Abstract
Objective
Rare mitochondrial mutations cause neurologic disease including ischemic stroke and MRI white matter changes. We investigated whether common mitochondrial genetic variants influence risk of sporadic ischemic stroke and, in patients with stroke, the volume of white matter hyperintensity (WMHV).
Methods
In this multicenter, mitochondrial genome-wide association study (GWAS), 2284 ischemic stroke cases and 1728 controls from the International Stroke Genetics Consortium were genotyped for 64 mitochondrial single nucleotide polymorphisms (SNPs). Imputation resulted in 144 SNPs, which were tested in each cohort and in meta-analysis for ischemic stroke association. A genetic score of all mitochondrial variants was also tested in association with ischemic stroke.
Results
No individual SNP reached adjusted significance in meta-analysis. A genetic score comprised of the summation of contributions from individual variants across the mitochondrial genome showed association with ischemic stroke in meta-analysis (OR = 1.13, p < 0.0001) with minimal heterogeneity (I2 = 0.00). This ischemic stroke score was robust to permutation, and was also associated with WMHV in 792 nested case individuals with ischemic stroke (p = 0.037).
Interpretation
In this mitochondrial GWAS of ischemic stroke, a genetic score comprised of the sum of all common variants in the mitochondrial genome showed association with ischemic stroke. In an independent analysis of a related trait, this same score correlated with WMHV in stroke cases. Despite this aggregate association, no individual variant reached significance. Substantially larger studies will be required to identify precise sequence variants influencing cerebrovascular disease.
Search Terms: mitochondria, stroke, genes, GWAS, white matter
INTRODUCTION
Rare diseases caused by gene mutations encoding mitochondrial proteins often result in neurologic manifestations, including seizures, optic atrophy, white matter MRI abnormalities, and ischemic stroke [1,2]. Because mitochondrial proteins are encoded within both the nuclear and mitochondrial genomes, mutations within either can result in mitochondrial disease, with resultant differences in inheritance patterns. Neurons are highly susceptible to such mutations because of their sensitivity to impaired mitochondrial energy metabolism [3].
Ischemic stroke is a heritable disease [4], and recent data suggest there may be a maternal inheritance pattern – the hallmark of a disorder related to mitochondrial DNA [5]. Traditional genome-wide association study (GWAS) approaches, which have focused on the nuclear genome, have included only limited coverage of sequence variation in the mitochondrial genome. Genome-wide coverage of common variation within the mitochondrial genome requires custom genotyping because the current generation of whole genome arrays does not provide complete coverage of common variation in the mitochondrial genome. While recent GWAS have identified promising loci where common autosomal genetic variants may alter the risk of ischemic stroke [6], no study has yet identified an association between individual common mitochondrial variants and ischemic stroke.
The mitochondrial genome predominantly encodes proteins functioning in the oxidative respiratory chain, thereby impacting energy metabolism [7]. Due to the high energy requirements of endothelial and neurologic tissues, we hypothesized that common variants in the mitochondrial genome may play a role in ischemic stroke, and that any variants that emerge may also influence the burden of white matter hyperintensity volume (WMHV), an established determinant of risk of stroke recurrence and degree of recovery from stroke [8,9].
To explore this hypothesis, we performed a multi-center mitochondrial GWAS (mtGWAS) of ischemic stroke with a panel of single nucleotide polymorphisms (SNPs) covering all common variation in the mitochondrial genome. Four centers within in the International Stroke Genetics Consortium (ISGC, http://www.strokegenetics.org) participated. Association testing was performed for all-cause ischemic stroke. A genetic score-based approach was used to combine effects of all mitochondrial SNPs in analysis, to assess the cumulative impact of multiple mitochondrial variants on respiratory chain function. This genetic score was further applied to WMHV as a biological extension to a related phenotype.
MATERIALS AND METHODS
Samples
Samples were contributed by the Massachusetts General Hospital Ischemic Stroke GWAS (MGH) (n = 1236) [10], the University of Cincinnati Greater Cincinnati and Northern Kentucky Stroke Study (GCNKSS) (n = 749) [11], the Jagiellonian University (Krakow, Poland) Stroke Study (JUSS) (n = 981) [12], and investigators participating in either the Ischemic Stroke Genetics Study or the Siblings With Ischemic Stroke Study (ISGS/SWISS) (n = 1046) [13,14] [Table 1]. All of these participating studies utilized a prospectively enrolled hospital-based case-control study design. Analysis of WMHV was restricted to ischemic stroke cases from individuals in the MGH and ISGS/SWISS datasets (n = 792). Individuals with T2-FLAIR sequences of sufficient quality for quantification on cranial MRIs obtained < 72 hours after admission for stroke were included in the WMHV analysis. All Institutional Review Boards approved the study, and all participants gave informed consent for data collection, genotyping, and analysis of genetic data.
Table 1. Demographic, clinical and radiographic characteristics, stratified by study.
| ISGS/SWISS | MGH | |||
|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |
| No. | 602 | 444 | 575 | 661 |
| Age (yrs.), mean (SD) | 65.7 (13.5) | 67.9 (14.8) | 65.6 (15.8) | 68.9 (9.3) |
| Sex (% male) | 53 | 59 | 57 | 51 |
| HTN (%) | 67 | 37 | 52 | 33 |
| DM II (%) | 23 | 11 | 26 | 18 |
| Smoking (% current) | 20 | 8.6 | 25 | 20 |
| No. WMH | 189 | -- | 603* | -- |
| WMH volume (cc) | 5.3 (2.6–11.1) | -- | 10.2 (4.8–22.9) | -- |
| TOAST Stroke Subtypes | ||||
| Cardioembolic | 150( 0.25) | -- | 230 (0.40) | -- |
| Large Artery | 114 (0.19) | -- | 104 (0.18) | -- |
| Small Vessel | 103 (0.17) | -- | 63 (0.11) | -- |
| Other Determined Etiology | 24 (0.04) | -- | 132 (0.23) | -- |
| Undetermined Etiology | 211 (0.35) | -- | 46 (0.08) | -- |
| GCNKSS | JUSS | |||
| Cases | Controls | Cases | Controls | |
| No. | 459 | 290 | 648 | 333 |
| Age (yrs.), mean (SD) | 70.4 (12.9) | 62.0 (14.9) | 64.2 (17.1) | 64.6 (15.0) |
| Sex (% male) | 52 | 47 | 51 | 49 |
| HTN (%) | 70 | 49 | 48 | 42 |
| DM II (%) | 36 | 16 | 17 | 22 |
| Smoking (% current) | 26 | 20 | 27 | 18 |
| TOAST Stroke Subtypes | ||||
| Cardioembolic | 105 (0.23) | -- | -- | -- |
| Large Artery | 69 (0.15) | -- | -- | -- |
| Small Vessel | 92 (0.20) | -- | -- | -- |
| Other Determined Etiology | 9 (0.02) | -- | -- | -- |
| Undetermined Etiology | 184 (0.40) | -- | -- | -- |
Individuals analyzed for white matter hyperintensity volume from MGH are greater than the total analyzed ischemic stroke cases because quality control procedures were implemented independently for the two cohorts.
All included individuals are of self-reported European or European-American ancestry. DM II = Type II Diabetes Mellitus, GCNKSS = Greater Cincinnati Northern Kentucky Stroke Study, HTN = Hypertension, ISGS/SWISS = Ischemic Stroke Genetic Study/Siblings with Ischemic Stroke Study, JUSS = Jagiellonian University Stroke Study, MGH = Massachusetts General Hospital Ischemic Stroke Genome-wide Association Study, SD = Standard Deviation, WMH = White Matter Hyperintensity
Patient selection and definitions
Case and control recruitment and phenotype ascertainment were performed in each cohort as described previously [10–14]. Cases were all consenting adults presenting to the emergency department of each participating institution for evaluation of acute stroke. For the purposes of this study, individuals were defined as ischemic stroke cases in the presence of (1) a radiographically proven infarct associated with an appropriate clinical syndrome, or (2) a fixed neurologic deficit persisting > 24 hours, consistent with a vascular event, without evidence of demyelination or non-vascular disease. All samples within the ISGS/SWISS, GCNKSS, and MGH datasets were assessed and Trial of Org 10172 in Acute Stroke Treatment (TOAST) stroke subtypes [15] were assigned by a neurologist. As in the original classification, infarct area, but not white matter burden, was considered in assigning TOAST subtype diagnosis. All controls were recruited from the same populations as the cases, and were confirmed to be free of stroke symptoms by interview and review of medical records. All clinical information, demographics, and comorbidities were abstracted prospectively by patient or proxy interview, and/or supplemented through medical chart review.
Genotyping of common mitochondrial variants
Common mitochondrial variants were genotyped according to a validated protocol, with selection of 144 SNPs capturing 100% of variants with MAF > 0.01 in Europeans [16]. A total of 64 tagging SNPs were selected to capture all common variation outside the hyper-variable D-loop, and genotyping was performed with use of a Sequenom platform (San Diego, CA, USA). Haplotype-based imputation was used to capture the remaining 80 variants (all r2 ≥ 0.8 with directly-genotyped SNPs). . All genetic analysis was performed using PLINK v1.07 [17].
Mitochondrial genomic quality control
Quality control of genotyped individuals included filters for missingness by individual > 0.1, missingness by SNP > 0.1, and minor allele frequency (MAF) < 0.005 [Supplementary Figure S1]. The MAF filter was set after empiric assessment of MAF in the current study population and in previously published data [16].
Haplogroup assignment and meta-analysis
European haplogroups (H1, H2, I, J, K, T, U, pre-HV, and W/X) were assigned according to previously published methods [18]. A total of 1074 complete sequences from the 10 most common European haplogroups were downloaded from mtDB (http://www.genpat.uu.se/mtDB/). From these sequences, the genotypes at tagging SNP loci were determined and used as predictors in a linear discriminant function analysis in R v2.10.0. The accuracy of prediction of haplogroup was determined using a bootstrap cross-validation approach, and determined to be >98.5%.
Population structure assessment and control
Only individuals of European ancestry were analyzed. Principal component analysis (PCA) has been identified as a robust means to control confounding by population stratification [19,20]. Population structure was assessed by performing PCA on all SNPs passing quality control. PCA was performed using the EIGENSOFT v3.0 software package [19] and confirmed using Multidimensional Scaling in PLINK v1.07. Principal components (PCs) 1 – 10 were extracted for each individual, and PCs were added until no additional reduction in mitochondrial genomic inflation factor (mtGIF) could be achieved (PC1–5 for all analyses) [Figure S2].
Study power
We determined statistical power for identification of association between analyzed variants and ischemic stroke at α = 0.00035 (144 independent tests). Power calculations specific to mitochondrial analysis were performed using a simulation-based method published in a prior study [18]. Power estimates were by Monte Carlo simulation, with power differences averaged over the observed distribution of MAF. Power was calculated for effect sizes of 1.2, using a haploid model with MAF 0.05, and disease prevalence of 0.03 [21]. Post-hoc power calculations were performed similarly, using assumptions of MAF and effect size based on data from meta-analysis results. Power estimation for the haplogroup-based meta-analysis was performed according to published methods using Monte Carlo permutation testing [22]. Because each haplogroup represents a variable proportion of the study population, detectable odds ratios at power 0.80 were assigned independently for each haplogroup, assuming α = 0.05.
Genetic association analysis by study cohort and meta-analysis
Genotype data were analyzed using an allele-based model, with odds ratio (OR) expressing the effect of the reference allele. All analyses included age, sex, and PC1–5 as covariates. Statistical significance was defined as Bonferroni-adjusted p-value for 144 independent tests. Association tests by study were combined in meta-analysis with a conservative, random-effects pooling method (DerSimonian-Laird), using R v2.10.0. SNPs that did not pass quality control filters in at least 3 studies were excluded, leaving 121 SNPs. Reference alleles were checked to ensure concordance, and MAFs were inspected by cohort.
Haplogroup-based analysis
European haplogroups were tested in a case-control logistic regression for ischemic stroke, using age and sex as covariates. Pooled results from each study were then used in a random-effects meta-analysis accounting for site. For this haplogroup-based analysis only, an additional cohort of 481 cases and 537 controls of European ancestry from the University of Maryland Genetics of Early Onset Stroke (GEOS) study were incorporated into the meta-analysis. These individuals had sufficient genotyping for haplogroup assignment, but lacked the comprehensive mitochondrial genotyping needed for the remainder of the study.
Genetic score analysis
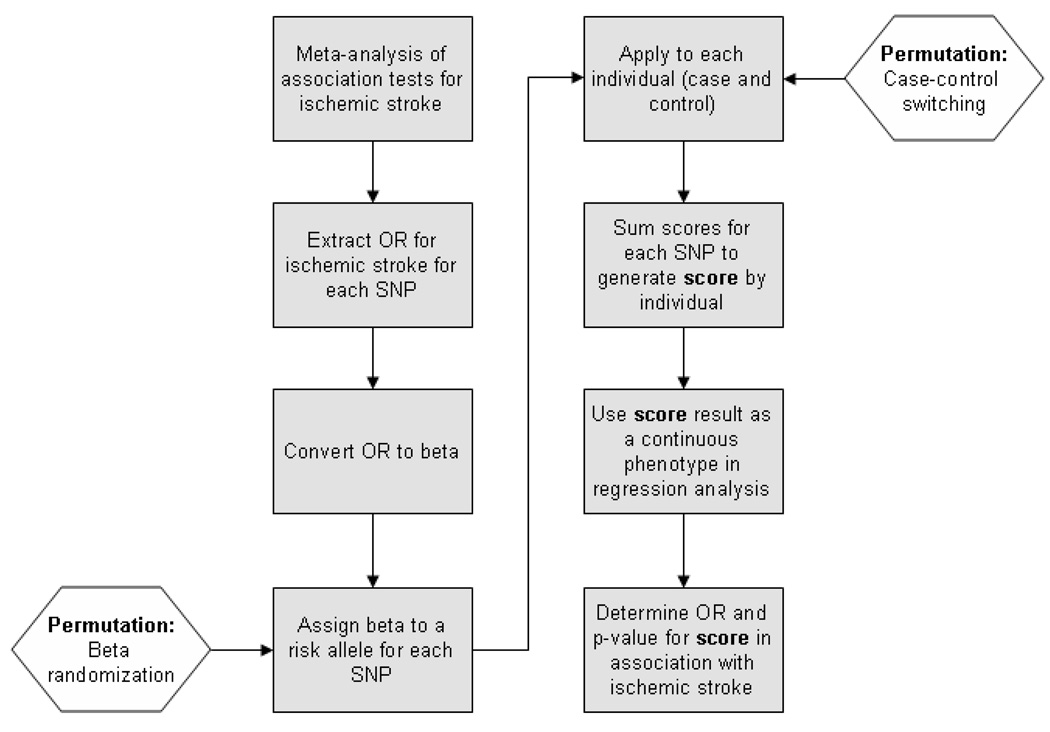
Combined effects of all mitochondrial SNPs were evaluated using a score-based method previously used to assess the cumulative effect of loci affecting lipid levels, risk for myocardial infarction, blood pressure, and schizophrenia [23–26]. Briefly, meta-analysis results were extracted for SNPs passing quality control filters in all 4 cohorts, resulting in 90 SNPs available across all 4 studies. The stroke risk conferred by each SNP (expressed as a beta coefficient) was then applied to each individual in the dataset, with the sum of all SNP beta coefficients resulting in the “score” for each individual [Figure 1]. This genetic score was then divided into quintiles and used as the dependent variable in a logistic regression for ischemic stroke, with age, sex, site, and PC1-PC5 used as covariates. Because only one test was performed in this analysis, Bonferroni correction was not necessary. Results from each cohort were meta-analyzed using the meta library for R v. 2.10.0. Power estimation for association with the genetic score was performed using R (http://www.r-project.org), v2.10.0, assuming an effect size of 1.20 (per risk score quintile) for ischemic stroke, and explanation of 1% of variance for WMHV.
Figure 1. Flowchart of genetic score-based analysis pipeline.
Boxes represent the steps for generation and application of the genetic score to test associations with ischemic stroke. Hexagons represent the locations where permutation algorithms can be applied to test the score generated against the null-hypothesis. OR = Odds Ratio, SNP = Single Nucleotide Polymorphism.
Permutation
To test the results of the genetic score analysis against the null hypothesis and limit the possibility of false positive association, two forms of permutation were employed using R v2.10.0. In the first method, case and control assignment was randomized in 10,000 permutations, betas and scores were re-calculated, and tests for association with the permuted case/control status were performed.
In the second method, the beta coefficients used to generate the score were assigned randomly to different SNPs in 10,000 iterations, scores were re-calculated, and these scores were then tested against the original case/control status. For both methods, the empiric p-value reported is defined as the number of times the observed test-statistic in permutation was superior to the test-statistic of the original dataset, divided by the number of permutations (10,000).
Quantification and analysis of WMHV
MRI scans performed within 72 hours of stroke onset were converted from DICOM to Analyze format using MRIcro software (University of Nottingham School of Psychology, Nottingham, UK, www.mricro.com) for computer-assisted determination of WMHV. All WMHV measurements were performed at MGH. Using MRIcro, a region-of-interest map of supratentorial WMHV was created by signal intensity thresholding followed by manual editing as necessary [27,28]. Axial T2 fluid attenuated inversion recovery (FLAIR) sequences were used to create the WMH maps. Areas of signal change from previous infarctions were not considered WMH and the corresponding brain regions were masked. In order to correct WMHV for head size we used the sagittal midline cross-sectional intracranial area (ICA) as a surrogate measure of the intracranial volume, according to a previously validated method [29]. The intracranial cavity was manually outlined on the two most mid-sagittal T1-weighted slices using MRIcro software, and the areas were averaged. All MRI measurements were performed centrally by readers blinded to clinical data, including functional outcome and TOAST subtype assignment.
The genetic score for each individual developed using the ischemic stroke phenotype was divided into quintiles to improve normality of distribution, and applied as a dependent variable in linear regression of WMHV. Additional covariates included age, sex, site, and PC1–5. Results were reported as a coefficient and p-value for association between the ischemic stroke genetic score and WMHV.
WMHV permutation
We performed permutation testing of the association between the ischemic stroke mitochondrial genetic score and WMHV through 10,000 permutations with randomization of the WMHV phenotype. Permutation testing was performed in R v2.10.0. Permutation empiric p-value is reported as the number of times the observed test-statistic in permutation was superior to the test-statistic of the original dataset, divided by the number of permutations (10,000).
RESULTS
Mitochondrial genetic quality control
5,912 individuals from the four cohorts had mitochondrial genotype information available. After quality control filtering, 2284 cases and 1728 controls remained for analysis of ischemic stroke. Filtering of mtGWAS data was applied to the initial pool of 64 SNPs, and after imputation, 109 – 132 SNPs remained for analysis within the four cohorts [Supplementary Figure S1]. Of these, 121 SNPs were common to at least 3 cohorts, and 90 SNPs were common to all four cohorts [Supplementary Table S1]. Population stratification was assessed by calculating the mtGIF for each study after incorporation of PCs [Supplementary Figure S2]. In pooled analysis of the 90 shared SNPs, the mtGIF was 1.00 after correction for PC1-PC5.
Statistical Power
Given our sample size, our study had power of 0.99 to detect an effect size of 2.0, given an expected MAF of 0.05 and disease prevalence of 0.03. Our power fell to 0.04 to detect effect sizes of 1.2. To achieve power of 0.8 to detect effect sizes of 1.2 at this MAF would require 13,638 cases with matching controls. For association between individual haplogroups and ischemic stroke, detectable odds ratios at power 0.80 varied from 1.18 for haplogroup H to 2.80 for haplogroup I as risk alleles, and 0.84 for haplogroup H to 0.36 to haplogroup I as protective alleles [Supplementary Table S2]. For analysis of WMHV, our genetic score analysis had 0.85 power to explain 1% of the variance in WMHV, assuming for WMHV α = 0.05.
Study results by cohort and meta-analysis
No individual SNP in any cohort met the pre-specified significance threshold of p < 0.00035. In meta-analysis of all cohorts, again no individual variant reached statistical significance [Table 2]. While there was significant heterogeneity (I2 > 40%) for a minority of SNPs (n = 14), none of the leading association results showed significant heterogeneity (all I2 < 40%). Accordingly, MAF concordance between cohorts was excellent (average Spearman correlation coefficient: 0.92, range 0.90 – 0.95, all p < 0.0001).
Table 2. Meta-analysis of leading SNPs in association with ischemic stroke.
Results for the SNPs with the lowest unadjusted p-values in meta-analysis of all four cohorts, after exclusion of SNPs genotyped adequately in fewer than 3 cohorts.
| SNP | OR | p | I2 | MAF | |||
|---|---|---|---|---|---|---|---|
| ISGS/SWISS | MGH | GCNKSS | JUSS | ||||
| mt930 | 0.81 | 0.03 | 0.00 | 0.059 | 0.045 | 0.046 | 0.043 |
| mt5147 | 0.80 | 0.03 | 0.00 | 0.057 | 0.045 | 0.046 | 0.043 |
| mt10915 | 1.35 | 0.08 | 0.00 | 0.012 | 0.013 | 0.017 | 0.013 |
| mt4580 | 0.83 | 0.11 | 0.34 | 0.032 | 0.032 | 0.039 | 0.028 |
| mt5426 | 0.80 | 0.13 | 0.00 | 0.015 | 0.018 | 0.017 | N/A |
| mt6221 | 0.83 | 0.15 | 0.00 | 0.015 | 0.020 | 0.012 | 0.019 |
| mt12633 | 1.33 | 0.15 | 0.38 | 0.030 | 0.020 | 0.019 | N/A |
| mt13879 | 0.78 | 0.15 | 0.00 | 0.009 | 0.013 | 0.010 | 0.010 |
| mt14233 | 0.86 | 0.18 | 0.00 | 0.095 | 0.072 | 0.086 | 0.075 |
| mt11812 | 0.86 | 0.18 | 0.00 | 0.095 | 0.072 | 0.086 | 0.077 |
GCNKSS = Greater Cincinnati Northern Kentucky Stroke Study, I2 = % of effect size attributable to meta-analysis heterogeneity, ISGS/SWISS = Ischemic Stroke Genetic Study/Siblings with Ischemic Stroke Study, JUSS = Jagiellonian University Stroke Study, MAF = Minor Allele Frequency, MGH = Massachusetts General Hospital Ischemic Stroke Genome-wide Association Study, N/A = SNP not available in this cohort, OR = Odds Ratio, SNP = Single Nucleotide Polymorphism
We tested all European haplogroups in meta-analysis (MGH, ISGS/SWISS, GCNKSS, JUSS, and GEOS cohorts) for association with ischemic stroke. No significant link between any particular haplogroup and ischemic stroke was identified [Table 3]. In particular, using our larger cohort and rigorous control for population stratification [20], we failed to replicate the findings of association with H1 and pre-HV haplogroups [30] and K haplogroup [31] as previously published. Using our power calculations [Supplementary Table S2], we had power > 0.99 to detect these associations at the effect sizes reported [30,31].
Table 3. Results from haplogroup association analysis for ischemic stroke.
| Haplogroup | Frequencies | OR | 95% CI | p | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MGH | ISGS-SWISS | GCNKSS | JUSS | GEOS | |||||||||
| Cases | Ctrls | Cases | Ctrls | Cases | Ctrls | Cases | Ctrls | Cases | Ctrls | ||||
| H | 0.40 | 0.41 | 0.41 | 0.42 | 0.43 | 0.42 | 0.36 | 0.36 | 0.40 | 0.42 | 0.98 | 0.88 – 1.09 | 0.65 |
| H1 | 0.13 | 0.15 | 0.14 | 0.15 | 0.15 | 0.14 | 0.12 | 0.11 | 0.15 | 0.15 | 0.97 | 0.91 – 1.04 | 0.41 |
| H2 | 0.27 | 0.25 | 0.26 | 0.28 | 0.28 | 0.27 | 0.24 | 0.25 | 0.25 | 0.27 | 1.05 | 0.90 – 1.23 | 0.52 |
| J | 0.09 | 0.09 | 0.10 | 0.09 | 0.09 | 0.10 | 0.08 | 0.08 | 0.10 | 0.09 | 1.02 | 0.99 – 1.04 | 0.15 |
| K | 0.07 | 0.06 | 0.06 | 0.06 | 0.06 | 0.07 | 0.05 | 0.06 | 0.05 | 0.06 | 1.03 | 0.90 – 1.20 | 0.68 |
| preHV | 0.05 | 0.06 | 0.05 | 0.06 | 0.07 | 0.04 | 0.06 | 0.07 | 0.07 | 0.06 | 0.86 | 0.82 – 1.08 | 0.39 |
| T | 0.09 | 0.09 | 0.08 | 0.09 | 0.08 | 0.10 | 0.09 | 0.09 | 0.10 | 0.08 | 0.97 | 0.86 – 1.10 | 0.66 |
| U | 0.15 | 0.15 | 0.14 | 0.15 | 0.15 | 0.16 | 0.19 | 0.18 | 0.14 | 0.16 | 1.00 | 0.81 – 1.22 | 0.99 |
| WX | 0.03 | 0.04 | 0.04 | 0.04 | 0.05 | 0.04 | 0.03 | 0.04 | 0.03 | 0.04 | 1.09 | 0.96 – 1.23 | 0.18 |
| I | 0.03 | 0.02 | 0.02 | 0.03 | 0.02 | 0.02 | 0.03 | 0.02 | 0.04 | 0.04 | 1.02 | 0.95 – 1.10 | 0.68 |
CI = confidence interval, Ctrls = controls, GCNKSS = Greater Cincinnati Northern Kentucky Stroke Study, GEOS = University of Maryland Genetics of Early Onset Stroke Study, ISGS/SWISS = Ischemic Stroke Genetic Study/Siblings with Ischemic Stroke Study, JUSS = Jagiellonian University Stroke Study, MGH = Massachusetts General Hospital Ischemic Stroke Genome-wide Association Study, OR = odds ratio
Post-hoc power calculation
In examination of all SNPs in meta-analysis, the median MAF was 0.032, with average effect sizes of 1.09 for risk alleles and 0.92 for protective alleles. We recalculated study power under these assumptions, both for risk and protective variants. Average study power for risk alleles was found to be 0.002, while power for protective alleles was 0.0017. To achieve power of 0.80 with this MAF and effect size would require over 80,000 cases with matching controls.
Mitochondrial genetic score results in ischemic stroke
We performed a genetic score-based analysis in order to assess the possible effects of multiple mitochondrial variants acting in concert to influence respiratory chain function. Using the effect sizes generated in meta-analysis for SNPs passing quality control filters in all cohorts [Supplementary Table S1], we computed genetic score values for all individuals [Figure 1]. These values were then used in logistic regression for association with ischemic stroke, both in each cohort as well as in meta-analysis of all cohorts [Figure 2, Table 4]. This genetic score analysis had power of 0.99 to detect an effect size of 1.20 for ischemic stroke per risk score quintile. Our mitochondrial genetic score was strongly associated with ischemic stroke in meta-analysis (OR 1.13, 95% confidence interval 1.06 – 1.20, p < 0.0001), and showed very little between-study heterogeneity (I2 = 0%). The odds ratios shown in Table 4 represent the risk of ischemic stroke for each quintile increase of the mitochondrial genetic score. This result was robust to permutation by both case-control switching (p = 0.0012) and beta randomization (p = 0.0023).
Figure 2. Forest plot of meta-analysis of association between mitochondrial genetic score and ischemic stroke.
GCNKSS = Greater Cincinnati Northern Kentucky Stroke Study, Het p-value = heterogeneity p-value, I2 = % of effect size attributable to meta-analysis heterogeneity, ISGS/SWISS = Ischemic Stroke Genetic Study/Siblings with Ischemic Stroke Study, JUSS = Jagiellonian University Stroke Study, MGH = Massachusetts General Hospital Ischemic Stroke Genome-wide Association Study
Table 4. Mitochondrial genetic score-based analysis by cohort and in meta-analysis.
| Study | Cases | Controls | OR | 95% CI | P* |
|---|---|---|---|---|---|
| MGH | 575 | 661 | 1.13 | 1.02 – 1.26 | 0.021 |
| ISGS/SWISS | 602 | 444 | 1.18 | 1.05 – 1.32 | 0.006 |
| GCNKSS | 459 | 290 | 1.10 | 0.95 – 1.28 | 0.189 |
| JUSS | 648 | 333 | 1.09 | 0.97 – 1.23 | 0.059 |
| Meta-Analysis | Cases | Controls | OR | 95% CI | p |
| Fixed Effects | 2284 | 1728 | 1.13 | 1.06 – 1.20 | < 0.0001 |
| Random Effects | 2284 | 1728 | 1.13 | 1.06 – 1.20 | < 0.0001 |
Unadjusted p-values shown.
CI = Confidence Interval, GCNKSS = Greater Cincinnati Northern Kentucky Stroke Study, ISGS/SWISS = Ischemic Stroke Genetic Study/Siblings with Ischemic Stroke Study, JUSS = Jagiellonian University Stroke Study, MGH = Massachusetts General Hospital Ischemic Stroke Genome-wide Association Study, OR = Odds Ratio. The OR shown for each study and in meta-analysis represent the risk of ischemic stroke associated with each quintile increase in the mitochondrial genetic score.
Mitochondrial ischemic stroke genetic score in WMHV
Using 792 ischemic stroke cases from the MGH and ISGS/SWISS cohorts, we determined whether the mitochondrial genetic score for ischemic stroke correlated with increasing WMHV. This independent case-only analysis demonstrated a significant (p = 0.037) association between the aggregated risk SNPs for ischemic stroke, and the quantity of WMHV, with a coefficient of 0.0665 (SE = 0.032). This coefficient represents the log-transformed WMHV attributable to each quintile of the mitochondrial ischemic stroke genetic score. Other predictors of WHMV in the regression included age (p < 0.001), and site (p < 0.001). To investigate the role of the site in the determination of WMHV, we inserted all TOAST subtype classifications into the regression model. None of the TOAST subtypes demonstrated a strong association (all p = NS), but the inclusion of these subtypes in the regression model reduced the significance of site in the association with WMHV, decreasing the p-value from < 0.001 to 0.02 with an effective halving of the effect size. The association between the ischemic stroke mitochondrial genetic score and WMHV was also robust to permutation via phenotype randomization, with p = 0.009.
DISCUSSION
Although our analysis was underpowered to detect associations for individual mitochondrial variants, our data provide evidence for an aggregate effect of common mitochondrial variation on risk of ischemic stroke. The score-based approach used to demonstrate this effect has been applied successfully to resolve genetic associations for lipid profiles, myocardial infarction, and blood pressure, as well as other complex traits [20–22]. While in these conditions the score was comprised of SNPs with genome-wide significance, in schizophrenia a score built from multiple non-significant SNPs with small effect sizes has been validated in multiple independent cohorts [26]. Our approach parallels that taken by the schizophrenia researchers, although we are not able to present replication in an independent data set. Our results were derived from meta-analysis of four independent cohorts, the results of which were all quite consistent with one another and without significant heterogeneity. Although this does not constitute formal replication, the correspondence in effect size and lack of between-study heterogeneity in these independent cohorts is suggestive of generalizability. In addition, the genetic score derived from these cohorts was also robust to two different forms of permutation, in which our results were tested repeatedly against the null hypothesis. Finally, we have applied our ischemic stroke genetic score to WMHV in an independent case-only sample, showing that this score comprised of mitochondrial variants that influence stroke risk also has an effect on the burden of white matter disease in individuals with ischemic stroke. This biologic extension to a related phenotype of ischemic stroke further strengthens the robustness of our findings.
We did not identify any SNP of large effect (OR > 2), despite sufficient power to detect such variants. Our analysis was underpowered to detect variants of small or moderate effects. Because we have not identified an association between ischemic stroke and any specific common variants, mitochondrial genetic testing for common alleles is not warranted at this time. Additionally, because our haplogroup-based analysis does not demonstrate a robust association between individual haplogroups and ischemic stroke, our results do not support haplogroup assignment for clinical prediction. However, future DNA sequencing studies building on our demonstration that common variants have an aggregate influence on risk of ischemic stroke may lead to discovery of individual rare variants with clinical implications for risk stratification in stroke.
This analysis provides concrete examples of the challenges particular to GWAS of mitochondrial variants. Autosomal GWAS studies often assume MAFs > 0.05 for common variants, while our observed median mitochondrial MAF was 0.032, with some variants displaying MAF as low as 0.005. These low observed MAFs greatly limit power to detect differences between groups, because larger sample sizes must be assembled to recruit rarer risk alleles. Furthermore, the mitochondrial genome is haploid, making it impossible for homozygous risk alleles to contribute additional signal in an additive genetic model. Finally, the low effect sizes seen in meta-analysis also limit power to detect novel SNP associations. These constraints altered our power estimates greatly in post hoc analysis.
The majority of the known genes in the mitochondrial genome subserve respiratory chain functions [7]. It has been demonstrated that multiple rare mitochondrial variants can work in concert to cause disease [32], possibly through collective impairment of the respiratory chain [33]. This provides a strong biologic rationale for the application of a genetic-score approach, as this technique allows assessment of the aggregate effect of all mitochondrial SNPs. Under the genetic score paradigm, SNPs that do not contribute to disease association only introduce random associations to the model, favoring the null hypothesis. For this reason, and because our genetic score was robust to two independent forms of permutation, our data provide strong support for a role of common mitochondrial variants in ischemic stroke. Of note, our genetic score was comprised of all available genetic variants, and was therefore an unbiased approach to disease association. The selection of individual candidates for inclusion in a genetic score could introduce biases that limit replicability.
Our mtGWAS was performed using an exhaustive panel of SNPs, encompassing all variation with MAF > 1% in individuals of European ancestry. While mitochondrial SNPs are available on commercially-available genotyping platforms, they do not capture all known common variation. Even with imputation, these platforms cannot achieve full coverage of all common variants, limiting the utility of pooling data from the exhaustive mitochondrial panel and standard GWAS platforms for analysis. Future mitochondrial association studies will require continued use of targeted exhaustive panels [16] to minimize risk of false negative results due to insufficient mitochondrial genome coverage.
Our study has limitations. Despite amassing a large, multi-center cohort of ischemic stroke cases and controls, we did not have power to detect individual SNP associations for the effect sizes and MAF observed in meta-analysis. To be adequately powered to identify associations with individual SNPs would require a cohort in excess of 80,000 cases with matching controls. Even introducing additional controls to our study to reach a 4:1 (control:case) ratio would only increase our power from 0.002 to 0.004. Our results cannot exclude a stroke subtype-specific effect for mitochondrial variants. Restricting sample size in sub-analysis of stroke subtypes would further erode power, decreasing the probability of detecting positive associations even for substantial subtype specificity. This study focused solely on inherited common variation in SNPs outside the D-loop. Mutations within the D-loop or insertion/deletion mutations would require a different study design to detect. We did not model heteroplasmy in our analysis, but this phenomenon would theoretically only erode power to detect associations for common inherited variants. Most importantly, we do not have an additional cohort of individuals in which to replicate our findings, and only after replication can our results be considered robust. While the fact that our results are robust to permutation supports our conclusions, our analysis is still based on a single data-set. However, the observation that the genetic score derived from ischemic stroke is also associated with WMHV in a nested case-only cohort represents a biologic extension of our results in what must be considered an independent phenotype assessed only in stroke cases. It is important to note that this result is insufficient to constitute a demonstration of an effect of specific common mitochondrial variants on WMHV. Rather, it shows that the common variants that increase the risk of ischemic stroke seem to also play some as-yet undefined role in the genetics of WMHV. Further research will be needed to independently assess the role of common mitochondrial variants in the phenotype of WMHV.
This large, multi-center mitochondrial GWAS of ischemic stroke was underpowered to detect individual variants. However, we have shown that some association does exist between common mitochondrial variants and stroke risk. Further studies are necessary to replicate our findings, and substantially larger sample sizes will be required to identify specific variants. It is possible that common variants in the mitochondrial genome conferring a small increase in stroke risk are in linkage with rare variants conferring much larger risk. Detection of these rare variants, through sequencing of the mitochondrial genome, may yield important new information on the role of mitochondria on ischemic stroke and other neurologic diseases.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to acknowledge the many study coordinators and technologists at all sites that assisted in the collection and analysis of samples for the current study. The Deane Institute for Integrative Study of Atrial Fibrillation and Stroke, Marriott Disease Risk and Regenerative Medicine Initiative Award in Individualized Medicine, Marriott Mitochondrial Fund, the Greater Cincinnati Foundation Grant, and the Myron and Jane Hanley Award in Stroke Research also contributed funding for this study. We would additionally like to acknowledge the GENEVA Consortium, including the CIDR Genotyping and Data Coordinating Center at the University of Washington, Seattle, for their support of the University of Maryland GEOS Study.
FUNDING AND SUPPORT
American Heart Association/Bugher Foundation Centers for Stroke Prevention Research (0775010N), Deane Institute for Integrative Study of Atrial Fibrillation and Stroke, Marriott Disease Risk and Regenerative Medicine Initiative Award in Individualized Medicine, Marriott Mitochondrial Fund, National Institute for Neurologic Disorders and Stroke (R01NS059727, R01NS42733, P50NS051343-04, K23NS064052, K24NS056207, R01NS042147, U54NS057405, R01NS039987, R01NS36695, R01NS30678), Greater Cincinnati Foundation Grant, Myron and Jane Hanley Award in Stroke Research, Fulbright Foundation, American Association of University Women, and the State Committee for Scientific Research (Poland) (2 PO5 CO1226).
Footnotes
Reprints: None Requested
STATEMENT OF CONTRIBUTION
Manuscript Preparation: Christopher D. Anderson (co-leader), Alessandro Biffi (co-leader), Jonathan Rosand. Data Acquisition: Rosanna Rahman, Natalia Rost, Yu-Ching Cheng, Lynelle Cortellini. Manuscript Revision: Rosanna Rahman, Lynelle Cortellini, Natalia Rost, Paul I.W. de Bakker, Daniel Woo, Owen A. Ross, Bradford B. Worrall, Robert D. Brown, Jr, Thomas G. Brott, Agnieszka Slowik, Jeremiasz M. Jagiella, Braxton D. Mitchell, John W. Cole, Steven J. Kittner, James F. Meschia, Joseph P. Broderick, Karen L. Furie, Steven M. Greenberg, Richa Saxena. Data Analysis: Christopher D. Anderson (co-leader), Alessandro Biffi (co-leader), Rosanna Rahman, Paul I.W. de Bakker, Yu-Ching Cheng, Richa Saxena. Study Management: Jonathan Rosand, Karen L. Furie, Joseph P. Broderick, Daniel Woo, Agnieszka Slowik, Bradford B. Worrall, Robert D. Brown, Jr, Thomas G. Brott, Braxton D. Mitchell, John W. Cole, Steven J. Kittner, James F. Meschia, Owen Ross.
REFERENCES
- 1.DiMauro S, Schon EA. Mitochondrial disorders in the nervous system. Annu Rev Neurosci. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- 2.Yu-Wai-Man P, Griffiths PG, Hudson G, Chinnery PF. Inherited mitochondrial optic neuropathies. J Med Genet. 2009;46(3):145–158. doi: 10.1136/jmg.2007.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholls DG. Oxidative stress and energy crises in neuronal dysfunction. Ann N Y Acad Sci. 2008;1147:53–60. doi: 10.1196/annals.1427.002. [DOI] [PubMed] [Google Scholar]
- 4.Flossmann E, Schulz UG, Rothwell PM. Systematic review of methods and results of studies of the genetic epidemiology of ischemic stroke. Stroke. 2004;35(1):212–227. doi: 10.1161/01.STR.0000107187.84390.AA. [DOI] [PubMed] [Google Scholar]
- 5.Touzé E, Rothwell PM. Sex differences in heritability of ischemic stroke: a systematic review and meta-analysis. Stroke. 2008;39(1):16–23. doi: 10.1161/STROKEAHA.107.484618. [DOI] [PubMed] [Google Scholar]
- 6.Ikram MA, Seshadri S, Bis JC, et al. Genomewide association studies of stroke. N Engl J Med. 2009;360(17):1718–1728. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lertrit P, Kapsa RM, Jean-Francois MJ, et al. Mitochondrial DNA polymorphism in disease: a possible contributor to respiratory dysfunction. Hum Mol Genet. 1994;3(11):1973–1981. doi: 10.1093/hmg/3.11.1973. [DOI] [PubMed] [Google Scholar]
- 8.Arsava EM, Rahman R, Rosand J, et al. Severity of leukoaraiosis correlates with clinical outcome after ischemic stroke. Neurology. 2009;72(16):1403–1410. doi: 10.1212/WNL.0b013e3181a18823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu JH, Lu CZ, Hong Z, et al. Extent of white matter lesions is related to acute subcortical infarcts and predicts further stroke risk in patients with first ever ischaemic stroke. J Neurol Neurosurg Psychiatry. 2005;76(6):793–796. doi: 10.1136/jnnp.2003.032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly PJ, Shih VE, Kistler JP, et al. Low Vitamin B6 but Not Homocyst(e)ine Is Associated With Increased Risk of Stroke and Transient Ischemic Attack in the Era of Folic Acid Grain Fortification. Stroke. 2003;34:e51–e54. doi: 10.1161/01.STR.0000071109.23410.AB. [DOI] [PubMed] [Google Scholar]
- 11.Broderick JP, Brott T, Kothari R, et al. Establishing an epidemiologic laboratory for stroke. Neuroepidemiology. 1996;15:19. [Google Scholar]
- 12.Turaj W, Slowik A, Wnuk M, Szczudlik A. Gender-related differences in diagnostic evaluation and outcome of ischemic stroke in Poland. Stroke. 2009;40(3):980–982. doi: 10.1161/STROKEAHA.108.528422. [DOI] [PubMed] [Google Scholar]
- 13.Meschia JF, Brott TG, Brown RD, Jr, et al. The Ischemic Stroke Genetics Study (ISGS) Protocol. BMC Neurol. 2003;3:4. doi: 10.1186/1471-2377-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meschia JF, Brown RD, Jr, Brott TG, et al. The Siblings With Ischemic Stroke Study (SWISS) protocol. BMC Med Genet. 2002;3:1. doi: 10.1186/1471-2350-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams HP, Jr, Bendixen BH, Kappelle LJ, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24:35–41. doi: 10.1161/01.str.24.1.35. [DOI] [PubMed] [Google Scholar]
- 16.Saxena R, de Bakker PI, Singer K, et al. Comprehensive association testing of common mitochondrial DNA variation in metabolic disease. Am J Hum Genet. 2006;79(1):54–61. doi: 10.1086/504926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 18.McRae AF, Byrne EM, Zhao ZZ, et al. Power and SNP tagging in whole mitochondrial genome association studies. Genome Res. 2008 Jun;18(6):911–917. doi: 10.1101/gr.074872.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reich D, Price AL, Patterson N. Principal component analysis of genetic data. Nat Genet. 2008;40(5):491–492. doi: 10.1038/ng0508-491. [DOI] [PubMed] [Google Scholar]
- 20.Biffi AB, Anderson CD, Nalls MA, et al. Principal component analysis for assessment of population stratification in mitochondrial medical genetics. Am J Hum Genet. 2010 doi: 10.1016/j.ajhg.2010.05.005. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feigin VL, Lawes CM, Bennett DA, Anderson CS. Stroke epidemiology: a review of population-based studies of incidence, prevalence, and case-fatality in the late 20th century. Lancet Neurol. 2003;2(1):43–53. doi: 10.1016/s1474-4422(03)00266-7. [DOI] [PubMed] [Google Scholar]
- 22.Samuels DC, Carothers AD, Horton R, Chinnery PF. The power to detect disease associations with mitochondrial DNA haplogroups. Am J Hum Genet. 2006;78(4):713–720. doi: 10.1086/502682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myocardial Infarction Genetics Consortium. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41(3):334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kathiresan S, Willer CJ, Peloso GM, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41(1):56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newton-Cheh C, Johnson T, Gateva V, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International Schizophrenia Consortium. Purcell SM, Wray NR, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurol ME, Irizarry MC, Smith EE, et al. Plasma {beta}-amyloid and white matter lesions in AD, MCI, and cerebral amyloid angiopathy. Neurology. 2006;66:23–29. doi: 10.1212/01.wnl.0000191403.95453.6a. [DOI] [PubMed] [Google Scholar]
- 28.Chen YW, Gurol M, Rosand J, et al. Progression of white matter lesions and hemorrhages in cerebral amyloid angiopathy. Neurology. 2006;67:83–87. doi: 10.1212/01.wnl.0000223613.57229.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nandigam K, Chen YW, Gurol ME, Rosand J, Greenberg SM, Smith EE. Intracranial area as a surrogate measure of intracranial volume. J Neuroimaging. 2007;17:74–77. doi: 10.1111/j.1552-6569.2006.00069.x. [DOI] [PubMed] [Google Scholar]
- 30.Rosa A, Fonseca BV, Krug T, et al. Mitochondrial haplogroup H1 is protective for ischemic stroke in Portuguese patients. BMC Med Genet. 2008;9:57. doi: 10.1186/1471-2350-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chinnery PF, Elliott HR, Syed A, Rothwell PM. Mitochondrial DNA haplogroups and risk of transient ischaemic attack and ischaemic stroke: a genetic association study. Lancet Neurol. 2010;9(5):498–503. doi: 10.1016/S1474-4422(10)70083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz-Pesini E, Mishmar D, Brandon M, et al. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303(5655):223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 33.Niemi AK, Moilanen JS, Tanaka M, et al. A combination of three common inherited mitochondrial DNA polymorphisms promotes longevity in Finnish and Japanese subjects. Eur J Hum Genet. 2005;13(2):166–170. doi: 10.1038/sj.ejhg.5201308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.