Abstract
Short-interfering RNAs (siRNAs) are common tools in molecular biology, however the development of RNAi-based therapeutics is limited by immunostimulatory and non-specific effects mediated by off-target RNA-binding proteins. PKR and ADAR1 are two proteins implicated in RNAi off-target effects, and share a common means of interaction with siRNAs through double-stranded RNA binding motifs (dsRBMs). Here we report the site-specific introduction of N2- propargyl 2-aminopurine into siRNAs and subsequent conversion to two bulky products via copper-catalyzed azide alkyne cycloaddition (CuAAC) with either Nazidoacetyl-mannosamine azide or N-ethylpiperidine azide. We observed position-specific effects on RNAi activity for modifications made to both the passenger and guide strands. These findings are rationalized in light of recent structural studies of components of the RNA-induced silencing complex (RISC) and RISC-loading complex (RLC). The most active siRNAs were assayed for binding affinity to the RNA-dependent protein kinase (PKR) and adenosine deaminase that acts on RNA 1 (ADAR1). PKR binding was significantly reduced by multiple modifications, regardless of size, and ADAR1 binding was reduced in a position and size-sensitive manner. Our findings present a strategy for designing siRNAs that reduce off-target dsRBM-protein binding while retaining native RNAi activity.
Introduction
Selective nuclease digestion of messenger RNAs inside living cells via the short interfering RNA (siRNA)-triggered RNA interference (RNAi) pathway has become a mainstay in molecular biology to study gene function and has exciting therapeutic potential (1). However, converting these valuable research tools into drugs presents challenges due to restrictions imposed by the natural RNA chemical structure. For instance, unmodified siRNAs are sensitive to nuclease degradation in serum (2), can suppress off-target genes (3), and often have poor cellular uptake and delivery to the desired site of action (4). In addition, administration of siRNAs has been shown to stimulate the immune system (5–7).
A principal source of the undesirable properties of siRNAs is their ability to bind to cellular proteins that are independent of the RNAi pathway (5). Several RNA-binding proteins have been implicated in the immunostimulatory effects of siRNAs, including toll-like receptors (TLRs) 3 (8, 9), 7 (10, 11) and 8 (12), and the RNA-dependent protein kinase (PKR) (7). Retinoic acid inducible gene 1 (RIG-1) and melanoma differentiation-associated gene 5 (MDA 5) are also capable of recognizing double-stranded RNA and can trigger immune responses, making them candidate off-target receptors for siRNAs (13, 14)
Recent studies highlight the need to regulate siRNA binding to double-stranded RNA binding motif (dsRBM) proteins, such as PKR and adenosine deaminases that act on RNA (ADARs) (7, 15, 16). The dsRBM is an evolutionarily conserved motif expressed in many proteins both on and off the RNAi pathway. The motif consists of ~70 amino acids and binds to duplex RNA in a largely sequence independent fashion (17–19). One dsRBM protein in particular, PKR, is well known for its role in siRNA-dependent immunostimulation and off-target effects (7, 15). Duplex RNA-dependent activation of PKR leads to phosphorylation of eIF2α, inhibition of translation initiation and other antiviral signaling events (20). Although siRNAs are shorter than ligands that trigger high PKR activity, lower level activation of PKR has been observed with siRNAs in vitro and when transfected into certain cell types (7, 15, 21, 22). A report by Donnely and coworkers described the PKR-dependent production of macrophage migratory inhibitory factor (MIF) and the resultant proliferation of breast cancer cells in response to anti-MIF siRNA transfection (15). These effects were induced by control siRNAs as well indicating they were sequence independent.
ADARs, RNA editing adenosine deaminases, also contain dsRBMs and have been shown to interact with substrates within the RNAi pathway (23, 24). The full-length isoform of ADAR1 (ADAR1p150) has been linked to decreased siRNA potency in mammalian cells, presumably due to the formation of high affinity siRNA • ADAR1p150 complexes within the cytoplasm (16). Additionally, dADAR has been shown to reduce RNAi efficiency in Drosophila (24) cell culture.
Chemical modification of the siRNA strands provides a solution to the undesired binding of cellular receptors and resultant off-target effects (25). To date, chemical modification has mainly been applied at the ribose and the phosphodiester backbone, and has focused on improving nuclease resistance, siRNA activity and eliminating immune responses. For example, inclusion of 2’-OMe, 2’-F, 2’-deoxy (DNA), locked ribose (LNA) and phosphorothioate modifications at specific sites in an RNA duplex protects against immune activation and improves in vivo persistence (26). Alternating 2’-OMe/2’-F modification also dramatically enhances silencing potency (27). Less is known about how structural changes to the nucleobases alter the characteristics of siRNAs. Reports indicate that incorporation of unnatural bases into siRNAs can alter the sequence specificity for the mRNA target (28), perturb duplex stability (29, 30) and increase serum stability (31, 32), but little information exists on the effects of base modifications towards off-pathway protein binding.
Our previous studies demonstrated that the base analog N2-benzyl-2’-deoxyguanosine is tolerated in the siRNA passenger strand with some loss of duplex stability and RNAi activity while also substantially reducing binding to PKR (22). Since dsRBM proteins recognize double-stranded RNA through minor groove contacts (33, 34), and the N2 substituent of guanosine is directed into the minor groove, this effect is most likely due to steric occlusion, as has been previously observed in non-siRNA substrates (35). Analogous to PKR, minor groove modifications are known to interrupt the binding of ADAR’s dsRBMs to RNA. This was demonstrated for ADAR2, a closely related member of the ADAR family (36), however, to date such modifications have not been analyzed in the context of siRNAs or for ADAR1.
Here we employ our recently developed 2-aminopurine substitution strategy (37) to modulate the minor groove structure at various sites in the passenger and guide strands of siRNAs. We demonstrate positional and structural dependent effects on RNAi activity and binding of the proteins PKR and ADAR1.
RESULTS AND DISCUSSION
Modular modification of the siRNA minor groove
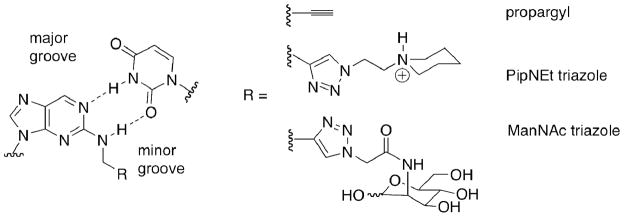
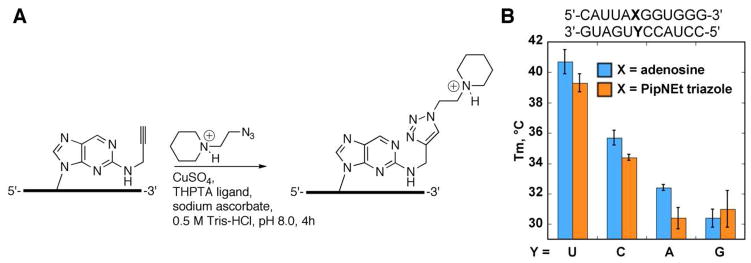
We tested three analogs of 2-aminopurine as replacements for adenosine in A•U pairs in siRNA. The analogs contain substituents on N2, which project into the minor groove of the RNA duplex, the site of recognition for dsRBM proteins (Figure 1) (33, 34). Replacement of adenosine with N2-propargyl-2-aminopurine ribonucleoside results in some loss of duplex stability (ΔTM = 4.5 °C per modification in a 12 mer duplex) but conserves base-pairing specificity, as previously reported (37). RNA strands containing this analog were further modified by copper-catalyzed azide-alkyne cycloaddition (CuAAC) (37) using two different azides. Reaction with N-azidoacetyl-D-mannosamine was chosen to generate a polar and neutral minor groove substituent more sterically demanding than propargyl (ManNAc triazole) (Figure 1). Also, a new analog was generated using an azide of N-ethylpiperidine, providing a large, positively charged minor groove substituent (PipNEt triazole) (Figure 1, Figure 2). We carried out the click reaction in aqueous conditions and purified the resultant triazole products by gel electrophoresis. This method provides an alternative to a previously described post-automated synthesis modification of the purine 2-position via convertible nucleosides (38), which requires strand deprotection after modification and longer reaction times. The advantages of click chemistry make it particularly desirable for the generation of modified siRNA libraries and to investigate siRNA structure activity relationships. Thermal denaturation studies either described previously (37) or reported here (Figure 2) indicate the two triazoles increase thermal stability relative to the propargyl modification to generate stable base pairs with uridine and effectively mimic adenosine’s base pairing specificity in duplex RNA.
Figure 1.
N2-modified 2-aminopurines employed in this study. The nucleobases replace adenosine in an A•U base pair, directing the N2 substituent into the minor grove of the siRNA duplex.
Figure 2.
Triazole-containing RNAs were generated from N2-propargyl 2-aminopurine-containing RNA via CuAAC reaction. A, generation of N-ethylpiperidine triazole RNA (PipNEt). B, Thermal denaturation analysis of the PipNEt triazole modification in a 12 mer duplex indicates that the stability and specificity of the modification is comparable to that of adenosine.
RNA interference with modified siRNAs
To evaluate the effect of the different nucleoside analogs on RNAi activity, we substituted adenosines in both the guide and passenger strands of an siRNA targeting the human caspase 2 message. We employed a commercial Renilla/firefly luciferase plasmid with the siRNA target sequence inserted into the 3’ UTR of the Renilla luciferase reporter. The efficiency of Renilla-directed RNAi was evaluated in HeLa cells. The results, showing Renilla luciferase activity normalized to nontargeted firefly luciferase activity, are shown in Figures 3 and 4.
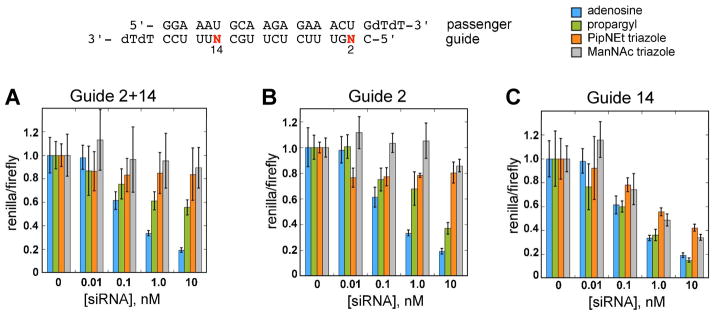
Figure 3.
Gene-specific RNAi activity for siRNAs containing guide strand modifications. A, 2 and 14 together, B, position 2 only and, C, position 14 only. Green, orange and gray bars correspond to propargyl, PipNEt triazole and ManNAz triazole modifications, respectively. Blue bars correspond to unmodified siRNA.
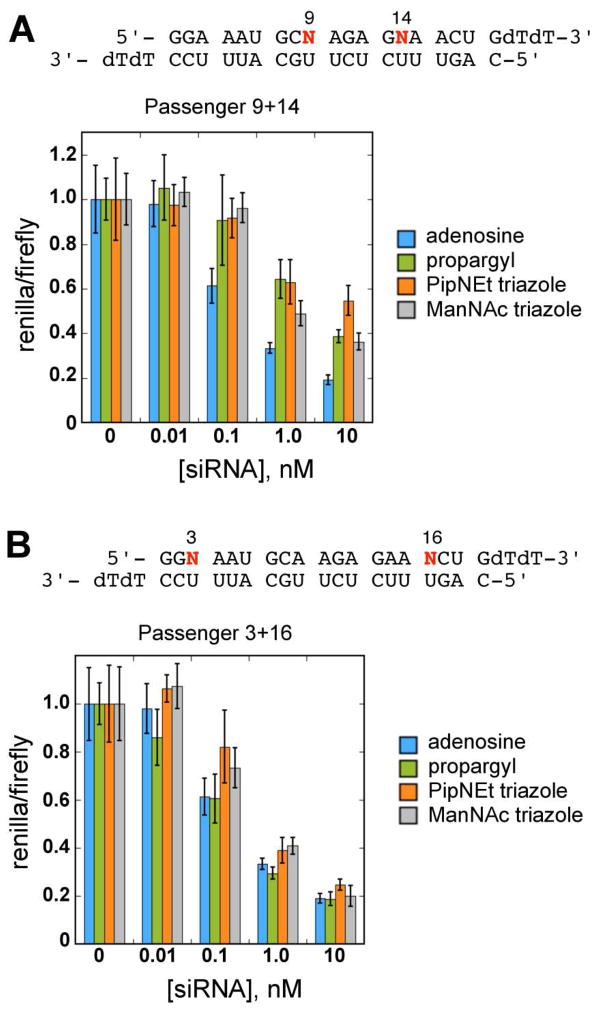
Figure 4.
Gene-specific RNAi activity for siRNAs containing passenger strand modifications. A, Positions 9 and 14 together and, B, positions 3 and 16 together. Green, orange and gray bars correspond to propargyl, PipNEt triazole and ManNAz triazole modifications, respectively. Blue bars correspond to unmodified siRNA.
The guide strand of the caspse 2 siRNA contains two adenosines (positions 2 and 14). We replaced these bases with 2-aminopurine analogs, in combination and individually (Figure 3, A–C). We found that replacing both adenosines (Guide 2+14) with any of the three analogs inhibited RNAi activity (Figure 3, A). The propargyl substitution had the least detrimental effect, giving 40% knockdown at 10 nM where > 80% was observed for the adenosine-containing control. The lower RNAi activity for the Guide 2+14 pattern is due primarily to changes at position 2, which falls within in the seed region (positions 2–8). Substitution of the adenosine at position 2 in the guide strand alone (Guide 2) with either of the two triazoles completely inhibits RNAi (Figure 3, B). Propargyl at this position retains activity, albeit at a slightly lower level (60% knockdown at 10 nM). The seed region guides target recognition by the active RNA-induced silencing complex (RISC), and is known to be sensitive to bulky modifications and modifications which dramatically increase or decrease the thermal stability (39). Our modifications do not dramatically alter thermal stability, however predictions based upon the crystal structures of a prokaryotic argonaute (Ago) protein bound to ssDNA guide and ssRNA target (40) (Figure 5, A) and an archaeal Piwi protein bound to siRNA-like duplexes (41) (Supplementary Figure 1) indicate that modification at position 2 could interfere with Ago binding. In these structures, the minor groove at position 2 is in close contact with amino acid residues of the PIWI/MID domains. A steric clash would occur upon the addition of bulky groups to the minor groove at this site. For example, in the prokaryotic Ago protein/guide complex, the 2-amino group of the guanosine nucleotide at position 2 in the guide is within 4.5 Å of three amino acid residues. The 2-amino group of guanosine in a G • C base pair is located in a similar relative spatial position to the 2-amino group of 2-aminopurine in a base pair with U, indicating introduction of substituents at N2 of 2-aminopurine would be predicted to cause a steric clash with these residues. Previous studies involving modifications made at position 2 of the guide strand are consistent with this observation. For instance, a single 2’-O-methoxyethyl ribose modification at position 2, which is also directed in to the minor groove, has been shown to reduce RNAi activity (42). However less sterically demanding 2’ modifications such as 2’-OMe (43) and 2’-F (44) are tolerated at this site. The observation that RNAi activity is dependent on minor groove structure at this position suggests interesting possibilities for temporal control of RNAi. For example, RNAi activity could be eliminated by installing a photocaging group either on the base or the sugar, and regained by in situ deprotection of the modifier (45).
Figure 5.
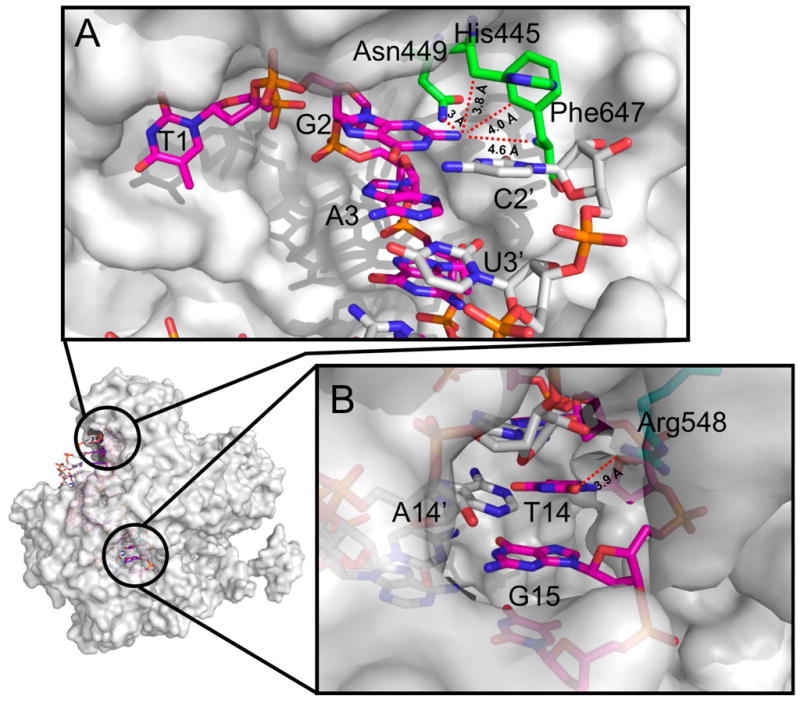
Crystal structure of T. thermophilus Ago(Asn478) in complex with 21 nt guide DNA (purple) and 19 nt target RNA (silver), showing amino acid residues (green) within 5 Å of the guide strand at A, N2 of guanosine 2 and, B, O2 of thymine 14. The structure suggests that minor groove modifications at position 2 would not be tolerated due to close protein contacts, but would be accommodated at position 14 due to a gap in the protein structure. PDB code: 3HK2(40).
Both of the triazole modifications are accommodated significantly better at position 14 of the guide strand than at position 2, and the corresponding propargyl modified siRNA has activity comparable to that of the unmodified siRNA (Figure 3, C). The prokaryotic Ago/duplex crystal structure indicates that position 14 of the guide strand is void of close protein contacts, suggesting that in RISC, modifications at this site would be directed into solvent (Figure 5, B). This is consistent with our observation that exceptionally bulky click products are tolerated at this position. Interestingly, 2’-O-methoxyethyl modification at position 14 has previously been reported to abolish RNAi activity (42). However, this is presumably due to the methoxyethyl residues interfering with catalysis, rather than due to steric demand, since the 2’-O-methoxyethyl modified guide strand retained affinity for Ago2. Taken together, these observations indicate a marked difference in tolerance between modification at the ribose and modification at the nucleobase at this particular position. This highlights an important and unforeseen advantage of base modifications, in that large minor groove substituents projecting from the base can be introduced at critical sites where modification of ribose is known to be detrimental to activity.
Guided by our previous work, in which we determined critical passenger strand modification sites for blocking dsRBM interactions (22), we employed a passenger strand modification pattern in which adenosines 9 and 14 are substituted together for 2-aminopurine analogs. We also chose a modification pattern in which adenosines 3 and 16 are substituted together, allowing us to evaluate the positional effects of modification (Figure 4). Both modification patterns lead to functional siRNAs. Modification in the middle of the passenger strand (Passenger 9+14) leads to a small but reproducible reduction in RNAi activity with each of the analogs tested (Figure 4, A). Importantly, however, we found that modification with all analogs near the ends of the passenger strand (Passenger 3+16) has no effect on RNAi activity (Figure 4, B). Ago2 has been shown to cleave the passenger strand during RISC activation in an Mg2+-dependent mechanism at the scissile phosphate of the passenger strand opposite position 10 of the guide (46, 47). In the 21-bp siRNA used in this study this corresponds to position 9 of the passenger strand. Previous studies have shown that 2’-OMe ribose modification at passenger position 9 substantially reduces RISC activity, presumably due to a disruption in magnesium ion binding in the active site (47). Modification of the base with large, bulky groups via the Passenger 9+14 modification pattern could have a similar effect, resulting in the observed mild inhibition of RNAi. An alternative explanation is that the Passenger 9+14 modification may impede RISC loading by inhibiting components of the RISC loading complex (RLC). In humans, TRBP and Dicer are involved in loading the siRNA guide strand into RISC (48–50). Both of these proteins contain dsRBM domains (51), and would therefore be expected to be sensitive to minor-groove modifications. It is possible that modifications near the middle of the duplex are capable of disrupting interactions with these critical dsRBMs.
Interestingly, the Passenger 3+16 pattern retained full RNAi capability despite the large triazole modifications, suggesting minimal recognition in the minor groove near the ends of the siRNA duplex during RISC loading. Recent studies of the RLC and RISC are consistent with this phenomenon. The process of RISC loading is still not fully understood, but involves the transfer of the siRNA from Dicer to Ago2 with the assistance of TRBP. A model of this process, supported by cryo-EM structures of the human Dicer/Ago2 complex, suggests an intermediate involved in the transfer of siRNA from Dicer to Ago2, in which both ends of the siRNA are simultaneously bound by the PAZ domains of Dicer and Ago2 (52–54). One would suspect that end modifications would disrupt this process by clashing with the PAZ domains, yet this does not appear to be the case. The crystal structure of a human Ago PAZ domain bound to an siRNA-like RNA shows that the minor groove near the ends of the duplex is largely free of protein contact (Supplementary Figure 2) (55, 56), indicating that large minor groove modifications would be tolerated in these positions, as we observe.
Considering the studies described above, RNAi experiments using siRNAs with 2-aminopurine derivatives replacing specific adenosines identified seven modified siRNAs with superior RNAi activity. Four of these duplexes had activity indistinguishable from the unmodified siRNA (Passenger 3+16 with propargyl, PipNEt triazole and ManNAc triazole along with Guide 14 with propargyl). Three of these duplexes showed small reductions in RNAi activity compared to the adenosine-containing control (Passenger 9+14 pattern with propargyl or ManNAc and Guide 14 with ManNAc).
Protein binding to modified siRNAs
For these seven most active duplexes, we tested the effects of the minor groove modifications on the binding of the proteins ADAR1 and PKR, two dsRBM proteins previously shown to interact with siRNAs and cause unwanted effects (vide supra). We generated corresponding siRNA duplexes with 5’-biotin labels on the unmodified strands and used affinity purification with magnetic streptavidin beads to isolate siRNA-binding proteins present in lysates from human cells grown in culture (21). The siRNA-binding proteins were then analyzed via Western blot, allowing us to evaluate the effect the modifications had on the binding to specific proteins of interest. For these studies, we used lysates from U87 human glioblastoma cells grown in the presence of interferon– α. The U87 cell line has been shown to up-regulate ADAR1 in response to interferon stimulation (57) and expresses PKR, making it a good a model to study the binding of siRNAs to dsRBM proteins.
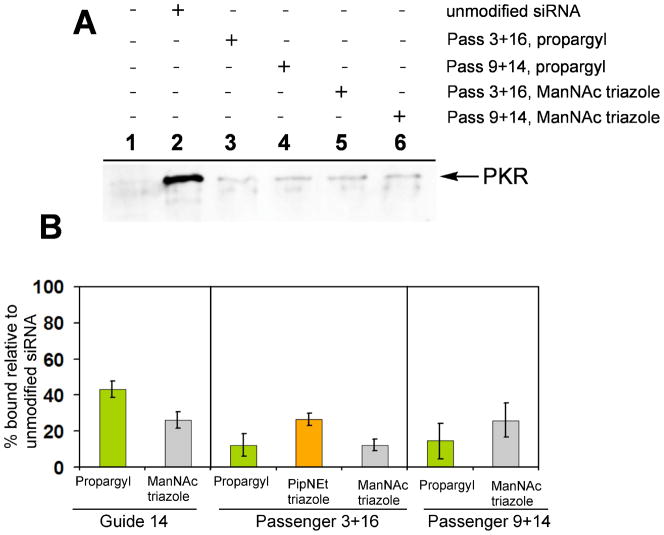
PKR was particularly sensitive to the minor groove modifications. Of the seven duplexes tested, only the duplex with the propargyl modification at guide position 14 retained levels of PKR binding greater than 30% (43 ± 5% of the unmodified control) (Figure 6A, B). Each of the remaining modified duplexes bound less than 30% of the PKR found in the control sample. Since binding is a prerequisite to PKR activation, we predict that the bulky minor groove modifications presented here will result in abatement of PKR activation. We have previously shown that that N2-benzyl-2’-deoxyguanosine incorporated at positions 9 and 14 of the passenger strand causes 75–80% reduction in maximal PKR activation (22). Here we report similar results with respect to PKR binding in cell lysates.
Figure 6.
A, Westen blot for PKR showing reduction in binding affinity as a result of propargyl modification (lanes 3 and 4) and ManNAc triazole modification (lanes 5 and 6) in the passenger strand, relative to unmodified siRNA (lane 2). B, Plot showing PKR binding (relative to unmodified siRNA) for the modified siRNAs that displayed the highest RNAi activity.
ADAR1 exists as two isoforms, cytoplasmic p150, and nuclear-localized p110. The two isoforms are distinguished by the presence of two Z-DNA binding domains at the N-terminal end of the p150 isoform (58). Both isoforms are retained on the streptavidin beads in an siRNA-dependent manner and the binding of both isoforms is reduced to similar extents by the chemical modifications (Figure 7). In contrast to PKR, ADAR1 binding was reduced to a lesser extent and was more sensitive to the sites and sizes of modifications. While modification of the guide strand at position 14 with propargyl showed minimal effect on the amount of ADAR1 retained, reduced binding was observed with the bulky ManNAc triazole at this position (51 ± 7% of the unmodified control for p150). This trend is also observed with the passenger strand modification patterns. The Passenger 3+16 pattern modified with propargyls bound 91 ± 15% of the ADAR1p150 bound by the control sample, and both the ManNAc and PipNEt triazoles were much more effective at preventing ADAR1 binding (< 40% for both forms). By comparison, the Passenger 3+16 pattern with propargyls reduced PKR binding to 12 ± 3% (Figure 6B). The increased tolerance to modified siRNA demonstrated by ADAR could be due to the presence of three dsRBMs compared to PKR’s two (59).
Figure 7.
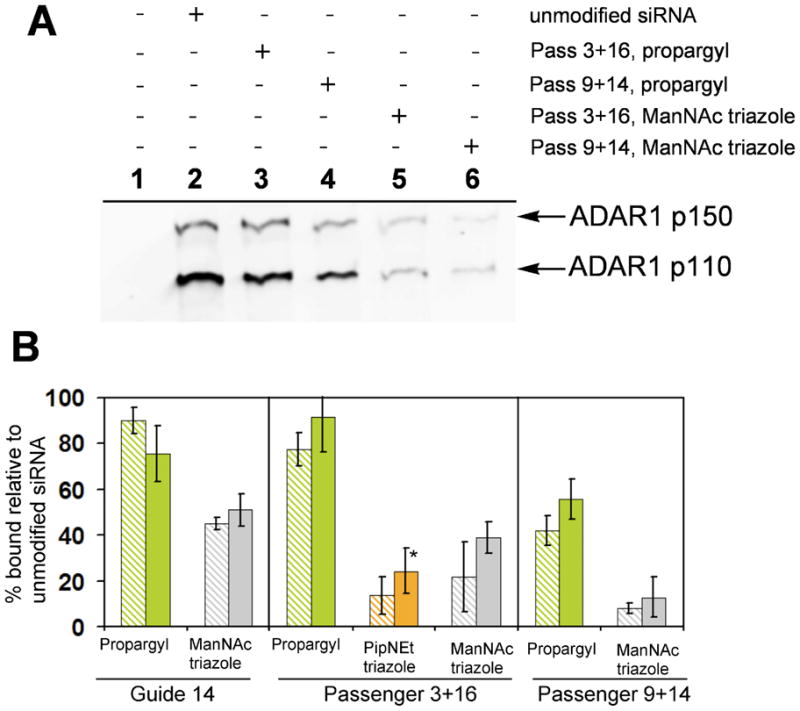
A, Westen blot for ADAR1 showing reduction in binding affinity as a result of propargyl modification (lanes 3 and 4) and ManNAc triazole modification (lanes 5 and 6) in the passenger strand, relative to unmodified siRNA (lane 2). B, Plot showing ADAR1 binding (relative to unmodified siRNA) for the modified siRNAs that displayed the highest RNAi activity. (Dashed bars: ADAR1p110, solid bars: ADAR1p150, *n = 2)
Modifications within the Passenger 9+14 pattern were more effective at blocking ADAR1 than corresponding modifications within the Passenger 3+16 pattern. When propargyls were incorporated in the Passenger 9+14 pattern, the ADAR1p150 binding was reduced to 56 ± 9%, significantly less than the propargyls in the Passenger 3+16 duplex. Additionally, the ManNAc triazole in the Passenger 9+14 pattern reduced ADAR1 binding to approximately 10% of the unmodified control siRNA. Since the Passenger 9+14 pattern contains modifications on opposite sides towards the middle of the duplex, while the Passenger 3+16 pattern features modifications on the same side and towards the ends of the duplex, these results suggest a potential binding configuration involving the placement of ADAR1’s dsRBMs on opposite sides of the duplex.
Overall, the Passenger 3+16 duplex containing the PipNEt triazole modification yielded the best combination of RNAi activity and dsRBM complex disruption. Its activity was indistinguishable from the unmodified siRNA and it blocked both PKR and ADAR1 binding to < 30% of the amount bound but the unmodified siRNA. The Passenger 9+14 duplex containing ManNAc triazoles was very effective at blocking both ADAR1 and PKR, however displayed slightly reduced RNAi activity.
These chemical modification strategies will prove useful in designing siRNAs refractory to confounding dsRBM proteins, thereby increasing gene-specific silencing activity. This will be realized in cells that express high levels of ADAR1, such as immune cells during inflammation (60), or cells that are known to be sensitive to siRNA-dependent PKR activation (15). Since the expression of both PKR and ADAR1p150 is stimulated by interferon (61, 62), the chemical modifications outlined here could also be particularly advantageous in the context of recent proposals for purposefully activating the immune system in combination with siRNA-directed gene knockdown (63, 64). This approach has potential in antiviral and antitumor therapy, where the induction of the innate immune response can be beneficial to clearance of the disease. Since expression levels of PKR and ADAR1p150 are elevated in an immunostimulatory environment, one would predict that increased gene-specific knockdown could be achieved by minimizing the binding of siRNAs to these receptors. As shown here, this can be achieved by incorporating bulky minor-groove modifications at key positions into the siRNAs.
In summary, we have described a method for varying the structure and position of minor groove-localized base substituents within siRNAs to block the binding to the off-pathway dsRBM proteins ADAR1 and PKR, while maintaining high RNAi activity. In general, minor-groove modifications are more detrimental to PKR than ADAR1 binding, however, the incorporation of bulky modifications (ManNAc and PipNEt triazoles) at two sites in the passenger strand of the duplex yielded siRNAs that displayed minimal binding to both of these proteins. The siRNAs modified towards the end of the passenger strand with the bulky modifications displayed activity indistinguishable from the unmodified duplex. In addition, bulky modifications were accommodated surprisingly well at position 14 of the guide strand. Our findings demonstrate the utility of base modifications in elucidating and blocking protein/siRNA interactions, and contribute to the growing portfolio of chemical modifications available for improving the properties of siRNAs.
METHODS
RNA oligonucleotides were synthesized using 5’-DMTrO, 2’-OTBDMS protected β-cyanoethyl phosphoramidites. CuAAC reaction between propargyl-containing RNAs and the relevant azides was performed in water or Tris-HCl, pH 8.0, using THPTA ligand (65), followed by PAGE purification. RNAi activity was measured in HeLa cells using a co-transfected dual-luciferase plasmid containing the target sequence (psiCHECK2, Promega) and the Dual Luciferase Assay Reporter system (Promega). siRNA protein binding was measured by incubating siRNA-conjugated magnetic streptavidin beads (Dynabeads M-280, Invitrogen) with lysate from U87 cells (treated with human interferon-α A (PBL Interferon Source)), followed by washing, elution, and western blotting for PKR and ADAR1. Full details of methods are provided in the Supporting Information.
Supplementary Material
Acknowledgments
P.A.B. acknowledges the National Institutes of Health for financial support in the form of grant R01-GM080784. We thank Cynthia Burrows and Arunkumar Kannan (University of Utah) for the gift of the psiCHECK2-caspase2 plasmid. Mass spectrometry was performed by Lieza M. Danan-Leon at the Mass Spectrometry Facility, Univerisity of California, Davis.
Supporting Information Available: This material is available free of charge via the Internet.
References
- 1.Kurreck J. RNA interference: from basic research to therapeutic applications. Angew Chem Int Ed. 2009;48:1378–1398. doi: 10.1002/anie.200802092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choung S, Kim YJ, Kim S, Park HO, Choi YC. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem Bioph Res Co. 2006;342:919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 3.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 4.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19:89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 6.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 7.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BRG. Activation of the Interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 8.Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, Albuquerque RJC, Yamasaki S, Itaya M, Pan YZ, Appukuttan B, Gibbs D, Yang ZL, Kariko K, Ambati BK, Wilgus TA, DiPietro LA, Sakurai E, Zhang K, Smith JR, Taylor EW, Ambati J. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kariko K, Bhuyan P, Capodici J, Weissman D. Small Interfering RNAs Mediate Sequence-Independent Gene Suppression and Induce Immune Activation by Signaling through Toll-Like Receptor 3. J Immunol. 2004;172:6545–6549. doi: 10.4049/jimmunol.172.11.6545. [DOI] [PubMed] [Google Scholar]
- 10.Gantier MP, Tong S, Behlke MA, Xu D, Phipps S, Foster PS, Williams BRG. TLR7 is involved in sequence-specific sensing of single-stranded RNAs in human macrophages. J Immunol. 2008;180:2117–2124. doi: 10.4049/jimmunol.180.4.2117. [DOI] [PubMed] [Google Scholar]
- 11.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, Fougerolles Ad, Endres S, Hartmann G. Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 12.Forsbach A, Nemorin JG, Montino C, Iler CM, Samulowitz U, Vicari AP, Jurk M, Mutwiri GK, Krieg AM, Lipford GB, Vollmer Jr. Identification of RNA Sequence Motifs Stimulating Sequence-Specific TLR8-Dependent Immune Responses. J Immunol. 2008;180:3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- 13.Marques JT, Devosse T, Wang D, Zamanian-Daryoush M, Serbinowski P, Hartmann R, Fujita T, Behlke MA, Williams BR. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 14.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Sousa CRE, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 15.Armstrong ME, Gantier M, Li LL, Chung WY, McCann A, Baugh JA, Donnelly SC. Small interfering RNAs induce macrophage migration inhibitory factor production and proliferation in breast cancer cells via a double-stranded RNA-dependent protein kinase-dependent mechanism. J Immunol. 2008;180:7125–7133. doi: 10.4049/jimmunol.180.11.7125. [DOI] [PubMed] [Google Scholar]
- 16.Yang W, Wang Q, Howell KL, Lee JT, Cho DC, Murray JM, Nishikura K. ADAR1 RNA Deaminase Limits Short Interfering RNA Efficacy in Mammalian Cells. J Biol Chem. 2005;280:3946–3953. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang KY, Ramos A. The double-stranded RNA-binding motif, a versatile macromolecular docking platform. FEBS J. 2005;272:2109–2117. doi: 10.1111/j.1742-4658.2005.04652.x. [DOI] [PubMed] [Google Scholar]
- 18.Beal PA. Duplex RNA-binding enzymes: Headliners from neurobiology, virology, and development. Chembiochem. 2005;6:257–266. doi: 10.1002/cbic.200400303. [DOI] [PubMed] [Google Scholar]
- 19.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded-RNA-binding motif: interference and much more. Nat Rev Mol Cell Bio. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 20.Li SD, Peters GA, Ding KY, Zhang XL, Qin J, Sen GC. Molecular basis for PKR activation by PACT or dsRNA. Proc Natl Acad Sci USA. 2006;103:10005–10010. doi: 10.1073/pnas.0602317103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Weinschenk T, Gua K, Schluesener HJ. siRNA Binding Proteins of Microglial Cells: PKR is an Unanticipated Ligand. J Cell Biochem. 2006;97:1217–1229. doi: 10.1002/jcb.20716. [DOI] [PubMed] [Google Scholar]
- 22.Puthenveetil S, Whitby L, Ren J, Kelnar K, Krebs JF, Beal PA. Controlling activation of the RNA-dependent protein kinase by siRNAs using site-specific chemical modification. Nucleic Acids Res. 2006;34:4900–4911. doi: 10.1093/nar/gkl464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Bio. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heale BS, Keegan LP, McGurk L, Michlewski G, Brindle J, Stanton CM, Caceres JF, O'Connell MA. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009;28:3145–3156. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shukla S, Sumaria CS, Pradeepkumar PI. Exploring chemical modifications for siRNA therapeutics: a structural and functional outlook. ChemMedChem. 2010;5:328–349. doi: 10.1002/cmdc.200900444. [DOI] [PubMed] [Google Scholar]
- 26.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, Vaish N, Zinnen S, Vargeese C, Bowman K, Shaffer CS, Jeffs LB, Judge A, MacLachlan I, Polisky B. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 27.Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Wanders L, Griffey RH, Swayze EE, Bhat B. Fully 2'-Modified Oligonucleotide Duplexes with Improved in Vitro Potency and Stability Compared to Unmodified Small Interfering RNA. J Med Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 28.Somoza A, Silverman A, Miller R. Steric effects in RNA interference: probing the influence of nucleobase size and shape. Chem Eur J. 2008;14:7978–7987. doi: 10.1002/chem.200800837. [DOI] [PubMed] [Google Scholar]
- 29.Sipa K, Sochacka E, Kazmierczak-Baranska J, Maszewska M, Janicka M, Nowak G, Nawrot B. Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA. RNA. 2007;13:1301–1316. doi: 10.1261/rna.538907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Somoza A, Chelliserrykattil J, Kool ET. The roles of hydrogen bonding and sterics in RNA interference. Angew Chem Int Ed. 2006;45:4994–4997. doi: 10.1002/anie.200601311. [DOI] [PubMed] [Google Scholar]
- 31.Terrazas M, Kool E. RNA major groove modifications improve siRNA stability and biological activity. Nucleic Acids Res. 2009;37:346–353. doi: 10.1093/nar/gkn958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia J, Noronha A, Toudjarska I, Li F, Akinc A, Braich R, Frank-Kamenetsky M, Rajeev KG, Egli M, Manoharan M. Gene Silencing Activity of siRNAs with a Ribo-difluorotoluyl Nucleotide. ACS Chem Biol. 2006;1:176–183. doi: 10.1021/cb600063p. [DOI] [PubMed] [Google Scholar]
- 33.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu HH, Henras A, Chanfreau G, Feigon J. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc Natl Acad Sci USA. 2004;101:8307–8312. doi: 10.1073/pnas.0402627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nallagatla S, Bevilacqua P. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephens OM, Haudenschild BL, Beal PA. The binding selectivity of ADAR2's dsRBMs contributes to RNA-editing selectivity. Chem Biol. 2004;11:1239–1250. doi: 10.1016/j.chembiol.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 37.Peacock H, Maydanovych O, Beal PA. N(2)-Modified 2-aminopurine ribonucleosides as minor-groove-modulating adenosine replacements in duplex RNA. Org Lett. 2010;12:1044–1047. doi: 10.1021/ol100019r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allerson C, Chen S, Verdine G. A chemical method for site-specific modification of RNA: the convertible nucleoside approach. J Am Chem Soc. 1997;119:7423–7433. [Google Scholar]
- 39.Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N, Babu BR, Hojland T, Abramov M, Van Aerschot A, Odadzic D, Smicius R, Haas J, Andree C, Barman J, Wenska M, Srivastava P, Zhou C, Honcharenko D, Hess S, Muller E, Bobkov GV, Mikhailov SN, Fava E, Meyer TF, Chattopadhyaya J, Zerial M, Engels JW, Herdewijn P, Wengel J, Kjems J. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lima WF, Wu H, Nichols JG, Sun H, Murray HM, Crooke ST. Binding and Cleavage Specificities of Human Argonaute2. J Biol Chem. 2009;284:26017–26028. doi: 10.1074/jbc.M109.010835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bramsen J, Laursen M, Nielsen A, Hansen T, Bus C, Langkjaer N, Babu B, Hojland T, Abramov M, Van Aerschot A, Odadzic D, Smicius R, Haas J, Andree C, Barman J, Wenska M, Srivastava P, Zhou C, Honcharenko D, Hess S, Muller E, Bobkov G, Mikhailov S, Fava E, Meyer T, Chattopadhyaya J, Zerial M, Engels J, Herdewijn P, Wengel J, Kjems J. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, Corey DR. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 45.Shah S, Rangarajan S, Friedman SH. Light-activated RNA interference. Angew Chem Int Ed. 2005;44:1328–1332. doi: 10.1002/anie.200461458. [DOI] [PubMed] [Google Scholar]
- 46.Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 47.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 48.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci USA. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gredell J, Dittmer M, Wu M, Chan C, Walton SP. Recognition of siRNA asymmetry by TAR RNA binding protein. Biochemistry. 2010;49:3148–3155. doi: 10.1021/bi902189s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sashital DG, Doudna JA. Structural insights into RNA interference. Curr Opin Struc Biol. 2010;20:90–97. doi: 10.1016/j.sbi.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lau PW, Potter CS, Carragher B, MacRae IJ. Structure of the human Dicer-TRBP complex by electron microscopy. Structure. 2009;17:1326–1332. doi: 10.1016/j.str.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang HW, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E. Structural insights into RNA processing by the human RISC-loading complex. Nat Struct Mol Biol. 2009;16:1148–1153. doi: 10.1038/nsmb.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.MacRae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 56.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang W, Wang Q, Kanes SJ, Murray JM, Nishikura K. Altered RNA editing of serotonin 5-HT2C receptor induced by interferon: implications for depression associated with cytokine therapy. Mol Brain Res. 2004;124:70–78. doi: 10.1016/j.molbrainres.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 58.Herbert A, Alfken J, Kim Y-G, Mian IS, Nishikura K, Rich A. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc Natl Acad Sci USA. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang JH, Luo XX, Nie YZ, Su YJ, Zhao QC, Kabir K, Zhang DX, Rabinovici R. Widespread inosine-containing mRNA in lymphocytes regulated by ADAR1 in response to inflammation. Immunology. 2003;109:15–23. doi: 10.1046/j.1365-2567.2003.01598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meurs E, Chong K, Galabru J, Thomas NSB, Kerr IM, Williams BRG, Hovanessian AG. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 62.George CX, Samuel CE. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Nat Acad Sci USA. 1999;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gantier M, Williams B. siRNA delivery not Toll-free. Nat Biotechnol. 2009;27:911–912. doi: 10.1038/nbt1009-911. [DOI] [PubMed] [Google Scholar]
- 64.Gantier M, Tong S, Behlke M, Irving A, Lappas M, Nilsson U, Latz E, McMillan NA, Williams B. Rational design of immunostimulatory siRNAs. Mol Ther. 2010;18:785–795. doi: 10.1038/mt.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hong V, Presolski SI, Ma C, Finn MG. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew Chem Int Ed. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.