Y, Liang Z, Zhang X, Choe M, Zhu A, Cho HT, Shin DM, Goodman MM, Chen ZG, Shim H. CXC chemokine receptor-4 antagonist blocks both growth of primary tumor and metastasis of head and neck cancer in xenograft mouse models. Cancer Res. 2007;67:7518–7524. doi: 10.1158/0008-5472.CAN-06-2263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.