Abstract
AtTSPO is a TspO/MBR domain-protein potentially involved in multiple stress regulation in Arabidopsis. As in most angiosperms, AtTSPO is encoded by a single, intronless gene. Expression of AtTSPO is tightly regulated both at the transcriptional and post-translational levels. It has been shown previously that overexpression of AtTSPO in plant cell can be detrimental, and the protein was detected in the endoplasmic reticulum (ER) and Golgi stacks, contrasting with previous findings and suggesting a mitochondrial subcellular localization for this protein. To ascertain these findings, immunocytochemistry and ABA induction were used to demonstrate that, in plant cells, physiological levels of AtTSPO colocalized with AtArf1, a mainly Golgi-localized protein in plant cells. In addition, fluorescent protein-tagged AtTSPO was targeted to the secretory pathway and did not colocalize with MitoTracker-labelled mitochondria. These results suggest that the polytopic membrane protein AtTSPO is cotranslationally targeted to the ER in plant cells and accumulates in the Trans-Golgi Network. Heterologous expression of AtTSPO in Saccharomyces cerevisiae, yeast devoid of TSPO-related protein, resulted in growth defects. However, subcellular fractionation and immunoprecipitation experiments showed that AtTSPO was targeted to mitochondria where it colocalized and interacted with the outer mitochondrial membrane porin VDAC1p, reminiscent of the subcellular localization and activity of mammalian translocator protein 18 kDa TSPO. The evolutionarily divergent AtTSPO appears therefore to be switching its sorting mode in a species-dependent manner, an uncommon peculiarity for a polytopic membrane protein in eukaryotic cells. These results are discussed in relation to the recognition and organelle targeting mechanisms of polytopic membrane proteins in eukaryotic cells.
Keywords: Abiotic stress, Arabidopsis, membrane protein, sorting, TSPO, yeast
Introduction
Eukaryotic cell interiors are subdivided into functionally distinct, membrane-bound subcellular compartments, each with a defined set of proteins required for its functioning. Each protein synthesized by the cell has to be correctly localized to perform its function. Knowing how newly synthesized proteins are targeted within cells is essential for understanding protein function, but also our knowledge of structure–function relationships of a given protein starts with identifying its subcellular localization. Concurrently with or shortly after their synthesis on ribosomes, α-helical polytopic membrane proteins are translocated across or asymmetrically integrated into distinct cellular membranes (Blobel, 1980). Some membrane proteins destined for the mitochondria have fewer hydrophobic TM (transmembrane) segments than those destined for the ER, and it is possible that the lower hydrophobicity is a critical determinant to escape the ER targeting mechanisms (Miyazaki et al., 2005).
Tryptophan-rich sensory protein/peripheral-type benzodiazepine receptor (TspO/MBR) proteins (referred to as TSPO hereafter) are membrane-anchored proteins found from Archaea to metazoans with few exceptions (reviewed in Gavish et al., 1999; Lacapere and Papadopoulos, 2003; Papadopoulos et al., 2006). Since their identification in the late 1970s (Braestrup et al., 1977) TSPO proteins have been the subject of intensive research to pinpoint their function, almost exclusively in animal cells. In mammals, TSPO1 is a widely expressed, evolutionarily conserved 18 kDa translocator protein localized primarily in the MOM (mitochondrial outer membrane), precisely enriched at the contact sites between the outer and inner mitochondrial membranes where it interacts structurally and functionally with the porin VDAC1 (McEnery et al., 1992; Veenman et al., 2008; Rampon et al., 2009, Rone et al., 2009). It is likely to possess a five-transmembrane structure, and it is endowed with several functions including cholesterol transport from the outer to the inner mitochondrial membrane, regulation of steroidogenesis, porphyrin transport, sensing of reactive oxygen species, and regulation of apoptosis (reviewed in Papadopoulos et al., 2006). However, a recently described divergent TSPO isoform from avians and mammals, TSPO2, which seems specific to erythropoetic cells, is localized to the endoplasmic reticulum (Fan et al., 2009).
TSPO-related proteins were recently identified and characterized in plants (Lindemann et al., 2004; Corsi et al., 2004; Frank et al., 2007; Guillaumot et al., 2009). Exhaustive analysis of the location of TSPO proteins both at the cellular and the tissue level is warranted to gain a better understanding of its biological role in plants. The Arabidopsis AtTSPO is a potential multiple abiotic stress regulator (Brady et al., 2007; Frank et al., 2007; Kleine et al., 2007; Dinneny et al., 2008; Kant et al., 2008; Guillaumot et al., 2009). The biological function of TSPO proteins in plant is not yet defined although their induction by the stress hormone abscisic acid (ABA) seems established (Frank et al., 2007; Kant et al., 2008; Guillaumot et al., 2009). It has previously been shown that overexpressed AtTSPO is targeted to the early secretory pathway in plant cells, and can be immunodetected in the ER and mainly in Golgi stacks (Guillaumot et al., 2009). These observations are in contrast with previous studies suggesting that AtTSPO is a mitochondrial protein (Lindemann et al., 2004). Towards the biochemical characterization and structure–function analyses, AtTSPO was heterologously expressed in Saccharomyces cerevisiae. In this report, evidence is presented that a physiologically relevant amount of AtTSPO is found solely in the secretory pathway in plant cells, colocalizing with the Golgi and Trans-Golgi Network (TGN) protein AtArf1. Unexpectedly, AtTSPO expressed in yeast is targeted to the MOM, suggesting a different sorting mechanism of the same polypeptide in yeast and plant cells.
Materials and methods
Plasmid construction and yeast transformation
Standard molecular biology protocols were followed and all genetic constructs were validated by sequencing. The full-length cDNA encoding AtTSPO was obtained from RIKEN (clones RAFL09-68-G14 and RAFL05-18-I12). To obtain AtTSPO-6×His, primers AtTSPO5′-5′ AAAATCTAGAATGGATTCTCAGGACATC and AtTSPO-6×His3′- 5′AAAACTCGAGTTAGTGATGGTGATGGTGATG were used, and for 6×His-AtTSPO the primers 5′-5′AAAATCTAGAATGCATCACCATCACCATCAC and AtTSPO3′-5′ AAAACTGAGTTATCACGCGACTGCAAGCTTTA were used to amplify AtTSPO cDNA. The PCR template was a plasmid containing either AtTSPO-TAP or TAP-AtTSPO (tandem affinity-tagged versions of AtTSPO in which the coding sequence of the protein is immediately extended at its C-terminus or N-terminus by a 6×Histidine tag (C Vanhee, H Batoko, unpublished data). These amplified PCR products were digested with XbaI/XhoI and cloned into the vector p426GAL1, opened with SpeI/XhoI. The generated plasmids were called p426GAL1-AtTSPO-6×His and p426GAL1-6×His-AtTSPO, respectively. All plasmids were amplified in Escherichia coli strain DH5α, purified, and further used to transform yeast cells. Yeast cells (CEN.PK113-7D) (Parrou et al., 1999) were transformed using the high-efficiency LiAc/single strand carrier DNA/PEG methods as described by Gietz and Woods (2002). The transformed yeast cells were grown for 4 d on solid SD selective medium made of yeast synthetic drop-out medium without uracil. The selected uracil auxotrophic clones were then grown in the presence of glucose or galactose as the carbon source and their proteins extract checked for the expression of AtTSPO by Western blotting (see below).
Yeast growth analysis
Yeast cells were routinely cultured in flasks with baffles, on an orbital shaker at 100 rotations per minute (rpm) at 30 °C in SD medium without uracil. Depending on the experiment either 2% glucose or 2% galactose were used as a carbon source. For solid medium, 2% (w/v) agar was added to the medium. Growth in liquid culture was followed by spectrophotometric absorbance measurements [optical density (OD) at 600 nm]. Experimental cultures were inoculated with log phase cells that were grown on SD medium without uracil and glucose as a carbon source, to obtain an initial OD of 0.2. The culture was incubated on a rotary shaker for 4 d and the OD was analysed at defined time points. The drop test on solid medium was carried out by spotting overnight cultures (OD600=0.3) and four 10-fold serial dilutions. The plates were incubated at 30 °C for 4–6 d and were monitored daily.
Subcellular fractionation
The subcellular fractionation was conducted according to Powers and Barlowe (1998), with some minor modifications. Briefly, cells were harvested by centrifugation in a JLA 9.1000 rotor Beckman-coulter, San Diego, USA for 5 min at 2000 g, and washed several times with cool sterile water. The cells were subsequently resuspended in spheroplasting buffer (0.7 M sorbitol, 10 mM TRIS/HCl pH 7.4, and 50 mM dithiothreitol) and washed once. The recovered pellet was resuspended in the spheroplasting buffer (2 ml of buffer g−1 of wet cells) and zymolyase was added (2.5 mg g−1 of wet cells) and incubated for 45 min at 30 °C. The mixture was centrifuged (5000 g for 5 min at 4 °C) to pellet the spheroplasts followed by at least three washes with the spheroplasting buffer. The recovered spheroplasts were resuspended in lysis buffer (10 mM HEPES pH 7.5, 12.5% sucrose, 1 mM MgCl2, and 1 mM PMSF) and disrupted by 20 strokes in a Dounce homogenizer. Cell debris were subsequently sedimented by centrifugation at 1500 g for 5 min at 4 °C in a C0560 rotor (Beckman-coulter, San Diego, USA) and the supernatant was applied on top of a discontinuous sucrose gradient (20–60% in 10 mM HEPES pH 7.5, 1 mM MgCl2, and 1 mM PMSF) and centrifuged for 2.5 h at 35 000 rpm (≈ 22 000 g) in a SW40Ti rotor (Beckman-coulter, San Diego, USA). Twelve fractions of about 700 μl were collected from the bottom of the tube using a peristaltic pump and analysed by SDS-PAGE and immunoblotting.
Antibodies and immunoblotting
The monoclonal antibody against VDAC1 was purchased from Invitrogen (Carlsbad, CA, USA), while the horseradish peroxidase-coupled anti-rabbit secondary antibodies were from Sigma-Aldrich and the anti-AtTSPO antibody was described previously (Guillaumot et al., 2009). Anti-Sna2 was generated by Renard et al. (2010), anti Gas1, anti Emp47, and anti Sec61 were a gift from H Riezman (Department of Biochemistry, Biozentrum, University of Basel). For Western blotting, anti-VDAC1 and anti-Sna2 were used at 1:1000, anti-Emp47 and anti-Sec61 were used at 1:2000, anti-CPY was used at 1:500 and anti-AtTSPO was used at 1:2000; anti-rabbit secondary antibody was used at 1:10 000 and anti-mouse secondary antibody at 1:10 000 (detection of anti-VDAC1). Total yeast proteins were prepared in 20 mM Na-phosphate buffer pH 7.8, 150 mM NaCl and 2% (w/v) DDM (dodecyl-β-D-maltoside). The extraction buffer was supplemented with 1 mM PMSF and 2 μg ml−1 of a protease inhibitors cocktail (leupeptin, aprotinin, antipain, pepstatin, chymostatin). Disruption of yeast cells was performed at room temperature in a Precellys24 apparatus (Bertin Technologies, Montigny le Bretonneux, France) using 500 μm glass beads. After homogenization, cell debris were pelleted by centrifugation for 5 min at 5000 g (bench top Eppendorf centrifuge) and the supernatant used immediately for protein quantification by the BCA method (Sigma, St Louis, USA) followed by SDS-PAGE or stored at –20 °C for subsequent use. Routinely, the proteins were electrophoresed on a 12% SDS-PAGE resolving gel in a Mini-Protean 3 apparatus (Bio-Rad, Hercules, USA). For standard Western blotting, after electrophoresis, the proteins were electrotransferred to a PVDF membrane (Millipore, Billerica, USA) using a semi-dry system (Bio-Rad) or by standard wet transfer (for quantification purposes) in 50 mM TRIS, 40 mM glycine, 0.0375% (w/v) SDS, and 10% methanol. The blot was blocked at room temperature for at least 45 min in 5% (w/v) low-fat dried milk dissolved in TRIS -buffered saline (TBS, 50 mM TRIS, 150 mM NaCl, pH 7.6) containing 0.5% (v/v) Tween 20 (saturation buffer). Primary antibody incubation was performed at 4 °C overnight in saturation buffer. After several washes of the blot with TBS containing 0.1% (v/v) Tween 20 and 0.5% (w/v) milk, secondary antibody incubation was performed at room temperature for 1–3 h, followed by Enhanced ChemiLuminescence (ECL) detection (Roche Diagnostics, Basel, Switzerland). The emitted signal was captured using a KODAK Image Station 4000R and, when required, relative band intensity was measured using KODAK 1D Image Analysis software (Eastman KODAK Company, Scientific Imaging Systems, Rochester, USA).
Immunocytochemistry and imaging
Arabidopsis cultured cells were grown in the dark as previously described by Guillaumot et al. (2009). The cells were subcultured for 3 d in fresh medium, 480 μl of cells were mixed with 500 μl paraformaldehyde (8% paraformaldehyde in phosphate buffer pH 7), and 20 μl dimethyl sulphoxide. The cells were fixed for 1 h at room temperature then washed three times in MEL buffer [CaCl2 4 mM, KCl 80 mM, mannitol 8% w/v, Na2HPO4/NaH2PO4 2 mM (pH 7), bovine serum albumin (BSA) 0.1% w/v] sterilized by filtration (0.22 μm) prior to use. The cells were pelleted by centrifugation at 1000 g in a swinging Eppendorf rotor (Beckman coulter, San Diego, USA) and digested for 5 min in digestion buffer [sorbitol 0.55 M, cellulose R10 0.6% (w/v), macerozyme 0.2% (w/v), pH 5.6] followed by three washes with MEL buffer. The cells were resuspended in PBS blocking buffer [per litre: NaCl 8 g, KCl 0.2 g, Na2HPO4 1.44 g, KH2PO4 0.24 g, pH 7.4 (HCl)], containing 1% BSA, 1% goat serum, 0.5% (v/v) Tween 20), and incubated for 2 h on a rotary wheel (15 rpm) at room temperature. Anti-AtArf1 was added at a final dilution of 1/50 and the cells incubated overnight at 4 °C followed by three washes with PBS blocking buffer. Texas red-coupled secondary anti-rabbit antibody was added (in PBS blocking buffer) at a final dilution of 1/500 and the cells incubated for 1 h at 37 °C in the dark. In case of double immunodetection, the cells were washed three times in PBS blocking buffer and then incubated for an additional 1 h at 37 °C with the affinity purified anti-AtTSPO antibody covalently coupled to FITC, at a final dilution of 1/50, followed by three more washes and mounting in the antifade medium Prolong™ containing DAPI (Invitrogen). The cells were imaged with a confocal microscope (Zeiss LSM 710) using an oil-immersion ×63 lens. Plant leaf cells expressing YFP-AtTSPO (Guillaumot et al., 2009) were infiltrated with 5 μM MitoTracker orange CMTMRos (Invitrogen) using a 1 ml syringe and imaged with a ×40 water-immersion lens. FITC was excited with the 488 nm argon multi-laser line and the emission beam 500–560 nm amplified; Texas red was excited with the 535 nm He–Ne line and the emission beam 600–680 nm amplified; DAPI was excited with the 405 nm diode laser and the emission beam 440–500 nm amplified; YFP was excited with the 514 nm argon multi-laser line and the emission beam 510–580 nm amplified. The Imaris™ software (Bitplane AG, Zurich, Switzerland) was used for image post-processing.
Purification of yeast microsomes
Yeast membrane fractions were prepared as in Goffeau and Dufour (1988), with the following modifications. Yeasts were grown in SD medium without uracil and 2% (w/v) glucose until an OD600 of 8, centrifuged for 5 min at 2000 g in a JLA 9.1000 rotor (Beckman coulter, San Diego, USA), washed twice with sterile distilled water, and resuspended in fresh SD growth medium containing 2% galactose. After overnight incubation at 30 °C the cells were harvested by centrifugation (for 5 min at 2000 g at 4 °C) and washed three times with ice-cold water. The recovered pellet was then resuspended (15 ml per 10 g of fresh weight) in MBS buffer (0.25 M sorbitol, 10 mM imidazole-NaOH pH 7.5, 2 mM MgCl2, 5 mM DTT, and the protease inhibitor cocktail as above) and the cells were disrupted for 2 min in the presence of 15 g of glass beads per 10 g of cells fresh weight by use of the braun mill (Braun Biotech; Melsungen, Germany). The remaining steps were as described by Goffeau and Dufour (1988). The resulting microsomal fraction was resuspended in MS buffer (10 mM imidazole-NaOH pH 7.5, 1 mM MgCl2, and the protease inhibitor cocktail as above), 1 ml aliquots were made and frozen in liquid nitrogen and stored at –80 °C. The protein concentration was determined by the BCA method using bovine serum albumin as the standard. For enrichment of the mitochondrial fraction, an acid precipitation of the microsomes was performed at pH 5.2, as described by Fuhrmann et al. (1976). The protein concentration of the microsomal fraction was brought to 5 mg ml−1 with MS buffer and 1 M acetic acid was added until the solution reached a pH of 5.2 and then the sample was centrifuged for 30 s at 6500 g. The pellet was next resuspended in MS buffer and the protein concentration was determined. The supernatant, obtained in the previous step, was adjusted to a pH of 7.5 with NaOH and subsequently centrifuged at 22 000 g for 40 min. The pellet, containing the plasma membrane-enriched fraction was resuspended in MS buffer followed by protein quantification as above.
Purification of histidine-tagged AtTSPO and mass spectrometry analysis
Microsomes were obtained as described above, resuspended in buffer A made of 20 mM Na-phosphate and 500 mM NaCl. Proteins solubilization was conducted according to Kanczewska et al. (2005), except that 0.1% polyoxyethylene 8-myristyl ether (C14E8) was used for stripping and 2% DDM was used for solubilizing detergent. The solubilized proteins were subsequently incubated for a minimum of 2 h at room temperature with buffer A containing 20 mM imidazole and prewashed Ni-sepharose high performance beads from GE healthcare (Little Chalfont, UK). Next, a batch purification was performed using increasing amounts of imidazole, in the washing buffer (20, 40, and 60 mM imidazole) made of a modified version of buffer A containing instead 0.05% (w/v) DDM. After the washing steps, Ni-NTA-bound proteins were eluted with 500 mM imidazole in the modified version of buffer A described above and analysed by SDS-PAGE followed by Western blotting as appropriate.
For mass spectrometry, the stained protein band was manually excised and in-gel tryptic digest performed using standard protocols. The peptides generated were mixed with 2 mg ml−1 of α-cyano-4-hydroxycinnamic acid matrix and MS and MS/MS spectra acquired using an Applied Bisosystems 4800 MALDI TOF/TOF™ Analyzer spectrometer. MS and MS/MS queries were performed using the Applied Biosystems GPS Explorer™ 3.6 software working with the Matrix Science Ltd MASCOT® Database search engine v2.1 (Boston, USA). The National Center for Biotechnology Information database restricted to yeasts was used. A 200 ppm precursor tolerance for MS spectra and a 0.1 Da fragment tolerance for MS/MS spectra were allowed.
Co-immunoprecipitation
In order to generate an immunocomplex, 10 μg of the monoclonal antibody against VDAC1, or 5 μg of anti-AtTSPO was mixed with 200 μg of protein (volume of 200 μl) and incubated at room temperature for 3 h on a rotating wheel (20 rpm). The immunocomplex was then isolated by the use 200 μl of protein-A sepharose 4 fast flow (GE healthcare) slurry, that was prewashed three times with the protein extraction buffer, and the mixture was incubated by rotating (20 rpm) overnight at 4 °C. The immunocomplexes attached to the beads were pelleted by centrifugation at 800 g for 2 min and the unbound proteins were washed off several times with the protein extraction buffer. The last wash was kept for further analysis and the immunocomplexes were eluted with Laemmli SDS-containing sample buffer and subsequently analysed by Western blotting.
Results
Expressed AtTSPO in Arabidopsis cultured cells colocalizes with the plant Golgi, TGN, and early endosome protein AtArf1
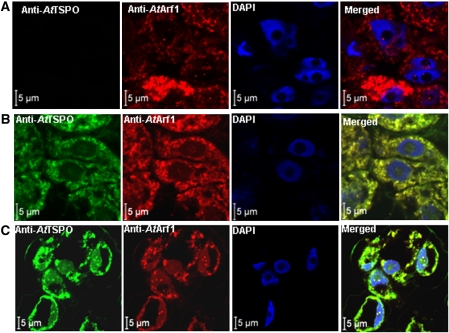
The experimental evidence for plant TSPO subcellular localization is scanty, most of the studies so far concluding to a mitochondrial localization (Corsi et al., 2004; Lindemann et al., 2004; Frank et al., 2007). The only direct evidence of the subcellular localization of a plant TSPO protein came from the studies by Lindemann et al. (2004) who used a polyclonal antibody against murine TSPO and transmission electron microscopy of Digitalis lanata leaf cells presumed to express a plant TSPO, and showed that the detected gold particles, thus the antigens, were in both chloroplasts and mitochondria (Lindemann et al., 2004). However, Western blot experiments suggested that the polyclonal antibody used cross-reacted with many plant proteins, which is not surprising since the primary sequence of plant and animal TSPO share not more than 25% identity. Recently, subcellular fractionation, immunocytochemistry and fluorescent protein tagging approaches were used and it was shown that AtTSPO is targeted to the secretory pathway in plant cells and can be detected in the ER and Golgi stacks (Guillaumot et al., 2009). However, most of these experiments were conducted with samples overexpressing the protein, since AtTSPO was not detectable in vegetative tissues from plants grown under unstressed conditions. In addition, the untagged protein was not colocalized directly with a genuine marker of the early secretory pathway as definitive proof of this unanticipated subcellular localization. To address these limitations, advantage was taken of the specific and affinity purified antibodies generated against AtTSPO, and the ABA-dependent induction of the protein in plant cells (Guillaumot et al., 2009). Arabidopsis cultured cells were probed with anti-AtTSPO and antibodies against the Arabidopsis thaliana ADP-ribosylation factor 1 (anti-AtArf1). AtArf1 can be detected in Golgi stacks in plant cells and its immunodetection has been used before to identify this organelle in plant cultured cells (Ritzenthaler et al., 2002; Xu and Scheres, 2005; Matheson et al., 2008). Figure 1 shows representative confocal images resulting from co-immunodetection of both AtTSPO and AtArf1. In Fig. 1A, the wild-type cells were probed with anti-AtArf1 followed by anti-AtTSPO and the nuclei are highlighted by DAPI (4',6-diamidino-2-phenylindole) staining. No AtTSPO could be detected in the wild-type cells (Fig. 1A, AtTSPO) consistent with previous studies showing that AtTSPO is undetectable in proteins extracted from wild-type Arabidopsis cultured cells grown under normal conditions (Guillaumot et al., 2009). A transgenic cell line was generated expressing N-terminally tagged AtTSPO with 6× histidine (6×His-AtTSPO) under the control of the 35S promoter. AtTSPO was immunodetected in these cells (Fig. 1B, AtTSPO) in punctuate structures with fluorescein isothiocyanate (FITC)-derived fluorescence also labelling the nuclear envelope. The signal from AtTSPO perfectly colocalized with that of AtArf1 (Fig. 1B, merged). It was checked that the pattern of the signal obtained with this sequential double immunodetection was consistent with those obtained separately with each antibody assessed alone (see Supplementary Fig. S1 at JXB online). The perfect colocalization of AtTSPO and AtArf1-derived signals suggest that the overexpressed 6×His-AtTSPO is mainly in the Golgi. None of the punctuate structures labelled by anti-AtTSPO was devoid of anti-AtArf1. Although unlikely, specific targeting of AtTSPO to the secretory pathway may result from mislocalization due to overexpression of a membrane protein as occasionally reported for other proteins (Sato et al., 1997; Lisenbee et al., 2003; Ma et al., 2006). To check this possibility, the endogenous protein was induced in cultured Arabidopsis wild-type cells by incubating them for 24 h in the presence of ABA, followed by immunodetection of AtTSPO and AtArf1. Figure 1C shows that both proteins were essentially detected in punctuate structures as in the transgenic overexpressing line, and the merged signals perfectly colocalized, suggesting that here also AtTSPO is a mainly Golgi-localized membrane protein. It is therefore concluded that, when expressed, AtTSPO is sorted to the ER and transported to Golgi stacks in plant cells through the secretory pathway. Thus, AtTSPO is an early secretory pathway (as defined by Robinson et al., 2007) membrane protein in plant cell.
Fig. 1.
AtTSPO colocalizes with AtArf1 in plant cells. (A) Wild-type cells were probed with anti-AtArf1 and Texas red-coupled anti-rabbit secondary antibody, then with anti-AtTSPO-coupled to FITC; the cells were mounted in antifade medium containing DAPI and imaged sequentially with a confocal microscope (Zeiss LSM 710) using an oil-immersion ×63 lens. No FITC signal could be detected in wild-type cells. (B) A transgenic cell line overexpressing 6×His-ATTSPO was treated as in (A), a FITC-derived signal could be seen (panel AtTSPO) and the merged image (panel merged) showed a perfect colocalization with the Texas red-derived signal. (C) Wild-type cells were pre-incubated in ABA (50 μM) before probing as in (A); the FITC-derived signal could be seen and perfectly colocalized with the Texas red-derived signal as in (B).
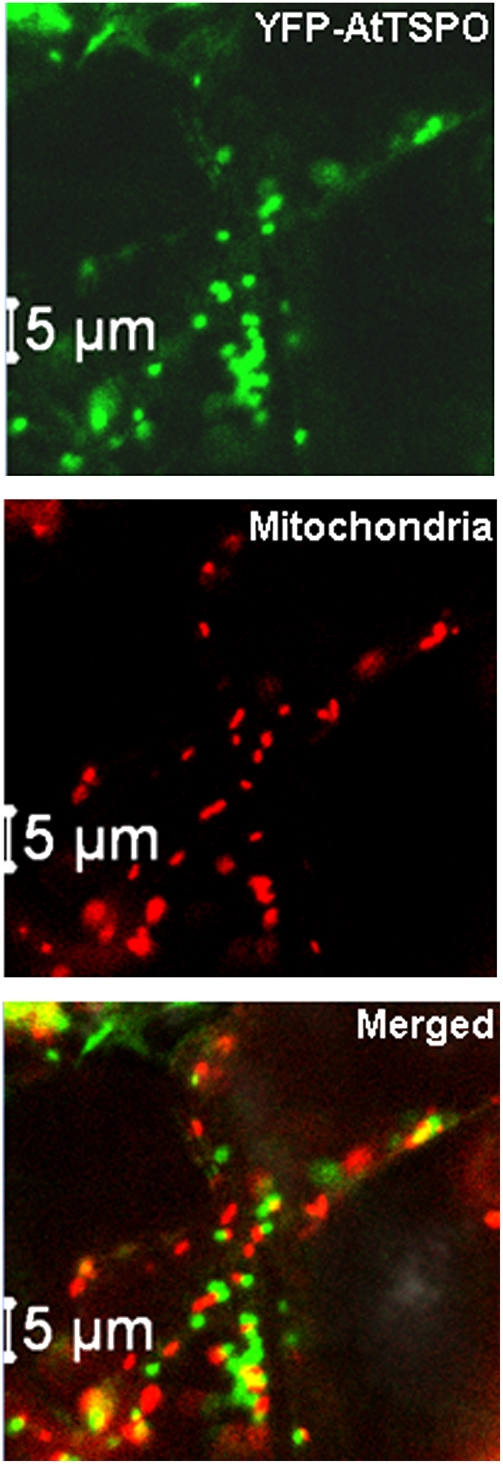
To check this conclusion independently, mitochondria in Arabidopsis transgenic plants overexpressing YFP-AtTSPO (Guillaumot et al., 2009) were stained with the mitochondria-specific dye MitoTracker. A representative confocal image of a leaf epidermal cell is shown in Fig. 2. The merged panel in Fig. 2 clearly demonstrates that YFP-AtTSPO and the MitoTracker signal did not colocalize (see also Supplementary Fig. S2 and the movie at JXB online). The software Imaris™ was used to analyse in detail the colocalization of the two signals in acquired time-lapse series. As shown in Supplementary Fig. S2 at JXB online, the two signals are on different organelles, with a Pearson's coefficient below 0.2 on average. A single eukaryotic translation product can occasionally be targeted to more than one organelle. These results clearly support the conclusion that AtTSPO is not, in addition to the organelles of the early secretory pathway, partially localized to the mitochondrion.
Fig. 2.
YFP-AtTSPO is not targeted to mitochondria in plant cells. Confocal images of YFP-AtTSPO and MitoTracker (panel mitochondria) imaged simultaneously as a time-lapse (see movies in the Supplementary data at JXB online); the two signals are not colocalizing as shown in the merged panel (see also Supplementary Fig. S2 at JXB online).
Expression of AtTSPO in S. cerevisiae reduces cell growth
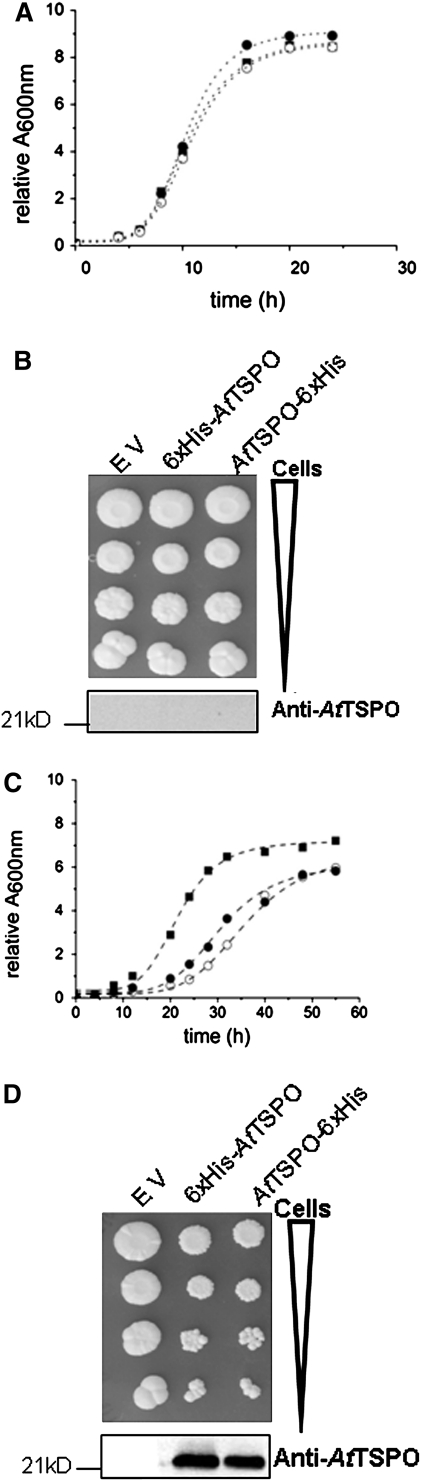
Functional and structural studies of membrane proteins usually require heterologous overexpression of the proteins in question (Wagner et al., 2006). S. cerevisiae is a common host in this respect, not only as a means to generate enough material for biochemical and structural studies, but also to search for potential protein partners. TspO/MBR domain-containing proteins are found from Archaea to metazoans with very few exceptions, notably the bakers yeast S. cerevisiae. However, the topology of animal TSPO1 was obtained after heterologous expression in yeast since the subcellular localization of the protein was conserved in yeast and mammalian cells (Bernassau et al., 1993; Joseph-Liauzun et al, 1998; Delavoie et al., 2003). For biochemical characterization of AtTSPO, the plant protein was expressed in yeast, driven by the inducible Gal1 promoter. To facilitate its affinity purification, a 6× histidines tag was added at the C-terminus (AtTSPO-6×His) or the N-terminus (6×His-AtTSPO). As a control, the yeast strain was also transformed with the empty vector (EV, no AtTSPO cDNA). The presence of AtTSPO in the yeast total protein extract was checked by Western blot using the affinity purified AtTSPO-specific antibodies (Guillaumot et al., 2009).
The growth of the EV strain was first compared with that of AtTSPO-6×His and 6×His-AtTSPO in the presence of glucose as the carbon source (Gal1 promoter repressed) or galactose (permissive conditions for AtTSPO expression). Growth was assessed by a time-course recording of the OD600 of the cells grown in liquid minimal synthetic medium with galactose (SDgal), or by a 10-fold dilution series of cells grown in liquid minimal medium with glucose (SDglu) then spotted on SDgal plates. The experiments were repeated at least three times and representative data are presented in Fig. 3. When glucose was used as the sole carbon source, growth of the three strains was similar in liquid culture (Fig. 3A) or after the serial dilution drop tests on plate (Fig. 3B). No AtTSPO could be detected in total protein extracts of AtTSPO-6×His and 6×His-AtTSPO (Fig. 3B, bottom panel). When glucose was replaced by galactose in the growth media, AtTSPO expression could be detected (Fig. 3D, lower panel). Interestingly, the relative growth of strains AtTSPO-6×His and 6×His-AtTSPO was substantially reduced as compared with the EV control (Fig. 3C, D). The size of the colonies generated by cells expressing AtTSPO-6×His or 6×His-AtTSPO was smaller as compared with those of the EV strain at each tested dilution (Fig. 3D), indicating clearly that expression of AtTSPO in S. cerevisiae reduces cell growth.
Fig. 3.
Expression of AtTSPO in yeast reduces cell growth. (A, C) Yeast strains (EV, 6×His AtTSPO, AtTSPO-6×His) growth monitored by OD600 nm absorbance either in SD-glucose medium (A) or SD-galactose medium (C). (B, D) The same yeast strains growth was also monitored through a serial dilution drop test; lower panels in (B) and (D) show the detection by Western blot of AtTSPO expression after growth in SD-glucose (B) or SD-galactose (D). EV, 6×His-AtTSPO, and AtTSPO-6×His stand for transgenic yeast transformed with, respectively, the empty vector, expression vector containing N-terminal tagged version of AtTSPO, and the expression vector containing C-terminal tagged version of AtTSPO. The filled squares are for the EV strain, the filled circles for 6×His-AtTSPO, and the open circles for AtTSPO-6×His.
AtTSPO is targeted to the mitochondria in yeast
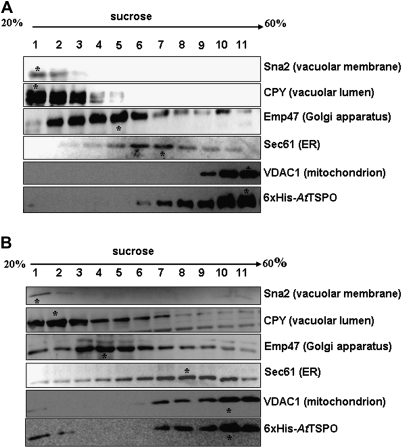
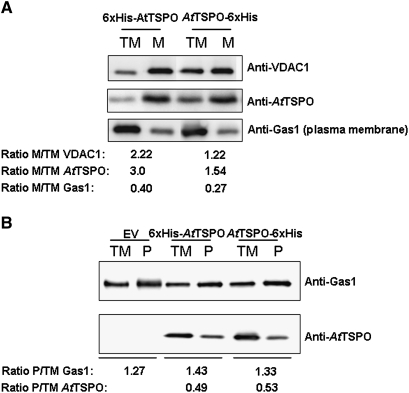
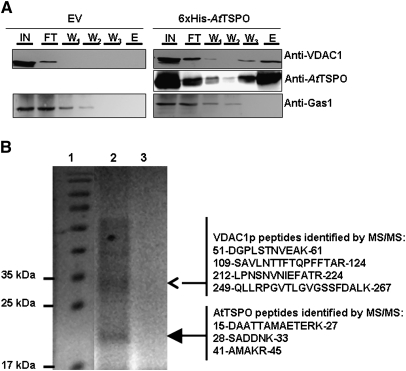
As shown above and in previous studies (Guillaumot et al., 2009) when expressed in plant cells, AtTSPO is targeted to the early secretory pathway. There was speculation that this subcellular localization was conserved in yeast as for the mammalian TSPO1. When expressed in yeast, mammalian translocator protein TSPO1 is targeted to the MOM (Bernassau et al., 1993; Joseph-Liauzun et al., 1998; Delavoie at al., 2003). To check the subcellular localization of AtTSPO expressed in yeast, subcellular fractionation experiments were conducted after 3 h and 12 h of AtTSPO induction. Total microsomes were separated from the cytosol and the former were used in a sucrose gradient to isolate fractions enriched in different subcellular membranes. Yeast Sna2p and CPY were used as a vacuolar membrane and a vacuolar lumen marker, respectively, Emp47p as Golgi membrane marker, Sec61p as ER membrane marker, and VDAC1p as the mitochondrial marker. AtTSPO was not detected in the soluble protein fraction (data not shown). Western blot analyses of the sucrose gradient-derived fractions are presented in Fig. 4. Figure 4A shows that 3 h after AtTSPO induction, the protein co-sedimented in the gradient with VDAC1p and both proteins peaked in fraction 11. Similar results were obtained at the 12 h time point (Fig. 4B), with some VDAC1p and AtTSPO also detected in the vacuolar membrane-enriched fractions (Fig. 4B). These results suggest that AtTSPO expressed in yeast is mainly targeted to the mitochondria and, with time, some of the protein may reach the vacuolar membrane. To confirm this observation, total microsomes (after 12 h induction) were submitted to acid-dependent precipitation resulting in plasma membrane-enriched and mitochondria-enriched fractions (Fuhrmann et al., 1976). These fractions were probed for the enrichment of the plasma membrane marker Gas1p and the mitochondria marker VDAC1p. Figure 5A shows that for the two AtTSPO expressing strains, the VDAC1p enrichment was concomitant with that of AtTSPO. By contrast, Fig. 5B shows that while GAS1p was enriched in plasma membrane fractions of all the samples analysed, AtTSPO was not. These results support the conclusion that VDAC1p and AtTSPO are colocalized in the same biological membrane, which is not the ER or the Golgi. Total microsomes from strain 6×His-AtTSPO were solubilized and subjected to nickel affinity chromatography. Figure 6A shows an example of affinity purification of 6×His-AtTSPO resulting in co-purification of some VDAC1p. Similar results were obtained for AtTSPO-6×His (data not shown). In control experiments, VDAC1p from the EV strain was not detected in the elution fraction (Fig. 6A, upper left panel), suggesting that VDAC1p retention by Ni-NTA requires AtTSPO. The eluted fraction was separated by SDS-PAGE and the polypeptides stained with colloidal blue. Two major bands were detected, one below 35 kDa (Fig. 6B open arrowhead) and the other around 20 kDa (Fig. 6B, filled arrowhead). These two stained polypeptide bands were manually excised, in-gel digested with trypsin, and the resulting peptides purified and subjected to mass spectrometry analysis (MALDI-TOF-TOF). The higher Mr band was identified as yeast VDAC1p (Fig. 6B) through four resulting MS/MS peptides (20% sequence coverage, MASCOT score 81) and the lower Mr band was identified as AtTSPO from three resulting MS/MS peptides (15% sequence coverage, MASCOT score 87). These results suggest that VDAC1p is the major co-purifying protein with AtTSPO.
Fig. 4.
Subcellular fractionation and membrane localization of 6×His-AtTSPO expressed in yeast. Lysed spheroplasts were separated on a discontinuous sucrose gradient and fractionation was performed by centrifugation. (A) Immunodetection of vacuolar markers Sna2 (membrane) and CPY (luminal), Golgi marker Emp47, ER marker Sec 61, mitochondria marker VDAC1, and AtTSPO from the 6×His-AtTSPO expressing strain, 3 h after induction. The asterisk indicates the sedimentation peak of each protein. (B) Immunodetection of vacuolar markers Sna2 (membrane) and CPY (luminal), Golgi marker Emp47, ER marker Sec 61, mitochondria marker VDAC1, and AtTSPO from the 6×His-AtTSPO expressing strain, 12 h after induction. (C) Immunodetection of vacuolar marker CPY, Golgi marker Emp47, ER marker Sec61, and mitochondria marker VDAC1 of the empty vector transformed strain, 12 h after induction.
Fig. 5.
Enrichment of AtTSPO in purified yeast microsomal fractions. TM stands for total microsomes; the ratio values >1 indicates an enrichment, and <1 indicates impoverishment with respect to the TM. (A) Immunoblot analysis of the mitochondria-enriched fraction (M) prepared from the strains expressing either the N-terminal tagged version of AtTSPO or the C-terminal tagged version of AtTSPO; anti-VDAC1 is used as a positive control. (B) Immunoblot analysis of the plasma membrane enriched fraction (P) prepared from the AtTSPO expressing yeast strains 6×His-AtTSPO and AtTSPO-6×His and the strain transformed with the empty vector, EV. Anti-Gas1 protein is used as a positive control for plasma membrane.
Fig. 6.
Yeast VDAC1p copurifies with 6×His-AtTSPO. The expressed 6×His-AtTSPO in yeast was solubilized from total microsomes and affinity purified using a Ni-NTA matrice, with microsomes from the yeast strain transformed with the empty vector (EV) used as control. (A) The input (IN), flowthrough (FT), the three successive washes (respectively W1, W2, and W3) and the elution fraction (E) were analysed by immunoblotting for the presence of AtTSPO, with VDAC1p and Gas1p as control membrane proteins. (B) The elution from 6×His-AtTSPO strain (lane 2) or the empty vector strain (lane 3) were separated by SDS-PAGE alongside Mr markers (lane 1); after blue colloidal staining of the polypeptides the main bands in lane 2 (arrowheads) were excised and in-gel tryptic-digested and the resulting peptides analysed by MALDI TOF/TOF; the identified peptide for each polypeptide are shown on the right with their start and end in the primary sequence of the MS/MS identified protein.
ATSPO expressed in yeast physically interacts with VDAC1p
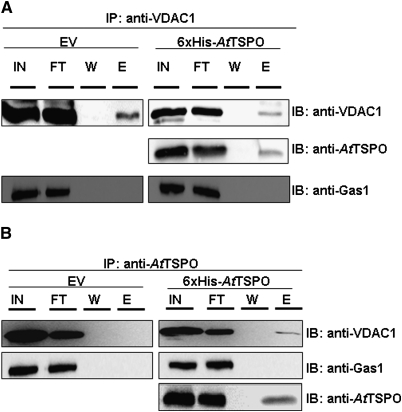
From the data presented above AtTSPO localization in yeast is reminiscent of that of mammalian TSPO1. Since mammalian TSPO1 can form oligomeric complexes with the porin VDAC (Papadopoulos et al., 1990, 1994, 1997), there was speculation that this may also be the case for AtTSPO expressed in yeast. A check was made for possible protein–protein interaction between AtTSPO and VDAC1p by co-immunoprecipitation. Figure 7A shows that when the solubilized microsomes from 6×His-AtTSPO strain were incubated with anti-VDAC1p antibodies, AtTSPO was detected in the eluted immuno-complex (Fig. 7A, second panel, lane E, elution), but the plasma membrane protein Gas1p was not. In a reversed experiment (Fig. 7B), immunoprecipitation with anti-AtTSPO antibodies resulted in co-immunoprecipitation of VDAC1p. VDAC1p was not detected in the elution fraction from the EV protein extract. Taken together, these results establish firmly that AtTSPO expressed in yeast is targeted to the MOM where it interacts with VDAC1p.
Fig. 7.
AtTSPO and VDAC1 interact physically. VDAC1 specific antibodies (A) or AtTSPO specific antibody (B) were used to generate an immunocomplex (IP) of their respective antigen from total protein extracts from yeast transformed with the empty vector (EV) or 6×His-AtTSPO. The immunocomplexes were coupled to protein A beads; the input (IN), flowthrough (FT), the last wash (W), and the eluted fraction (E) were analysed by immunoblotting (IB). To verify the specificity of the interactions, a check was made for the presence of Gas1p, a plasma membrane protein, in the immunocomplexes.
Discussion
In this study, direct immunolocalization was used to show that, in plant cells, AtTSPO, an Arabidopsis polytopic membrane protein, is sorted to the early secretory pathway and colocalizes with the small GTPase AtArf1. AtTSPO induction by ABA was exploited to probe physiologically relevant levels of the protein, and the results confirmed previous findings (Guillaumot et al., 2009) and demonstrated unambiguously the subcellular localization of AtTSPO in plant cells as an early secretory pathway membrane protein. However, the same variant of the polypeptide 6×His-AtTSPO which is cotranslationally targeted to the secretory pathway in plant cells was post-translationally sorted to the MOM in yeast cells, suggesting that the recognition and targeting of this α-helical membrane protein by the translocon in both eukaryotic systems was different.
The subcellular localization of plant TSPO has been previously studied to some extent with conflicting results. Using a polyclonal antibody against animal TSPO, Corsi et al. (2004) identified a 30–36 kDa cross-reacting protein in potato tuber extracts that they considered to be potato TSPO. Subcellular fractions of potato tuber were probed with this antibody and resulted in no immunoreactivity for chloroplast-enriched or mitochondria-enriched fractions, while an antigen was detected in nuclear preparations, suggesting an ER localization of the detected protein. Lindemann et al. (2004) showed binding of TSPO-cognate ligands to mitochondrial fractions from various plant species, and to heterologously expressed and purified AtTSPO. In addition, the authors used anti-murine TSPO antibodies and immunodetected by Western blot polypeptides of various sizes in these plant materials. The same antibodies immunogold-labelled antigens in mitochondria and thylakoid membranes of D. lanata leaf cells, suggesting that, at least in this species, the supposed TSPO protein(s) may be localized in the mitochondria and the chloroplasts. The moss Physcomitrella patens encodes five TSPO-like proteins and one of these, PpTSPO1 fused to GFP, could be detected in organelles morphologically resembling the mitochondria in P. patens protoplasts (Franck et al., 2007). Whether all these P. patens TSPO-like proteins are targeted to the same subcellular compartment is not yet known. TspO/MBR domain-containing proteins are fairly conserved between kingdoms. The primary sequence of plant and animal TSPO appear to share less than 25% identity (Lindemann et al., 2004; Frank et al., 2007; Guillaumot et al., 2009). The antibodies used by Corsi et al. (2004) and Lindemann et al. (2004) cross-reacted with various polypeptides from plant cell or with an antigen, supposedly a plant TSPO, whose size is questionable and intriguing. It is not clear whether an antibody raised against animal TSPO can cross-react with a genuine plant TSPO protein. Angiosperm TSPO seems to be transcriptionally regulated, the expression being at least ABA-dependent. In the absence of stress or ABA treatment, AtTSPO was not detected in Arabidopsis vegetative tissues, consistent with all the transcriptome data available to date. Whether D. lanata TSPO is constitutively expressed in leaf cells awaits experimental evidence. AtTSPO was detected neither in a purified mitochondrial fraction nor in a chloroplast fraction from a transgenic plant line constitutively expressing the protein (Guillaumot et al., 2009). Data presented in Figs 1 and 2 confirms that AtTSPO is only detected in the ER and Golgi stacks in plant cells. As a regulator of retrograde trafficking between the Golgi and the ER, the small GTPase AtArf1 temporally localize to Golgi cisterna. However, previous studies showed a colocalization of YFP-AtTSPO with ST-GFP (Guillaumot et al., 2009). ST-GFP is a predominantly trans-most Golgi cisterna and TGN marker in plant cells. Furthermore, recent evidence suggests that AtArf1 localizes predominantly to the TGN (Stierhof and El Kasmi, 2010) and the TGN is now considered to be an independent and endocytic compartment of plant cells, transiently associating with Golgi stacks (Viotti et al., 2010). Therefore, AtTSPO may be targeted to the Golgi and predominantly accumulate in the TGN and/or the early endosomes, reflecting the perfect colocalization of YFP-AtTSPO and ST-GFP (Guillaumot et al., 2009). It may be that the angiosperms’ lineage of this protein family evolved to perform additional or different functions in plants. In contrast to animal cells, AtTSPO is not a housekeeping gene since knockout mutants are viable under normal growth conditions.
One argument so far in favour of plant TSPO being mitochondrial proteins is mainly based on the well-established subcellular localization of mammalian TSPO1, and the general assumption that related proteins in other eukaryotes should be targeted to the same organelle, although there is no evidence so far that members of this fairly conserved family of proteins are involved in the same physiological activities. However, Fan et al. (2009) recently characterized an avian and mammalian erythroblast-specific translocator protein, TSPO2, localized in the ER and the nuclear membrane but not in the mitochondria. AtTSPO is not more related to TSPO2 than it is to TSPO1 (see Supplementary Fig. S3 at JXB online).
How structurally similar membrane proteins or even the same protein are targeted to either the ER or the mitochondria is a currently intriguing topic in cell biology (Miyazaki et al., 2005; Ma and Taylor, 2008; Balss et al., 2008). The subcellular localization of TSPO1 is conserved after heterologous expression in yeast. The topology of TSPO1, with five α-helical transmembrane domains and a cytoplasmic C-terminus, was first studied in vivo in yeast (Joseph-Liauzun et al., 1998). Whether the subcellular localization of TSPO2 is also conserved in yeast is not known. Unexpectedly, it is shown clearly here that AtTSPO expressed is yeast is sorted and targeted to the MOM, where it interacts with the yeast porin VDAC1p, a situation reminiscent of TSPO1 in animal cell and yeast. Why is AtTSPO mis-sorted in yeast? Indeed AtTSPO seems to switch its sorting mode from cotranslational ER integration in plant cells to post-translational mitochondrial import in yeast, suggesting that, in yeast, the SRP-RNC (signal recognition particle–ribosome nascent chain complex) failed to identify and integrate the α-helical transmembrane domains of AtTSPO in the ER membrane. Sorting and orderly insertion of an α-helical polytopic membrane protein in the ER starts with the recognition of the first transmembrane domain. Some membrane proteins destined to the mitochondria have fewer hydrophobic transmembrane segments than those destined to the ER, and it is possible that the lower hydrophobicity is a critical determinant to escape the ER targeting mechanisms (Miyazaki et al., 2005). A thorough quantitative assessment of the transmembrane helix recognition and insertion efficiency revealed that it depends on a ‘biological’ hydrophobicity scale, the apparent free energy of the amino acid residues (ΔGapp) (Hessa et al., 2005, 2007). The predicted first transmembrane of TSPO1 is far less hydrophobic than that of AtTSPO (predicted ΔGapp of +2.672 and –0.660, respectively) (see Supplementary Fig. S4 at JXB online) suggesting that TSPO1 is more likely to escape the translocon-mediated ER insertion than AtTSPO. Targeting and import of TSPO1 to MOM require the helix terminating Schellman motif (between predicted TM segments 2 and 3) and the C-terminal cholesterol-binding domain (Rone et al., 2009). The Schellman motif appears not to be conserved in AtTSPO, neither is the cholesterol binding domain, suggesting that, in yeast, other structural determinants are conducive of the targeting and insertion of AtTSPO in the MOM. AtTSPO contains a plant-specific N-terminal hydrophilic, highly charged (net charge +4) extension. It is known that some hydrophobic mitochondrial membrane proteins can escape the cotranslational targeting to the ER because of an N-terminal hydrophilic extension (Miyazaki et al., 2005). The addition of 6×His to this already charged peptide did not affect the targeting since the untagged protein also localized in the mitochondria in yeast (C Vanhee and H Batoko, unpublished data). It is possible that this N-terminal extension in AtTSPO inhibits cotranslational ER insertion in yeast but not in plant cells as shown for ABCme (Miyazaki et al., 2005). As described for TSPO1, it is possible that AtTSPO is integrated in the MOM through the solely TOM70-dependent pathway (Otera et al., 2007). It is shown that in yeast MOM, AtTSPO physically interacts with VDAC1p, although AtTSPO predicted topology may be different from that of TSPO1 (see Supplementary Table 1 at JXB online). It may be that the TspO/MBR domains fold is conserved and responsible for porin-TSPO interaction described in bacteria and eukaryotes, although TSPO proteins form a heterogeneous family of membrane proteins.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. Summary of number of transmembrane domains in human TSPO1 and TSPO2, Arabidopsis AtTSPO and Vitis vinifera VvTSPO using different transmembrane (TM) prediction software.
Supplementary Fig. S1. Anti-AtArf1 and anti-AtTSPO probing of plant cells.
Supplementary Fig. S2. YFP-AtTSPO and Mitotracker-labelled mitochondria do not colocalize.
Supplementary Fig. 3. ClustalW alignment of TSPO proteins.
Supplementary Fig. 4. Helical wheel projection of TSPO proteins.
Supplementary Material
Acknowledgments
We thank Drs JL Parrou, J François, and H Riezman for sharing materials or reagents; A Errachid and the imaging platform IMABIOL for assistance. This work was supported by the Inter-University Attraction Poles Program–Belgian Science Policy, the Communauté Française de Belgique–Action de Recherche Concertée grant no. ARC-0510-329, the Fonds de la Recherche Fondamentale Collective project no. 2.4523.09. MD and HB are Research Associate of the Fonds de la Recherche Scientifique-FNRS.
References
- Balss J, Papatheodorou P, Mehmel M, et al. Transmembrane domain length of viral K+channels is a signal for mitochondria targeting. Proceedings of the National Academy of Sciences, USA. 2008;105:12313–12318. doi: 10.1073/pnas.0805709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braestrup C, Albrechtsen R, Squires RF. High densities of benzodiazepine receptors in human cortical areas. Nature. 1977;269:702–704. doi: 10.1038/269702a0. [DOI] [PubMed] [Google Scholar]
- Bernassau JM, Reversat JL, Ferrara P, Caput D, Lefur G. A 3D model of the peripheral benzodiazepine receptor and its implication in intra mitochondrial cholesterol transport. Journal of Molecular Graphics. 1993;11:236–244. doi: 10.1016/0263-7855(93)80003-a. [DOI] [PubMed] [Google Scholar]
- Blobel G. Intracellular protein topogenesis. Proceedings of the National Academy of Sciences, USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- Corsi L, Avallone R, Geminiani E, Cosenza F, Venturini I, Baraldi M. Peripheral benzodiazepine receptors in potatoes (Solanum tuberosum) Biochemical and Biophysical Research Communications. 2004;313:62–66. doi: 10.1016/j.bbrc.2003.11.089. [DOI] [PubMed] [Google Scholar]
- Delavoie F, Li H, Hardwick M, Robert JC, Giatzakis C, Peranzi G, Yao ZX, Maccario J, Lacapere JJ, Papadopoulos V. In vivo and in vitro peripheral-type benzodiazepine receptor polymerization: Functional significance in drug ligand and cholesterol binding. Biochemistry. 2003;42:4506–4519. doi: 10.1021/bi0267487. [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320:942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- Fan JJ, Rone MB, Papadopoulos V. Translocator protein 2 is involved in cholesterol redistribution during erythropoiesis. Journal of Biological Chemistry. 2009;284:30484–30497. doi: 10.1074/jbc.M109.029876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank W, Baar KM, Qudeimat E, Woriedh M, Alawady A, Ratnadewi D, Gremillon L, Grimm B, Reski R. A mitochondrial protein homologous to the mammalian peripheral-type benzodiazepine receptor is essential for stress adaptation in plants. The Plant Journal. 2007;51:1004–1018. doi: 10.1111/j.1365-313X.2007.03198.x. [DOI] [PubMed] [Google Scholar]
- Furhmann GF, Boehm C, Theuvenet APR. Sugar transport and potassium permeability in yeast plasma membrane vesicles. Biochimica et Biophysica Acta-Biomembranes. 1976;433:583–596. doi: 10.1016/0005-2736(76)90283-2. [DOI] [PubMed] [Google Scholar]
- Gavish M, Bachman I, Shoukrun R, Katz Y, Veenman L, Weisinger G, Weizman A. Enigma of the peripheral benzodiazepine receptor. Pharmacological Reviews. 1999;51:629–650. [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Guide to Yeast Genetics and Molecular and Cell Biology. 2002;Pt B 350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Dufour JP. Plasma membrane ATPase from the yeast Saccharomyces cerevisiae. Methods in Enzymology. 1988;157:528–533. doi: 10.1016/0076-6879(88)57101-x. [DOI] [PubMed] [Google Scholar]
- Guillaumot D, Guillon S, Deplanque T, Vanhee C, Gumy C, Masquelier D, Morsomme P, Batoko H. The Arabidopsis TSPO-related protein is a stress and abscisic acid-regulated, endoplasmic reticulum-Golgi-localized membrane protein. The Plant Journal. 2009;60:242–256. doi: 10.1111/j.1365-313X.2009.03950.x. [DOI] [PubMed] [Google Scholar]
- Heinrich SU, Mothes W, Brunner J, Rapoport TA. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- Joseph-Liauzun E, Delmas P, Shire D, Ferrara P. Topological analysis of the peripheral benzodiazepine receptor in yeast mitochondrial membranes supports a five-transmembrane structure. Journal of Biological Chemistry. 1998;273:2146–2152. doi: 10.1074/jbc.273.4.2146. [DOI] [PubMed] [Google Scholar]
- Kanczewska J, Marco S, Vandermeeren C, Maudoux O, Rigaud JL, Boutry M. Activation of the plant plasma membrane H+-ATPase by phosphorylation and binding of 14-3-3 proteins converts a dimer into a hexamer. Proceedings of the National Academy of Sciences, USA. 2005;102:11675–11680. doi: 10.1073/pnas.0504498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant P, Gordon M, Kant S, Zolla G, Davydov O, Heimer YM, Chalifa-Caspi V, Shaked R, Barak S. Functional-genomics-based identification of genes that regulate Arabidopsis responses to multiple abiotic stresses. Plant, Cell and Environment. 2008;31:697–714. doi: 10.1111/j.1365-3040.2008.01779.x. [DOI] [PubMed] [Google Scholar]
- Kleine T, Kindgren P, Benedict C, Hendrickson L, Strand A. Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of arabidopsis to high irradiance. Plant Physiology. 2007;144:1391–1406. doi: 10.1104/pp.107.098293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacapere JJ, Papadopoulos V. Peripheral-type benzodiazepine receptor: structure and function of a cholesterol-binding protein in steroid and bile acid biosynthesis. Steroids. 2003;68:569–585. doi: 10.1016/s0039-128x(03)00101-6. [DOI] [PubMed] [Google Scholar]
- Lindemann P, Koch A, Degenhardt B, Hause G, Grimm B, Papadopoulos V. A novel Arabidopsis thaliana protein is a functional peripheral-type benzodiazepine receptor. Plant and Cell Physiology. 2004;45:723–733. doi: 10.1093/pcp/pch088. [DOI] [PubMed] [Google Scholar]
- Lisenbee CS, Karnik SK, Trelease RN. Overexpression and mislocalization of a tail-anchored GFP redefines the identity of peroxisomal ER. Traffic. 2003;4:491–501. doi: 10.1034/j.1600-0854.2003.00107.x. [DOI] [PubMed] [Google Scholar]
- Ma B, Cui ML, Sun HJ, Takada K, Mori H, Kamada H, Ezura H. Subcellular localization and membrane topology of the melon ethylene receptor CmERS1. Plant Physiology. 2006;141:587–597. doi: 10.1104/pp.106.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YL, Taylor SS. A molecular switch for targeting between endoplasmic reticulum (ER) and mitochondria: conversion of a mitochondria-targeting element into an ER-targeting signal in DAKAP1. Journal of Biological Chemistry. 2008;283:11743–11751. doi: 10.1074/jbc.M710494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson LA, Suri SS, Hanton SL, Chatre L, Brandizzi F. Correct targeting of plant ARF GTPases relies on distinct protein domains. Traffic. 2008;9:103–120. doi: 10.1111/j.1600-0854.2007.00671.x. [DOI] [PubMed] [Google Scholar]
- McEnery MW, Snowman AM, Trifiletti RR, Snyder SH. Isolation of the mitochondrial benzodiazepine receptor: association with the voltage-dependent anion channel and the adenine-nucleotide carrier. Proceedings of the National Academy of Sciences, USA. 1992;89:3170–3174. doi: 10.1073/pnas.89.8.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki E, Kida Y, Mihara K, Sakaguchi M. Switching the sorting mode of membrane proteins from cotranslational endoplasmic reticulum targeting to posttranslational mitochondrial import. Molecular Biology of the Cell. 2005;16:1788–1799. doi: 10.1091/mbc.E04-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Taira Y, Horie C, Suzuki Y, Suzuki H, Setoguchi K, Kato H, Oka T, Mihara K. A novel insertion pathway of mitochondrial outer membrane proteins with multiple transmembrane segments. Journal of Cell Biology. 2007;179:1355–1363. doi: 10.1083/jcb.200702143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos V, Amri H, Boujrad N, et al. Peripheral benzodiazepine receptor in cholesterol transport and steroidogenesis. Steroids. 1997;62:21–28. doi: 10.1016/s0039-128x(96)00154-7. [DOI] [PubMed] [Google Scholar]
- Papadopoulos V, Baraldi M, Guilarte TR, et al. Translocator protein (18 kDa): new nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends in Pharmacological Sciences. 2006;27:402–409. doi: 10.1016/j.tips.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Papadopoulos V, Boujrad N, Ikonomovic MD, Ferrara P, Vidic B. Topography of the Leydig-cell mitochondrial peripheral-type benzodiazepine receptor. Molecular and Cellular Endocrinology. 1994;104:R5–R9. doi: 10.1016/0303-7207(94)90061-2. [DOI] [PubMed] [Google Scholar]
- Papadopoulos V, Mukhin AG, Costa E, Krueger KE. The peripheral-type benzodiazepine receptor is functionally linked to Leydig-cell steroidogenesis. Journal of Biological Chemistry. 1990;265:3772–3779. [PubMed] [Google Scholar]
- Parrou JL, Enjalbert B, Plourde L, Bauche A, Gonzalez B, Francois J. Dynamic responses of reserve carbohydrate metabolism under carbon and nitrogen limitations in Saccharomyces cerevisiae. Yeast. 1999;15:191–203. doi: 10.1002/(SICI)1097-0061(199902)15:3<191::AID-YEA358>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Powers J, Barlowe C. Transport of Axl2p depends on Erv14p, an ER-vesicle protein related to the Drosophila cornichon gene product. Journal of Cell Biology. 1998;142:1209–1222. doi: 10.1083/jcb.142.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampon C, Bouzaffour M, Ostuni MA, Dufourcq P, Girard C, Freyssinet JM, Lacapere JJ, Schweizer-Groyer G, Vriz S. Translocator protein (18 kDa) is involved in primitive erythropoiesis in zebrafish. FASEB Journal. 2009;23:4181–4192. doi: 10.1096/fj.09-129262. [DOI] [PubMed] [Google Scholar]
- Renard HF, Demaegd D, Guerriat B, Morsomme P. Efficient ER exit and vacuole targeting of yeast Sna2p require two tyrosine-based sorting motif. Traffic. 2010;11:931–946. doi: 10.1111/j.1600-0854.2010.01070.x. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler C, Nebenfuhr A, Movafeghi A, Stussi-Garaud C, Behnia L, Pimpl P, Staehelin LA, Robinson DG. Reevaluation of the effects of brefeldin A on plant cells using tobacco bright yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. The Plant Cell. 2002;14:237–261. doi: 10.1105/tpc.010237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DG, Herranz MC, Bubeck J, Pepperkok R, Ritzenthaler C. Membrane dynamics in the early secretory pathway. Critical Reviews in Plant Sciences. 2007;26:199–225. [Google Scholar]
- Rone MB, Liu J, Blonder J, Ye XY, Veenstra TD, Young JC, Papadopoulos V. Targeting and insertion of the cholesterol-binding translocator protein into the outer mitochondrial membrane. Biochemistry. 2009;48:6909–6920. doi: 10.1021/bi900854z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Sato M, Nakano A. Rer1p as common machinery for the endoplasmic reticulum localization of membrane proteins. Proceedings of the National Academy of Sciences, USA. 1997;94:9693–9698. doi: 10.1073/pnas.94.18.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierhof YD, EL Kasmi F. Strategies to improve the antigenicity, ultrastructure preservation and visibility of trafficking compartments in Arabidopsis tissue. European Journal of Cell Biology. 2010;89:285–297. doi: 10.1016/j.ejcb.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Veenman L, Shandalov Y, Gavish M. VDAC activation by the 18 kDa translocator protein (TSPO), implications for apoptosis. Journal of Bioenergetics and Biomembranes. 2008;40:199–205. doi: 10.1007/s10863-008-9142-1. [DOI] [PubMed] [Google Scholar]
- Viotti C, Bubeck J, Stierhof YD, et al. Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. The Plant Cell. 2010;22:1344–1357. doi: 10.1105/tpc.109.072637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Bader ML, Drew D, de Gier JW. Rationalizing membrane protein overexpression. Trends in Biotechnology. 2006;24:364–371. doi: 10.1016/j.tibtech.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Xu J, Scheres B. Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. The Plant Cell. 2005;17:525–536. doi: 10.1105/tpc.104.028449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.