Abstract
Transgenic tomato plants (Solanum lycopersicum L.) with reduced mRNA levels of AUXIN RESPONSE FACTOR 7 (SlARF7) form parthenocarpic fruits with morphological characteristics that seem to be the result of both increased auxin and gibberellin (GA) responses during fruit growth. This paper presents a more detailed analysis of these transgenic lines. Gene expression analysis of auxin-responsive genes show that SlARF7 may regulate only part of the auxin signalling pathway involved in tomato fruit set and development. Also, part of the GA signalling pathway was affected by the reduced levels of SlARF7 mRNA, as morphological and molecular analyses display similarities between GA-induced fruits and fruits formed by the RNAi SlARF7 lines. Nevertheless, the levels of GAs were strongly reduced compared with that in seeded fruits. These findings indicate that SlARF7 acts as a modifier of both auxin and gibberellin responses during tomato fruit set and development.
Keywords: AUXIN RESPONSE FACTOR, fruit development, gibberellin (GA), parthenocarpy, tomato
Introduction
At the end of flower development, the cell division activity in the ovary stops when it has reached its mature size. After a few days, the flower will abscise unless successful completion of pollination and fertilization occur and cell division activity resumes (Gillaspy et al., 1993). In tomato (Solanum lycopersicum), this period of cell division continues for 10–14 d. During the following 6–7 weeks, cell division activity is low and fruit growth mainly depends on cell expansion (Mapelli et al., 1978; Bünger-Kibler and Bangerth, 1982; Gillaspy et al., 1993). At the end of this cell-expansion period, the fruit has reached its final size and will start to ripen (Gillaspy et al., 1993). These changes require tight regulation both at the level of gene activity and translation, which is mediated by phytohormones such as auxin and gibberellin (GA). Application of either of these two hormones on tomato ovaries leads to the formation of seedless (parthenocarpic) fruit, without the need for pollination and fertilization (Gustafson, 1937, 1960; Wittwer et al., 1957; Bünger-Kibler and Bangerth, 1982; Serrani et al., 2007a). Furthermore, the auxin and GA contents in the ovary were shown to increase upon pollination and fertilization (Mapelli et al., 1978; Sjut and Bangerth, 1982; Koshioka et al., 1994). However, to date, the molecular mechanisms by which these hormones regulate fruit set and development are still poorly understood, although recent advances have shed some light on their regulatory role.
Despite the fact that application of either auxin or GA can trigger tomato fruit development, there are several indications that each of these hormones has a specific role in fruit development. Treatments of unpollinated ovaries with auxins stimulated cell division for an extended period, resulting in the formation of fruits with a higher number of pericarp cells. By contrast, the pericarp of GA-induced tomato fruits contained fewer cells but with a volume, on average, larger than the cells of control fruits. The application of GA together with auxin, resulted in the formation of fruits in which the number of pericarp cells and cell size were similar to that in seeded fruits (Bünger-Kibler and Bangerth, 1982; Serrani et al., 2007a), suggesting that both hormones are required for normal fruit development. In accordance, the expression of both auxin and GA response genes appeared to be up-regulated within 48 h after pollination. Interestingly, none of the auxin signalling genes seemed to be influenced by GA treatment of unpollinated ovaries (Vriezen et al., 2008), suggesting that auxin acts prior to or independently of GA. Serrani et al. (2008) showed that auxin-induced fruit development was significantly reduced by the simultaneous application of GA biosynthesis inhibitors, suggesting that the effect of auxin is mediated by GA. This hypothesis was supported by the transcript levels of the GA biosynthesis genes, copalyldiphosphate synthase (SlCPS) and GA 20-oxidases, SlGA20ox1, SlGA20ox2, and SlGA20ox3 which are all up-regulated after pollination (Rebers et al., 1999; Serrani et al., 2007b), but also in auxin-treated ovaries (Serrani et al., 2008). In addition, the transcript level of SlGA2ox2 encoding a GA inactivating GA 2-oxidase was found to be lower, also leading to higher levels of active GA (Serrani et al., 2008). These data indicate that auxin and GA action successively regulate tomato fruit development. In Arabidopsis thaliana and Pisum sativum (pea), auxin may act as an early post-pollination signal, which originates in the ovules upon successful fertilization that, in turn, stimulates GA biosynthesis. Subsequently, these GAs are transported to the surrounding tissues and trigger fruit development (Ozga and Reinecke, 2003; Dorcey et al., 2009). Thus, in order to unravel the molecular mechanisms that regulate fruit set and development, it is important to understand the cross-talk between these two hormones.
Previously, a functional analysis was performed of Solanum lycopersicum AUXIN RESPONSE FACTOR 7 (SlARF7), the transcript level of which was found to be high in the unpollinated mature tomato ovary, but decreased within 48 h after pollination or after auxin application (De Jong et al., 2009b). Transgenic plants with decreased SlARF7 mRNA levels formed parthenocarpic fruits, indicating that SlARF7 may act as a negative regulator of fruit set. These fruits displayed characteristics that seemed to be the result of both increased auxin and GA responses during fruit growth. Here, a more detailed analysis of these transgenic lines is presented, which establishes that SlARF7 is indeed affecting the signalling response pathways of auxin and GA and is part of the cross-talk between these two hormones. The silencing of SlARF7 affected part of the auxin signalling response pathway, and resulted in enhanced GA signalling. However, the levels of GA were strongly reduced, suggesting that SlARF7 also acts as a modifier of the GA response during the early stages of tomato fruit development.
Materials and methods
Plant materials and growth conditions
Tomato plants (Solanum lycopersicum L. cv. Moneymaker) were grown on soil under standardized greenhouse conditions during spring, with a daily temperature regime of 20–25 °C (day) and 15–18 °C (night). The photoperiod was extended to 16 h by low-intensity light supplied by high-pressure sodium lamps (600 W, Philips, http://www.philips.com).
All analyses were performed on ovaries and fruits from wild-type and the third generation of RNAi SlARF7 lines 4 and 6, these were the two transgenic lines which had only a fruit phenotype (De Jong et al., 2009b). The tissues were collected at several stages of development: unpollinated ovaries at pre-anthesis, corresponding to the stage at which wild-type flowers were emasculated (3 d before anthesis); unpollinated ovaries at anthesis; 3–4 mm, 5–6 mm, 7–8 mm, and 9–10 mm fruits, corresponding to approximately 6, 8, 10, and 12 DAP, respectively. Fruit size was used as a marker for developmental stage, as fruit set in the transgenic plants was changed, making it difficult to determine the same stage of development. All collected tissues were frozen in liquid N2 and stored at –80 °C until RNA or GA extraction.
Real-time quantitative PCR
Total RNA was extracted from the frozen tomato plant tissues using a NucleoSpin® RNA plant kit (Macherey-Nagel, http://www.macherey-nagel.com) and was treated with RNase-free DNase I (Fermentas, http://www.fermentas.com). The total DNA-free RNA (400 ng) was used as a template for cDNA synthesis (iScript™ cDNA synthesis kit, Bio-Rad, http://www.bio-rad.com). For real-time quantitative PCR, 5 μl of 25-fold diluted cDNA were used in a 25 μl PCR reaction containing 400 nM of each primer and 12.5 μl iQ™ SYBR Green Supermix (Bio-Rad). The PCR reactions were performed in a 96-well iCycler (Bio-Rad), with a temperature programme starting with 3 min at 95 °C, then 40 cycles of 15 s at 95 °C and 45 s at 60 °C. At the end, the melting temperature of the product was determined to verify the specificity of the amplified fragment. The sequences of the primers (see Supplementary Table S1 at JXB online) used for real-time quantitative PCR were obtained from Serrani et al. (2008), De Jong et al. (2009b), and Dr A Czerednik (Radboud University, http://www.ru.nl), or designed with a computer program (Beacon Designer 5.01, Premier Biosoft International, http://www.premierbiosoft.com).
Quantification of gibberellins
The GAs were quantified following the protocol described in Fos et al. (2000). In short, aliquots (2 g) of frozen material were extracted with 80% (v/v) methanol. After removing the organic phase, the water fraction was partitioned against ethyl acetate, and purified by QAE-Sephadex chromatography and C18 cartridges. Subsequently, the GAs were separated by reverse phase HPLC chromatography (4 μm C18 column, 15 cm long, 3.9 mm inner diameter; NovaPak; Millipore, http://millipore.com), and appropriate fractions were grouped for further analysis. After methylation and trimethylsilylation, the GAs were quantified by GC-SIM, using a gas chromatograph (model 5890; Hewlett-Packard, http://www.hp.com) coupled to a mass-selective detector (model 5971A; Hewlett-Packard). The concentrations of GAs in the extracts were determined with the calibration curves methodology, using the internal standards [17,17-2H]GA1, [17,17-2H]GA4, [17,17-2H]GA8, [17,17-2H]GA19, [17,17-2H]GA20, [17,17-2H]GA29, [17,17-2H]GA34, [17,17-2H]GA44, and [17,17-2H]GA53 (purchased from Dr L Mander, Australian National University, http://www.anu.edu.au) that were added to the extracts.
Microscopy
Tissues were fixed in a 2% (v/v) glutaraldehyde, 0.1 M phosphate buffer pH 7.2 solution overnight at 4 °C. Subsequently, the tissues were dehydrated in 100% (v/v) ethanol and embedded in Spurr's resin (Agar Scientific, http://www.agarscientific.com). Sections of 1 μm were stained with a toluidine blue solution (0.1% in 1% borax) and viewed under a Leitz Orthoplan microscope (Leica Microsystems, http://www.leica-microsystems.com). The micrographs were made with a Leica digital camera (model DFC 420C; Leica Microsystems), while applying shading correction with the Leica Application Suite software (Leica Microsystems).
Quantification methodology of cell area and number of dividing cells
The micrographs of the pericarp were optimized for further analysis by applying stitching and levelling in Adobe Photoshop CS3 (Adobe, http://www.adobe.com). Subsequently, the micrographs were subdivided into the inner and outer epidermal layer, endocarp, mesocarp, and exocarp. The endocarp was defined as the inner 1 or 2 cell layers of the pericarp, excluding the inner epidermal layer. The mesocarp included all the cell layers in between the endocarp and exocarp. The exocarp was defined as the outer 4–6 cell layers of the pericarp, excluding the outer epidermal layer. So-called exocarp cells in the layer closest to the mesocarp with an area of more than 450 μm2 were re-defined as mesocarp cells. The vascular bundles, intercellular cavities, and outer-tissue regions were masked manually. Then, the programs ImageJ (Rasband, 1997-2009, http://rsb.info.nih.gov/ij/) and Adobe Photoshop CS3 (Adobe) were used to create a binary outline of the cell walls and cell contents to measure the morphometric features automatically. Briefly, macros, inspired by G Polder's approach (http://www.biometris.wur.nl) were compiled in ImageJ to carry out routine operations, such as colour thresholding, conversion to binary image, dilation, erosion, filling holes, and skeletonization. The resulting raw skeleton image was superimposed over the original image and corrected semi-automatically for edge errors and false bifurcations using Adobe Photoshop CS3 (Adobe) utilities. The corrected skeleton image was then reintroduced into ImageJ and measurements on cell number and calibrated cell area were performed (detailed information on the macros is available on request). Recent mitotic divisions were defined as the incomplete separation of daughter cells, when the cell plate just has reached the perimeter of the parent cell. In total, two different regions per fruit and five fruits per line have been analysed.
Results
Microscopic analysis of early fruit development of wild-type and transgenic fruits
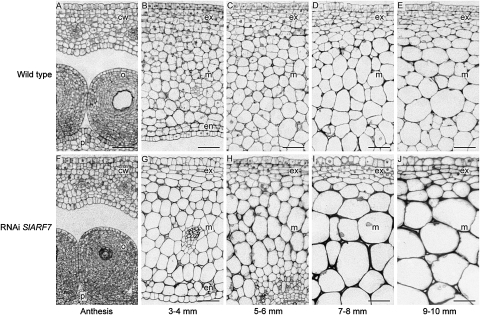
A previous study showed that transgenic plants, in which SlARF7 transcript levels were reduced by an RNA interference (RNAi) approach, produced parthenocarpic fruits (De Jong et al., 2009b). The characteristics of these fruits suggest that SlARF7 is involved in the hormonal regulatory mechanisms of both auxin and GA during the early stages of fruit development. One of these characteristics was the thick pericarp of the transgenic fruits compared with the pericarp of wild-type developing fruits of 3 cm in diameter, resulting in smaller locules. This thick pericarp was the result of increased cell expansion, while no obvious difference in cell number was observed (De Jong et al., 2009b). In this study, the pericarp of both wild-type and transgenic fruits was analysed during the early stages of fruit development. In general, the pericarp is composed of parenchyma cells that are differentiated into three layers: the endocarp, mesocarp, and exocarp (Gillaspy et al., 1993). These fruits were collected before and during the early stages of fruit development, ranging from unpollinated ovaries collected at anthesis to fruits that were 10 mm in diameter (Fig. 1). At anthesis, there were no clear differences between the pericarp of wild-type and transgenic ovaries (Fig. 1A, F), but in 3–4 mm fruits, corresponding to 6 d post anthesis (DPA), the difference in cell size between the pollinated wild-type fruits and parthenocarpic transgenic fruit became visible. At this stage, the cells in the exocarp were dividing, while most cells in the mesocarp and endocarp had already started to expand. Nevertheless, the cells in the mesocarp and endocarp of the transgenic fruits were bigger compared with the cells in wild-type fruits (Fig. 1B, G). This difference in cell size became even more apparent during the next stages: 5–6 mm, 7–8 mm, and 9–10 mm fruits, corresponding to approximately 8, 10, and 12 DPA, respectively (Fig. 1C–E, H–J). Detailed analysis of the pericarp cells from 5–6 mm fruits (Table 1) showed that, in the transgenic fruits, the average cell area of the exocarp cells, located at the outer 4–6 cell layers of the pericarp, and the average cell area of the mesocarp cells were significantly increased compared with that of wild-type fruits (P <0.05, Student's t test). The analysis of the pericarp cells from these fruits also showed that the number of dividing cells was significantly decreased in both exocarp and mesocarp of the transgenic fruits, compared with the number of dividing cells in the pericarp of wild-type fruits (P <0.05, Student's t test). This aspect was also manifested by the cell division planes, visible in the mesocarp of 3–4 mm and 5–6 mm wild-type fruits, but absent in the mesocarp of transgenic fruits (Fig. 1B, C, G, J). However, the fact that no difference in cell number was observed in the later stages of fruit development (De Jong et al., 2009b), suggests that the rate of cell division in the RNAi SlARF7 fruits might be reduced but also that the period in which cell division takes place is prolonged, resulting in a similar number of cells as in wild-type fruits at the later stages of fruit development.
Fig. 1.
Microscopic analysis of the pericarp during early fruit development of wild-type and RNAi SlARF7 fruits. (A) Micrograph of an unpollinated wild-type ovary at anthesis. (B) Micrograph of pollinated wild-type tomato fruit, 3–4 mm in diameter. The cells in the exocarp are dividing, while the cells in the mesocarp and endocarp mainly expand. (C) Micrograph of the exocarp and mesocarp of a wild-type tomato fruit, 5–6 mm in diameter. The cells in the endocarp look similar to the mesocarp cells (not shown). At this stage, cell divisions also occur in the mesocarp. (D) Micrograph of the exocarp and mesocarp of a wild-type tomato fruit, 7–8 mm in diameter. The fruit mainly grows through cell expansion. (E) Micrograph of the exocarp and mesocarp of a wild-type tomato fruit, 9–10 mm in diameter. (F) Micrograph of an ovary from the RNAi SlARF7-4 line at anthesis. At this stage, there are no clear differences between wild-type and transgenic ovaries. (G) Micrograph of a parthenocarpic fruit from the RNAi SlARF7-4 line, 3–4 mm in diameter. The cells in the mesocarp and endocarp are already bigger compared with those in wild-type fruit. (H) Micrograph of the exocarp and mesocarp of a fruit from the RNAi SlARF7-4 line, 5–6 mm in diameter. At this and the following two stages, the pericarp mainly grows through cell expansion of the mesocarp cells. (I) Micrograph of the exocarp and mesocarp of a fruit from the RNAi SlARF7-4 line, 7–8 mm in diameter. (J) Micrograph of the exocarp and mesocarp of a fruit from the RNAi SlARF7-4 line, 9–10 mm in diameter. Bars 50 μm; cw, carpel wall; o, ovules; pl, placenta; ex, exocarp; m, mesocarp; en, endocarp.
Table 1.
Quantification of cell area and number of dividing cells in the pericarp of wild-type and transgenic fruits
| Line | Exocarp |
Mesocarp |
||
| Mean area (μm2) | % Dividing cells | Mean area (μm2) | % Dividing cells | |
| Wild type | 105±6.6 (n=1051) | 74±2.1 | 391±18.8 (n=2519) | 41±4.2 |
| RNAi SlARF7-4 | 131±4.3 (n=842) | 61±4.1 | 997±135.4 (n=1110) | 7±1.2 |
| RNAi SlARF7-6 | 141±19.5 (n=857) | 43±8.0 | 810±216.2 (n=1052) | 7±2.0 |
Wild-type and transgenic fruits, 5–6 mm in diameter, were used for the quantification of cell area (in μm2) and the percentage of dividing cells in the exocarp and mesocarp. The total number of cells that has been analysed from each line is indicated (n). The data represent the means ±standard error of five fruits. For all measurements, the differences between wild-type and transgenic lines were statistically significant (P <0.05, Student's t test).
The effect of SlARF7 silencing on transcript levels of cell division- and cell expansion-related genes
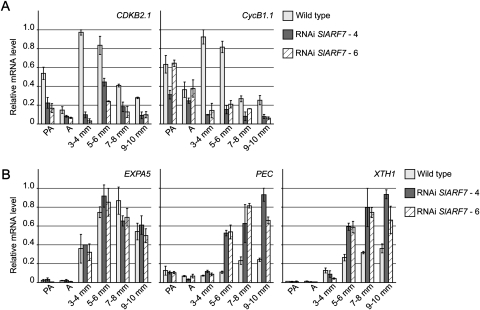
To support the histological data, the ovaries and fruits collected for microscopic analysis, were also used for total RNA extraction to analyse the transcript levels of several genes involved in cell division and expansion. Expression analyses by real-time quantitative PCR showed that, in the wild type, mRNA levels of the cell cycle genes SlCDKB2.1 and SlCyclinB1.1 increased after pollination, and subsequently decreased during the later stages of fruit development, as previously described by Joubès et al. (2000, 2001). In the transgenic lines, however, the transcript levels of SlCDKB2.1 were reduced (Fig. 2A). In the wild type, the transcript levels of this gene were highest at the 3–4 mm stage, while in the transgenic lines, transcript levels reached their maximum at the 5–6 mm stage, but this peak was still less than 50% of that found at a similar stage in the wild type. Also the transcript levels of SlCyclinB1.1 were strongly reduced in the transgenic lines, with the exception of the levels at the pre-anthesis and anthesis stage.
Fig. 2.
Transcript levels of cell cycle and cell division-related genes in developing wild-type and transgenic fruits. (A) Relative mRNA levels of cell cycle genes SlCDKB2.1 and SlCyclinB1.1 in ovaries from wild-type and RNAi SlARF7-4 and SlARF7-6 lines, collected 3 d before anthesis, the pre-anthesis stage at which wild-type flowers were emasculated (PA), at anthesis (A), and in pollinated wild-type or parthenocarpic transgenic fruits that were 3–4 mm, 5–6 mm, 7–8 mm, and 9–10 mm in diameter. (B) Relative mRNA levels of cell expansion genes encoding for an expansin precursor (SlEXPA5), pectate lyase (SlPEC), and an endo-xyloglucan transferase (SlXTH1). Standard errors are indicated (n=2).
In the wild type, transcript levels of cell expansion-related genes encoding for an expansin precursor (SlEXPA5), a pectate lyase (SlPEC), and an endo-xyloglucan transferase (SlXTH1) increased after pollination, confirming the results of Vriezen et al. (2008), who showed that pollination induces several genes involved in cell expansion. Moreover, that study showed that some of these genes, like the SlPEC and SlXTH1, are also induced by GA application. In the transgenic lines, the relative mRNA levels of SlEXPA5 were similar to those in the wild type. However, at the stages from 5–10 mm, the expression of SlPEC and SlXTH1 was much higher than in wild type (Fig. 2B).
Altogether, these expression data support the findings of the microscopic analysis, indicating that once fruit development is initiated in the RNAi SlARF7 lines, cell division activity was de-regulated, whereas cell expansion was increased as compared to wild type.
The effect of SlARF7 silencing on transcript levels of auxin-related genes
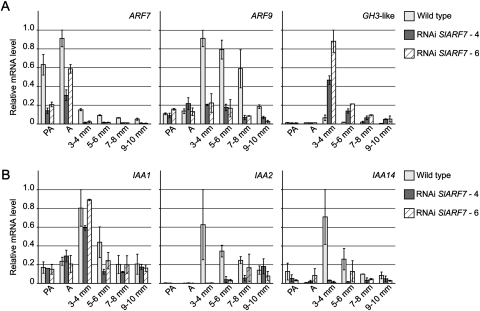
It was previously reported that auxin stimulates cell division (Bünger-Kibler and Bangerth, 1982; Serrani et al., 2007a). Therefore, the reduced cell division activity in the RNAi SlARF7 lines indicates that the auxin response is also diminished, although the morphological characteristics of the fruits, such as the heart-like shape and the formation of pseudoembryos that were also found in fruits with high levels of auxin (Pandolfini et al., 2002; Serrani et al., 2007a), suggest the opposite. To investigate this apparent contradiction further, transcript levels of auxin-related genes were analysed. SlARF9, the putative orthologue of Arabidopsis ARF9, was found by Vriezen et al. (2008) to be up-regulated after pollination or auxin application, with an exactly opposite expression pattern to that of SlARF7 (Fig. 3A), but was not induced by GA application (Serrani et al., 2008; Vriezen et al., 2008). In this study, the high SlARF9 transcript level at wild-type stage 3–4 mm confirmed that SlARF9 indeed is induced by pollination. In the subsequent stages, its expression slowly decreased (Fig. 3A). Interestingly, SlARF9 expression was not induced in the growing parthenocarpic fruits of the RNAi SlARF7 lines, suggesting that the auxin signalling pathway was not induced in these fruits. However, the expression of SlGH3-like was strongly up-regulated in the transgenic lines, compared with the expression in wild-type fruits (Fig. 3A; De Jong et al., 2009b). SlGH3-like is the tomato homologue of the Arabidopsis auxin-responsive gene GH3.6 encoding an IAA-amido synthethase (Staswick et al., 2005). In accordance, the expression of SlGH3-like was also induced in unpollinated ovaries treated with auxin (data not shown), possibly as negative feedback on high auxin levels. These findings indicate that a part of the auxin signalling pathway is enhanced in the RNAi SlARF7 lines. This bifurcation in the auxin signalling transduction was further supported by the analysis of the transcript levels of genes encoding the Aux/IAA repressors IAA1, IAA2, and IAA14. Previous studies have shown that SlIAA2 and SlIAA14 are induced by pollination, and that the expression of these genes, including SlIAA1, is induced by auxin-application to unpollinated ovaries (Serrani et al., 2008; Vriezen et al., 2008). Accordingly, the transcript levels of these Aux/IAA's were high in 3–4 mm wild-type fruits, and subsequently decreased at the later stages of fruit development. In the transgenic lines, the expression pattern of IAA1 was similar to that in the wild type, whereas the expression of IAA2 and IAA14 was strongly reduced (Fig. 3B).
Fig. 3.
Transcript levels of auxin-related genes in developing wild-type and transgenic fruits. (A) Relative mRNA levels of SlARF7, SlARF9, and SlGH3-like in ovaries and fruits collected from wild-type and RNAi SlARF7-4 and SlARF7-6 lines. (B) Relative mRNA levels of Aux/IAA genes SlIAA1, SlIAA2, and SlIAA14. Standard errors are indicated (n=2).
Quantification of GA content in wild-type and transgenic fruits
The fruits of the RNAi SlARF7 lines displayed several characteristics that were also found in GA-induced fruits, the most clear of which was an increase in cell expansion. To determine if these characteristics resulted from an increase in GA content, the concentrations of GA53, GA44, GA19, GA20, GA29, GA1, and GA8, GAs from the early 13-hydroxylation pathway, and GA9, GA4, and GA34, GAs from the non-13-hydroxylation pathway were quantified. To exclude the GAs that are present in the fertilized ovules of wild-type fruit, which are absent in the unfertilized ovules of the parthenocarpic fruits, only pericarp tissues of 9–10 mm fruits were collected.
The pericarp of wild-type fruits contained much lower concentrations of GA19, whereas the concentrations of GA20, GA29, and GA1 were much higher compared with concentrations in the pericarp of the RNAi SlARF7 fruits (Table 2). Also, the concentrations of GA4 and GA34 were higher in wild-type fruits compared with those in transgenic fruits. Concentrations of GA53 and GA44, the precursors of GA19, and concentrations of GA9, the precursor of GA4 could not be quantified in any of the samples.
Table 2.
Quantification of the gibberellin content of the pericarp from wild-type and transgenic fruits
| Line | GA19 | GA20 | GA29 | GA1 | GA8 | GA4 | GA34 |
| Wild type | <0.05 | 2.88±0.30 | 3.26±0.11 | 1.50±0.02 | 1.77±0.04 | 1.20±0.13 | 1.46±0.13 |
| RNAi SlARF7-4 | 0.50±0.05 | 0.13±0.02 | 0.31±0.03 | 0.43±0.15 | <0.05 | 0.12±0.01 | 0.32±0.04 |
| RNAi SlARF7-6 | 0.46±0.04 | 0.27±0.04 | 0.35±0.04 | 0.60±0.10 | <0.05 | <0.05 | 0.40±0.05 |
Gibberellin (GA) content (ng g−1 fresh weight) of the pericarp from pollinated wild-type fruits and parthenocarpic RNAi SlARF7 fruits, 9–10 mm in diameter. Data for GA9, GA44, and GA53 are not included because they could not be quantified in any of the samples. The data represent the means ±standard error of two or three biological replicates.
The effect of SlARF7 silencing on transcript levels of gibberellin-related genes
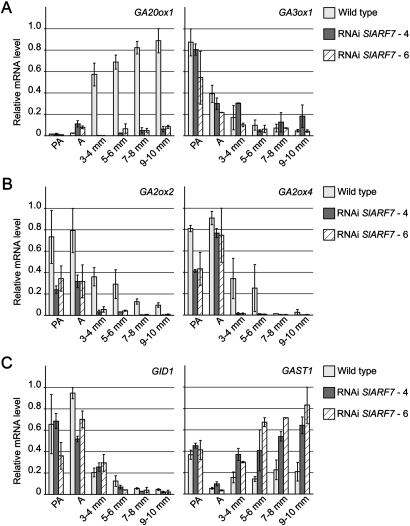
Although the content of bioactive GA is reduced in the RNAi SlARF7 lines, it is possible that their phenotype is caused by an increased GA response. The GA response is tightly regulated by the negative feedback of GA on its own biosynthesis (Hedden and Kamiya, 1997; Yamaguchi and Kamiya, 2000). Therefore, the expression of GA genes, involved in GA biosynthesis, and metabolism were analysed. The GA level is particularly limited by the last steps of its biosynthesis. Bioactive GAs are produced by GA 20-oxidases that convert GA12 and GA53 to GA9 and GA20, respectively, the precursors of the bioactive forms (Hedden and Phillips, 2000). Subsequently GA9 and GA20 are converted to GA1 and GA4 by 3β-hydroxylation. In wild-type plants, the transcript levels of SlGA20ox1, a GA 20-oxidase gene, strongly increased after pollination, but not in developing unpollinated fruits of the RNAi SlARF7 lines (Fig. 4A), corresponding to the reduced concentration of active GA in the latter. Notwithstanding, the expression pattern of the SlGA3ox1 gene, encoding a 3β-hydroxylase, was similar in both wild-type and transgenic lines.
Fig. 4.
Transcript levels of gibberellin-related genes in developing wild-type and transgenic fruits. (A) Relative mRNA levels of GA-biosynthesis genes SlGA20ox1 and SlGA3ox1 in ovaries and fruits collected from wild-type and RNAi SlARF7-4 and SlARF7-6 lines. (B) Relative mRNA levels of GA-inactivating genes SlGA2ox2 and SlGA2ox4. (C) Relative mRNA levels of GA-signalling genes SlGID1 and SlGAST1. Standard errors are indicated (n=2).
The content of active GA is not only controlled at the level of biosynthesis, but also by inactivation. This step is executed by GA 2-oxidases, that convert the active GA4 and GA1 to the inactive GA34 and GA8, respectively, and which are known to be induced by high levels of bioactive GAs (Sponsel and Hedden, 2004). Figure 4B shows that during fruit development the mRNA levels of SlGA2ox2 and -4, encoding GA 2-oxidases, were lower in the transgenic lines than in young wild-type fruits, suggesting a reduced feedback regulation by GAs, in this case corresponding to the reduced GA concentration in transgenic fruits.
In addition, the expressions of two GA-response genes that are not related to GA biosynthesis were analysed; SlGID1, a putative GA receptor in tomato (Ueguchi-Tanaka et al., 2007) and SlGAST1 (Shi et al., 1992), both induced in wild-type plants by GAs (Serrani et al., 2008). The pattern of the GID1 mRNA levels, decreasing after anthesis and during fruit development, was comparable between wild-type and transgenic plants. However, the activity of the GAST1 gene was significantly higher in the SlARF7 silenced plants (Fig. 4C).
In all, these findings suggest that the silencing of SlARF7 not only induced part of the auxin response pathway, but also part of that of GA.
Discussion
Successful completion of the fruit developmental programme depends on the action of phytohormones such as auxin and GA. These hormones are considered to play an important role in the initiation of fruit development, and in the co-ordination of cell division and expansion during the early stages of this process (Gillaspy et al., 1993). However, fruit set is not only regulated by positive growth factors, but also by negative regulators (Vivian-Smith et al., 2001). One of these regulators is Solanum lycopersicum AUXIN RESPONSE FACTOR 7 (SlARF7), as our previous work demonstrated that transgenic tomato plants with decreased SlARF7 mRNA levels produced seedless (parthenocarpic) fruits (De Jong et al., 2009b). The morphological characteristics of these fruits, such as the heart-like shape, the presence of pseudoembryos, the thick pericarp, and the empty locules, seemed to be the result of both increased auxin and GA responses during fruit growth. Here, a more detailed analysis of these SlARF7 RNA interference (RNAi) lines is described. This analysis provides more insight into the role of SlARF7 as a modulator of tomato fruit development, and its putative function in the regulation of the auxin and GA signalling response pathways in the fruit.
Fruit morphology
One of the characteristics of the parthenocarpic transgenic fruits was the thick pericarp compared with that of wild-type fruits, caused by an increase in cell expansion (De Jong et al., 2009b), particularly in the mesocarp and endocarp. In general, the cells in the exocarp are smaller than the mesocarp and endocarp cells, because this is the tissue in which new cell layers arise due to periclinal cell divisions. In the wild type, these divisions are completed within 5 DPA, while random orientated cell divisions occur in the pericarp up to 20 DPA (Cheniclet et al., 2005). However, analysis of young fruits demonstrated that, in the transgenic lines, the number of cell divisions was strongly reduced. This reduction in cell division activity was confirmed by the decreased transcript levels of the cell cycle-related genes CDKB2.1 and CyclinB1.1. Interestingly, the parthenocarpic fruit development of the tomato mutant pat, seemed to be initiated by the precocious onset of cell divisions in the pericarp (Mapelli et al., 1978; Mazzucato et al., 1998). Similarly, in the pat3/pat4 mutant, the expression of cell cycle-related genes was not reduced at the stage of anthesis (Pascual et al., 2009). Also in the parthenocarpic transgenic lines in which the negative auxin response regulator SlIAA9 was down-regulated using an antisense approach, cell cycle-related genes were already activated at anthesis, independent of pollination and fertilization (Wang et al., 2009). However, transgenic lines in which SlDELLA, a repressor of GA signalling, was silenced using the antisense approach, formed parthenocarpic fruits in which the number of cell divisions was decreased and cell size was increased (Martí et al., 2007), as in the RNAi SlARF7 fruits. Based on these results, Martí et al. (2007) generated a model for the role of SlDELLA in early fruit development, suggesting that, in the parthenocarpic transgenic lines, the phase of auxin-regulated cell division was bypassed and fruit growth mainly depended on cell expansion. Furthermore, the locular tissue in fruits of both RNAi SlARF7 and antisense SlDELLA lines barely developed, which might be due to a reduced cell division activity as well. These findings are similar to those of Bünger-Kibler and Bangerth (1982) and Serrani et al. (2007a), who showed that the pericarp of GA-induced tomato fruits contained fewer cells but with a larger volume than the cells of seeded fruits, and that no jelly was formed resulting in empty locules. Vriezen et al. (2008) compared the gene expression profiles of pollinated ovaries and GA-treated ovaries. The cell cycle-associated genes were more strongly induced by pollination than by GA treatment, while most genes involved in cell expansion were induced by both pollination and GA treatment. However, some of these genes, such as SlEXPA5, appeared to be more highly expressed after pollination than after GA application, whereas other genes, such as SlPEC and SlXTH1, had a higher level of expression after GA application (Vriezen et al., 2008). Interestingly, in the RNAi SlARF7 lines, the expression levels of SlPEC and SlXTH1 were higher during early fruit development than in the wild type, while transcript levels of SlEXPA5 were similar to those in the wild type, also indicating the presence of a strong GA signal.
Hence, both morphological and molecular analyses displayed similarities between GA-induced fruits, fruits of the antisense SlDELLA lines and fruits formed by the RNAi SlARF7 lines, supporting the hypothesis that in the RNAi SlARF7 fruits, the GA response is increased (De Jong et al., 2009b), and that normally SlARF7 moderates the GA response during the early stages of fruit development.
Hormone biosynthesis and signalling
In addition to the GA-related phenotype, the fruits formed by the RNAi SlARF7 lines also displayed features that might be related to high levels of auxin, such as the heart-like shape, which was also found in other transgenic fruits with high auxin levels (Pandolfini et al., 2002), and the formation of seed-like structures that resemble the pseudoembryos found in auxin-induced fruit (Asahira et al., 1967; Serrani et al., 2007a). Moreover, the auxin-related phenotype of the RNAi SlARF7 fruits was supported by the increased expression of the auxin-inducible GH3-like (De Jong et al., 2009b). Nevertheless, the expression of several other auxin signalling-related genes, such as SlARF9, SlIAA2, and SlIAA14, known to be induced by pollination or auxin application (Serrani et al., 2008; Vriezen et al., 2008), were not induced in fruits of the RNAi SlARF7 lines. This implies that the auxin content within the parthenocarpic fruit did not increase during early development as it normally does after pollination and fertilization (Mapelli et al., 1978; Sjut and Bangerth, 1982), although the increased expression of the GH3-like gene would suggest otherwise. Interestingly, the transcript levels of the auxin-inducible SlIAA1 did increase during the early stages of parthenocarpic fruit development. These levels were similar to those found in wild-type fruits, just like the transcript levels of SlIAA9 (data not shown). These results suggest that SlARF7 may regulate only part of the auxin signalling pathway involved in tomato fruit set and development, and that the expression of other auxin-related genes, such as SlARF9, SlIAA2, and SlIAA14, requires pollination and fertilization signals to be activated, as previously suggested by Pascual et al. (2009).
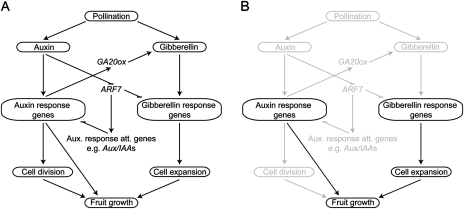
Successful completion of pollination and fertilization not only induces an increase in auxin content, but also causes the GA content to increase rapidly during the first 10 DPA (Mapelli et al., 1978; Koshioka et al., 1994; Serrani et al., 2008). The phenotypic similarities between GA-induced fruits and RNAi SlARF7 fruits suggest that the GA content in the transgenic fruit could be higher compared with that in wild-type fruit. However, the GA content of the transgenic fruits was actually lower than in the wild type, except for GA19, the concentrations of which appeared to be higher in both transgenic lines. These results suggest that GA19 could not be converted to GA20 due to the reduced expression of SlGA20ox1. The reduction of SlGA20ox1 expression in the RNAi SlARF7 fruits could be the result of feedback regulation, when the GA-related phenotype of the transgenic fruits might not be due to increased GA biosynthesis, but due to increased GA signalling. Also in parthenocarpic antisense SlDELLA fruits, SlGA20ox mRNA levels were strongly reduced (Martí et al., 2007). However, if the reduction of SlGA20ox1 expression in the RNAi SlARF7 lines was the result of feedback regulation, one would also expect feedback-related expression of SlGA3ox1, SlGA2ox2, SlGA2ox4, and SlGID1, unless the feedback mechanism of the GA 20-oxidase genes is differently regulated. Alternatively, the SlGA20ox1 gene may be part of the auxin signalling pathway that requires pollination and fertilization to be activated, corresponding to the data of Serrani et al. (2008) who showed that the expression of SlGA20ox1 depends on auxin. The GA-related phenotype together with the increased expression of the GA-response gene SlGAST1 in both transgenic lines shows that, with the silencing of SlARF7, part of the GA signalling is enhanced. These findings indicate that SlARF7 not only acts as a modulator of the auxin response, but also as a modulator of the GA response. Normally, SlARF7 transcript levels are reduced after pollination and fertilization (Vriezen et al., 2008; De Jong et al., 2009b). Reduction of SlARF7 transcript levels by an RNAi approach may release the repression of the auxin and GA signalling pathways that are imposed by SlARF7 independently of pollination and fertilization, resulting in the partial activation of these pathways and thus in parthenocarpic fruit growth. Hence, the fertilization-dependent step of the auxin signalling transduction pathway may be bypassed, which might be necessary to initiate cell division activity and stimulate GA biosynthesis (Fig. 5).
Fig. 5.
Model for SlARF7 function in tomato fruit set and development. (A) After pollination, the levels of both auxin and GA increase, resulting in the activation of auxin and GA response genes, which, in turn, will trigger fruit growth by regulating cell division and cell expansion (adapted from De Jong et al., 2009a). However, these pathways do not act independently of each other. Our data suggest that after pollination the auxin response genes stimulate GA biosynthesis through the transcriptional activation of GA 20-oxidases (GA20ox). Furthermore, SlARF7 acts as a negative regulator of the auxin and GA signalling pathways. After fertilization, transcript levels of SlARF7 decrease and both pathways are released from repression. (B) In the RNAi SlARF7 lines, SlARF7 transcript levels are reduced independently of fertilization. This reduction causes the partial activation of the auxin and GA signalling pathways (black), resulting in parthenocarpic fruit growth. Due to the silencing of SlARF7, the pollination- and fertilization-dependent activation of the auxin and GA pathways is bypassed (grey).
The work presented here contributes to the understanding of the cross-talk between the signalling pathways of auxin and GA in early fruit development. Recent studies (Vriezen et al., 2008; Pascual et al., 2009; Wang et al., 2009; Xiao et al., 2009) have shown that the hormones ethylene and abscisic acid also play an important role in the regulation of tomato fruit set and development, demonstrating the complexity of the network that regulates these processes. The analysis of differences in gene expression between seeded fruits and parthenocarpic fruits from mutant or transgenic lines already has provided new insights into this regulatory network, but further work will be necessary to understand the precise relationship between these different plant hormones.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table 1. Primer sequences used for real-time quantitative PCR analysis of cell cycle and cell division-related genes, genes involved in the auxin response, genes from GA metabolism, and GA response of tomato.
Supplementary Material
Acknowledgments
We are grateful to Mrs T Sabater (Universidad Politécnica de Valencia-CSIC, Spain) for her help with the GA analysis. We also like to thank Dr Elisabeth Pierson (Radboud University, the Netherlands) for her help on the analysis of the micrographs, Dr A Czerednik (Radboud University, the Netherlands) for the primers of the cell cycle related genes, and Dr I Rieu (Radboud University, the Netherlands) for critical reading of the manuscript.
References
- Asahira T, Takeda Y, Nishio T, Hirabayashi M, Tsukamoto Y. Studies on fruit development in tomato. I. Ovule development and content of diffusible auxin- and gibberellin-induced parthenocarpic tomato fruits in relation to their development. Memoirs of the Research Institute for Food Science, Kyoto University. 1967;28:47–74. [Google Scholar]
- Bünger-Kibler S, Bangerth F. Relationship between cell number, cell size and fruit size of seeded fruits of tomato (Lycopersicon esculentum Mill.), and those induced parthenocarpically by the application of plant growth regulators. Plant Growth Regulation. 1982;1:143–154. [Google Scholar]
- Cheniclet C, Rong WY, Causse M, Frangne N, Bolling L, Carde J, Renaudin J. Cell expansion and endoreduplication show a large genetic variability in pericarp and contribute strongly to tomato fruit growth. Plant Physiology. 2005;139:1984–1994. doi: 10.1104/pp.105.068767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong M, Mariani C, Vriezen WH. The role of auxin and gibberellin in tomato fruit set. Journal of Experimental Botany. 2009a;60:1523–1532. doi: 10.1093/jxb/erp094. [DOI] [PubMed] [Google Scholar]
- De Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH. The Solanum lycopersicum Auxin Response Factor 7 (SlARF7) regulates auxin signalling during tomato fruit set and development. The Plant Journal. 2009b;57:160–170. doi: 10.1111/j.1365-313X.2008.03671.x. [DOI] [PubMed] [Google Scholar]
- Dorcey E, Urbez C, Blázquez MA, Carbonell J, Perez-Amador MA. Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis. The Plant Journal. 2009;58:318–332. doi: 10.1111/j.1365-313X.2008.03781.x. [DOI] [PubMed] [Google Scholar]
- Fos M, Nuez F, García-Martínez JL. The gene pat-2, which induces natural parthenocarpy, alters the gibberellin content in unpollinated tomato ovaries. Plant Physiology. 2000;122:471–480. doi: 10.1104/pp.122.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W. Fruits: a developmental perspective. The Plant Cell. 1993;5:1439–1451. doi: 10.1105/tpc.5.10.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson FG. Parthenocarpy induced by pollen extracts. American Journal of Botany. 1937;24:102–107. [Google Scholar]
- Gustafson FG. Influence of gibberellic acid on setting and development of fruits in tomato. Plant Physiology. 1960;35:521–523. doi: 10.1104/pp.35.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Kamiya Y. Gibberellin biosynthesis: enzymes, genes and their regulation. Annual Review of Plant Physiology and Plant Molecular Biology. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL. Gibberellin metabolism: new insights revealed by the genes. Trends in Plant Science. 2000;5:523–530. doi: 10.1016/s1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- Joubès J, Lemaire-Chamley M, Delmas F, Walter J, Hernould M, Mouras A, Raymond P, Chevalier C. A new C-type cyclin-dependent kinase from tomato expressed in dividing tissues does not interact with mitotic and G1 cyclins. Plant Physiology. 2001;126:1403–1415. doi: 10.1104/pp.126.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J, Walsh D, Raymond P, Chevalier C. Molecular characterization of the expression of distinct classes of cyclins during the early development of tomato fruit. Planta. 2000;211:430–439. doi: 10.1007/s004250000306. [DOI] [PubMed] [Google Scholar]
- Koshioka M, Nishijima T, Yamazaki H, Liu Y, Nonaka M, Mander LN. Analysis of gibberellins in growing fruits of Lycopersicon esculentum after pollination or treatment with 4-chlorophenoxyacetic acid. Journal of Horticultural Science. 1994;69:171–179. [Google Scholar]
- Mapelli S, Frova C, Torti G, Soressi GP. Relationship between set, development and activities of growth regulators in tomato fruits. Plant and Cell Physiology. 1978;19:1281–1288. [Google Scholar]
- Martí C, Orzáez D, Ellul P, Moreno V, Carbonell J, Granell A. Silencing of DELLA induces facultative parthenocarpy in tomato fruits. The Plant Journal. 2007;52:865–876. doi: 10.1111/j.1365-313X.2007.03282.x. [DOI] [PubMed] [Google Scholar]
- Mazzucato A, Taddei AR, Soressi GP. The parthenocarpic fruit (pat) mutant of tomato (Lycopersicon esculentum Mill.) sets seedless fruits and has aberrant anther and ovule development. Development. 1998;125:107–114. doi: 10.1242/dev.125.1.107. [DOI] [PubMed] [Google Scholar]
- Ozga JA, Reinecke DM. Hormonal interactions in fruit development. Journal of Plant Growth Regulation. 2003;22:73–81. [Google Scholar]
- Pandolfini T, Rotino GL, Camerini S, Defez R, Spena A. Optimisation of transgene action at the post-transcriptional level: high quality parthenocarpic fruits in industrial tomatoes. BMC Biotechnology. 2002;2:1. doi: 10.1186/1472-6750-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual L, Blanca JM, Cañizares J, Nuez F. Transcriptomic analysis of tomato carpel development reveals alterations in ethylene and gibberellin synthesis during pat3/pat4 parthenocarpic fruit set. BMC Plant Biology. 2009;9:67. doi: 10.1186/1471-2229-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS. NIo Health. Bethesda, Maryland: USA; 1997-2009. ImageJ. [Google Scholar]
- Rebers M, Kaneta T, Kawaide H, Yamaguchi S, Yang Y, Imai R, Sekimoto H, Kamiya Y. Regulation of gibberellin biosynthesis genes during flower and early fruit development of tomato. The Plant Journal. 1999;17:241–250. doi: 10.1046/j.1365-313x.1999.00366.x. [DOI] [PubMed] [Google Scholar]
- Serrani JC, Fos M, Atarés A, García-martínez JL. Effect of gibberellin and auxin on parthenocarpic fruit growth induction in the cv. micro-tom of tomato. Journal of Plant Growth Regulation. 2007a;26:211–221. [Google Scholar]
- Serrani JC, Ruiz-Rivero O, Fos M, García-Martínez JL. Auxin-induced fruit-set in tomato is mediated in part by gibberellins. The Plant Journal. 2008;56:922–934. doi: 10.1111/j.1365-313X.2008.03654.x. [DOI] [PubMed] [Google Scholar]
- Serrani JC, Sanjuán R, Ruiz-Rivero O, Fos M, García-Martínez JL. Gibberellin regulation of fruit set and growth in tomato. Plant Physiology. 2007b;145:246–257. doi: 10.1104/pp.107.098335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Gast RT, Gopalraj M, Olszewski NE. Characterization of a shoot-specific, GA3-and ABA regulated gene from tomato. The Plant Journal. 1992;2:153–159. [PubMed] [Google Scholar]
- Sjut V, Bangerth F. Induced parthenocarpy: a way of changing the levels of endogenous hormones in tomato fruits (Lycopersicon esculentum Mill.).1. Extractable hormones. Plant Growth Regulation. 1982;1:243–251. [Google Scholar]
- Sponsel V, Hedden P. Gibberellin biosynthesis and inactivation. In: Davies PJ, editor. Plant hormones: biosynthesis, signal transcuction, action! Dordrecht: Kluwer Academic Publishers; 2004. pp. 63–94. [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. The Plant Cell. 2005;17:616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M. Gibberellin receptor and its role in gibberellin signalling in plants. Annual Review of Plant Biology. 2007;58:183–198. doi: 10.1146/annurev.arplant.58.032806.103830. [DOI] [PubMed] [Google Scholar]
- Vivian-Smith A, Luo M, Chaudhury A, Koltunow AM. Fruit development is actively restricted in the absence of fertilization in Arabidopsis. Development. 2001;128:2321–2331. doi: 10.1242/dev.128.12.2321. [DOI] [PubMed] [Google Scholar]
- Vriezen WH, Feron R, Maretto F, Keijman J, Mariani C. Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytologist. 2008;177:60–76. doi: 10.1111/j.1469-8137.2007.02254.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, Latché A, Pech J, Fernie AR, Bouzayen M. Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. The Plant Cell. 2009;21:1428–1452. doi: 10.1105/tpc.108.060830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer SH, Bukovac MJ, Sell HM, Weller LE. Some effects of gibberellin on flowering and fruit setting. Plant Physiology. 1957;32:39–41. doi: 10.1104/pp.32.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Radovich C, Welty N, Hsu J, Li D, Meulia T, van der Knaap E. Integration of tomato reproductive developmental landmarks and expression profiles, and the effect of SUN on fruit shape. BMC Plant Biology. 2009;9:49. doi: 10.1186/1471-2229-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y. Gibberellin biosynthesis: its regulation by endogenous and environmental signals. Plant and Cell Physiology. 2000;41:251–257. doi: 10.1093/pcp/41.3.251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.