Abstract
The modulation of primary nitrogen metabolism by water deficit through ABA-dependent and ABA-independent pathways was investigated in the model legume Medicago truncatula. Growth and glutamate metabolism were followed in young seedlings growing for short periods in darkness and submitted to a moderate water deficit (simulated by polyethylene glycol; PEG) or treated with ABA. Water deficit induced an ABA accumulation, a reduction of axis length in an ABA-dependent manner, and an inhibition of water uptake/retention in an ABA-independent manner. The PEG-induced accumulation of free amino acids (AA), principally asparagine and proline, was mimicked by exogenous ABA treatment. This suggests that AA accumulation under water deficit may be an ABA-induced osmolyte accumulation contributing to osmotic adjustment. Alternatively, this accumulation could be just a consequence of a decreased nitrogen demand caused by reduced extension, which was triggered by water deficit and exogenous ABA treatment. Several enzyme activities involved in glutamate metabolism and genes encoding cytosolic glutamine synthetase (GS1b; EC 6.3.1.2.), glutamate dehydrogenase (GDH3; EC 1.4.1.1.), and asparagine synthetase (AS; EC 6.3.1.1.) were up-regulated by water deficit but not by ABA, except for a gene encoding Δ1-pyrroline-5-carboxylate synthetase (P5CS; EC not assigned). Thus, ABA-dependent and ABA-independent regulatory systems would seem to exist, differentially controlling development, water content, and nitrogen metabolism under water deficit.
Keywords: Abscisic acid, glutamate, Medicago truncatula, nitrogen metabolism, seedlings, water deficit
Introduction
Young seedlings and plants have developed complex cellular signalling mechanisms to sense and to respond to unfavourable environmental situations (Yamaguchi-Shinozaki and Shinozaki, 2006; Nakashima et al., 2009). Such responses can be achieved by distinct physiological and metabolic adjustments, mediated by different hormones. The phytohormone abscisic acid (ABA) has been reported to play a central role in controlling growth and development throughout the plant life cycle, and specifically in the adaptation to environmental constraints (Finkelstein et al., 2002; Hirayama and Shinozaki, 2007). ABA is synthesized from carotenoids (C40) in higher plants by an indirect pathway, in which the oxidative cleavage of cis-epoxycarotenoids catalysed by 9-cis-epoxycarotenoid dioxygenase (NCED) is the main regulatory step in the biosynthesis of ABA (Nambara and Marion-Poll, 2005). In vegetative plant organs, ABA is rapidly synthesized and accumulates under different environmental stresses (e.g. salt, cold, and drought), and responses mediated by this hormone lead to the induction of complex tolerance mechanisms to osmotic stress (Shinozaki and Yamaguchi-Shinozaki, 2007). Many of the cellular components involved in ABA reception and downstream signal transduction have been identified (for reviews see Finkelstein et al., 2002; Himmelbach et al., 2003). Several ABA receptors have been reported (Shen et al., 2006; Liu et al., 2007), which were later partly questioned (McCourt and Creelman, 2008; Pennisi, 2009). Recently, two independent research groups have demonstrated that regulators of type 2C protein phosphatases (PP2Cs) function as ABA receptors (Ma et al., 2009; Park et al., 2009).
Generally, studies on the molecular responses to drought stress have revealed numerous changes in the expression of genes (Bray, 2004; Yamaguchi-Shinozaki and Shinozaki, 2006). Use of ABA-deficient and ABA-insensitive mutants has indicated the existence of both ABA-dependent and ABA-independent regulatory systems (cis- and trans-acting factors) governing the expression of drought-induced genes (Shinozaki and Yamaguchi-Shinozaki, 2000; Takahashi et al., 2004; Yamaguchi-Shinozaki and Shinozaki, 2006). Since it has been recognized that post-transcriptional events control the pattern of expression of genes (Gallie and Bailey-Serres, 1997), ABA may act not only as a signal molecule at the transcriptional, but also at the post-transcriptional level (Liu and Hill, 1995). Recent integrated transcriptomic and metabolomic analyses have revealed some new ABA-regulated molecular mechanisms of metabolic networks occurring under drought stress in Arabidopsis thaliana (Urano et al., 2009).
Stress conditions induce modifications in many metabolic pathways, such as glycolysis, photosynthesis, and carbohydrate and nitrogen metabolism (Martinez et al., 2005; Debouba et al., 2007). The expression of several genes that are involved in drought-induced metabolite accumulation, such as sugars (trehalose, sorbitol, and mannitol), amino acids (proline and asparagine), and amines (glycine betaine and polyamines), have been shown to increase as part of the tolerance to abiotic stresses (Seki et al., 2007). Among these, free amino acid accumulation appears to be of major importance in the protection of plants (Good and Zaplachinski, 1994; Rhodes et al., 1999), but how amino acid accumulation is regulated during water deficit or after exogenous ABA application is still poorly understood. Glutamate plays a central role in amino acid metabolism (Forde and Lea, 2007) and is positioned at a ‘cross-road’ between carbon and nitrogen metabolism in higher plants. Glutamate is (i) the product of ammonia assimilation through the reaction catalysed by glutamine synthetase (GS, EC 6.3.1.2.), which exists in cytosolic and chloroplastic isoforms (GS1 and GS2, respectively) co-operating with glutamate synthase [GOGAT, EC 1.4.1.13. (NADH-GOGAT) or EC 1.4.7.1. (Fd-GOGAT)], (ii) the product and substrate for aminating and deaminating glutamate dehydrogenase (GDH, EC 1.4.1.2.), (iii) the preferred precursor for proline synthesis through Δ1-pyrroline-5-carboxylate synthetase (P5CS; EC not assigned) activity, and (iv) both the amino donor and acceptor of many different reversible aminotransferase reactions, such as alanine aminotransferase and aspartate aminotransferase. The combined actions of these different glutamate pathways (GS/GOGAT and GDH) within plants may be required and/or be involved in the homeostatic control of glutamate concentrations (Glevarec et al., 2004; Masclaux-Daubresse et al., 2006; Limami et al., 2008).
The present work aimed to improve the understanding of the metabolic changes in seedlings during the initial stages of water deficit. Using Medicago truncatula as a model for other grain legume crops, the work was focused on ABA-dependent and ABA-independent responses of nitrogen metabolism related to glutamate. As previous experiments have shown that seedling growth and amino acid metabolism were more active in continuous dark than under diurnal conditions (E Planchet et al., unpublished data), the experiments described in this paper were performed only in darkness for up to 72 h. Water deficit has been induced by polyethylene glycol (PEG), a non-ionic molecule and a non-permeating osmolyte which has been reported to maintain uniform water potential in a way similar to soil drying (Larher et al., 1993; Lu and Neumann, 1998; Verslues et al., 1998) and to lead to an increase in ABA content in the shoots of alfalfa seedlings (Luo et al., 1992). To our knowledge, this is the first differential investigation into glutamate metabolism in M. truncatula seedlings either under water deficit, or following exogenous ABA application.
Materials and methods
Germination and seedling growth conditions
Seeds of M. truncatula cv. Paraggio were germinated in darkness at 21 °C in Petri dishes (diameter 9 cm) on Whatman paper soaked with 4 ml sterile deionized water for 46 h. Afterwards, seedlings were transferred for 72 h in Petri dishes on to polyethylene glycol 8000 solution (PEG; Sigma-Aldrich, St Quentin Fallavier, France) at –0.25 MPa (in order to mimic a mild water deficit), or on to a 10 μM solution of abscisic acid (ABA; Sigma-Aldrich) and were maintained in darkness (at 21 °C). As a control, well-watered seedlings were used. Into each Petri dish, 30 seedlings were grown. Seedlings were harvested at different times (0, 24, 48, and 72 h) following the onset of the three treatments. Cotyledons were discarded and axes (hypocotyl plus radicle) were immediately frozen in liquid nitrogen before being stored at –80 °C. The fresh weight was also determined for each time point and the dry weight was measured after 48 h of desiccation in an oven at 80 °C. Specific water content was calculated as the difference between fresh weight and dry weight as a function of axis length.
ABA quantitative assay
Freeze-dried samples were homogenized in powder and weighted (100 mg). Forty ng of a standard ABA-d4 (Euriso-Top SA, France) were then added as internal tracers for recovery and analytical purposes. Organs were extracted with 2 ml of acetone/water/acetic acid (80/19/1, by vol.) vigorously shaken for 1 min, sonicated for 1 min at 25 Hz, and then centrifuged (8230 g, 4 °C, 15 min). The supernatants were collected, and the pellets were re-extracted with 2 ml of the same extraction solution, then vigorously shaken (1 min) and sonicated (1 min; 25 Hz). Following centrifugation, the two supernatants were pooled and dried. The dry extract was dissolved in 200 μl acetonitrile/water (50/50; v/v), filtered, and submitted to analysis by HPLC-electrospray-MSMS (HPLC-ESI-MSMS). The compounds were introduced in the ESI source using a Waters 2695 separation module (Alliance; Waters, Milford, MA, USA) equipped with a Waters 2487 dual UV detector. Separation was achieved on a reverse-phase column (Uptisphere C18, 5 μm, 150×2 mm i.d, Interchrom) using a flow-rate of 0.15 ml min−1 and a binary gradient: (A) acetonitrile 0.5% acetic acid (v/v) and (B) acetic acid 0.5% (v/v). The solvent gradient was programmed as following: 0–5 min 20% A, 5–15 min 65% A, 15–20 min 100% A, before returning to the initial composition at 30 min. The analyses were performed on a Waters Quattro LC triple quadrupole mass spectrometer (Waters) operating in a Multiple Reaction Monitoring (MRM) scanning mode. Relevant instrumental parameters were set as follows: capillary 2.75 kV (negative mode), extractor 2 V, source block and desolvation gas temperatures 120 °C and 350 °C, respectively. Nitrogen was used to assist the nebulization and the desolvation (250 l h−1 and 450 l h−1, respectively), argon was used as collision gas at 3.5×10−3 mbar. The parameters used for MRM quantification of ABA-d4 and ABA in positive mode were: cone potential 16 V, collision energy 10 eV for two transitions used m/z 267>251 and 265>247, respectively. For a five μl injection volume of sample prepared with 40 ng internal standard and reconstituted in 200 μl of 50/50 acetonitrile/H2O (v/v), the limit of detection (LOD) and limit of quantification (LOQ) were calculated from calibration curve and sample using the Quantify module of MassLynx (version 4.1 software). The respective LOD and LOQ are 1 and 3 pg mg−1 dry mass. Measurements were repeated three times with independent biological samples.
Quantitative real-time RT-PCR
For total RNA isolation, frozen axes were crushed in liquid nitrogen with a mortar and pestle. The homogenous powder was treated with TRI Reagent® (Ambion, Austin, TX, USA) according to the manufacturer's protocol. cDNAs were obtained by retrotranscription of 2 μg of total RNA using 200 units of RT-MMLV (Promega, Madison, WI, USA), 2 μg of random primer (Invitrogen, Breda, The Netherlands) in the presence of 40 U of a recombinant RNasin® ribonuclease inhibitor (Promega). The reaction mixture was incubated for 1 h at 37 °C in a total volume of 50 μl.
PCR was performed on a light cycler ABI Prism 7000 SDS (Applied Biosystems, Foster city, CA, USA). Each reaction was performed with 3 μl of a 1/2 (v/v) dilution of the first cDNA strands using a SYBR Green PCR Master Mix (Applied Biosystems), following the manufacturer's instructions, with 200 nM of each primer in a total reaction of 25 μl. The primer sequences for the amplification of each gene are given in Table 1. The sequences of genes were obtained from the Medicago Gene Index (http://compbio.dfci.harvard.edu/tgi; see also Limami et al., 2008). The PCR-reaction was incubated for 2 min at 50 °C and 10 min at 95 °C followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. The specificity of the PCR amplification procedure was checked with a heat-dissociation protocol (from 65 °C to 95 °C) after the final cycle of PCR. Each incubation was carried out with three biological replicates using a duplicated PCR reaction for determining Ct values. mRNA amounts were calculated in copy numbers, and the results are expressed as a ratio [treatment versus control (H2O)].
Table 1.
Primer sequences for real-time quantitative PCR amplification
| Gene | Forward primer sequence (5′–3′) | Reverse primer sequence (5′–3′) |
| GS2 | 5′-GTTATTGGTTATGAGATGAATGCACAT | 5′-TGCAACTCTGTCCATACCATATGC |
| GS1b | 5′-ACCACCATTCTCTGGAAACCAT | 5′-ACAATGCATGTGTGTGTTTTATAGCA |
| NADH-GOGAT | 5′-CAGGTGACTCTCGGCGTG | 5′-TCTTTGGTGAGGTAGCTGTCAACT |
| GDH3 | 5′-CATAAGAGTAATATGAGCAACCATTCTTG | 5′-GAAATAGACGTATCCACATTAAAGAAATGA |
| AS | 5′-CAACATGGTGACTGCTTTTTGG | 5′-TCCCCAGAAGCAGGATCAAC |
| P5CS | 5′-GAGAGGGAACGGCCAAGTG | 5′-TTGCAGATCCTTGTGTGTATAAATCA |
| NCED5 | 5′-GGACGGTTTTATGCATGATCCT | 5′-TGGCAAAGTCATGCATCATGAT |
Amino acid extraction and analysis by HPLC
Total amino acids were extracted from axes in 96% (v/v) ethanol for 1 h at 4 °C. After centrifugation (9800 g, 4 °C, 15 min), the ethanol fraction was removed and the same process was then repeated with deionized water. The ethanol and water fractions were combined. After evaporation of the extract under vacuum, organic residues were dissolved in deionized water and extracted with the same volume of chloroform. After centrifugation (12 000 g, 4 °C, 15 min), an aqueous phase containing amino acids was vacuum dried. The amino acids were then redissolved in deionized water. Samples were passed through a nylon syringe filter (0.45 μm) and then analysed by HPLC (Waters Corporation, Massachusetts, USA). The amino acids were determined by the Waters AccQTag method which uses Waters AccQFluor Reagent (patent pending) to derivatize the amino acids. The reagent is a highly reactive compound, 6-aminoquinolyl-N-hydroxy-succinimidyl carbamate (AQC), which forms stable derivatives with primary and secondary amino acids including proline. Derivatives were separated by reverse-phase (C18 column) HPLC and quantified by fluorescence detection. Each sample was analysed over 1 h. The amounts of all the amino acids determined by HPLC were summed up for the determination of the total amount of amino acids. Measurements were repeated twice with independent biological samples.
Enzyme in vitro assays
Glutamine synthetase (GS) and glutamate dehydrogenase (GDH) were extracted from frozen seedling plant material in 2 ml of phosphate buffer (Na2HPO4/KH2PO4; 10 mM, pH 7.5) supplemented with 1% (w/v) polyvinylpolypyrrolidone. The homogenate was centrifuged (15 000 g, 4 °C, 15 min) and the supernatant was used as the crude extract. GS activity was measured using 120 μl of the crude extract in a reaction mixture containing TRIS-HCl buffer (50 mM, pH 7.6) supplemented with MgSO4 (20 mM), glutamate (80 mM), hydroxylamine (6 mM), EDTA (4 mM), and ATP (8 mM). The reaction was stopped after 30 min at 37 °C by the addition of FeCl3 reagent and centrifuged. γ-glutamyl hydroxamate (GHM) formed from hydroxylamine and glutamate was determined colorimetrically at 540 nm after complexation with FeCl3.
Aminating- and deaminating-GDH activities were assayed by following the oxidation of NADH and the reduction of NAD+ at 340 nm, respectively. Aminating-GDH activity was measured using 20 μl of the crude extract in a reaction mixture containing TRIS-HCl buffer (100 mM, pH 8) supplemented with CaCl2 (1 mM), 2-oxoglutarate (13 mM), (NH4)2SO4 (50 mM) and NADH (0.25 mM). For deaminating-GDH activity, the reaction mixture contained TRIS-HCl buffer (100 mM, pH 9) supplemented with CaCl2 (1 mM), glutamate (33 mM), and NAD+ (0.25 mM).
Results
Induction of NCED gene expression and ABA accumulation by water deficit
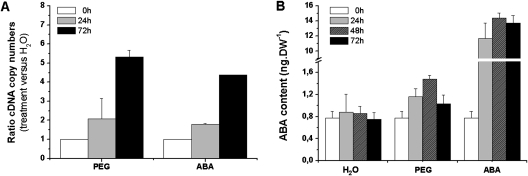
The expression of a NCED (9-cis-epoxycarotenoid dioxygenase) gene of A. thaliana has been shown to control the endogenous content of ABA under water deficit conditions (Iuchi et al., 2001). In order to show that, in the present system, the ABA biosynthesis pathway in M. truncatula seedlings was activated by water deficit (PEG), quantitative RT-PCR analysis of the content of NCED5 mRNA and determination of ABA content were carried out. Transcription of the NCED5 gene was induced by PEG (Fig. 1A). Treatment of seedlings with ABA (10 μM) also induced the expression of NCED5 at least 4-fold in the axis after 72 h (Fig. 1A). The endogenous ABA content was increased transiently under PEG and after ABA application, reaching a maximum after 48 h (Fig. 1B). This ABA concentration was almost 2-fold higher under water deficit than under normal conditions, but to a lesser extent than after ABA application.
Fig. 1.
Relative expression of the NCED5 gene and ABA content in Medicago truncatula seedlings under water deficit induced by PEG or after ABA treatment. After 46 h of germination (T=0 h), seedlings were incubated with PEG (–0.25 MPa) or with ABA (10 μM) for 72 h. As a control, seedlings were well-watered. (A) NCED5 transcript abundance of treated dark-grown seedlings was plotted as the relative expression (ratio) of the non-stressed control seedlings at the indicated time points. (B) Determination of ABA content in axis during 72 h according the treatments. Means ±SE, n=3.
Morphological adaptations in response to water deficit or ABA
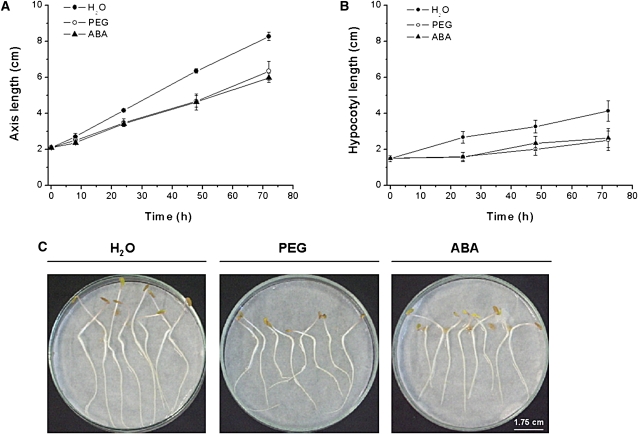
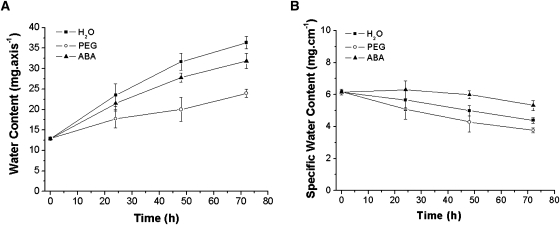
In response to water deficit (PEG) or to exogenous ABA, the axis length was reduced in comparison to the water treated controls (Fig. 2A, C). This decrease was mainly due to a reduction in the hypocotyl length (Fig. 2B). Whilst both PEG and ABA caused almost a 35% inhibition of axis extension, the water content was only weakly affected by exogenous ABA, but strongly reduced by PEG (Fig. 3A). The specific water content of the axis (expressed as mg cm−1) was lower under PEG treatment than the control, whereas ABA induced a significantly higher specific water content compared with the control (Fig. 3B).
Fig. 2.
Effects of ABA or water deficit on growth of Medicago truncatula seedlings. All treatments were the same as in Fig. 1. The axis (A) and hypocotyl lengths (B) were measured at the indicated time points. Each datum point represents the mean of three independent experiments ±SE. (C) Pictures of seedlings were taken after 72 h according the treatments.
Fig. 3.
Interaction of water deficit and ABA on water relations on Medicago truncatula seedlings. All treatments were the same as in Fig. 1. Water content (expressed as mg axis−1) and specific water content (expressed as mg cm−1) of treated and untreated axes were measured at the indicated time points. Each value represents the mean ±SE of three independent experiments.
Modifications of amino acid metabolism by PEG or ABA
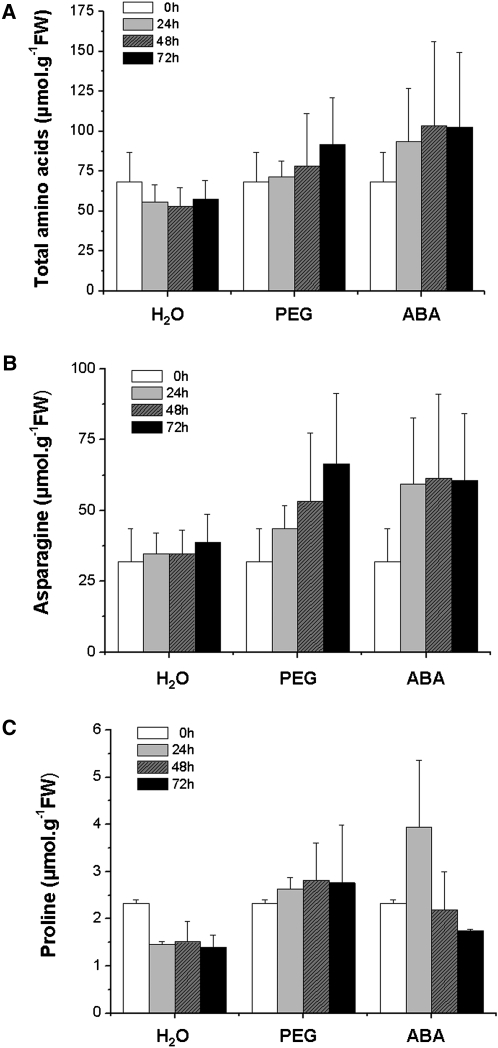
In order to analyse the ABA-dependent regulation of metabolic changes in response to water deficit, the free amino acid (AA) were quantified during the different treatments. As shown in Fig. 4A, the total AA content of well-watered seedlings was relatively constant during the 72 h period (57 μmol g−1 FW), with a slight trend towards a decrease. PEG and ABA induced an increase in the total free AA content of axes (55% and 69% at 72 h compared with the water treatment, respectively).
Fig. 4.
Total free amino acid, asparagine and proline contents of Medicago truncatula seedlings subjected to exogenous ABA or water deficit. All treatments were the same as in Fig. 1. Measurements were made at the indicated time points and expressed as μmol g−1FW. Each datum point is the mean of two independent experiments.
The analysis of individual amino acids showed that asparagine was the major amino acid present in axes, accounting for more than 60% of the total free AA content (Table 2; Fig. 4B). The asparagine content increased steadily during the incubation period with PEG over 72 h, whilst with ABA, the asparagine content increased only for the first 24 h, after which it remained constant (Fig. 4B). The proline content of axes increased until 48 h and remained stable after 72 h following incubation with PEG, whereas the application of ABA induced a transient increase in proline, with a maximum at 24 h (Fig. 4C). Although the proline content of axes did vary following the application of PEG and ABA, it represented only 2.9% of the total free amino acids after 72 h of PEG treatment. Several other amino acids showed a rapid accumulation in response to PEG (water deficit) and ABA, increasing by an average of 1.5–3-fold over that of the control (Table 2). The continuous reduction of serine content under normal conditions over 72 h was slowed down under both PEG application and exogenous ABA. Similar results were observed for alanine concentration, except with ABA treatment where the reduced content was accentuated. Whereas the cysteine content remained more or less constant under normal conditions over 72 h, PEG and ABA induced, respectively, an increase and a decrease of cysteine content. The concentration of free ammonium decreased slightly during the onset of PEG stress (in comparison to the control) and increased with ABA.
Table 2.
Major individual amino acid composition of axes subjected to treatment with PEG or ABA
| H2O |
PEG |
ABA |
||||||||
| 0 h | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | |
| Alanine | 4.23±0.87 | 2.13±0.19 | 1.34±0.19 | 1.04±0.08 | 2.98±0.14 | 1.99±0.56 | 1.45±0.29 | 3.62±1.07 | 1.11±0.23 | 0.87±0.16 |
| Aspartate | 1.05±0.15 | 0.44±0.07 | 0.22±0.05 | 0.19±0.03 | 0.64±0.04 | 0.46±0.10 | 0.50±0.05 | 0.47±0.07 | 0.35±0.11 | 0.39±0.19 |
| Cysteine | 2.30±0.59 | 2.18±0.42 | 2.36±0.46 | 2.91±0.12 | 2.75±0.29 | 3.39±1.21 | 4.44±0.33 | 4.03±1.68 | 1.26±1.26 | 1.99±1.49 |
| GABA | 0.23±0.10 | 0.35±0.08 | 0.28±0.15 | 0.07±0.02 | 0.42±0.05 | 0.42±0.14 | 0.12±0.03 | 0.24±0.21 | 0.24±0.14 | 0.16±0.05 |
| Glutamine | 6.00±0.33 | 0.81±0.12 | 0.33±0.05 | 0.22±0.09 | 1.23±0.62 | 0.69±0.69 | 0.05±0.05 | 1.97±0.50 | 0.23±0.23 | 0.64±0.54 |
| Glycine | 2.09±0.53 | 1.26±0.19 | 0.82±0.37 | 0.78±0.21 | 1.15±0.47 | 0.47±0.04 | 0.45±0.04 | 1.08±0.32 | 1.09±0.08 | 0.45±0.09 |
| Histidine | 2.03±0.62 | 1.70±0.34 | 1.60±0.23 | 1.69±0.32 | 2.09±0.26 | 1.65±1.09 | 1.42±0.12 | 2.57±0.70 | 2.27±0.06 | 1.68±0.20 |
| Isoleucine | 1.94±0.42 | 1.43±0.27 | 1.37±0.27 | 0.97±0.60 | 1.87±0.16 | 1.94±0.70 | 2.20±0.31 | 2.55±0.96 | 1.93±0.64 | 1.83±0.28 |
| Leucine | 2.22±0.40 | 1.04±0.16 | 0.76±0.16 | 0.57±0.20 | 1.54±0.03 | 1.26±0.47 | 1.13±0.29 | 1.28±0.00 | 1.08±0.35 | 0.61±0.04 |
| Methionine | 0.24±0.05 | 0.13±0.01 | 0.12±0.00 | 0.14±0.02 | 0.18±0.02 | 0.18±0.01 | 0.28±0.06 | 0.18±0.01 | 0.13±0.00 | 0.08±0.03 |
| Phenylalanine | 2.18±0.59 | 1.74±0.38 | 2.25±0.31 | 2.84±0.02 | 2.18±0.36 | 2.65±1.00 | 3.07±1.27 | 3.29±1.36 | 1.98±0.23 | 1.86±1.15 |
| Serine | 5.82±1.10 | 4.07±1.18 | 3.30±1.29 | 3.50±0.86 | 5.21±1.06 | 4.86±2.33 | 4.90±0.56 | 5.78±2.03 | 4.56±1.37 | 4.14±0.46 |
| Threonine | 2.24±0.55 | 1.88±0.28 | 1.75±0.20 | 1.73±0.17 | 2.34±0.14 | 1.53±1.53 | 2.21±0.58 | 2.10±0.09 | 1.86±0.09 | 0.95±0.49 |
| Tyrosine | 0.61±0.07 | 0.36±0.00 | 0.29±0.06 | 0.26±0.09 | 0.47±0.06 | 0.38±0.02 | 0.33±0.04 | 0.72±0.20 | 0.42±0.20 | 0.24±0.09 |
| Ammonium | 1.16±0.31 | 1.11±0.04 | 0.93±0.08 | 1.24±0.07 | 0.88±0.17 | 0.59±0.29 | 0.75±0.32 | 1.52±0.19 | 1.23±0.14 | 1.57±0.24 |
The amino acid contents are expressed as μmol g−1 FW. The asparagine and proline contents are shown in Fig. 4. Each datum point is the mean of two independent experiments ±SE.
Changes in the activities of nitrogen metabolism enzymes related to glutamate
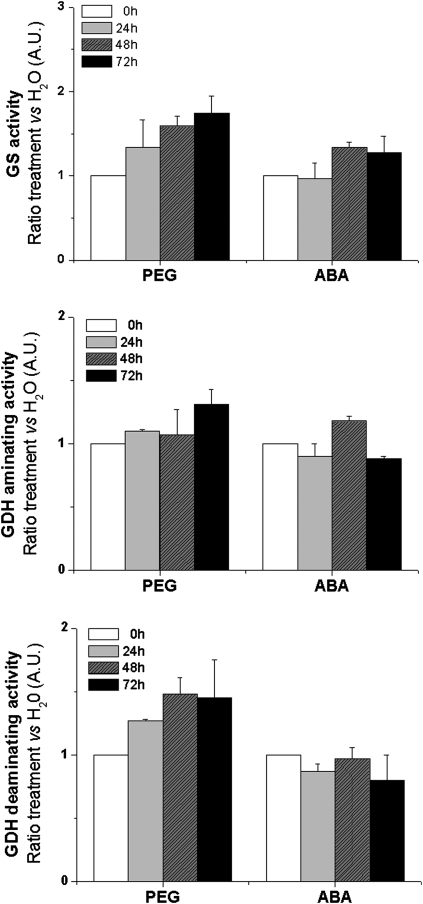
Changes in metabolite pools often reflect modified activities of involved enzymes. In order to check the post-transcriptional effects of treatments with PEG or ABA, enzymes of primary nitrogen metabolism such as GS (cytosolic GS1 and chloroplastic GS2) and GDH (aminating and deaminating) were measured in vitro in crude organ extracts of M. truncatula seedlings (Fig. 5). GS, GDH aminating, and GDH deaminating activities all increased to varying extents, in response to PEG treatment (water deficit). By contrast, only GS increased to a small extent following incubation with exogenous ABA, whereas the two GDH activities remained relatively constant.
Fig. 5.
Changes in the activities of nitrogen metabolism enzymes related to glutamate according stress treatments. Glutamine synthetase (GS) and glutamate dehydrogenase (GDH) aminating/deaminating activities were expressed as ratio (treatment versus H2O). Seedlings were exposed 3 d to ABA (10 μM) or PEG (–0.25 MPa). Each datum point is the mean of three independent experiments ±SE.
Expression of nitrogen metabolism genes related to glutamate
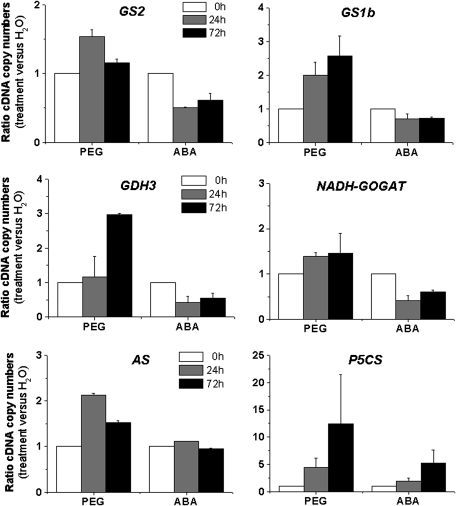
The intricate interplay between ABA and water deficit in the transcriptional regulation of genes encoding enzymes of primary nitrogen metabolism remains unclear. The pattern of expression of the following genes, GS1b, NADH-GOGAT, asparagine synthetase (AS), and Δ1-pyrroline-5-carboxylate synthetase (P5CS), was monitored by quantitative RT-PCR (Fig. 6) in seedlings submitted to water deficit (PEG) or to exogenous ABA. The majority of the monitored transcripts were up-regulated by PEG treatment, including cytosolic GS1b, which encodes the predominant GS isoenzyme in the root cortex (Carvalho et al., 2000), GDH3, NADH-GOGAT, AS, and P5CS (12-fold higher than the control after 72 h). In contrast to the PEG treatment, exogenous ABA slightly decreased the expression of GS1b, GS2, GDH3, and NADH-GOGAT. Conversely, the expression of P5CS increased by exogenous ABA was 6-fold higher after 72 h. The expression of AS was not affected by exogenous ABA.
Fig. 6.
Changes in expression of genes related to glutamate metabolism. Total RNA was isolated from axes of Medicago truncatula subjected to H2O (control), PEG (–0.25 MPa) or ABA (10 μM). Each RNA sample was used for quantitative RT-PCR analyses. The cDNA copy numbers for glutamine synthetase (GS1b and GS2), glutamate dehydrogenase (GDH3), NADH-GOGAT, asparagine synthetase (AS), and Δ1-pyrroline-5-carboxylate synthetase (P5CS) gene were expressed as ratio (treatment versus H2O). Each datum point is the mean of three independent experiments ±SE.
Discussion
The aim of this work was to study the changes in primary nitrogen metabolism in young seedlings during the initial stages of water deficit in the model legume M. truncatula, with a focus on a differentiation between ABA-dependent and ABA-independent pathways. The induction by PEG of the expression of NCED5, the gene that controls the first step of the ABA biosynthetic pathway, and of ABA accumulation supports the hypothesis that ABA synthesis was activated in M. truncatula seedlings. The expression of NCED5 was also induced by exogenous ABA, suggesting a positive feedback regulation by ABA. These results are supported by other studies in root tissues of A. thaliana dedicated to the regulation of AtNCED3, the major NCED gene in Arabidopsis involved in the regulation of ABA contents under drought (Iuchi et al., 2001) or salt stress (Barrero et al., 2006).
ABA-dependent and ABA-independent rearrangement of development and amino acid metabolism
Water deficit (PEG) reduced the extension and the specific water content of axes. As exogenous ABA caused similar changes of extension, a conclusion could be that the water deficit-induced reduction of extension was mediated by endogenous ABA. It is very likely that this reduction of development was the consequence of ABA-induced inhibition of cell expansion as already observed in the radicles of post-germinating M. truncatula seeds (Gimeno-Gilles et al., 2009). Contrary to PEG treatment, ABA increased the specific water content in the axis, indicating that the water deficit-induced reduction of the specific water content was not mediated by endogenous ABA.
Since exogenous ABA triggered an increase in the content of total free amino acids, a suggestion would be that the similar response observed under PEG treatment was mediated via ABA. Although the increase in AA contents has been reported in other species in response to various stresses such as salt, drought, mineral deficiencies, heavy metals or pathogen attack (for reviews see Azevedo et al., 2006; Lea et al., 2007), the role of this accumulation in stress tolerance is still poorly understood. A role in the control of osmotic homeostasis has been proposed for specific amino acids such as asparagine or proline under osmotic stresses related either to drought or salt (Lea et al., 2007; Verbruggen and Hermans, 2008). In the present work, the apparent non-relationship between the water deficit-induced AA accumulation and the specific water content would not be in favour of an osmotic role. However, this consideration does not take into account the possibility that the increase in asparagine (the major amino acid accumulated) may be restricted to physiologically active subcellular compartments, such as the cytosol, plastids, and mitochondria, which would result in much greater concentration changes. Alternatively, the stress-induced amino acid accumulation could be a mechanism to provide the cells with the precursors of several compounds known to be involved in biotic and abiotic stress responses (Sanchez et al., 2008), such as polyamines or metabolites of secondary metabolism (phenylpropanoids, flavonoids, alkaloids, and phenolic compounds). The storage of amino acids could also be considered as useful in anticipation of the return to non-stressful conditions. This idea has been put forward for the expression of genes under stress whose transcription products function during the recovery period (Drew, 1997). Finally, amino acid accumulation could also be just a consequence of (i) the decrease in nitrogen demand caused by reduced growth triggered by stress, (ii) the reduced rate of protein synthesis which often occurs under stressful conditions, and at the same time (iii) the increased rate of proteolysis which could be occurring in the cotyledons.
A central role of glutamate metabolism in response to water deficit or ABA
Evidence has emerged that glutamate is an important signalling molecule in higher plants (for a review see Forde and Lea, 2007). Glutamate is synthesized by the ammonium assimilating pathway catalysed by the GS/GOGAT cycle. Glutamate is also regenerated as a by-product of asparagine synthesis by asparagine synthetase (AS). Degradation of glutamate is catalysed by the GDH-deaminating activity. This oxidative glutamate deamination sustains carbohydrate metabolism by contributing to the regeneration of the respiratory substrates (α-ketoglutarate), ammonium, and NADH as reducing power (Aubert et al., 2001). The replenishment of the glutamate pool is also required to produce protective metabolites, since the glutamate pathway is the predominant route for proline biosynthesis in plants during conditions of osmotic stress (Armengaud et al., 2004; Szekely et al., 2008).
Recently, an integrated metabolome and transcriptome analysis of A. thaliana suggested that dehydration stress induces ABA accumulation, which regulates global metabolic networks and changes in amino acid contents (Urano et al., 2009). Interestingly, the authors found that dehydration-induced galactinol and raffinose accumulation was regulated by ABA-independent pathways. While such a global metabolic analysis confers a general overview of the metabolic networks, it might overlook some important details, which become obvious only in a narrow-scale analyses focusing on a limited number of genes, enzymes, and metabolites.
In the present work, it was observed that PEG-induced proline and asparagine accumulation was correlated with induced expression of the P5CS and AS genes. As P5CS expression was induced by exogenous ABA and AS expression was not affected by this hormone, changes in proline content under water deficit could be controlled at the transcriptional level via an ABA-dependent pathway, while changes in asparagine content under water deficit could also be controlled at the transcriptional level, but through an ABA-independent pathway.
The PEG-induced activities of GS and deaminating-GDH in extracts correlated with the water-deficit-induced expression of the genes encoding these enzymes (GS2, GS1b, and GDH3). Exogenous ABA did not stimulate either these enzyme activities or the expression of the genes, suggesting that these enzymes were induced by water deficit through an ABA-independent pathway. The involvement of GDH in drought tolerance has been shown previously with the expression of bacterial gdhA genes in transgenic plants of maize (Lightfoot et al., 2007) or Nicotiana tabacum (Mungur et al., 2006).
The concentrations of the metabolites (glutamine and ammonium) were not correlated with the variations in measured enzyme activities. The pools of these metabolites probably undergo a high turnover, as they are located at the main crossroads between the pathways of ammonium assimilation and amino acid transamination and deamination. These results are consistent with work on A. thaliana, which showed the existence of regulatory processes of metabolite content independent of transcript abundance (Gibon et al., 2006) during cold acclimation (Kaplan et al., 2007).
Although ABA was not required for the regulation of the glutamate metabolism genes, supplementary control mechanisms interacting with elements involved in ABA regulation should exist. Exploring the putative central role of glutamate in water-deficit tolerance, through ABA signalling, could lead to more information on changes of nitrogen metabolism under adverse environmental conditions.
Acknowledgments
The authors are grateful to Peter J Lea for critical reading of the article and English language correction, and to Werner M Kaiser for helpful discussions.
Glossary
Abbreviations
- AA
amino acid
- ABA
abscisic acid
- AS
asparagine synthetase
- GDH
glutamate dehydrogenase
- GOGAT
glutamate synthase
- GS
glutamine synthetase
- NCED
9-cis-epoxycarotenoid dioxygenase
- P5CS
Δ1-pyrroline-5-carboxylate synthetase
- PEG
polyethylene glycol
References
- Armengaud P, Thiery L, Buhot N, Grenier-de March G, Savouré A. Transcriptional regulation of proline biosynthesis in Medicago truncatula reveals developmental and environmental specific features. Physiologia Plantarum. 2004;120:442–450. doi: 10.1111/j.0031-9317.2004.00251.x. [DOI] [PubMed] [Google Scholar]
- Aubert S, Bligny R, Douce R, Gout E, Ratcliffe RG, Roberts JK. Contribution of glutamate dehydrogenase to mitochondrial glutamate metabolism studied by 13C and 31P nuclear magnetic resonance. Journal of Experimental Botany. 2001;52:37–45. [PubMed] [Google Scholar]
- Azevedo RA, Lancien M, Lea PJ. The aspartic acid metabolic pathway, an exciting and essential pathway in plants. Amino Acids. 2006;30:143–162. doi: 10.1007/s00726-005-0245-2. [DOI] [PubMed] [Google Scholar]
- Barrero JM, Rodríguez PL, Quesada V, Piqueras P, Ponce MR, Micol JL. Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant, Cell and Environment. 2006;29:2000–2008. doi: 10.1111/j.1365-3040.2006.01576.x. [DOI] [PubMed] [Google Scholar]
- Bray EA. Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. Journal of Experimental Botany. 2004;55:2331–2341. doi: 10.1093/jxb/erh270. [DOI] [PubMed] [Google Scholar]
- Carvalho H, Lima L, Lescure N, Camut N, Salema R, Cullimore J. Differential expression of the two cytosolic glutamine synthetase genes in various organs of. Medicago truncatula. Plant Science. 2000;159:301–312. doi: 10.1016/s0168-9452(00)00360-5. [DOI] [PubMed] [Google Scholar]
- Debouba M, Maâroufi-Dghimi H, Suzuki A, Ghorbel MH, Gouia H. Changes in growth and activity of enzymes involved in nitrate reduction and ammonium assimilation in tomato seedlings in response to NaCl stress. Annals of Botany. 2007;99:1143–1151. doi: 10.1093/aob/mcm050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MC. Oxygen deficiency and root metabolism: Injury and acclimation under hypoxia and anoxia. Annual Review of Plant Physiology and Plant Molecular Biology. 1997;48:223–250. doi: 10.1146/annurev.arplant.48.1.223. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. The Plant Cell. 2002;14:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG, Lea PJ. Glutamate in plants: metabolism, regulation and signalling. Journal of Experimental Botany. 2007;58:2339–2358. doi: 10.1093/jxb/erm121. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Bailey-Serres J. Eyes off transcription! The wonderful world of post-transcriptional regulation. The Plant Cell. 1997;9:667–673. doi: 10.1105/tpc.9.5.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R, Stitt M. Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biology. 2006;7 doi: 10.1186/gb-2006-7-8-r76. R76 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno- Gilles C, Lelièvre E, Viau L, Malik-Ghulam M, Ricoult C, Niebel A, Leduc N, Limami AM. ABA-mediated inhibition of germination is related to the inhibition of genes encoding cell-wall biosynthetic and architecture: modifying enzymes and structural proteins in Medicago truncatula embryo axis. Molecular Plant. 2009;2:108–119. doi: 10.1093/mp/ssn092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glevarec G, Bouton S, Jaspard E, Riou MT, Cliquet JB, Suzuki A, Limami AM. Respective roles of the glutamine synthetase/glutamate synthase cycle and glutamate dehydrogenase in ammonium and amino acid metabolism during germination and post-germinative growth in the model legume Medicago truncatula. Planta. 2004;219:286–297. doi: 10.1007/s00425-004-1214-9. [DOI] [PubMed] [Google Scholar]
- Good AG, Zaplachinski ST. The effects of drought stress on free amino acid accumulation and protein synthesis in Brassica napus. Physiologia Plantarum. 1994;90:9–14. [Google Scholar]
- Himmelbach A, Yang Y, Grill E. Relay and control of abscisic acid signaling. Current Opinion in Plant Biology. 2003;6:470–479. doi: 10.1016/s1369-5266(03)00090-6. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends in Plant Science. 2007;12:343–351. doi: 10.1016/j.tplants.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. The Plant Journal. 2001;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Sung DY, Zhao W, Popp M, Porat R, Guy CL. Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. The Plant Journal. 2007;50:967–981. doi: 10.1111/j.1365-313X.2007.03100.x. [DOI] [PubMed] [Google Scholar]
- Larher F, Leport L, Petrivalsky M, Chappart M. Effectors for the osmoinduced proline response in higher plants. Plant Physiology and Biochemistry. 1993;31:911–922. [Google Scholar]
- Lea PJ, Sodek L, Parry MAJ, Shewry PR, Halford NG. Asparagine in plants. Annals of Applied Biology. 2007;150:1–26. [Google Scholar]
- Lightfoot DA, Mungur R, Ameziane R, et al. Improved drought tolerance of transgenic Zea mays plants that express the glutamate dehydrogenase gene (gdhA) of. E. coli. Euphytica. 2007;156:103–116. [Google Scholar]
- Limami AM, Glevarec G, Ricoult C, Cliquet JB, Planchet E. Concerted modulation of alanine and glutamate metabolism in young Medicago truncatula seedlings under hypoxic stress. Journal of Experimental Botany. 2008;59:2325–2335. doi: 10.1093/jxb/ern102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JH, Hill RD. Post-transcriptional regulation of bifunctional α-amylase/subtilisin inhibitor expression in barley embryos by abscisic acid. Plant Molecular Biology. 1995;29:1087–1091. doi: 10.1007/BF00014980. [DOI] [PubMed] [Google Scholar]
- Liu X, Yue Y, Li B, Nie Y, Li W, Wu WH, Ma L. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science. 2007;315:1712–1716. doi: 10.1126/science.1135882. [DOI] [PubMed] [Google Scholar]
- Lu Z, Neumann PM. Water-stressed maize, barley and rice seedlings show species diversity in mechanisms of leaf growth inhibition. Journal of Experimental Botany. 1998;49:1945–1952. [Google Scholar]
- Luo M, Liu JH, Mohapatra S, Hill RD, Mohapatra SS. Characterization of a gene family encoding abscisic acid- and environmental stress-inducible proteins of alfalfa. Journal of Biological Chemistry. 1992;267:15367–15374. [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Martinez JP, Kinet JM, Bajji M, Lutts S. NaCl alleviates polyethylene glycol-induced water stress in the halophyte species Atriplex halimus L. Journal of Experimental Botany. 2005;56:2421–2431. doi: 10.1093/jxb/eri235. [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Reisdorf-Cren M, Pageau K, Lelandais M, Grandjean O, Kronenberger J, Valadier MH, Feraud M, Jouglet T, Suzuki A. Glutamine synthetase–glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink source nitrogen cycle in tobacco. Plant Physiology. 2006;140:444–456. doi: 10.1104/pp.105.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCourt P, Creelman R. The ABA receptors: we report you decide. Current Opinion in Plant Biology. 2008;11:474–478. doi: 10.1016/j.pbi.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Mungur R, Wood AJ, Lightfoot DA. Water potential is maintained during water deficit in Nicotiana tabacum expressing the Escherichia coli glutamate dehydrogenase gene. Plant Growth Regulation. 2006;50:231–238. [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiology. 2009;149:88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annual Review in Plant Biology. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. Stressed out over a stress hormone. Science. 2009;324:1012–1013. doi: 10.1126/science.324_1012. [DOI] [PubMed] [Google Scholar]
- Rhodes D, Verslues PE, Sharp RE. Role of amino acids in abiotic stress resistance. In: Singh BK, editor. Plant amino acids: biochemistry and biotechnology. New York: Marcel Dekker; 1999. pp. 319–356. [Google Scholar]
- Sanchez DH, Lippold F, Redestig H, Hannah MA, Erban A, Krämer U, Kopka J, Michael K, Udvardi MK. Integrative functional genomics of salt acclimatization in the model legume Lotus japonicus. The Plant Journal. 2008;53:973–987. doi: 10.1111/j.1365-313X.2007.03381.x. [DOI] [PubMed] [Google Scholar]
- Seki M, Umezawa T, Urano K, Shinozaki K. Regulatory metabolic networks in drought stress responses. Current Opinion in Plant Biology. 2007;10:296–302. doi: 10.1016/j.pbi.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Shen YY, Wang XF, Wu FQ, et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443:823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Current Opinion in Plant Biology. 2000;3:217–223. [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Székely G, Abrahám E, Cséplo A, et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. The Plant Journal. 2008;53:11–28. doi: 10.1111/j.1365-313X.2007.03318.x. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Seki M, Ishida J, et al. Monitoring the expression profiles of genes induced by hyperosmotic, high salinity, and oxidative stress and abscisic acid treatment in Arabidopsis cell culture using a full-length cDNA microarray. Plant Molecular Biology. 2004;56:29–55. doi: 10.1007/s11103-004-2200-0. [DOI] [PubMed] [Google Scholar]
- Urano K, Maruyama K, Ogata Y, et al. Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. The Plant Journal. 2009;57:1065–1078. doi: 10.1111/j.1365-313X.2008.03748.x. [DOI] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C. Proline accumulation in plants: a review. Amino Acids. 2008;35:753–759. doi: 10.1007/s00726-008-0061-6. [DOI] [PubMed] [Google Scholar]
- Verslues PE, Ober ES, Sharp RE. Root growth and oxygen relations at low water potentials. Impact of oxygen availability in polyethylene glycol solutions. Plant Physiology. 1998;116:1403–1412. doi: 10.1104/pp.116.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]