Abstract
Endosialin/TEM1 is predominantly expressed on neovasculature, thus ideally suited for diagnostic, targeted-imaging and -therapy of cancer. To isolate TEM1-specific affinity reagents, we thought to screen a recombinant antibody (scFv) library derived from the repertoire of a patient with thrombotic thrombocytopenic purpura (TTP), as autoimmune disorders may produce self-reactive specificities. The yeast-display scFv library was constructed by homologous recombination of the TTP patient repertoire originally expressed on M13 bacteriophage in the novel vector pAGA2 for yeast-display expression. The TTP yeast-display library (10^9 members) was screened by magnetic and flow sorting with human TEM1 recombinant protein. A pool of yeast-display scFv able to detect 2 nM of TEM1 was obtained and transformed into yeast-secreted scFv by homologous recombination using the novel p416 BCCP vector for yeast secretion of biotinylated scFv. Anti-TEM1 yeast-secreted scFv were independently validated in vitro by flow cytometry analysis and ELISA assays, then in vivo biotinylated in N-termini to produce biobodies. Biobody-78 bound specifically to Endosialin/TEM1-expressing ovarian tumor in vivo, with functional stability over 48 hrs. Our results suggest that our novel paired display-secretory yeast libraries can serve as an ideal platform for the rapid isolation of high affinity reagents, and that anti-TEM1 biobody-78 can be used for in vitro assays including flow cytometry analysis, as well as in vivo for targeted-imaging and -therapy of cancer.
Keywords: Tumor vascular markers, ovarian cancer, yeast-display scFv, biobody, flow cytometry, in vivo targeting
Introduction
The growth of solid tumors beyond a diameter of 1–2 mm critically depends on the formation of a supporting stroma of newly formed blood vessels (Folkman, 1985). Tumor endothelial cells, stromal fibroblasts (activated fibroblast or myofibroblasts) and/or vascular pericytes acquire a phenotype different from that of normal stromal cells (Rettig et al., 1992; Christian et al., 2008) and express Tumor Vascular Markers (TVM). TVM provide attractive targets for antibody-based tumor diagnosis and therapy (St Croix et al., 2000; Marty et al., 2006; Teicher, 2007; Rouleau et al., 2008) due to i) the relative stability of TVM-expressing cells comparing to tumor cells; ii) neovasculature essential function for tumor maintenance, as demonstrated by the widespread necrosis of solid tumor after destruction of their blood vessels (Hinnen and Eskens, 2007); iii) neovasculature leaky capillaries that permit circulating antibodies and antibody conjugates to easily access TVM.
Endosialin/Tumor Endothelial Marker 1 (TEM1 or CD248) is a TVM and a type I transmembrane protein which comprises an 80.9 kDa protein core modified by extensive sialylated O-linked glycosylation that gives rise to an approximately 175 kDa mature glycoprotein (Christian et al., 2001). Endosialin/TEM1 was originally discovered by an anti-fibroblast monoclonal antibody (FB5) as a glycoprotein expressed by the pericytes and myofibroblasts associated with tumor vasculature (MacFadyen et al., 2005; Christian et al., 2008; Rouleau et al., 2008) as well as by tumor-associated vascular endothelial cells in various human cancers (Rettig et al., 1992; Davies et al., 2004; Rmali et al., 2005; Becker et al., 2008), including ovarian cancer (Conejo-Garcia et al., 2005). Endosialin/TEM1 plays a unique role in tumor progression as a promoting factor of tumor angiogenesis (Bagley et al., 2008), proliferation, migration and metastasis through interaction with matrix proteins such as fibronectin, collagen type I and IV (Tomkowicz et al., 2007) and Mac-2 BP/90K (Becker et al., 2008). Importantly, mice without functional Tem1 gene present a striking reduction in tumor growth, invasiveness, and metastasis after tumor transplantation to abdominal sites (Nanda et al., 2006). Taken together, these results suggest that targeting Endosialin/TEM1 for diagnostic and/or therapy could provide a valuable strategy against cancer.
Isolation of antigen-specific antibodies has been achieved through a variety of methods, including screening of phage- and yeast-display recombinant antibody (scFv) libraries (Vaughan et al., 1996; Feldhaus et al., 2003; Paschke, 2006; Scholler et al., 2006; Bergan et al., 2007; Scholler et al., 2008a; Scholler et al., 2008b). Yeast-display recently emerged as an efficient alternative strategy due to the advantages it offers over prokaryotic systems, including faster and more controlled flow cytometry-based selection compared to solid phase panning (Feldhaus et al., 2003; Bergan et al., 2007); a highly efficient sampling of the immune antibody repertoire (Bowley et al., 2007); post-translational modifications (glycosylation) due to the eukaryotic expression; and absence of scFv-induced growth bias because scFv are not displayed during the amplification step, when yeast multiply. Yet, transfer of scFv from displayed to secreted forms has often resulted in loss of antigen specificity and/or affinity, requiring additional time-consuming and costly steps, including in vitro maturation of scFv sequence and/or recloning of scFv fused to immunoglobulin (Ig) constant regions. Mechanisms underlying the loss of scFv function include changes in scFv conformation and post-translational modifications, due to the use of different expression systems for displayed and secreted forms.
We sought to generate a highly efficient system for high-throughput identification of antigen-specific affinity reagents. Because patients with autoimmune disorders produce large variety of antibodies, we hypothesized that a library derived from an autoimmune patient could contain high affinity antibodies against various antigens, including tumor vascular markers. We also hypothesized that only one expression system (Saccharomyces cerevisiae) for both scFv display and secretion could eliminate changes in scFv post-translational modifications, while keeping the advantages of an eukaryotic system for the expression of high-affinity antibodies, and that conformational changes would be minimized during the shift from displayed to secreted scFv forms if both displayed and secreted scFv were modified only at the N-terminus, which binds to the yeast surface or to secondary reagents, respectively. To test our hypotheses, we generated a 1×109 yeast display scFv library by homologous recombination of our new vector pAGA2 with scFv amplified from a phage-display scFv library derived from a patient with Thrombotic Thrombocytopenic Purpura (TTP), an autoimmune system disease related to the production of autoantibodies against coagulation factors (Siegel, 2008). We screened the TTP yeast-display scFv library in two steps using two complementary yeast systems of expression that permit to express yeast-display and yeast-secreted scFv with the same post-translational modifications while minimizing conformational changes. We identified several Endosialin/TEM1-specific scFvs using human TEM1 recombinant protein, including one with affinity in the nanomolar range, that were further transformed in biobodies (Scholler et al., 2006). Antigen-specific binding of anti-TEM1 scFvs and biobodies were characterized in vitro by flow cytometry analysis and ELISA assay, and in vivo by injection in an orthotopic mouse model of ovarian cancer. The anti-TEM1 biobody of highest affinity was able to bind specifically to both murine and human Endosialin/TEM1 and to target Endosialin/TEM1-expresser tumor cells in vivo, paving the way to the development of novel anti-angiogenesis targeted-theranostics.
Experimental Materials and Methods
Development of companion vectors for yeast-display and yeast-secretion of N-terminal biotinylated scFv
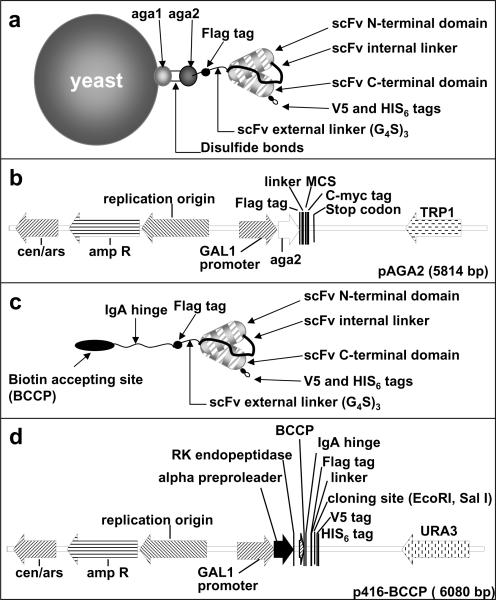
The pAGA2 vector for yeast-display (fig. 1a–b) was derived from shuttle vector p414 GAL1 (Mumberg et al., 1994) (the kind gift of Martin Funk, IMT, Philipps-Univ. Marburg, Germany). The pAGA2 multiple cloning site (MCS) was engineered as follows: the first site Nhe1 was inserted after a FLAG tag and a (G4S)3 linker sequence. The second site EcoR1 was part of a stop codon that is removed when cDNA is inserted in frame in the cloning site. The third site Xho1 was inserted just 5' to the c-myc tag, out of frame with the FLAG tag, to insure that both tags could be expressed only in the presence of correctly inserted cDNA. pAGA2 full insert sequence is described in supplementary methods.
Figure 1. Companion vectors for yeast-display and yeast-secreted scFv expression.
a–b: pAGA2 vector for yeast-display. The shuttle vector p414 GAL1 that allows galactose-inducible expression in presence of uracil was modified to include Nhe1, EcoR1 and Xho1 restriction enzyme sites matching the cloning sites of its companion vector p416 BCCP (d). The multiple cloning site (MCS) was engineered by inserting a Nhe1 site after the FLAG tag and (G4S)3 linker sequence; an EcoR1 site, also part of a stop codon that is removed when cDNA are inserted in frame in the cloning site; and a Xho1 site, inserted directly before the c-myc tag, out of frame with the FLAG tag, to insure that both tags would be expressed only in presence of correctly inserted cDNA. c–d: p416-BCCP for yeast-secretion. The shuttle vector p416 GAL1 that allows for galactose-inducible secretion in the presence of tryptophane was linearized by BamH1 and Xho1 and co-transformed in yeast with a purified PCR product encoding alpha preproleader and RK endopeptidase sequences followed by biotin accepting site (BCCP), IgA hinge, FLAG tag, (G4S)3 linker, cloning site with stop codon, and V5-HIS tags.
To construct the p416 BCCP vector, the companion vector of pAGA2 for the secretion of N-terminal biotinylated scFv (fig. 1c–d), the shuttle vector p416 GAL1 (Mumberg et al., 1994) (the kind gift of Martin Funk) was linearized by BamH1 and Xho1, and co-transformed in yeast with a PCR product encoding alpha preproleader and RK endopeptidase sequences fused to a biotin accepting site (BCCP), IgA hinge, FLAG tag, (G4S)3 linker, cloning site with a stop codon, and V5-HIS tags. p416 BCCP full insert sequence is described in supplementary methods.
Generation of rhTEM1-GST and rGST recombinant proteins and of TEM1-transduced cell lines
hTEM1 cDNA (NM_020404) was a kind gift from Dr. Ballmer-Hofer (Paul Scherrer Institut, Switzerland). The 1113 bp fragment corresponding to nucleotides 75–1187 of NM_020404 was cloned into the BamH1 site of the Glutathione S-transferase Gene Fusion Vector pGEX-2TK (Life Science, Piscataway, NJ) to obtain hTEM1-pGEX-2TK plasmid. hTEM1-pGEX-2TK and the control vector pGEX-2TK were transformed into E. coli BL21-CodonPlus(DE3)-RIPL (Stratagene, Cedar Creek, Texas) to produce human TEM1 recombinant protein fused to GST (rhTEM1-GST) and GST recombinant protein (rGST). Transformants were inoculated into fresh 2YT medium and incubated at 37°C on an orbital shaker (200 rpm) overnight. Each sample was then inoculated into 500 mL of fresh medium at a dilution of 1:50, and incubated in a shaking incubator at 37°C until the OD600 was 0.8. Isopropyl β-d-1-thiogalactopyranoside (IPTG) (Qiagen, Valencia, CA) was then added to a final concentration of 1 mM for the induction of expression at 25°C for 6 h. Bacterial cells were collected by centrifugation, lyzed by Bugbuster (Novagen, Gibbstown, NJ) and sonicated according to the manufacturer's instructions. Glutathione Sepharose 4B columns were equilibrated with phosphate buffered saline (PBS, pH 7.4) and lysate supernatant samples were loaded at a flow rate of approximately 1 mL/min.
The columns were then washed with three column-volumes of PBS. Finally, 50 mM Tris-HCl buffer (pH 7.4) containing 20 mM glutathione was used to elute the recombinant protein that was further purified with Mono Q 5/50 GL (GE Healthcare) with 20 mM Tris buffer (pH 6.8). Purified recombinant proteins were analyzed by SDS-PAGE (4–15% separation gel). Yields of rhTEM1-GST and rGST proteins were approximately 1 mg and 35 mg per liter of culture, respectively. Recombinant proteins were biotinylated using the EZ-Link Sulfo-NHS-Biotin-Reagents kit (Pierce, Rockford, IL) according to the manufacturer's instructions and dialyzed against PBS.
The full length hTEM1 cDNA was cloned into MluI/PacI-digested lentiviral plasmid vector pHRSIN-GFP (Hasegawa et al., 2006) to generate pHRSIN-TEM1 vector that expresses human TEM1 but not GFP. The corresponding lentivirus was generated by transient transfection of HEK293T cells with calcium phosphate. Conditioned medium containing viral vectors was harvested 24 and 48 hours after transfection, filtered (0.45 μm) and frozen at −80°C until use. MS1, a murine pancreatic islet endothelial cell line transformed by SV40 (ATCC number CRL-2279™), H5V, a murine heart endothelial cell line transformed by Polyoma middle-size T antigen (PmT) (Garlanda et al., 1994) and SKOV3, a human ovarian carcinoma cell line (Chan et al., 1988) (2 × 104 per 24-well plate) were transduced using 500 μL viral supernatants, and expression of hTEM1 was confirmed by RT-PCR (supplementary Fig. 4 and methods), Western blot and flow cytometry analysis using a rabbit anti-TEM1 polyclonal antibody (kindly provided to GC by Morphotek, Inc, Malvern, PA) (data not shown).
Yeast transformation
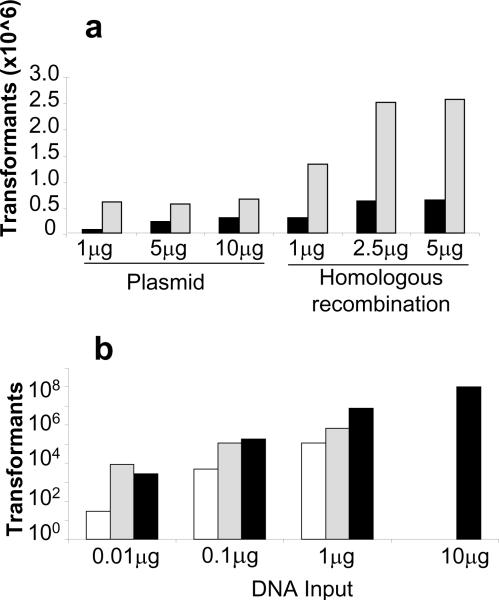
Table 1 summarizes the procedure for optimization of the preparation of chemical competent cell and transformation. Optimization was conducted in two steps, first by calibrating competent cell preparation and transformation, and then by optimizing DNA input. Condition 2 was best for chemical transformation of both EBV100 (Table 1) and YVH10 (data not shown), thus was applied for the transformation of both stains with intact plasmid or with combinations of linearized vector and insert for homologous combination (Fig. 2a). YVH10 chemical transformation yielded 106 transformants per microgram of DNA by homologous recombination, which was unexpectedly higher than transformation with intact plasmid (Fig. 2a, gray bars). However, for EBY100 yeast the chemical transformation efficiency was low (Fig. 2a, black bars). Thus, we developed an alternate protocol for EBY100 transformation using electroporation (Table 2). While electroporation of YVH10 resulted in low yield and was not pursued (data not shown), condition G (Table 2) gave the highest yield of transformants for EBV100, producing 1×108 transformants per electroporation with 10 μg of linearized vector and insert combined (Fig. 2b). Thus, both YVH10 and EBV100 yeast strains are more efficiently transformed by homologous recombination than by intact plasmids. YVH10 is best transformed by chemical transformation with heat shock, while EBV100 is best transformed by electroporation, and at levels compatible with generation of scFv libraries.
Table 1. Calibration of chemical transformations.
EBY100 cells in log phase were treated with five different conditions, as indicated, and transformed with one microgram of DNA plasmid.
| Conditions | Yeast resuspension | Yeast washes w/ water & LIAC/TE | Yeast incubation w/ vector + insert | Heat Shock at 42°C | Yeast recovery in YPED | Transformants per μg of DNA |
|---|---|---|---|---|---|---|
| 1 | Chilled in ice 60 min | 4°C | w/o DMSO | 30 min | 2h | 3 × 10^4 |
| 2 | Room temperature | 4°C | w/o DMSO | 30 min | 2h | 8.1 × 10^4 |
| 3 | Room temperature | 4°C | w/ DMSO | 30 min | 2h | 0.5 × 10^4 |
| 4 | Room temperature | Room temperature | w/o DMSO | 30 min | 2h | 0.7 × 10^4 |
| 5 | Room temperature | Room temperature | w/ DMSO | 15 min | 2h | 1 × 10^4 |
Figure 2. Optimization of yeast transformations for EBY100 and YVH10 yeast strains.
a: Chemical transformations. Chemical transformation of EBY100 (black bars) vs. YVH10 (grey bars) using condition 2 described in table 1, with different concentrations of intact DNA plasmids (1μg, 5μg and 10μg, as indicated) or of combinations of linearized plasmid and insert for homologous recombination (1μg, 2.5μg and 5μg of linearized vector combined with 3-fold molar excess of scFv fragments, as indicated). The number of transformants x106 is plotted on the Y axis. b: Electroporations. All experiments were performed in 0.2cm cuvettes, in 50μl volume, with settings of 1.5kV, 25mF and 200Ω. Electroporation of EBY100 using condition G described in table 2, with different concentrations of linearized vector (white bars) or intact DNA plasmid (grey bars) or combination of linearized plasmid and insert (black bars) for homologous recombinations (as indicated). The number of transformants per transformation is plotted on the Y axis.
Table 2. Calibration of electroporation.
All experiments were performed in 0.2cm cuvettes, in 50μl volume, with settings of 1.5kV, 25mF and 200Ω. EBY100 cells in log phase were treated with fifteen different conditions, as indicated, and transformed with one microgram of DNA plasmid.
| Conditions | DTT conc. | Buffer | Sorbitol conc. | Shaking Time | Shaking Temperature | Transformants per μg of DNA |
|---|---|---|---|---|---|---|
| A | 25 mM | 15 min | 30°C | 0.05 × 10^6 | ||
| B | 25 mM | 30 min | 30°C | 0.05 × 10^6 | ||
| C | 25 mM | 10 mM Tris | 30 min | 30°C | 0.23 × 10^6 | |
| D | 25 mM | 20 mM Hepes | 30 min | 30°C | 0.14 × 10^5 | |
| E | 25 mM | 1M | 30 min | 30°C | 0.01 × 10^5 | |
| F | 25 mM | 1M | 60 min | 4°C | 0.46 × 10^6 | |
| G | 25 mM | 20 mM Hepes | 0.6M | 30 min | 30°C | 1.15 × 10^6 |
| H | 25 mM | 20 mM Hepes | 1M | 30 min | 30°C | 0.74 × 10^6 |
| 1 | 25 mM | LiAc-TE | 0.6M | 30 min | 30°C | 0.69 × 10^6 |
| J | 25 mM | LiAc-TE | 0.6M | 60 min | 4°C | 0.44 × 10^5 |
| K | 25 mM | LiAc-TE | 1M | 30 min | 30°C | 0.1 × 10^5 |
| L | 25 mM | LiAc-TE | 1M | 60 min | 4°C | 0.14 × 10^5 |
| M | 25 mM | LiAc-TE | 30 min | 30°C | 0.37 × 10^5 | |
| N | 25 mM | LiAc-TE | 30 min | 4°C | 0.05 × 10^6 | |
| 0 | 25 mM | LiAc-TE | 60 min | 4°C | 0.17 × 10^6 |
Shuffling of phage-display scFv library into yeast
ScFvs initially cloned in the M13 phage display vector pComb3X (Scripps Research Institute, La Jolla, CA) (Barbas et al., 2001–07) were rescued by PCR from a phagemid DNA preparation. The primers were designed to bind to the original phage library scFv 5' and 3' end sequences, as well as to promote homologous recombination with the yeast-display vector pAGA2. Since the scFv constructs comprised light chain (kappa or lambda) variable regions (VL's) followed by a GS-rich linker peptide followed by γ heavy chain variable region sequences (VH's), forward primers annealed to the 5' ends of VL's and reverse primers annealed to the conserved 5' end of the γ1,2,3,4 CH1 domain of the IgG heavy chain. The primer sequences are listed in the supplementary methods.
PCR conditions for scFv amplification were: 94°C for 5 min followed by 25 cycles of 94°C 1 min, 55°C 1 min and 72°C 1 min, and a final extension of 7 min at 72°C. PCR products were then purified by electrophoresis using the Qiaquick Gel Extraction Kit (Invitrogen, Carlsbad, CA). pAGA2 vector for yeast display was linearized with NheI and XhoI and purified using the Qiaquick PCR Purification Kit (Invitrogen, Carlsbad, CA). EBY100 competent cells were prepared for electroporation according to the condition G (Table 2), and co-transfected with purified PCR products and pAGA2 linearized vector. Transfected yeast cells were finally expanded in SD-CAA medium (Scholler et al., 2006) at 30°C at 200 rpm until saturation. Ten-fold serial dilutions of the transfected yeast were cultured on SD-CAA plates for calculation of the library's size.
The library's diversity and gap repair efficiency were evaluated by sequencing and flow cytometry analysis, respectively. In brief, 30 individual clones were randomly selected, induced in the medium SGR-CAA (Scholler et al., 2006), and assessed for scFv expression using the c-myc tag expressed only by yeast displaying scFv. Plasmid DNA was extracted from these clones using MasterPure Yeast DNA Purification Kit (Epicentre Biotechnologies, Madison, WI) and scFv gene fragments were amplified for sequencing with primers flanking the gap repair sites. PCR amplification primers are: pAGA2-scFv-For: 5'-ccgtactctttgtcaacgac-3' and pAGA2-scFv-Rev: 5'-ttaaagccttcgagcgtccc-3'. Sequencing primers are: pAGA2-seq-For: 5'- gggaaggcaatgcaagga g-3' and pAGA2-seq-Rev: 5'- tgcgtacacgcgtctgtacag-3'.
Antibodies
Yeast-display scFv expression was detected with anti-c-myc mouse monoclonal antibody (mAb), 9E10 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and Alexa488 F(ab')2 fragment of goat anti-mouse IgG (H+L) (488 anti-IgG) (Invitrogen, Carlsbad, CA) or PECy5 goat F(ab')2 anti-mouse IgG(H+L) (PE-Cu5 anti-IgG) (Cedarlane Laboratories Limited, Burlington, NC). Biotinylated antigen binding to yeast-display scFv was detected with goat anti-biotin-FITC (Abcam, Cambridge, MA) or streptavidin-PE (BD Pharmingen, San Jose, CA). ScFv binding to cell lines were detected with APC-conjugated anti-V5 mouse mAb (AbD Serotec, Raleigh, NC) (APC anti-V5) and scFv binding to plastic-immobilized antigen was detected by HRP-conjugated mouse anti-V5 mAb (AbD Serotec) (HRP-anti-V5). Biobody binding to TEM1-expresser cells was detected with APC-labeled streptavidin (eBioscience, San Diego, CA). Confocal microscopy was performed with anti-SV40 Tag mAb (Santa-Cruz Biotech, Santa Cruz, CA) detected by goat anti-mouse IgG1k-Alexa 488 mAb (488 anti-IgG1k) (Invitrogen, Carlsbad, CA), and rhodamine-labeled streptavidin (Invitrogen).
Identification of anti-TEM1 scFv
The TTP yeast-display scFv library was first screened by magnetic and flow sorting for anti-TEM1 scFv using progressively decreasing concentrations of human TEM1 recombinant protein fused to a GST tag (rhTEM1-GST) (supplementary fig. 1), as previously described (Scholler et al., 2006; Bergan et al., 2007). cDNAs encoding selected yeast-display scFv were PCR amplified, purified and transformed into yeast-secreted scFv. Briefly, the library was magnetically enriched once for scFvs that bound to 20 nM of biotinylated rhTEM1-GST, twice for scFvs that bound to 6 nM of biotinylated rhTEM1-GST, and then magnetically depleted for the scFv that bound to 60 nM of control biotinylated rGST. Selected yeast-display scFvs were flow sorted for cmyc/TEM1 double positive clones. Progressively decreasing concentrations of rhTEM1-GST were further used during the screening (from 2nM to 40 pM). DNA plasmids were extracted from yeast-display scFv that bound to 400 pM of rhTEM1-GST but not to 2nM of rGST (supplementary fig. 1b–d), and scFv fragments were amplified using primers allowing homologous recombination with the yeast secretion vector p416-BCCP. The primers used were: Forward shuffling primer: 5'-ggttctggtggtggaggttctggtggtggtggatctg-3'; Reverse shuffling primer: 5'-gagaccgaggagagggttagggataggcttaccgtcgaccaagtcttcttcagaaataagctt-3'. ScFv fragments and linearized p416-BCCP were co-transfected into YVH10 by chemical transformation. Soluble scFv screening for specific binding to TEM1 was performed by ELISA with yeast supernatants of 470 random transformants in Maxisorp ELISA plates (Nunc, Rochester, NY) coated with 0.8 μg/ml of rhTEM1-GST vs. rGST. Recombinant proteins were plastic-immobilized in carbonate-bicarbonate buffer 0.5 M, pH 9.6 (Fisher Scientific, Pittsburgh, PA) ON at 4°C. Wells were then blocked with 5% dry milk in PBS (PBMS) (Biorad, Hercules, CA) for 2 hours at RT with gentle agitation and incubated with yeast supernatants diluted 1:1 with PBSM at RT for 1h. After three washes with PBS supplemented with 0.05% Tween (PBST) (Biorad), scFv binding to immobilized proteins was detected with HRP anti-V5. Colorimetric signals were developed with TMB substrate solution (KPL, Inc., Gaithersburg, MD), quenched with sulfuric acid (KPL, Inc) and read at 450 nm on a Fluoroskan Ascent FL (Thermo Fisher Scientific, Pittsburgh, PA). Approximately 50% of the colonies (232/470) gave a colorimetric signal higher than the average background plus 3 standard deviations. No cross-reactivity with rGST control protein was detected. Sequencing of 40 scFv identified five unique clones that were then produced and Ni-purified as previously described (Scholler et al., 2006).
Measurement of scFv affinity by ELISA
To assess scFv-78 affinity maxisorp ELISA plates were coated with rhTEM1-GST at two-fold decreasing concentrations from 0.4 to 0.05 μ/ml, in carbonate-bicarbonate buffer. After blocking with PBSM, wells were incubated with ten-fold serial dilutions of scFv-78, starting from 1 μM. ScFv binding to immobilized proteins was detected with HRP anti-V5. Colorimetric signals were developed as previously described. The same procedure was followed for the other scFvs, but using 2 and 1 μg/ml of coated rhTEM1-GST and three-fold serial dilutions of scFv, starting at 2 μM.
Flow cytometry analysis
Analysis of scFv expression by yeast was performed as previously described (Scholler et al., 2006). Briefly, binding of anti-TEM1 scFv and biobodies to TEM1-expresser mammalian cells was evaluated using various human or murine cell lines expressing TEM1 endogenously (HEK-293T, MOV1 and 2H11) or stably transduced with pHRSINTEM1 (hTEM1-MS1, hTEM1-H5V and hTEM1-SKOV3). MS1 (SV40-transformed murine endothelial cells), H5V (PmT-transformed murine endothelial cells) and SKOV3 (human ovarian carcinoma cell) lines were used as negative or weakly positive control cell lines, in accordance with previous published findings reporting Endosialin/TEM1 to be expressed by mouse immortalized endothelial cells and various tumors, including carcinomas and sarcomas (Dolznig et al., 2005; Teicher, 2007). Anti-TEM1 scFvs were preincubated for 30 min at RT with APC-anti-V5 at a ratio 1:1 and anti-TEM1 biobodies were preincubated with APC-labeled streptavidin at a ratio of 1:4. A non-relevant scFv was used as a negative control for binding.
Orthotopic mouse model of ovarian cancer
MOV-1 mouse ovarian cancer cell line was derived from an ovarian cancer that spontaneously arose in female transgenic mice that express the transforming region of SV40 under control of the Mullerian inhibitory substance type II receptor gene promoter (Tg-MISIIR-TAg) (Connolly et al., 2003). MOV-1 cell line expresses SV40 antigen and TEM1. To emulate ovarian cancer mouse ovarian cancer cells, MOV1 were orthotopically injected in the ovarian bursa of NOD-Scid-γcnull (NSG) mice (Shultz et al., 2000). Four month-old multiparious females were anesthetized according to the protocol approved by the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC). A dorso-lateral incision on right caudal portion of the animal dorsum was made. The retroperitoneum was dissected to expose the left ovary using the forceps to grasp, retract, position, and secure the organ for injection. Five million MOV1 cells were injected in the ovarian bursa in a volume of 20 μl of PBS using an insulin syringe. Retroperitoneum wounds were closed, animals were administered antibiotics and fluids, and tumor growth was monitored by in vivo imaging.
Analysis of in vivo distribution of anti-TEM1 biobodies by confocal microscopy
Anti-TEM1 biobody-78 of high affinity was injected intravenously (IV) 3 weeks after tumor cell implantation. As control for the Enhanced Permeability and Retention (EPR) effect (Maeda and Matsumura, 1989; Greish, 2007), we used the anti-TEM1 biobody-137 of low affinity in the same conditions. Spleen, liver, kidney and ovaries were harvested 24 or 48 hours after biobody injection and preserved in frozen tissue matrix OCT compound (Tissue-Tek, Sakura Finetek USA). Slides of 5 mm thickness were cut from frozen sections, air dried 1 hr at RT and fixed by immersion in cold 100% acetone 5 min. After 2 washes in PBS, slides were blocked for endogenous biotin by pre-treatment with Avidin/Biotin Blocking solution (avidin-skim milk 0.001% in PBS). Anti-TEM1 biobody binding was detected with rhodamine-conjugated streptavidin. MOV1 tumor cells derived from a Tg-MISIIR-TAg tumor express SV40 and thus could be detected with anti-SV40 Tag antibody (2 μg/ml) for 30 min at RT, followed by 1 μg/ml Alexa 488 goat anti-mouse IgG1κ for 30 min at RT. Slides were incubated with 1:2000 diluted DAPI (Invitrogen) for 30 min at RT to visualize the nuclei. Fluorescent signals were acquired by confocal analysis (Zeiss LSM 510META NLO) at 63× magnification
Results
Generation of companion vectors for yeast-display and yeast-secretion of N-terminal biotinylated scFv
To overcome the loss of antigen specificity and/or affinity due to scFv transfer from cell surface display to secreted, we developed two complementary vectors, pAGA2 and p416 BCCP, which permit scFv to be displayed (Fig. 1a–b) or secreted (Fig. 1c–d) by the same expression system (S. cerevisiae) and through engineering at the same domain (N terminus), thus with similar post-translational modifications and conformation. In pAGA2 vector, scFv are fused at the N-terminus to Aga2, to permit yeast display (Fig. 1 a–b). ScFv expressed by p416 BCCP are soluble and fused at the N-terminus to an enzymatically biotinylable domain (BCCP) separated from the scFv functional site by a flexible IgA hinge (Fig. 1 c–d). The BCCP domain is biotinylated in vivo by a biotin ligase expressed by yeast mating partners, and the presence of the IgA hinge minimizes scFv conformational changes when the biotinylated BCCP binds to immobilized or labeled streptavidin (Scholler et al., 2006). The vectors pAGA2 (Fig. 1a–b) and p416 BCCP (Fig. 1c–d) permit galactose-inducible expression in the presence of uracil or tryptophane, respectively, which minimizes growth bias (Mumberg et al., 1994).
Construction and validation of TTP yeast-display scFv library
The M13 phage-display human scFv library derived from a patient with thrombotic thrombocytopenic purpura (TTP), a coagulation system disorder caused by autoantibodies to the metalloprotease ADAMTS13, was previously reported (Siegel, 2008). ScFv gene segments from the TTP phage display library were rescued by PCR from a phagemid DNA preparation, and co-transformed with pAGA2 linearized vector by electroporation in EBV100 yeast strain to allow homologous recombination and cell surface display. The diversity of the resulting yeast-display scFv library was validated by the sequencing of 20 randomly selected clones that demonstrated a gap repair rate of 95% (data not shown). The correct display of scFv at the yeast cell surface was assessed by flow cytometry through the detection of a c-myc tag fused to the scFv C-terminus. Nine out of 24 randomly selected clones displayed scFvs on their surface after induction (supplementary Fig. 2, clones a,g,i,k,o,p,r,u,w), which was consistent with the expression ratio of phage and yeast libraries previously reported (Vaughan et al., 1996; Blaise et al., 2004).
Identification of anti-TEM1 scFv
Targeting TEM1 with antibody or antibody-conjugated reagents such as an immunotoxin, isotope, or nanoparticles, is a promising approach for both diagnosis and therapy. To identify recombinant antibodies directed against TEM1, the TTP yeast-display library was enriched for scFv that bind to recombinant human TEM1-GST protein (rhTEM1-GST), first by magnetic sorting then by flow sorting, and finally depleted by magnetic sortings for the scFv that bound to rGST control protein. Magnetic sorting is based on one parameter of selection only (capture of antigen-binding yeast), thereby providing a rapid means for robust enrichment of antigen-specific scFv. However, to prevent the selection of yeast that non-specifically bind to antigens, a flow sorting step using two parameters of selection was included. Yeast that both bound to TEM1-1 and expressed cmyc tag due to the display of scFv at their surface were sorted and their cDNA rescued for amplification with primers allowing homologous recombination with p416 BCCP vector. Resulting transformants secreted scFv enriched for TEM1-specific binders.
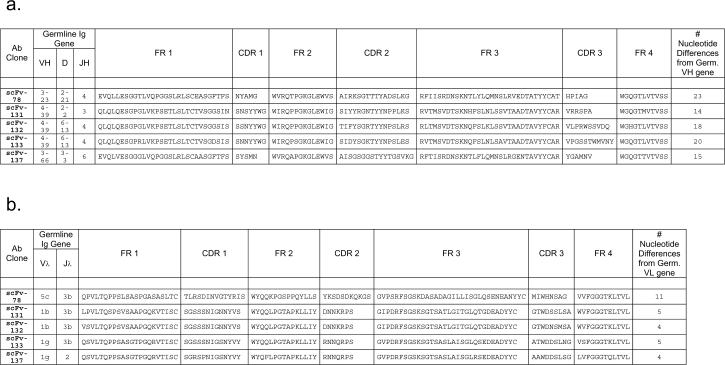
Four hundred and seventy yeast-secreting scFv were tested by ELISA detection assays for their ability to detect rhTEM1-GST. Almost 50% (232/470) of the soluble scFv bound to rhTEM1-GST but not to rGST. Anti-TEM1 soluble scFv were classified into two affinity categories: “high” when the optical density (OD) by detection ELISA was greater than 0.5, and “low” when lower than 0.5. Twenty scFv of each category were sequenced. The high OD group contained only one sequence (scFv-78) and three clones presented each one point mutation in the VL; #6 had a point mutation in FR1 that changed a serine in leucin; #8 a point mutation in FR3 changing a glycin in valine; #10 a point mutation in CDR2 changing a leucine in methionine. The low OD group included four different scFvs (scFv-131; scFv-132; scFv-133; scFv-137) (Figure 3). Alignment of scFv heavy and light chain variable region sequences to a database of human immunoglobulin germline sequences (V Base Directory of Human V Gene Sequences (Tomlinson et al., 1996)) indicated that the heavy chain variable region sequences used VH3- and VH4-family-encoded gene products and showed extensive somatic mutation particularly in the CDR regions as though they evolved during an antigen-driven immune response (Fig. 3a). The light chain sequences used by all 5 anti-TEM1 scFv's were lambda and were minimally mutated from their most likely VL and JL germline genes (Fig. 3b).
Figure 3. Germline immunoglobulin gene usage and predicted amino acid sequences of anti-TEM1 scFv.
a. Anti-TEM1 heavy chain variable regions. The number of nucleotide differences from germline VH is tabulated to the right of each sequence. In general, D segments showed very poor homology with known D genes so mutations were not scored in these regions. FR (framework region) and CDR (complementarily determining region) designations as per Kabat. (Kabat et al., 1991) b. Anti-TEM1 light chain variable regions. The number of nucleotide differences from germline VL is tabulated to the right of each sequence.
Characterization and in vitro validation of anti-TEM1 scFvs and biobodies by ELISA assays and flow cytometry analysis
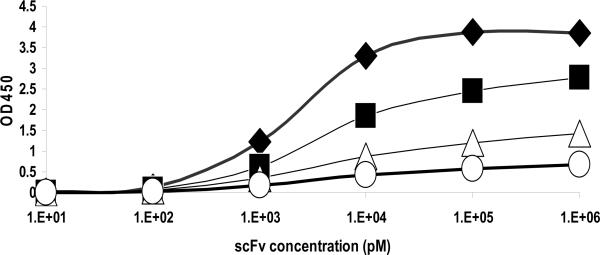
ELISA provides a convenient way to evaluate the affinity of an antibody (Beatty et al., 1987). Because a scFv has only one binding site for antigen, the affinity can be calculated by the equation of Kd=2[Ab']t−[Ab]t, derived from the Beatty's equation Kaff=1/2(2[Ab']t−[Ab]t), where [Ab']t refers to scFv concentration at half the maximal OD (OD50) for rhTEM1-GST-coated wells at half concentration, and [Ab] refers to scFv concentration at OD50 for rhTEM1-GST-coated wells at whole concentration. The calculated Kd of the five anti-TEM1 scFvs with distinct sequences were 4.3+/− 1.5 nM for scFv-78 (Fig. 4), 148 nM for scFv-132, 218 nM for scFv-133, 682 nM for scFv-131, and 4.4 μM for scFv-137, respectively (supplementary Fig. 3). R2 of the curve fittings for all ELISAs were above 0.99. ScFv were then biotinylated on the BCCP site in N-termini by yeast mating with biotin ligase-bearing yeast to produce biobodies, as described by Scholler et al. (Scholler et al., 2006), and expressed at a yield of 10 mg per liter.
Figure 4. Measurement of scFv-78 affinity by ELISA.
Kd was calculated by the equation Kd=2[Ab']t-[Ab]t, where [Ab']t refers to the scFv concentration at OD50 for the half concentration rhTEM1 coated wells while [Ab]t refers to the scFv concentration at OD50 for one concentration rhTEM1-GST coated wells. The calculated Kd for scFv-78 was 5.8 nM using antigen concentrations of 0.4 μg/ml (diamonds) and 0.2 μg/ml (squares), 4.5 nM using antigen concentrations of 0.2 μg/ml and 0.1 μg/ml (open triangles), or 2.8 nM using antigen concentration of 0.1 μg/ml and 0.05 μg/ml (open circles). ELISAs were performed in duplicate in two independent experiments. Lines are fitted using the antibody-antigen reaction equation.
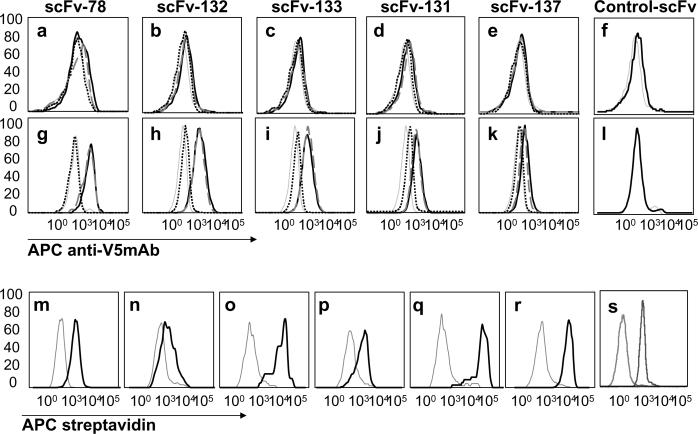
Flow cytometry showed that all anti-TEM1 scFvs bound to the human TEM1-transfected mouse endothelial cell line hTEM1-MS1 but not to wild-type MS1 cells, while binding could be blocked with rhTEM1 but not with rGST (Fig. 5a–l). The remainder of the study was performed with the high affinity anti-TEM1 scFv-78 and low affinity scFv-137 after targeted biotinylation, resulting in biobody-78 and biobody-137, respectively. Biobody-78 strongly bound to cell lines transduced with human TEM1 (Fig. 5o–q) and cells expressing high levels of endogenous human (Fig. 5m) as well as mouse (Fig. 5r,s) TEM1. Biobody-78 also bound to cell lines that express moderate levels of endogenous mouse or human TEM1, such as H5V (Fig. 5n) and SKOV3 (Fig. 5p), respectively. TEM1 mRNA expression level was verified by qRT-PCR in all cell lines (supplementary Fig. 4).
Figure 5. Analysis of anti-TEM1 scFv binding by flow cytometry.
Wild type (a–f) and hTEM1-transfected (g–l) microvascular endothelial cells of murine pancreatic origin (Mile-Sven, MS1 cells, American Type Culture Collection, Manassas, VA) were incubated with five different anti-TEM1 scFv (as indicated) at 10 nM for scFv-78 and at 1mM for scFv-132, scFv-133, scFv-131, scFv-137, and an irrelevant control scFv. ScFv binding to cell surface was detected with APC-labeled anti-V5 (solid black line). Blocking conditions were performed in presence of 20 nM of rhTEM1-GST (small dotted line) or 100 nM of rGST control protein (dashed line) for scFv78 and 1 μM of rhTEM1-GST (smalldotted line) or 1 μM of rGST control protein (dashed line) for scFv-131, -132, -133 and -137. As a negative control, cells were incubated with APC anti-V5 mAb only (grey line). m–s: Targeted biotinylated ScFv-78 (biobody-78) was incubated with TEM1-endogenous expresser embryonic kidney 293 cells (HEK293) (m); wild type heart endothelial mouse cells (H5V) (n); hTEM1-transfected H5V cells (o); wild type human ovarian cancer cells (SKOV3) (p); hTEM1-transfected SKOV3 cells (q); mouse TEM1-endogenous expresser endothelial cells (2H11) (r), and mouse TEM1-endogenous expresser ovarian cancer cells (MOV-1) (s). Biobody-78 binding was detected by 30 nM of APC-labeled streptavidin (black line). As a negative control, cells were incubated with APC-labeled streptavidin only (grey line).
In vivo validation of anti-TEM1 scFvs
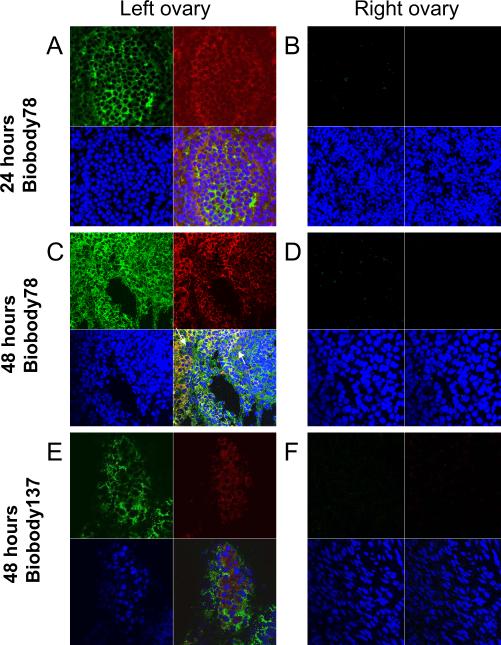
To test the ability of anti-TEM1 scFvs to recognize target in vivo, immunodeficient mice were transplanted orthotopically in the left ovarian bursae with MOV1 ovarian cancer cells, which express mouse TEM1 (supplementary Fig. 4B). Groups of mice were injected IV with a bolus of either the high-affinity biobody-78 or the low affinity biobody-137 (50 μg/mouse) to control for the EPR effect. Animals were euthanized after 24 or 48 hours, and spleens, livers, kidneys and ovaries were collected to monitor biobody distribution by confocal microscopy. Anti-TEM1 biobody-78 was detected in kidneys 24 hours after IV injection (supplementary Fig. 5C upper and lower right panels), but was cleared after 48 hours (supplementary Fig. 5F upper and lower right panels). Both anti-TEM1 biobody-78 and biobody-137 specifically localized to MOV1 cells implanted in the left ovaries after 48 hours (Fig. 6 A,C,E, upper and lower right panels) but not in the contralateral normal ovaries (Fig. 6 B,D,F) or in the other normal mouse organs (spleen, liver and kidney) (supplementary Fig. 5, as indicated). The signal generated by the biobody-137 was weaker than the signal from biobody-78 (compare Fig. 6C with Fig. 6E) which demonstrated a correlation between biobody affinity and tumor binding, and suggested that biobody-78 binding to the tumor was both independent of the EPR effect and antigen-specific.
Figure 6. Biodistribution of anti-TEM1 biobodies in the ovaries.
Mice were first implanted in the left ovary with MOV1 cells, IV-injected three weeks later with anti-TEM1 biobody-78 (A–D) or biobody-137 (E,F), and sacrificed 24 (A,B) or 48 hours (C,F) later. Upper left panel: The presence of tumor cells in the ovaries was detected with 2 μg/ml of anti-SV40 mAb followed by 1 μg/ml of alexa-488 goat anti-mouse IgG2a. Upper right panel: Comparative binding of anti-TEM1 biobodies to the left ovary (A,C,E) or right ovary (B,D,F) were detected by staining with 1 μg/ml of rhodamine-conjugated streptavidin. Lower left panel: The presence of nuclei was detected by DAPI staining. Lower right panel: Merged images of all three conditions were assessed by confocal microscopy (63×).
Discussion
We isolated high affinity reagents (scFv and biobodies) that bound to both mouse and human Endosialin/TEM1, using a high throughput yeast-based platform and a two-step screening method. Anti-TEM1 scFv and biobodies were validated for flow cytometry use, ELISA detection assays and in vivo targeting. The affinity of our novel anti-TEM1 biobody-78 was in the nanomolar range which permitted to generate high mean fluorescence intensities (MFI) by flow cytometry analysis at very low concentration (10 nM, Fig. 5), which favorably compared to the anti-TEM1 scFv previously published by Marty and colleagues (Marty et al., 2006). In addition and in contrast to the other available anti-TEM1 antibodies, our novel anti-TEM1 biobody-78 binds to both mouse and human TEM1, which is invaluable for preclinical model systems. Our screening strategy was based on cell sorting of yeast-display scFv, combined to ELISA screening and flow cytometry analysis of yeast-secreted biotinylated scFv (biobodies), using a comprehensive set of vectors to shuttle scFv without loss of function from displayed to secreted, labeled forms.
High affinity anti-TEM1 human scFv and biobodies were isolated from an yeast-display scFv library derived from a patient with TTP, an autoantibody-mediated disorder (Siegel, 2008). Given that patients with autoimmune disorders have lost some degree of immune tolerance, we thought that an immune library derived from such a patient could contain high affinity antibodies against various self-antigens. The transfer of the TTP scFv library from phage-display to yeast-display permitted a highly efficient initial selection by cell sorting (Bowley et al., 2007). Although here we used a phage library as starting material because it was previously shown to comprise high affinity clones against vascular targets (Siegel, 2008), we do not foresee phage display as a necessary initial step in the development of yeast scFv libraries. Furthermore, although the library derived from a patient with autoimmune disorder exhibited high affinity antibody clones against TEM1, it is possible that patients with cancer also exhibit high affinity clones against tumor-associated self-antigens (Anderson and LaBaer, 2005; Tan and Zhang, 2008; Qiu and Hanash, 2009), including antigens up-regulated by tumor vasculature.
Our new vector for yeast-display, pAGA2, prevents the cell surface expression of c-myc in the absence of scFv insertion in frame with the cloning site, which permits efficient positive and negative selections by cell sorting. We also generated a companion vector for yeast-secretion of scFv, p416-BCCP. The combined use of pAGA2 and p416-BCCP vectors permit consistency both in the expression systems used for scFv surface display or secretion (yeast), and in the scFv orientation (attachment to the N-termini when displayed at the yeast cell surface as well as when soluble and bound to streptavidin), which minimizes post-translational and conformational changes from displayed to secreted forms. In addition, the transfer from display to secreted forms can be simply achieved by yeast homologous recombination with an optimized protocol permitting high yields of transformation. Finally, yeast secreting soluble scFv cloned into p416 BCCP can be mated with biotin-ligase bearing yeast (Scholler et al., 2006) to produce diploid yeast that secrete N-termini biotinylated scFv (biobodies) directly in the yeast culture medium. Biobodies are inexpensive to produce, can be retrieved by any streptavidin-coated surfaced directly from the yeast culture medium or purified through their HIS6 tag, and are detectable with labeled streptavidin.
Conclusion
The foreseeable clinical and laboratory applications of fully human scFv and especially biobodies are quite diverse. We describe here for the first time a biobody (biobody-78) that can detect both human and mouse Endosialin/TEM1 for flow cytometry analysis and ELISA applications, and that is stable in vivo with intact functional binding 48 hours after IV injection (Fig. 6C upper and lower right panels), permitting confocal analysis of harvested tissues. Therefore, biobody-based targeting of tumor targets in vivo for the purpose of imaging or to deliver therapeutic payloads is now possible. Antibody-conjugated reagents such as an immunotoxins, immunoisotopes, or immunonanoparticles are high promising approaches for diagnosis and/or therapy. Indeed, the versatility of biobodies allows for modular design of both imaging and therapeutic tools, thus paving the way for the development of personalized paired imaging and therapy, or theranostic, tools. We believe that we resolved several limitations existing presently in the field of scFv antibody development and we provide the first evidence of a high throughput paired yeast platform that can lead to the rapid isolation of functional high affinity biotinylated scFv for the development of theranostics. To our knowledge, biobody-78 is the only high affinity antibody of human origin directed against both human and murine Endosialin/TEM1, which will greatly facilitate Endosialin/TEM1-based preclinical targeted imaging and therapeutic studies.
Supplementary Material
Acknowledgments
We acknowledge Yi Cheng and Lindsay Bergan for excellent technical assistance, John Facciaponte for help with the qRT-PCR assays, Carmine Carpenito for the preparation of lentiviruses from the pTurboFP635-C vector and Ryan D. Wychowanec for flow sorting. The project described was first supported by the Pacific Ovarian Cancer Research Consortium, Award Number P50 CA083636 from the National Cancer Institute (Nicole Urban) and the Canary Foundation (Nicole Urban). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institute of Health. The project described was also supported by The Institute for the Translational Medicine and Therapeutics (CA016520/NIH) (Nathalie Scholler), the Claneil Foundation (George Coukos, Nathalie Scholler), the Alliance for Cancer Gene Therapy (George Coukos), the Ovarian Cancer Research Foundation (George Coukos), and the National Heart, Lung, and Blood Institute (P50-HL81012) (Don. L. Siegel)
Abbreviations
- scFv
single chain variable fragments
- vs.
versus
- pM
picomolar
- nM
nanomolar
- rhTEM1-GST
recombinant human protein Endosialin/TEM1 fused to GST tag
- rGST
recombinant GST tag
- RT
room temperature
- ON
overnight
- PBS
phosphate buffered saline
- PBSM
PBS supplemented with 5% dry milk
- PBST
PBS supplemented with 0.05% Tween
- mAb
mouse monoclonal antibody
- g/ml
microgram per ml
- IV
intra venous.
Appendix
AZ designed and performed optimizations of yeast transfection, transferred the phage-display scFv library to yeast-display form, screened the yeast-display library, identified anti-TEM1 scFv, validated them in vitro and contributed to manuscript preparation. SNC established MOV1 cell line and the orthotopic model of ovarian cancer, performed in vivo experiments and confocal analysis and contributed to manuscript preparation. CL generated hTEM1-pGEX-2TK and pHRSIN-TEM1, transduced the cell lines, and produced and purified rhTEM1-GST and rGST. GC supervised CL and contributed to project conception and design and manuscript preparation. DLS constructed the phage-displayed scFv library and contributed to the design of scFv transfer from phage to yeast and to manuscript preparation. NS generated pAGA2 and p416-BCCP vectors, contributed to the design of scFv transfer from phage to yeast, supervised AZ and SNC, and wrote the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests statement The authors declare no competing financial interests.
References
- Anderson KS, LaBaer J. The sentinel within: exploiting the immune system for cancer biomarkers. J Proteome Res. 2005;4:1123–33. doi: 10.1021/pr0500814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley RG, Rouleau C, St Martin T, Boutin P, Weber W, Ruzek M, Honma N, Nacht M, Shankara S, Kataoka S, Ishida I, Roberts BL, Teicher BA. Human endothelial precursor cells express tumor endothelial marker 1/endosialin/CD248. Mol Cancer Ther. 2008;7:2536–46. doi: 10.1158/1535-7163.MCT-08-0050. [DOI] [PubMed] [Google Scholar]
- Barbas CF, III, Burton DR, Scott JK, Silverman GJ. Phage Display: A laboratory Manual. Cold Spring Harbor Laboratory Press; 2001–07. [Google Scholar]
- Beatty JD, Beatty BG, Vlahos WG. Measurement of monoclonal antibody affinity by non-competitive enzyme immunoassay. J Immunol Methods. 1987;100:173–9. doi: 10.1016/0022-1759(87)90187-6. [DOI] [PubMed] [Google Scholar]
- Becker R, Lenter MC, Vollkommer T, Boos AM, Pfaff D, Augustin HG, Christian S. Tumor stroma marker endosialin (Tem1) is a binding partner of metastasis-related protein Mac-2 BP/90K. Faseb J. 2008;22:3059–67. doi: 10.1096/fj.07-101386. [DOI] [PubMed] [Google Scholar]
- Bergan L, Gross JA, Nevin B, Urban N, Scholler N. Development and in vitro validation of anti-mesothelin biobodies that prevent CA125/Mesothelin-dependent cell attachment. Cancer Lett. 2007;255:263–74. doi: 10.1016/j.canlet.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Blaise L, Wehnert A, Steukers M, van den Beucken T, Hoogenboom H, Hufton S. Construction and diversification of yeast cell surface displayed libraries by yeast mating: application to the affinity maturation of Fab antibody fragments. Gene. 2004;342:211–8. doi: 10.1016/j.gene.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Bowley DR, Labrijn AF, Zwick MB, Burton DR. Antigen selection from an HIV-1 immune antibody library displayed on yeast yields many novel antibodies compared to selection from the same library displayed on phage. Protein Eng Des Sel. 2007;20:81–90. doi: 10.1093/protein/gzl057. [DOI] [PubMed] [Google Scholar]
- Chan HS, Bradley G, Thorner P, Haddad G, Gallie BL, Ling V. A sensitive method for immunocytochemical detection of P-glycoprotein in multidrug-resistant human ovarian carcinoma cell lines. Lab Invest. 1988;59:870–5. [PubMed] [Google Scholar]
- Christian S, Ahorn H, Koehler A, Eisenhaber F, Rodi HP, Garin-Chesa P, Park JE, Rettig WJ, Lenter MC. Molecular cloning and characterization of endosialin, a C-type lectin-like cell surface receptor of tumor endothelium. J Biol Chem. 2001;276:7408–14. doi: 10.1074/jbc.M009604200. [DOI] [PubMed] [Google Scholar]
- Christian S, Winkler R, Helfrich I, Boos AM, Besemfelder E, Schadendorf D, Augustin HG. Endosialin (Tem1) is a marker of tumor-associated myofibroblasts and tumor vessel-associated mural cells. Am J Pathol. 2008;172:486–94. doi: 10.2353/ajpath.2008.070623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conejo-Garcia JR, Buckanovich RJ, Benencia F, Courreges MC, Rubin SC, Carroll RG, Coukos G. Vascular leukocytes contribute to tumor vascularization. Blood. 2005;105:679–81. doi: 10.1182/blood-2004-05-1906. [DOI] [PubMed] [Google Scholar]
- Connolly DC, Bao R, Nikitin AY, Stephens KC, Poole TW, Hua X, Harris SS, Vanderhyden BC, Hamilton TC. Female mice chimeric for expression of the simian virus 40 TAg under control of the MISIIR promoter develop epithelial ovarian cancer. Cancer Res. 2003;63:1389–97. [PubMed] [Google Scholar]
- Davies G, Cunnick GH, Mansel RE, Mason MD, Jiang WG. Levels of expression of endothelial markers specific to tumour-associated endothelial cells and their correlation with prognosis in patients with breast cancer. Clin Exp Metastasis. 2004;21:31–7. doi: 10.1023/b:clin.0000017168.83616.d0. [DOI] [PubMed] [Google Scholar]
- Dolznig H, Schweifer N, Puri C, Kraut N, Rettig WJ, Kerjaschki D, Garin-Chesa P. Characterization of cancer stroma markers: in silico analysis of an mRNA expression database for fibroblast activation protein and endosialin. Cancer Immun. 2005;5:10. [PubMed] [Google Scholar]
- Feldhaus MJ, Siegel RW, Opresko LK, Coleman JR, Feldhaus JM, Yeung YA, Cochran JR, Heinzelman P, Colby D, Swers J, Graff C, Wiley HS, Wittrup KD. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat Biotechnol. 2003;21:163–70. doi: 10.1038/nbt785. [DOI] [PubMed] [Google Scholar]
- Folkman J. Tumor angiogenesis. Adv Cancer Res. 1985;43:175–203. doi: 10.1016/s0065-230x(08)60946-x. [DOI] [PubMed] [Google Scholar]
- Garlanda C, Parravicini C, Sironi M, De Rossi M, Wainstok de Calmanovici R, Carozzi F, Bussolino F, Colotta F, Mantovani A, Vecchi A. Progressive growth in immunodeficient mice and host cell recruitment by mouse endothelial cells transformed by polyoma middle-sized T antigen: implications for the pathogenesis of opportunistic vascular tumors. Proc Natl Acad Sci U S A. 1994;91:7291–5. doi: 10.1073/pnas.91.15.7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greish K. Enhanced permeability and retention of macromolecular drugs in solid tumors: a royal gate for targeted anticancer nanomedicines. J Drug Target. 2007;15:457–64. doi: 10.1080/10611860701539584. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Pham L, O'Connor MK, Federspiel MJ, Russell SJ, Peng KW. Dual therapy of ovarian cancer using measles viruses expressing carcinoembryonic antigen and sodium iodide symporter. Clin Cancer Res. 2006;12:1868–75. doi: 10.1158/1078-0432.CCR-05-1803. [DOI] [PubMed] [Google Scholar]
- Hinnen P, Eskens FA. Vascular disrupting agents in clinical development. Br J Cancer. 2007;96:1159–65. doi: 10.1038/sj.bjc.6603694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat EA, Wu TT, Perry HM, K.S. G, Foeller C. Sequences of Proteins of Immunological Interest. In: NIH, editor. Sequences of Proteins of Immunological Interest. ed 5 Bethesda, MD: 1991. [Google Scholar]
- MacFadyen JR, Haworth O, Roberston D, Hardie D, Webster MT, Morris HR, Panico M, Sutton-Smith M, Dell A, van der Geer P, Wienke D, Buckley CD, Isacke CM. Endosialin (TEM1, CD248) is a marker of stromal fibroblasts and is not selectively expressed on tumour endothelium. FEBS Lett. 2005;579:2569–75. doi: 10.1016/j.febslet.2005.03.071. [DOI] [PubMed] [Google Scholar]
- Maeda H, Matsumura Y. Tumoritropic and lymphotropic principles of macromolecular drugs. Crit Rev Ther Drug Carrier Syst. 1989;6:193–210. [PubMed] [Google Scholar]
- Marty C, Langer-Machova Z, Sigrist S, Schott H, Schwendener RA, Ballmer-Hofer K. Isolation and characterization of a scFv antibody specific for tumor endothelial marker 1 (TEM1), a new reagent for targeted tumor therapy. Cancer Lett. 2006;235:298–308. doi: 10.1016/j.canlet.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–8. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda A, Karim B, Peng Z, Liu G, Qiu W, Gan C, Vogelstein B, St Croix B, Kinzler KW, Huso DL. Tumor endothelial marker 1 (Tem1) functions in the growth and progression of abdominal tumors. Proc Natl Acad Sci U S A. 2006;103:3351–6. doi: 10.1073/pnas.0511306103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschke M. Phage display systems and their applications. Appl Microbiol Biotechnol. 2006;70:2–11. doi: 10.1007/s00253-005-0270-9. [DOI] [PubMed] [Google Scholar]
- Qiu J, Hanash S. Autoantibody profiling for cancer detection. Clin Lab Med. 2009;29:31–46. doi: 10.1016/j.cll.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Rettig WJ, Garin-Chesa P, Healey JH, Su SL, Jaffe EA, Old LJ. Identification of endosialin, a cell surface glycoprotein of vascular endothelial cells in human cancer. Proc Natl Acad Sci U S A. 1992;89:10832–6. doi: 10.1073/pnas.89.22.10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rmali KA, Puntis MC, Jiang WG. Prognostic values of tumor endothelial markers in patients with colorectal cancer. World J Gastroenterol. 2005;11:1283–6. doi: 10.3748/wjg.v11.i9.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau C, Curiel M, Weber W, Smale R, Kurtzberg L, Mascarello J, Berger C, Wallar G, Bagley R, Honma N, Hasegawa K, Ishida I, Kataoka S, Thurberg BL, Mehraein K, Horten B, Miller G, Teicher BA. Endosialin protein expression and therapeutic target potential in human solid tumors: sarcoma versus carcinoma. Clin Cancer Res. 2008;14:7223–36. doi: 10.1158/1078-0432.CCR-08-0499. [DOI] [PubMed] [Google Scholar]
- Scholler N, Garvik B, Quarles T, Jiang S, Urban N. Method for generation of in vivo biotinylated recombinant antibodies by yeast mating. J Immunol Methods. 2006;317:132–43. doi: 10.1016/j.jim.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholler N, Gross JA, Garvik B, Wells L, Liu Y, Loch CM, Ramirez AB, McIntosh MM, Lampe PD, Urban N. Use of cancer-specific yeast-secreted in vivo biotinylated recombinant antibodies for serum biomarker discovery. J Transl Med. 2008a;6:41. doi: 10.1186/1479-5876-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholler N, Lowe KA, Bergan LA, Kampani AV, Ng V, Forrest RM, Thorpe JD, Gross JA, Garvik BM, Drapkin R, Anderson GL, Urban N. Use of Yeast-Secreted In vivo Biotinylated Recombinant Antibodies (Biobodies) in Bead-Based ELISA. Clin Cancer Res. 2008b;14:2647–55. doi: 10.1158/1078-0432.CCR-07-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz LD, Lang PA, Christianson SW, Gott B, Lyons B, Umeda S, Leiter E, Hesselton R, Wagar EJ, Leif JH, Kollet O, Lapidot T, Greiner DL. NOD/LtSz-Rag1null mice: an immunodeficient and radioresistant model for engraftment of human hematolymphoid cells, HIV infection, and adoptive transfer of NOD mouse diabetogenic T cells. J Immunol. 2000;164:2496–507. doi: 10.4049/jimmunol.164.5.2496. [DOI] [PubMed] [Google Scholar]
- Siegel DL. Translational applications of antibody phage display. Immunol Res. 2008;42:118–3. doi: 10.1007/s12026-008-8044-y. [DOI] [PubMed] [Google Scholar]
- St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C, Vogelstein B, Kinzler KW. Genes expressed in human tumor endothelium. Science. 2000;289:1197–202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- Tan EM, Zhang J. Autoantibodies to tumor-associated antigens: reporters from the immune system. Immunol Rev. 2008;222:328–40. doi: 10.1111/j.1600-065X.2008.00611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teicher BA. Newer vascular targets: endosialin (review) Int J Oncol. 2007;30:305–12. [PubMed] [Google Scholar]
- Tomkowicz B, Rybinski K, Foley B, Ebel W, Kline B, Routhier E, Sass P, Nicolaides NC, Grasso L, Zhou Y. Interaction of endosialin/TEM1 with extracellular matrix proteins mediates cell adhesion and migration. Proc Natl Acad Sci U S A. 2007;104:17965–70. doi: 10.1073/pnas.0705647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson IM, Williams SC, Corbett SJ, Cox JPL, Winter G. Base Directory of Human V Gene Sequences. 1996 [Google Scholar]
- Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, McCafferty J, Hodits RA, Wilton J, Johnson KS. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol. 1996;14:309–14. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.