Abstract
Diacylglycerol acyltransferases (DGATs) catalyse the final step of the triacylglycerol (TAG) biosynthesis of the Kennedy pathway. Two major gene families have been shown to encode DGATs, DGAT1 (type-1) and DGAT2 (type-2). Both genes encode membrane-bound proteins, with no sequence homology to each other. In this study, the molecular cloning and characterization of a type-2 DGAT cDNA from olive is presented. Southern blot analysis showed that OeDGAT2 is represented by a single copy in the olive genome. Comparative transcriptional analysis revealed that DGAT1 and DGAT2 are developmentally regulated and share an overall overlapping but distinct transcription pattern in various tissues during vegetative growth. DGAT2 is highly expressed in mature or senescing olive tissues. In flowers, the expression of DGAT1 was almost undetectable, while DGAT2 transcripts accumulated at the later stages of both anther and ovary development. Differential gene regulation was also detected in the seed and mesocarp, two drupe compartments that largely differ in their functional roles and mode of lipid accumulation. DGAT1 appears to contribute for most of the TAG deposition in seeds, whereas, in the mesocarp, both DGAT1 and DGAT2 share an overlapping expression pattern. During the last stages of mesocarp growth, when TAGs are still accumulating, strong up-regulation of DGAT2 but a marked decline of DGAT1 transcript levels were detected. The present results show overlapping gene expression for olive DGATs during mesocarp growth, with a more prominent implication of DGAT2 in floral bud development and fruit ripening.
Keywords: Diacylglycerol acyltransferase, gene expression, in situ hybridization, olive development, triacylglycerols
Introduction
A number of plants accumulate large amounts of triacylglycerols (TAGs) in their seeds as storage reserves for germination and seedling development. Key points in the accumulation of TAGs are the early events of fatty acid biosynthesis and the last and critical events of TAG synthesis (Bao and Ohlrogge, 1999; Jako et al., 2001; Weselake, 2005; Lung and Weselake, 2006). There are few fruit crops that deposit most of the oil in the mesocarp tissues to attract animals for seed dispersal. Among them, olive is of predominant economic importance because its oil is ideal for direct consumption. It is therefore of great importance to elucidate the key-points in the olive oil biosynthesis pathway and storage. Such knowledge could speed up the breeding programmes aimed at selecting clones with superior fatty acid composition and is also essential for selecting high oil-yielding genotypes more efficiently and rapidly, thus improving decision-making processes. Nevertheless, the molecular basis of gene regulation underlying olive oil production is far from complete. There is a significant amount of information concerning the regulation of several genes involved in fatty acid synthesis and modification (Hatzopoulos et al., 2002; Doveri and Baldoni, 2007; Banilas and Hatzopoulos, 2009), but much less is known about the cellular mechanisms governing the transfer of fatty acids into storage TAGs, not only in olive but generally in plants (Shockey et al., 2006).
TAG biosynthesis is principally accomplished by membrane-bound enzymes that operate in the endoplasmic reticulum through the glycerol-3-phosphate or the so-called Kennedy pathway (Kennedy, 1961; Browse and Somerville, 1991). The first step in the process involves the acylation of glycerol-3-phosphate (GP) at the sn-1 position to produce lysophosphatidic acid (LPA) by GP acyltransferase (GPAT). LPA is further acylated at the sn-2 position by LPA acyltransferase (LPAT) resulting in the formation of phosphatidic acid (PA). PA is dephosphorylated to produce diacylglycerol (DAG), which is further acylated to produce TAG by diacylglycerol acyltransferase (DGAT), the only enzyme in the pathway that is thought to be exclusively committed to TAG synthesis. Inasmuch as DGAT catalyses the final and most critical step for TAG synthesis, it has been suggested that it may constitute a rate-limiting factor in TAG bioassembly in developing seeds (Ichihara et al., 1988; Jako et al., 2001; Weselake, 2005; Lung and Weselake, 2006). However, TAGs could also be produced via the transfer of acyl groups from phospholipids to diacylglycerols, an acyl-CoA-independent reaction catalysed by the enzyme phospholipid:diacylglycerol acyltransferase (PDAT) (Dahlqvist et al., 2000; Stahl et al., 2004; Zhang et al., 2009).
TAGs are not only produced in seeds or mesocarps. Both TAG accumulation and DGAT activity have been reported in several other organs, such as flowers, developing siliques, germinating seeds, young seedlings, and senescing leaves of Arabidopsis (Zou et al., 1999; Kaup et al., 2002), and in stems, flowers, roots, and leaves of tobacco (Zhang et al., 2005). Based on those observations, it has been suggested that TAG may also be implicated in physiological roles other than as a carbon or energy source (Lu and Hills, 2002; Lu et al., 2003).
Two major unrelated gene families have been shown to encode DGATs, namely DGAT1 (type-1) and DGAT2 (type-2) both of which are ER-localized. DGAT1 genes have been cloned from several plant species, including olive (Giannoulia et al., 2000). DGAT2 genes have been cloned from diverse eukaryotes, including the oleaginous fungus Mortierella ramanniana (Lardizabal et al., 2001), human (Cases et al., 2001), and the plant species Arabidopsis (Lardizabal et al., 2001), castor bean (Kroon et al., 2006), and tung tree (Shockey et al., 2006). A third member of the DGAT family (type-3), highly unrelated to the previously reported, was identified in peanut that possesses a cytosolic localization (Saha et al., 2006).
Accumulating data suggest that DGAT activity may have a substantial effect on carbon flow into seed oil of Brassica napus (Perry et al., 1999; Weselake et al., 2008), Arabidopsis thaliana (Katavic et al., 1995; Zou et al., 1999; Jako et al., 2001), and maize (Zheng et al., 2008). In an attempt to gain further insight into the role(s) of DGATs in plant lipid biosynthesis, it is shown here that DGAT2 is highly expressed in mature or senescent olive tissues. The expression patterns of DGAT1 and DGAT2 during drupe development and in several other organs/tissues of the olive tree indicated that genes are differentially regulated to fulfil the needs for TAG accumulation at certain points of growth and development.
Materials and methods
Plant material
Leaves, buds, flowers, and drupes at different developmental stages were harvested from ‘Koroneiki’, an oil olive (Olea europaea L.) cultivar, grown in a natural environment at the Agricultural University of Athens (37°58' N, 23°46' E). Ovaries and anthers were dissected from flowers, while primary roots, hypocotyls, cotyledons, and shoot tips were dissected from seedlings grown in a growth chamber at 23 °C under a 16 h photoperiod. Samples were immediately frozen in liquid nitrogen and stored at –80 °C for RNA and DNA extractions.
RNA extraction and RT reactions
Total RNA was isolated from different olive tissues by a phenol:chloroform extraction procedure as described previously (Haralampidis et al., 1998). The concentration of total RNA was determined spectrophotometrically and verified following agarose gel electrophoresis by ethidium bromide staining. Total RNA was treated with RNase-free DNAse I (Promega) and 2 μg were used as a template in first-strand cDNA synthesis using Superscript™ II RNase H- Reverse Transcriptase (Invitrogen) following the manufacturer's instructions. Unless otherwise stated, the first-strand cDNA was primed off by the poly-A tail with the reverse transcription primer T17XHO (5′-GTCGACCTCGAGTTTTTTTTTTTTTTTTT-3′).
cDNA cloning
To amplify a central fragment of olive diacylglycerol acyltansferase type- 2 cDNA (OeDGAT2), the degenerate primers FOR1 (5′-C C/T TA C/T GT A/C/T TT C/T GG A/G/T TATGA A/G/ CC-3′) and REV2 (5′-CC A/G/C/T ACCACCAC A/G TG A/C/G/ AT A/T GG-3′) were designed for conserved regions of orthologous genes from GenBank. A cDNA amount corresponding to 150 ng of total RNA from mesocarp tissue at 25 weeks after flowering (WAF) was used as a template in PCR together with 200 μM of each dNTP, 20 pmol of each primer, 2 U of DNA polymerase (Expand™ High Fidelity, Boehringer, Mannheim) and 1× PCR buffer (provided by the manufacturer of the enzyme). Amplification was achieved in a thermal cycler (Model PTC-200, MJ Research, Waltham, MA) during 35 cycles of the following program: 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 45 s. PCR products were gel extracted (QIAquick® Gel Extraction Kit), cloned into pGEM-T Easy (Promega) and sequenced by Macrogen Inc. (Seoul, Korea). That sequence was used to design primers for amplification of the 3′ and 5′ ends by standard RACE PCR methods (Frohman et al., 1990). For 3′-RACE, the first-strand cDNA was primed off with the T17XHO primer and the amplification was achieved by using in reverse the specific forward primer 3aDG2 (5′-TAACACCAGCAACGAGGAAG-3′) followed by the nested primer 3bDG2 (5′-ATGGGCAAACCTCTGGTTC-3′). For 5′-RACE, the first-strand cDNA was synthesized by the reverse specific primer 5zDG2 (5′-TTTCCCACTAGGCTTCCAC-3′), and after dA-tailing the amplification was achieved using the nested primers 5bDG2 (5′-AATGCCTCCTTGACTCCCCCT-3′) and 5eDG2 (5′-TAGAACACAGCAGTACTAGCG-3′) in combination with T17XHO. The final full-length sequence was amplified using the primers 5UTRDGAT2 (5′-TCCCATCTACCAATTTCACACTC-3′) and 3UTRDGAT2 (5′-GCGTTTGTTATCTGCAGCTATTC-3′), designed from the 5′- and 3′-UTRs.
Sequence analysis
Nucleotide and deduced amino acid sequences were identified by the NCBI BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/). Prediction of open reading frame (ORF) and molecular weight estimation of the deduced polypeptide (GenBank accession number: GU357635) were made by the EditSeq program (version 3.88). Sequence comparisons were made by the MegAlign program (DNASTAR Inc., London UK). Transmembrane regions were predicted by the TMHMM Server ver. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and subcellular localization of the deduced polypeptide by PSORT (http://psort.ims.u-tokyo.ac.jp/form.html) and TargetP (http://www.cbs.dtu.dk/services/TargetP/) algorithms. Amino acid multiple alignments were made with the ClustalW program (http://www.ebi.ac.uk/clustalw/) under default parameters. A phylogenetic tree was constructed using the Neighbor–Joining algorithm included in the ClustalW program and visualized by TreeView, version 1.6.6.
DNA gel blot analysis
Genomic DNA was extracted from olive leaves by the CTAB method (Murray and Thomson, 1980) and 5 μg were digested with the restriction endonucleases EcoRI, HindIII or EcoRI/HindIII. DNA was then fractionated on an 0.8% agarose gel, transferred to nylon membrane, probed with a 32P-labelled OeDGAT2 cDNA fragment of 482 bp, corresponding to the 3'-RACE, and washed under high stringency conditions at 65 °C (Church and Gilbert, 1984).
RNA gel blot and semi-quantitative RT-PCR
For RNA gel blot analysis (Sambrook et al., 1989), total RNA (10 μg per lane, standardized by spectrophotometric analysis and gel electrophoresis of equivalent amounts of rRNA), was separated on denaturing 1.4% formaldehyde-agarose gel, transferred onto nitrocellulose membranes without treatment, probed and washed as described for DNA gel blot hybridization. For semi-quantitative RT-PCR analysis, the OeDGAT2-specific primers 5UTRDGAT2 and 5eDG2, corresponding to the 5′ end of the gene, or the OeDGAT1-specific primers 3DGAT (5′-TTGGCTGAATATATTAGCGGAACTTC-3′) and SDGAT (5′-CTCATCATAAAAATGTCCACATCC-3′), corresponding to the 3′ end of the gene, were used. As a control, part of the coding region of the olive β-tubulin gene was amplified with the primers NTUB1 (5′-CCGGTACAAAGCGACAATGAT-3′) and NTUB2 (5′-AGGGGATGGGAAGACAGAGAAAGT-3′). The PCRs were performed using equal amounts of templates and gene-specific or β-tubulin primers and carried out for different numbers of cycles in order to optimize reproducibility.
Microscopy and in situ hybridization
Slides of fixed and paraffin-embedded tissue from young expanding leaves, floral buds (1.5–2.0 mm in length), anthers, and ovaries of buds (3.0 mm in length) or drupes at an early stage of development (9 WAF) were prepared as described previously by Banilas et al. (2007). Sense and anti-sense RNA probes were generated by using the T7 or SP6 RNA promoter of pGEM T-easy vector (Promega), in which the 5′-end OeDGAT2 or the 3′-end OeDGAT1 cDNA fragment was cloned. The riboprobes were labelled with digoxigenin (DIG)-UTP (Roche) by run-off transcription using T7 and SP6 RNA polymerases (Takara Bio Inc.) according to the manufacturer's instructions. In situ hybridization was performed as described by Poghosyan et al. (1999). Signal was detected through the alkaline phosphatase-catalysed precipitation of BCIP/NBT. Sites of positive hybridization signals were detected as blue/violet regions using bright-field microscopy (Olympus BX50).
Results
Cloning and sequence analysis of OeDGAT2 cDNA
Based on conserved amino acid sequences of different type-2 DGATs, degenerate oligonucleotide primers were designed to amplify a central fragment of the homologous gene in olive. PCR employing cDNA from mesocarp tissue generated a fragment of 496 bp. BLAST searches of both nucleotide and deduced amino acid sequences predicted this fragment to be a central part of a type-2 DGAT gene. The full-length cDNA was cloned by conducting 3′- and 5′-RACE PCRs. Sequence comparisons of the 3′- and 5′-ends with the central part of the gene showed that the overlapping regions match perfectly. Based on the above sequence data, primers were designed from the 5′- and 3′-UTRs and the full-length cDNA was amplified, cloned, and sequenced, revealing 100% identity to the expected sequence.
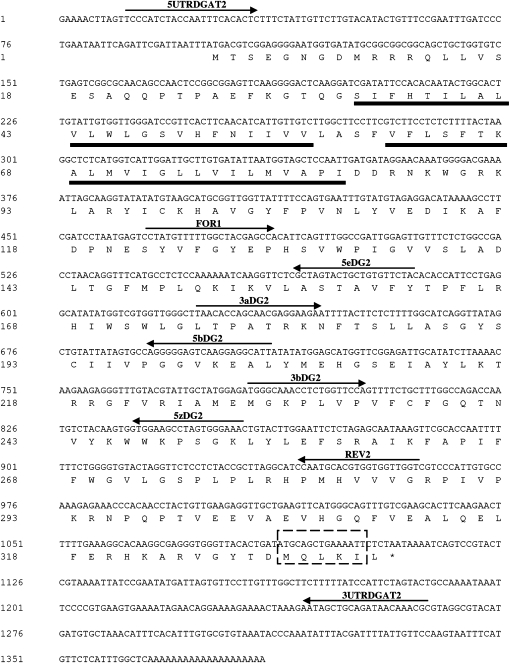
The full-length OeDGAT2 cDNA consisted of a 100 nt 5′-UTR, a 277 nt 3′-UTR, and a putative 1008 nt ORF (Fig. 1), encoding a predicted polypeptide of 335 amino acid residues with a calculated molecular mass of 37.9 kDa and an isolectric point of 9.6. The encoded polypeptide contains two putative transmembrane domains close to the N-terminus, at amino acid residues 34–56 and 61–83, as predicted by the TMHMM server (Fig. 1). Hydrophobicity plot analysis of other type-2 DGAT proteins from both plants and mammals is consistent with the presence of at least one membrane spanning domain close to the N-terminus (Shockey et al., 2006; Kroon et al., 2006). As opposed, hydropathy plots of various type-1 DGAT proteins, including OeDGAT1 (Giannoulia et al., 2000), consist of 9–10 putative transmembrane domains and a relatively hydrophilic N-terminus (Lung and Weselake, 2006). By using two different algorithms (PSORT and TargetP), no plastidial or other signal transit peptide was predicted at the N-terminal region. At the C-terminus a sequence motif was detected (Fig. 1) that resembles the recently described endoplasmic reticulum (ER) retrieval motif -Φ-X-X-K/R/D/E-Φ-COOH, where –Φ- are large hydrophobic amino acid residues (McCartney et al., 2004). A similar motif seems to be highly conserved in both DGAT1 and DGAT2 polypeptides of different plant species (Kroon et al., 2006) including tung tree (Shockey et al., 2006).
Fig. 1.
Nucleotide sequence and deduced amino acid sequence, in single-letter code, of the olive type-2 diacylglycerol acyltransferase cDNA (OeDGAT2). The stop codon is denoted by an asterisk. Numbers represent the position of nucleotides and amino acids. Arrows indicate the position of primers used. A predicted ER-retrieval motif is denoted with a discontinuous lined box at the protein C-terminus. The amino acid sequences corresponding to the predicted transmenbrane domains are underlined.
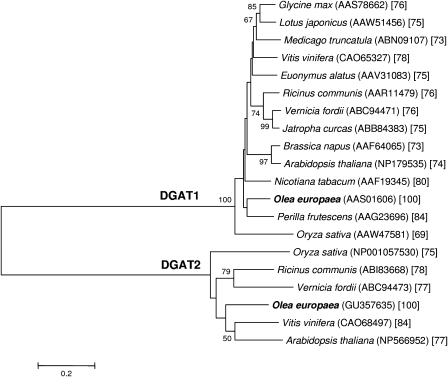
BLAST search showed high similarities of the predicted OeDGAT2 amino acid sequence to type-2 DGATs from other plants, like grapevine (70% identity and 84% similarity) and Arabidopsis (61% and 77%, respectively). No homology (8% identity, 19% similarity) was shared between OeDGAT2 and its type-1 isoform from olive (Giannoulia et al., 2000). To elucidate phylogenetic relationships of OeDGAT2, the deduced amino acid sequence was aligned with other plant type-2 and type-1 DGATs and a phylogenetic tree was constructed. The Neighbor–Joining tree was composed of two distinct branches representing the two types of DGAT families, with OeDGAT1 and OeDGAT2 being clustered within their expected groups (Fig. 2).
Fig. 2.
Phylogenetic relationships among deduced amino acid sequences of OeDGAT1, OeDGAT2, and other plant diacylglycerol acyltransferases (DGATs). The tree was constructed according to the Neighbor–Joining algorithm. GenBank accession numbers are given in parentheses. Numbers at branch points are bootstrap percentages derived from 1000 replicates. Only values ≥50% are presented. Numbers within brackets correspond to % amino acids sequence homologies between olive and other species within each group.
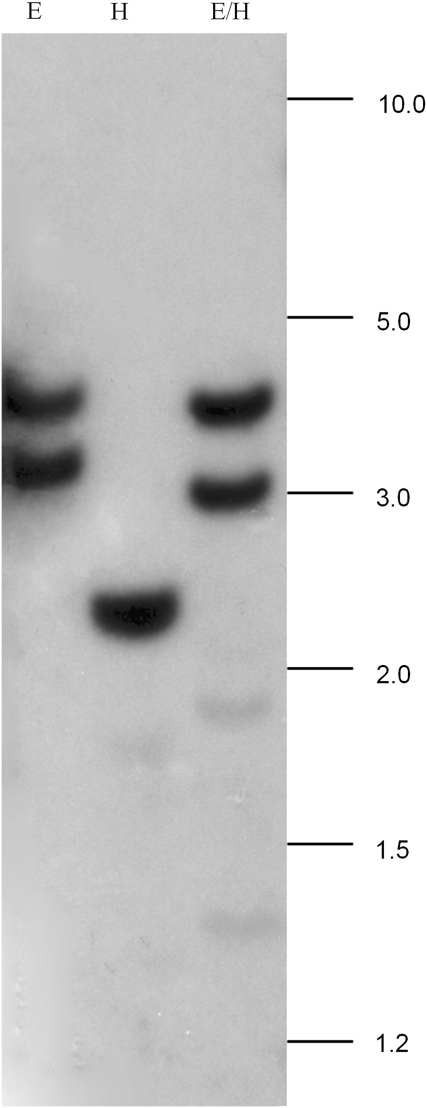
To determine the copy number of OeDGAT2 within the olive genome, genomic DNA was digested with EcoRI, HindIII or EcoRI/HindIII and separated on an agarose gel. Within the target sequence there is a unique EcoRI restriction site, but there are no restriction sites for HindIII (data not shown). Single digest with EcoRI revealed two hybridizing bands, suggesting the presence of a single gene within the olive genome (Fig. 3). This was also confirmed with the HindIII digestion of gDNA. A single hybridizing band was detected (Fig. 3). Double digest also confirmed that the olive genome contains a single copy of a type-2 DGAT gene.
Fig. 3.
Southern analysis of olive genomic DNA digested with EcoRI (E), EcoRI/HindIII (E/H), or HindIII (H) and probed with a cDNA fragment of OeDGAT2. Hybridization was performed at 65 °C (0.745 M Na+). Blot was subjected to high stringency wash conditions (3× in 0.21 M Na+ and 3× in 0.075 M Na+ at 65 °C). Numbers indicate molecular size markers in kb.
DGAT1 and DGAT2 are differentially regulated during olive drupe development
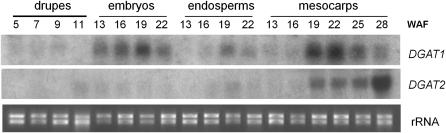
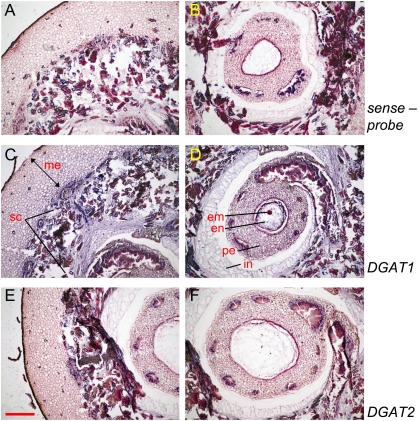
To determine whether the pattern of mRNA synthesis of the type-2 OeDGAT gene shares any similarities to that of type-1 OeDGAT, the expression profile of the two genes was analysed in different tissues. The temporal expression and developmental accumulation of olive type-1 and -2 DGAT transcripts were investigated during olive fruit development. Total RNA was isolated from embryos, endosperms, and mesocarps at different times and analysed by RNA gel blotting. At the very early stages of drupe development (5–11 WAF) RNA was isolated from intact drupes, since zygotic embryos at early globular and heart stages could not be excised without injury. Embryos at the early torpedo, early mid-torpedo, mid-torpedo, and late torpedo stages (13, 16, 19, and 22 WAF, respectively) were dissected out from the surrounding endosperm and collected. As it is shown in Fig. 4, both OeDGATs are developmentally regulated. At the early stages of drupe development (5–11 WAF), low levels of mRNA accumulation were observed for DGAT1. The DGAT2 mRNA was not detected in drupes from 5–9 WAF, while transcripts were just detectable at 11 WAF. In situ mRNA hybridization on 7 WAF drupes revealed that DGAT1 is expressed in both seed and mesocarp tissues of the drupe, confirming the above results. Consistent with RNA blotting results, no hybridization signal was observed for the DGAT2 anti-sense probe (Fig. 5). The expression of DGAT1 occurred throughout the cell types of the drupe. A prominent staining was observed in the mesocarp, the developing seed coat tissues and the perisperm. However, globular embryos showed high levels of expression (Fig. 5). The higher expression level of the DGAT1 gene observed in globular embryos than in the mesocarp (Fig. 5) was also apparent during the later stages of drupe growth (Fig. 4).
Fig. 4.
Northern blot analysis of DGAT1 and DGAT2 expressions during olive fruit development. Total RNA was extracted from drupes, embryos, endosperms, and mesocarps at different developmental stages. Drupes lanes: 5 (1–2 mm in diameter drupes), 7 (3–4 mm), 9 (5–8 mm), and 11 (9–11 mm) weeks after flowering (WAF). Embryo lanes: 13 (early torpedo stage), 16 (early-mid torpedo), 19 (mid-late torpedo), and 22 (late torpedo) WAF. Endosperm lanes: 13, 16, 19, and 22 WAF. Mesocarp lanes: 13, 16, 19, 22, 25, and 28 WAF. Equivalent amounts of rRNA were loaded onto a gel and stained with ethidium bromide to evaluate equal loading in each lane (lower panel).
Fig. 5.
Localization of OeDGAT1 and OeDGAT2 transcripts in young drupes at 7 weeks after flowering (WAF). Longitudinal sections were processed for in situ hybridization with DIG-labelled antisense RNA probes of OeDGAT1 (C, D) and OeDGAT2 (E, F). Negative controls were included by using two respective DIG-labelled riboprobes sense probes. Only representatives using OeDGAT1 sense-probe are presented here (A, B). DGAT1 expression was detected in mesocarp (me), the developing seed coat tissues (sc), the perisperm (pe), and the globular embryo (em), while the expression was relatively lower in endosperm (en) and the integuments (in). Sites of positive hybridization signals are shown as blue/violet regions. Scale bar represents 300 μm.
RNA gel blotting (Fig. 4) showed relatively high DGAT1 transcript accumulation in embryos, starting as early as the early torpedo stage (13 WAF). The signal increased gradually at the later stages, peaked at 19 WAF (mid-torpedo stage), and finally declined substantially at 22 WAF. A similar bell-shaped accumulation pattern was observed in endosperms, with a peak at 19 WAF, but the signal was less intense throughout the time-course. The respective expression of DGAT2 had a different pattern of mRNA accumulation, nevertheless the signal was weak throughout the time-course. Higher expression could be detected during the early- to mid-torpedo embryos. In endosperms, a transient slight increase at 19 WAF was also detected (Fig. 4). At the early stages of mesocarp development (13 and 16 WAF), the expressions of both DGAT1 and DGAT2 were very low, almost undetectable (Fig. 4). At 19 WAF, however, both transcript levels increased rapidly, particularly for DGAT1 that reached almost maximum levels. DGAT1 transcripts peaked at 22 WAF and thereafter decreased gradually until the end of the time-course. At 28 WAF the OeDGAT1 transcript levels were very low. As opposed, transcription of DGAT2 remained at rather low levels, similar to that detected at 19 WAF through 25 WAF. Notably, a sharp up-regulation was observed at 28 WAF, the late maturity stage. During that time (28 WAF), the olive drupes are starting to turn in colour from green to purplish-black. The results show that the two types of DGATs have distinct developmental regulation of gene expression in olive drupes.
Tissue-specific expression patterns in various reproductive or vegetative organs
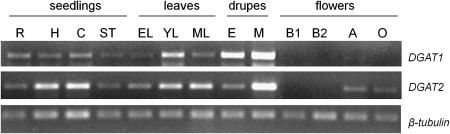
To uncover whether this differential regulation of the two DGATs was also apparent in other parts of the plant, their expression patterns were compared in other than drupe tissues. Semi-quantitative RT-PCR analysis was carried out with total RNA extracted from various seedling parts (roots, hypocotyls, cotyledons, and shoot tips), leaves at different developmental stages (expanding, young, mature), floral buds (1.5 and 2.0 mm in length), anthers and ovaries of open flowers (2.5–3.0 mm in length). For comparison reasons, two samples from RNA gel blotting, i.e. embryos at 13 WAF and mesocarps at 22 WAF, were also included in the analysis. The results obtained from RT-PCR analysis (Fig. 6) were similar to the ones observed in the Northern blot analysis (Fig. 4). DGAT1 expression level in embryos was almost half of the mRNA accumulation detected in mesocarp, while DGAT2 mRNA transcripts in the mesocarp were at much higher levels than those of embryos (Figs 4, 6).
Fig. 6.
Semi-quantitative RT-PCR expression analysis of olive DGAT1 and OeDGAT2 genes. First-strand cDNAs were synthesized from total RNA extracted from roots (R), hypocotyls (H), cotyledons (C), and shoot tips (ST) of olive seedlings; expanding (EL), young (YL), and mature (ML) leaves; embryos (E) at 13 weeks after flowering (WAF) and mesocarps (M) at 22 WAF; flower buds 1.0 mm (B1) and 2.0 mm (B2) in length; anthers (A) and ovaries (O) of flowers 2.5–3.0 mm in length. To ensure equal amounts of template, olive β-tubulin was used as a reference gene.
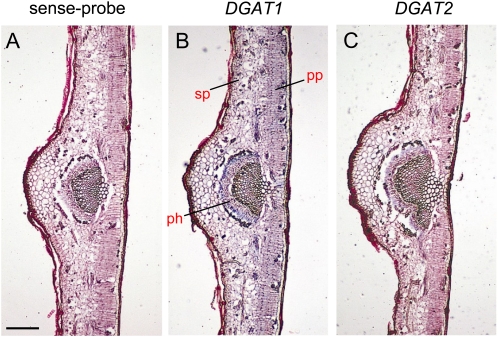
Both DGATs were transcribed in all the vegetative tissues examined (Fig. 6). DGAT1 transcript levels were very low in all seedling parts and relatively constant, as compared to embryo and mesocarp tissues. Root, hypocotyl, and cotyledons had similar levels of DGAT1 mRNAs. By contrast, relatively high DGAT2 expression was recorded in both cotyledons and hypocotyls. Different expression patterns between the two genes were also observed during leaf development. The expression of DGAT1 was almost undetectable in expanding leaves, peaked at the young stage, and was strongly down-regulated in mature leaves (Fig. 6). The DGAT1 mRNA accumulation in young leaves was the highest among the vegetative tissues. The expression of DGAT2 in expanding leaves was low, as per DGAT1, increased during the young stage and remained at similar high levels at maturity. In situ mRNA hybridization analysis on expanding leaves revealed that DGAT1 was expressed in almost every cell type including palisade and spongy parenchyma cells. However, the respective signal for DGAT2 was most prominent in phloem cells of the vascular tissue of veins (Fig. 7). The expression level of DGAT2 in mature leaves was similar to that of the cotyledons, but nevertheless lower than that of the 28 WAF mesocarp (Fig. 6).
Fig. 7.
In situ localization of DGAT1 and DGAT2 transcript accumulation in transverse sections of expanding olive leaves. A representative negative control using the OeDGAT1 sense-probe is presented here (A). Signals of DGAT1 expression are prominent in almost every cell type including palisade parenchyma (pp), spongy parenchyma (sp), and phloem (ph) of the central vein, while DGAT2 mRNA is mostly localized to the phloem tissue of the vascular bundle. Sites of positive hybridization signals are shown as blue/violet regions. Scale bar represents 50 μm.
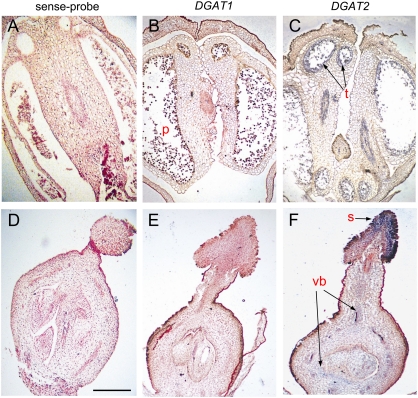
Distinct pattern of expression between DGAT1 and DGAT2 genes was also observed in flower tissues. Based on RT-PCR analysis, no transcripts were detected for DGAT1 and DGAT2 genes in young floral buds (1.5 or 2.0 mm). At later stages of bud development (2.5–3.0 mm) relatively weak signals corresponding to DGAT2 were observed both in anthers and ovules, while no transcripts were detected for DGAT1 (Fig. 6). The levels of DGAT2 mRNAs in both anthers and ovules were comparable with those observed in root tissues. In situ hybridization verified the above results and further showed that DGAT2 transcripts are largely localized in the tapetum cells and vascular bundles of anthers and mostly in the stigma and the vascular bundles of ovaries (Fig. 8). The above results show a prominent differential spatial and temporal gene expression of DGAT1 and DGAT2 in most organs/tissues examined.
Fig. 8.
In situ localization of OeDGAT1 and OeDGAT2 transcript accumulation in longitudinal sections of anthers (A–C) and pistils (D–F) from buds of about 2.5 mm in length. A representative negative control using the OeDGAT2 sense-probe is presented (A, D). DGAT1 exhibited no labelling or negligible background reaction in pollen grains (p) (B) or in ovaries (E). DGAT2 transcripts are largely localized in the tapetum cells (t) of anthers (C) and in the stigma (s) and the vascular bundles (vb) of ovaries (F). Sites of positive hybridization signals are shown as blue/violet regions. Scale bar represents 500 μm.
Discussion
Although several type-1 DGATs have been isolated and characterized from various eukaryotes, to date, the physiological functions of type-2 DGATs have been much less determined (Shockey et al., 2006). In order to understand the mechanisms of TAG biosynthesis in olive better, a novel DGAT cDNA was cloned from olive drupe mesocarp. The potential contribution of type-1 and -2 DGATs in olive TAG biosynthesis were investigated through tissue-specific transcriptional analysis in various vegetative and reproductive organs, at different times during development. DGAT activity exerts strong control over flux in the Kennedy pathway of oleaginous seeds and in olive tissue cultures as well (Ramli et al., 2005), therefore particular emphasis was given to the temporal regulation of olive DGATs during drupe development. In olive fruit, TAGs are formed and stored in both the mesocarp and the seed, two drupe compartments that have different physiological functions and roles and also display differences in the mode of TAG accumulation. Storage TAGs in seeds are proposed to provide energy for germination. They are present in small (0.5–2 mm diameter) subcellular oil bodies completely covered by oleosins to prevent them from coalescence (Hsieh and Huang, 2004). By contrast, TAGs in the mesocarp have no such clear physiological role for the plant per se, but may attract animals for seed dissemination. The fleshy olive mesocarp possesses much larger (about 30 mm diameter) lipid particles of TAGs, which are devoid of surface oleosins (Murphy, 2001; Giannoulia et al., 2007). Accumulation of TAG in olive seeds is relatively fast, compared with the mesocarp, being completed within a relatively short period (Sanchez, 1994). Although massive TAG storage in seeds starts at about 11 WAF, coinciding with endocarp lignification, DGAT1 transcripts were present as early as 5 WAF, albeit at low levels. By contrast, DGAT2 transcripts were almost undetectable until 11 WAF, pointing to a principal role of DGAT1 in early TAG accumulation in olive drupes, especially in the seed.
As the drupe grows further, the rate of oil synthesis in seed tissues accelerates reaching a plateau at about 22 WAF (Sanchez, 1994). The pattern of oil deposition in seeds correlates well with DGAT1 regulation both in embryo and endosperm, whereas the relative expression of DGAT2 was barely detectable. The bell-shaped expression pattern of DGAT1 coincides well with the relative expression of the olive oleosin gene in seed tissues (Giannoulia et al., 2007). Similarly, in oilseed species, a transient increase of DGAT activity occurs at the stage of active oil accumulation, but when the lipid content reaches plateau the activity decreases markedly (Tzen et al., 1993; Weselake et al., 1993). Taken together, the present results suggest a prominent role of DGAT1 in seed olive oil accumulation compared with DGAT2. This is in contrast to oleogenic seed crops that contain unusual fatty acids, where DGAT2 may play a more central role than DGAT1 in oil production (Yu et al., 2006; Shockey et al., 2006; Burgal et al., 2008; Li et al., 2010).
Olive oil does not contain unusual fatty acids and olive is one of the few exceptions of commercially important oil-producing crops in that most of the oil is produced in the mesocarp. Oil accumulation in the mesocarp follows a typical sigmoidal curve (Sanchez, 1994). The major proportion of oil generally starts to accumulate at 16–19 WAF and reaches a plateau at about 28 WAF. However, the pattern of accumulation may vary due to environmental conditions, different agricultural practices and/or the olive variety (Connor and Fereres, 2005). Both DGAT1 and DGAT2 share an overlapping expression pattern from 19–25 WAF, suggesting that they probably function together at those stages. However, following maximal mRNA levels at 22 WAF, DGAT1 transcription declined substantially. As opposed to DGAT1, a sharp up-regulation of DGAT2 was detected at 28 WAF reaching by far its maximum mRNA accumulation in the time-course. As this is also the case in planta, the present data suggest that DGAT2 predominates at the onset of ripening. Although most of TAG accumulation in olives takes place before the onset of ripening, it has been demonstrated that oil cultivars, like ‘Koroneiki’ in this study, have much longer oil-filling periods than those of table cultivars (Garcia-Martos and Mancha, 1992; Farinelli et al., 2002). Indeed, as shown previously, the transcripts of stearoyl-ACP desaturase, the responsible enzyme for oleate production, accumulates as the growth of the mesocarp proceeds, reaching maximum levels at 28 WAF (Haralampidis et al., 1998). It is still unclear why DGAT2 expression overwhelms DGAT1 at the late stages of mesocarp growth. DGAT2 implication in altering the fatty acid composition of TAGs is unlikely here, because there are not sufficient structural changes in olive oil composition during ripening (Gutierrez et al., 1999; Ayton et al., 2001; Cossignani et al., 2001). Some plant DGATs exhibit a broad acyl-CoA preference as revealed by specificity and selectivity studies (Lung and Weselake, 2006; Shockey et al., 2006). Therefore, it is plausible that the acyl composition of TAG in oilseeds is predominantly dictated by the availability of specific DAG and acyl-CoA pools (Kamisaka et al., 1997; Lung and Weselake, 2006; Shockey et al., 2006). The higher proportion of oleoyl-CoA throughout olive fruit growth dictates a higher incorporation of oleic acid, which is consistent with analytical data for the fatty acid composition of olive oils (Sanchez and Harwood, 2002).
It is believed that the increase in lipid droplet size in olive mesocarpic cells is facilitated through the coalescence of smaller lipid bodies (Rangel et al., 1997). Concomitantly, during the olive fruit-ripening phase, oil biosynthesis continues together with an increase in dry matter, albeit at a slower rate than in previous phases (Conde et al., 2008). The mechanism of lipid-droplet formation and the growth in size in eukaryotes is largely unknown (Guo et al., 2009). Recent data revealed that DGAT2, but not DGAT1, may be dynamically associated with lipid droplets and mitochondrial compartments, promoting active synthesis and storage of TAGs in oleate-treated mammalian (COS-7) cells (Stone et al., 2009). It is unknown as yet whether the plant DGAT2 enzyme may have a similar function on lipid droplet enlargement in olive mesocarp cells. In any case, present data suggest that olive DGAT2 is a key player in the late production of mesocarp oil, the time of lipid droplet enlargement. By contrast, in both embryos and endosperm the oil bodies remain extremely small (Ross et al., 1993) and in those tissues the DGAT2 expression is almost negligible. This raises the possibility that OeDGAT2 may be useful for breeding programmes aimed at high oil-yielding olive genotypes through marker-assisted selection or by genetic engineering. In Arabidopsis, DGAT1 and PDAT1 have overlapping functions in TAG biosynthesis (Zhang et al., 2009). PDAT may also contribute to TAG biosynthesis in olive callus cultures (Hernandez et al., 2008). Nevertheless, the activity of this enzyme as measured in subcellular fractions of olive calli was rather low, indicating that DGAT exerts significant flux control over TAG formation in olives (Ramli et al., 2005; Hernandez et al., 2008).
Both DGAT genes were expressed in all seedling organs examined, with mRNA accumulation of DGAT2 being the highest in the cotyledons and hypocotyls. To our knowledge, this is the first time that the induction of DGAT2 has been reported in seedling tissues. Transcriptional analysis in Arabidopsis seedlings has revealed high mRNA levels of type-1 DGAT during the post-embryonic stages in both cotyledons and hypocotyls (Lu et al., 2003) and DGAT1 activity has been previously correlated with de novo TAG synthesis in the cotyledons of castor seedlings (He et al., 2006). It is likely that excess sugar production is channelled into TAGs for internal use or for storage to prevent osmotic effects (He et al., 2006). In the latter case, glyoxysomes and other machineries may convert degraded lipids into sugar for export to non-senescing tissues (Pracharoenwattana and Smith, 2008).
Huang et al. (2009) showed that oil bodies, primarily composed of steryl esters and triacylglycerols, were abundant in the Physcomitrella photosynthetic vegetative gametophyte. In this study, relatively high levels of transcription of both OeDGATs were detected in olive leaves, where the regulation of expression was clearly developmentally regulated. Transcripts of both DGATs and, possibly, the accompanied neutral lipid accumulation, start as the leaf reaches its final size, but is still young. Cross-sections of young leaves followed by Sudan III staining and microscopic observation revealed the presence of lipid droplets throughout the mesophyll but mostly close to the epidermis and the vascular tissue (data not shown). Accumulating data, as stated above, suggest that DGAT genes also play roles other than its ‘classical’ role to synthesize TAGs in the storage organs (Zhang et al., 2009). The present results point to a differential contribution of each DGAT gene in various organs in a temporal-related manner. This notion was further justified in flowers. OeDGAT2, but not OeDGAT1, was highly expressed in the tapetum cells of anthers and in different ovule tissues. Neither DGAT expression was detected in floral buds while, in actively dividing cells like shoot tips and young seedling roots, both genes were down-regulated. DGAT2 expression in tapetum cells is in concert with the deposits of TAGs (Murphy, 2001). Taken together, the high levels of DGAT2 expression in leaves and in vascular and tapetum cells of flowers, point to its prominent role in the synthesis of TAGs in those organs possibly for later lipid mobilization and sugar transport.
In conclusion, both DGAT1 and DGAT2 share overlapping but distinct transcription patterns during vegetative and reproductive growth, suggesting that they are differentially regulated in a developmental and cellular manner. They probably have similar functions but they also serve different purposes. Distinct expression patterns of DGAT1 and DGAT2 were observed between the seed and mesocarp, with DGAT1 contributing most of the TAG deposition in seeds, reflecting the large differences in the mode of TAG accumulation between the two fruit compartments. Important differences between the expression profiles of the two genes were also apparent during drupe ripening. Our findings in this study provide evidence that DGAT2 may be a key mediator of higher oil yields in ripening mesocarps, where oil droplets increase in size and TAGs are still accumulating. This important finding, however, remains to be further justified by measuring the respective enzymatic activities. Apart from the ripening mesocarp, DGAT2 may also serve for the accumulation of TAGs in senescing organs, like cotyledons and flower tissues, possibly for further lipid mobilization or other internal uses.
Glossary
Abbreviations
- DAG
diacylglycerol
- DGAT
diacylglycerol acyltransferase
- GP
glycerol-3-phosphate
- GPAT
glycerol-3-phosphate acyltransferase
- LPA
lysophosphatidic acid
- LPAT
lysophosphatidic acid acyltransferase
- PA
phosphatidic acid
- PDAT
phospholipid:diacylglycerol acyltransferase
- TAG
triacylglycerol
References
- Ayton J, Mailer RJ, Robards K. Changes in oil content and composition of developing olives in a selection of Australian cultivars. Australian Journal of Experimental Agriculture. 2001;41:815–821. [Google Scholar]
- Banilas G, Nikiforiadis A, Makariti I, Moressis A, Hatzopoulos P. Discrete roles of a microsomal linoleate desaturase gene in olive identified by spatiotemporal transcriptional analysis. Tree Physiology. 2007;27:481–490. doi: 10.1093/treephys/27.4.481. [DOI] [PubMed] [Google Scholar]
- Banilas G, Hatzopoulos P. Developmental perplexity of oil biosynthesis gene expression in olive. In: Berti L, Maury J, editors. Advances in olive resources. Kerala, India: Transworld Research Network; 2009. pp. 1–22. [Google Scholar]
- Bao X, Ohlrogge J. Supply of fatty acid is one limiting factor in the accumulation of triacylglycerol in developing embryos. Plant Physiology. 1999;120:1057–1062. doi: 10.1104/pp.120.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J, Somerville C. Glycerolipid synthesis: biochemistry and regulation. Annual Review of Plant Physiology and Plant Molecular Biology. 1991;42:467–506. [Google Scholar]
- Burgal J, Shockey J, Lu C, Dyer J, Larson T, Graham I, Browse J. Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnology Journal. 2008;6:819–831. doi: 10.1111/j.1467-7652.2008.00361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases S, Stone SJ, Zhou P, Yen E, Tow B, Lardizabal KD, Voelker T, Farese RV., Jr Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. Journal of Biological Chemistry. 2001;276:38870–38876. doi: 10.1074/jbc.M106219200. [DOI] [PubMed] [Google Scholar]
- Church G, Gilbert W. Genomic sequencing. Proceedings of the National Academy of Sciences, USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor DJ, Fereres E. The physiology of adaptation and yield expression in olive. Horticultural Reviews. 2005;31:155–229. [Google Scholar]
- Conde C, Delrot S, Geros H. Physiological, biochemical and molecular changes occurring during olive development and ripening. Journal of Plant Physiology. 2008;165:1545–1562. doi: 10.1016/j.jplph.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Cossignani L, Simonetti MS, Damiani P. Structural changes of triacylglycerol and diacylglycerol fractions during olive drupe ripening. European Food Research and Technology. 2001;212:160–164. [Google Scholar]
- Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S. Phospholipid: diacylglycerol acyltransferase: an enzyme that catalyses the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proceedings of the National Academy of Sciences, USA. 2000;97:6487–6492. doi: 10.1073/pnas.120067297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doveri S, Baldoni L. Olive. In: Kole C, editor. Genome mapping and molecular breeding in plants, fruits and nuts. Vol. IV. Berlin, Heidelberg: Springer-Verlag; 2007. pp. 253–264. [Google Scholar]
- Farinelli D, Boco M, Tombesi A. Intensity and growth period of the fruit components of olive varieties. Acta Horticulturae. 2002;586:607–610. [Google Scholar]
- Frohman MA. RACE: rapid amplification of cDNA ends. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. London: Academic Press; 1990. pp. 28–38. [Google Scholar]
- Garcia-Martos JM, Mancha M. Evoluciσn de la biosνntesis de lνpidos durante la maduraciσn de las variedades de aceituna ‘Picual’ y ‘Godal’. Grasas y Aceites. 1992;43:277–280. [Google Scholar]
- Giannoulia K, Banilas G, Hatzopoulos P. Oleosin gene expression in olive. Journal of Plant Physiology. 2007;164:104–107. doi: 10.1016/j.jplph.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Giannoulia K, Haralampids K, Poghosyan Z, Murphy DJ, Hatzopoulos P. Differential expression of diacylglycerol acyltransferase (DGAT) genes in olive tissues. Biochemical Society Transactions. 2000;28:695–697. [PubMed] [Google Scholar]
- Guo Y, Cordes KR, Farese RV, Jr, Walther TC. Lipid droplets at a glance. Journal of Cell Science. 2009;122:749–752. doi: 10.1242/jcs.037630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez F, Jimenez B, Ruνz A, Albi MA. Effect of olive ripeness on the oxidative stability of virgin olive oil extracted from the varieties Picual and Hojiblanca and on the different components involved. Journal of Agricultural and Food Chemistry. 1999;44:121–127. doi: 10.1021/jf980684i. [DOI] [PubMed] [Google Scholar]
- Haralampidis K, Milioni D, Sanchez J, Baltrusch M, Heinz E, Hatzopoulos P. Temporal and transient expression of stearoyl-ACP carrier protein desaturase gene during olive fruit development. Journal of Experimental Botany. 1998;49:1661–1669. [Google Scholar]
- Hatzopoulos P, Banilas G, Giannoulia K, Gazis F, Nikoloudakis N, Milioni D, Haralampidis K. Breeding, molecular markers and molecular biology of the olive tree. European Journal of Lipid Science and Technology. 2002;104:574–586. [Google Scholar]
- He X, Chen GQ, Lin JT, McKeon TA. Diacylglycerol acyltransferase activity and triacylglycerol synthesis in germinating castor seed cotyledons. Lipids. 2006;41:281–285. doi: 10.1007/s11745-006-5098-2. [DOI] [PubMed] [Google Scholar]
- Hernandez ML, Guschina IA, Martínez-Rivas JM, Mancha M, Harwood JL. The utilization and desaturation of oleate and linoleate during glycerolipid biosynthesis in olive (Olea europaea L.) callus cultures. Journal of Experimental Botany. 2008;59:2425–2435. doi: 10.1093/jxb/ern121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K, Huang AHC. Endoplasmic reticulum, oleosins, and oils in seeds and tapetum cells. Plant Physiology. 2004;136:3427–3434. doi: 10.1104/pp.104.051060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Chung CI, Lin YC, Hsing YI, Huang AH. Oil bodies and oleosins in Physcomitrella possess characteristics representative of early trends in evolution. Plant Physiology. 2009;150:1192–1203. doi: 10.1104/pp.109.138123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihara K, Takahashi T, Fujii S. Diacylglycerol acyltransferase in maturing safflower seeds: its influences on the fatty acid composition of triacylglycerol and on the rate of triacylglycerol synthesis. Biochimica et Biophysica Acta. 1988;958:125–129. doi: 10.1016/0005-2760(88)90253-6. [DOI] [PubMed] [Google Scholar]
- Jako C, Kumar A, Wei YD, Zou JT, Barton DL, Giblin EM, Covello PS, Taylor DC. Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiology. 2001;126:861–874. doi: 10.1104/pp.126.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisaka Y, Mishra S, Nakahara T. Purification and characterization of diacylglycerol acyltransferase from the lipid body fraction of an oleaginous fungus. Journal of Biochemistry. 1997;121:1107–1114. doi: 10.1093/oxfordjournals.jbchem.a021702. [DOI] [PubMed] [Google Scholar]
- Katavic V, Reed DW, Taylor DC, et al. A role for diacylglycerol acyltransferase during leaf senescence. Plant Physiology. 2002;129:1616–1626. doi: 10.1104/pp.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EP. Biosynthesis of complex lipids. Federation Proceedings. 1961;20:934–940. [PubMed] [Google Scholar]
- Kroon JT, Wei W, Simon WJ, Slabas AR. Identification and functional expression of a type 2 acyl-CoA:diacylglycerol acyltransferase (DGAT2) in developing castor bean seeds which has high homology to the major triglyceride biosynthetic enzyme of fungi and animals. Phytochemistry. 2006;67:2541–2549. doi: 10.1016/j.phytochem.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Lardizabal KD, Mai JT, Wagner NW, Wyrick A, Voelker T, Hawkins DJ. DGAT2 is a new diacylglycerol acyltransferase gene family: purification, cloning, and expression in insect cells of two polypeptides from Mortierella ramanniana with diacylglycerol acyltransferase activity. Journal of Biological Chemistry. 2001;276:38862–38869. doi: 10.1074/jbc.M106168200. [DOI] [PubMed] [Google Scholar]
- Li R, Yu K, Hildebrand DF. DGAT1, DGAT2, and PDAT expression in seeds and other tissues of epoxy and hydroxy fatty acid accumulating plants. Lipids. 2010;45:145–157. doi: 10.1007/s11745-010-3385-4. [DOI] [PubMed] [Google Scholar]
- Lu CL, de Noyer SB, Hobbs DH, Kang J, Wei Y, Krachtus D, Hills MJ. Expression pattern of diacylglycerol acyltransferase-1, an enzyme Involved in triacylglycerol biosynthesis, in Arabidopsis thaliana. Plant Molecular Biology. 2003;52:31–41. doi: 10.1023/a:1023935605864. [DOI] [PubMed] [Google Scholar]
- Lu C, Hills MJ. Arabidopsis mutants deficient in diacylglycerol acyltransferase display increased sensitivity to abscisic acid, sugars, and osmotic stress during germination and seedling development. Plant Physiology. 2002;129:1352–1358. doi: 10.1104/pp.006122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung SC, Weselake RJ. Diacylglycerol acyltransferase: a key mediator of plant triacylglycerol synthesis. Lipids. 2006;41:1073–1088. doi: 10.1007/s11745-006-5057-y. [DOI] [PubMed] [Google Scholar]
- McCartney AW, Dyer JM, Dhanoa PK, Kim PK, Andrews DW, McNew JA, Mullen RT. Membrane-bound fatty acid desaturases are inserted co-translationally into the ER and contain different ER retrieval motifs at their carboxy termini. The Plant Journal. 2004;37:156–173. doi: 10.1111/j.1365-313x.2004.01949.x. [DOI] [PubMed] [Google Scholar]
- Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants, and microorganisms. Progress in Lipid Research. 2001;40:325–438. doi: 10.1016/s0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- Murray MG, Thomson WF. Rapid isolation of high weight plant DNA. Nucleic Acids Research. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry HJ, Bligny R, Gout E, Harwood JL. Changes in Kennedy pathway intermediates associated with increased triacylglycerol synthesis in oil-seed rape. Phytochemistry. 1999;52:799–804. [Google Scholar]
- Poghosyan Z, Haralampidis K, Martsinkovskaya AI, Murphy DJ, Hatzopoulos P. Developmental regulation and spatial expression of a plastidial fatty acid desaturase from Olea europaea. Plant Physiology and Biochemistry. 1999;37:109–119. [Google Scholar]
- Pracharoenwattana I, Smith SM. When is a peroxisome not a peroxisome? Trends in Plant Science. 2008;13:522–525. doi: 10.1016/j.tplants.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Ramli US, Salas JJ, Quant PA, Harwood JL. Metabolic control analysis reveals an important role for diacylglycerol acytransferase in olive but not in oil palm lipid accumulation. FEBS Journal. 2005;272:5764–5770. doi: 10.1111/j.1742-4658.2005.04964.x. [DOI] [PubMed] [Google Scholar]
- Rangel B, Platt KA, Thomson WW. Ultrastructural aspects of the cytoplasmic origin and accumulation of oil in olive fruit (Olea europaea) Physiologia Plantarum. 1997;101:109–114. [Google Scholar]
- Ross JHE, Sanchez J, Millan F, Murphy DJ. Differential presence of oleosins in oleogenic seed and mesocarp tissues in olive (Olea europaea) and avocado (Persea americana. Plant Science. 1993;93:203–210. [Google Scholar]
- Saha S, Enugutti B, Rajakumari S, Rajasekharan R. Cytosolic triacylglycerol biosynthetic pathway in oilseeds: molecular cloning and expression of peanut cytosolic diacylglyerol acyltransferase. Plant Physiology. 2006;141:1533–1543. doi: 10.1104/pp.106.082198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanchez J. Lipid photosynthesis in olive fruit. Progress in Lipid Research. 1994;33:97–104. doi: 10.1016/0163-7827(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Sanchez J, Harwood JL. Biosynthesis of triacylglycerols and volatiles in olives. European Journal of Lipid Science and Technology. 2002;104:564–573. [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM. Tung (Vernicia fordii) DGAT1 and DGAT2 possess different affinities for eleostearic acid-containing substrates and are localized to different subdomains of the endoplasmic reticulum. The Plant Cell. 2006;18:2294–2313. doi: 10.1105/tpc.106.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl U, Carlsson AS, Lenman M, Dahlqvist A, Huang B, Banas W, Banas A, Stymne S. Cloning and functional characterization of a phospholipid:diacylglycerol acyltransferase from Arabidopsis. Plant Physiology. 2004;135:1324–1335. doi: 10.1104/pp.104.044354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SJ, Levin MC, Zhou P, Han J, Walther TC, Farese RV., Jr The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. Journal of Biological Chemistry. 2009;284:5352–5361. doi: 10.1074/jbc.M805768200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzen JTC, Cao YZ, Laurent P, Ratnayake C, Huang AHC. Lipids, proteins, and structure of seed oil bodies from diverse species. Plant Physiology. 1993;101:267–276. doi: 10.1104/pp.101.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weselake RJ. Storage lipids. In: Murphy DJ, editor. Plant lipids: biology, utilization, and manipulation. Oxford: Blackwell Publishing; 2005. pp. 162–225. [Google Scholar]
- Weselake RJ, Pomeroy MK, Furukawa TL, Golden JL, Little DB, Laroche A. Developmental profile of diacylglycerol acyltransferase in maturing seeds of oilseed rape and safflower and microspore-derived cultures of oilseed rape. Plant Physiology. 1993;102:565–571. doi: 10.1104/pp.102.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weselake RJ, Shah S, Tang M, et al. Metabolic control analysis is helpful for informed genetic manipulation of oilseed rape (Brassica napus) to increase seed oil content. Journal of Experimental Botany. 2008;59:3543–3549. doi: 10.1093/jxb/ern206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, McCracken CTJ, Li R, Hildebrand DF. Diacylglycerol acyltransferases from Vernonia and Stokesia prefer substrates with vernolic acid. Lipids. 2006;41:557–566. doi: 10.1007/s11745-006-5005-x. [DOI] [PubMed] [Google Scholar]
- Zhang FY, Yang MF, Xu YN. Silencing of tobacco causes a reduction in seed oil content. Plant Science. 2005;169:689–694. [Google Scholar]
- Zhang M, Fan J, Taylor DC, Ohlrogge JB. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. The Plant Cell. 2009;21:3885–3901. doi: 10.1105/tpc.109.071795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Allen WB, Roesler K, et al. A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nature Genetics. 2008;40:367–372. doi: 10.1038/ng.85. [DOI] [PubMed] [Google Scholar]
- Zou J, Wei Y, Jako C, Kumar A, Selvaraj G, Taylor DC. The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. The Plant Journal. 1999;19:645–653. doi: 10.1046/j.1365-313x.1999.00555.x. [DOI] [PubMed] [Google Scholar]