Abstract
In Arabidopsisfloral meristems are specified on the periphery of the inflorescence meristem by the combined activities of the FLOWERING LOCUS T (FT)–FD complex and the flower meristem identity gene LEAFY. The floral specification activity of FT is dependent upon two related BELL1-like homeobox (BLH) genes PENNYWISE (PNY) and POUND-FOOLISH (PNF) which are required for floral evocation. PNY and PNF interact with a subset of KNOTTED1-LIKE homeobox proteins including SHOOT MERISTEMLESS (STM). Genetic analyses show that these BLH proteins function with STM to specify flowers and internodes during inflorescence development. In this study, experimental evidence demonstrates that the specification of flower and coflorescence meristems requires the combined activities of FT–FD and STM. FT and FD also regulate meristem maintenance during inflorescence development. In plants with reduced STM function, ectopic FT and FD promote the formation of axillary meristems during inflorescence development. Lastly, gene expression studies indicate that STM functions with FT–FD and AGAMOUS-LIKE 24 (AGL24)–SUPPRESSOR OF OVEREXPRESSION OF CONTANS1 (SOC1) complexes to up-regulate flower meristem identity genes during inflorescence development
Keywords: Coflorescence, development, floral transition, flower specification, homeobox, inflorescence, shoot apical meristem
Introduction
The shoot apical meristem (SAM) is the site at which organs, meristems, and structures are produced such as leaves, axillary meristems (AMs), and internodes (Sablowski, 2007; Barton, 2009; Bleckmann and Simon, 2009; Dodsworth, 2009). The continuous growth and development displayed by shoots is dependent upon the ability of the meristem to maintain an intricate balance between the perpetuation of stem cells in the central apical zone and the organogenic mechanisms that specify lateral organs and meristems on the periphery (Bennett and Leyser, 2006; Sablowski, 2007; Barton, 2009; Bleckmann and Simon, 2009; Dodsworth, 2009).
Specific members of the KNOTTED1-like HOMEOBOX (KNOX) family of transcription factors regulate SAM function during plant development (Hake et al., 2004; Scofield and Murray, 2006; Hay and Tsiantis, 2009). In addition, Class I KNOX proteins regulate leaf dissection in a subset of plants (Champagne and Sinha, 2004; Barkoulas et al., 2008). Null alleles of knotted1 (kn1) and shoot meristemless (stm) produce terminal shoots comprised of cotyledons and, in some cases, a leaf or two, in maize and Arabidopsis, respectively (Barton and Poethig, 1993; Vollbrecht et al., 2000). In Cardamine hirsuta, RNAi lines directed against the orthologue of STM also produce a terminal shoot phenotype (Hay and Tsiantis, 2006). Interestingly, an allele of stm called gorgon causes an increase in the size of the SAM indicating that STM regulates stem cell homeostasis (Takano et al., 2010). STM and kn1 also act to regulate reproductive patterning events as plants with decreased levels of these KNOX genes alter flower patterning, branching, as well as internode growth (Clark et al., 1996; Endrizzi et al., 1996; Kerstetter et al., 1997; Kanrar et al., 2006; Scofield et al., 2007; Yu et al., 2009; Takano et al., 2010).
KNOX proteins interact with members of the BELL1-like HOMEODOMAIN (BLH) proteins (Hake et al., 2004). In Arabidopsis, BLH proteins regulate developmental pathways that control plant architecture, organ specification, and phase change (Hamant and Pautot, 2010). For example, two paralogous BLH proteins, PENNYWISE (PNY: also known as BLH9, BELLRINGER, REPLUMLESS, and VAAMANA) and POUND-FOOLISH (PNF), are essential for floral evocation, internode patterning, and specification of AMs during inflorescence development (Byrne et al., 2003; Roeder et al., 2003; Smith and Hake, 2003; Bao et al., 2004; Bhatt et al., 2004; Smith et al., 2004; Cole et al., 2006; Rutjens et al., 2009). In addition, genetic studies indicate that BLH proteins ARABIDOPSIS THALIANA HOMEOBOX1 (ATH1), PNY, and PNF function with STM to maintain meristem maintenance patterning events during shoot development (Byrne et al., 2003; Bhatt et al., 2004; Kanrar et al., 2006; Rutjens et al., 2009). Therefore, Class I KNOX function is modulated through the interaction with specific BLH proteins, which co-ordinate meristem maintenance and shoot patterning events throughout development.
Shoot and organ architecture are modified and altered as plants transition through each phase of development (Poethig, 2003). The floral transition is a major developmental phase change in which flower inductive cues produced in the leaves converge at the SAM to mediate the transition from vegetative to inflorescence development (Kobayashi and Weigel, 2007; Turck et al., 2008; Zeevaart, 2008). FLOWERING LOCUS T (FT) functions as a mobile photoperiodic signal that moves from the leaves to the SAM to promote flowering (Kobayashi and Weigel, 2007; Turck et al., 2008; Zeevaart, 2008). In the SAM, FT associates with the b-ZIP transcription factor, FD, to promote floral evocation and flower meristem specification (Pnueli et al., 2001; Abe et al., 2005; Wigge et al., 2005). Moreover, recent studies in tomato not only demonstrate the mobile flowering function of FT, but also show that FT modifies leaf morphology and meristem activity in conjunction with auxin and TERMINAL FLOWER1 (TFL1), respectively (Shalit et al., 2009).
In Arabidopsis, flower specification involves the activation of flower meristem identity genes during AM development (Liu et al., 2009). Flower specification is controlled in part by LEAFY (LFY), which is induced by multiple flowering time pathways (Nilsson et al., 1998; Blazquez and Weigel, 2000; Schmid et al., 2003; Eriksson et al., 2006). Two MADS box transcription factors, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) and AGAMOUS-LIKE 24 (AGL24) function together to activate LFY directly in response to floral inductive cues (Lee et al., 2008; Liu et al., 2008). In turn, LFY positively regulates APETALA1 (AP1) directly and through a cascade of late flower meristem identity genes (Bowman et al., 1993; Schultz and Haughn, 1993; Parcy et al., 1998; Liljegren et al., 1999; Wagner et al., 1999; William et al., 2004; Saddic et al., 2006). Once activated, AP1 maintains LFY expression, creating a positive feed-back loop, which functions to maintain flower meristem identity (Bowman et al., 1993; Schultz and Haughn, 1993; Liljegren et al., 1999). The FT–FD complex also functions to specify flower meristem identity by directly activating AP1 (Ruiz-García et al., 1997; Abe et al., 2005; Wigge et al., 2005). In addition, the FT–FD complex indirectly regulates LFY by positively regulating SOC1 (Abe et al., 2005; Moon et al., 2005; Wigge et al., 2005; Yoo et al., 2005; Searle et al., 2006).
Recent studies showed that the expression of LFY and AP1 requires PNY and PNF (Smith et al., 2004; Kanrar et al., 2008). Moreover, the flower specification function of FT is dependent upon PNY and PNF (Kanrar et al., 2008). Lastly, the interplay between PNY/PNF and the floral specification integrators LFY and FT is not only crucial for floral determination but this network also regulates the formation of coflorescence meristems. Given the interplay between PNY/PNF and FT together with the genetic and biochemical studies showing that STM–PNY and STM–PNF act to specify floral meristems, the relationship between STM and FT–FD was examined during inflorescence development. In this study, genetic analyses showed that the specification of coflorescence and floral meristems requires both STM and FT/FD function during inflorescence development. Surprisingly, a role for FT in meristem maintenance and carpel development was identified. Based on gene expression studies, it is proposed that STM functions with FT–FD and AGL24–SOC1 for the activation of flower meristem identity genes.
Materials and methods
Genetic analyses
The Arabidopsis plants used in this study were grown under long-day growth conditions: 16/8 h light/dark cycle at 22 °C. Genetic studies were performed to analyse the inflorescence phenotypes resulting from combining stm-10 with ft-2 and fd-3 mutants, in the Columbia ecotype (Koornneef et al., 1991; Abe et al., 2005; Wigge et al., 2005; Kanrar et al., 2006). The ft-2 and fd-3 mutants are likely null alleles, while stm-10 is a weak allele in which a stop codon is located in the first helix of the homeodomain (Koornneef et al., 1991; Kardailsky et al., 1999; Abe et al., 2005; Wigge et al., 2005; Kanrar et al., 2006).
In order to determine the genetic relationship between FT and STM, ft-2 was backcrossed into the Columbia ecotype three times (Kanrar et al., 2008). FT and STM are located on chromosome one, separated by approximately 12.6 Mb. Pollen from stm-10 was crossed to ft-2 and F2 seed was collected from F1 plants derived from this cross. Because FT and STM are linked, the F3 seed from all F2 ft-2 plants was collected individually. Subsequently, the F3 ft-2 plants were screened for the stm-10-like phenotypes. Seed derived from ft-2 STM/stm-10 parental plants were used to characterize the inflorescence phenotypes of ft-2 stm-10 plants, which segregated 25% of the time.
The genetic relationship between STM and FD was determined by transferring pollen from stm-10 to the carpels of fd-3 mutants. F1 plants were self-pollinated and the resulting F2 seed was planted out. 298 F2 plants were scored: ∼1/16 fd-3 stm-10 plants, ∼3/16 fd-3 plants, ∼3/16 stm-10 plants, and ∼9/16 wild-type plants. Genotype determination via PCR was used to verify fd-3 homozygous plants. Seed collected from F2 fd-3 plants were screened for the fd-3 stm-10 plants, which segregated 25% of the time. The progeny derived from the F3 fd-3 STM/stm-10 plants were used to characterize the fd-3 stm-10 inflorescence phenotypes.
To determine how ectopic FT or FD alter reproductive patterning events in stm-10, 35S:FT and 35S:FD was crossed with stm-10. All F1 plants flowered early and displayed the 35S:FT or 35S:FD phenotypes. The F1 plants were self-pollinated and seed from these crosses were planted. Because 35S:FT and 35S:FD plants are resistant to the herbicide, glufosinate, the resulting F2 plants were screened for: (i) stm-10 like plants that flowered early and (ii) were resistant to the herbicide basta. To characterize the 35S:FT stm-10 and 35S:FD stm-10 inflorescences further, F3 35S:FT and 35S:FD plants that segregated for the stm-10 phenotype were identified.
To determine the fold change in number of cauline leaves/coflorescences produced in ft-2 and fd-3, the average number of cauline leaves produced by ft-2 or fd-3 was divided by the average number of cauline leaves initiated in wild-type (note: in wild-type, ft-2 and ft-3 plants, all cauline leaves contained a coflorescence shoot in its leaf axil). The fold change in the number of cauline leaves produced by ft-2 stm-10 and fd-3 stm-10 was calculated by dividing the average number of cauline leaves initiated in ft-2 stm-10 and fd-3 stm-10 by the average number of cauline leaves formed in stm-10 inflorescence shoots.
In situ hybridization
The expression patterns of AP1, LFY, AGL24, and SOC1 transcripts were examined in wild-type, fd-3 and stm-10 inflorescence apices as well as in the non-flower producing shoot tips of fd-3 stm-10. The pKY89 vector containing the AP1 cDNA lacking the MADS domain was a gift provided by Dr Xuemei Chen. The AP1 UTP-digoxigenin anti-sense probe was synthesized using the SP6 RNA polymerase (Promega, Madison). Using the T7 RNA polymerase (Promega, Madison), the LFY UTP-digoxigenin anti-sense probe was synthesized from the pDW122 vector (Weigel et al., 1992). For localization of AGL24 and SOC1 transcripts, primers were designed and used to PCR amplify gene-specific sequences for these MADS-box genes. Primer sequences for SOC1 were SOC1-F (CTTATGAATTCGCCAGCTCC) and SOC1-R (GAAATAATACGACTCACTATAGGGACTCTAGAGAGGCAAGTGTAAGAACATAG). AGL24 primer sequences were AGL24-F (CTCCAGCTCAAGAATGAGAGAC) and AGL24-R (GAAATAATACGACTCACTATAGGGACTCATTCCCAAGATGGAAGCCCAAGC). The T7 RNA polymerase was used to synthesize the UTP-digoxigenin anti-sense SOC1 and AGL24 probes [note: the reverse (R) primer contains the T7 promoter (underlined)]. Plant fixation, sectioning, and mRNA in situ hybridization were performed as described previously (Jackson, 1991; Chuck et al., 2002).
Results
The combined functions of FT, FD, and STM are crucial for flower formation
In Arabidopsis, inflorescence architecture is, in part, dependent upon developmental patterning events that regulate the formation and identities of AMs (Benlloch et al., 2007; Prusinkiewicz et al., 2007). In wild-type plants, the SAM initiated 2–4 (average=3.2) coflorescence meristems subtended by cauline leaves during the initial stages of inflorescence development (Fig. 1A) (Table 1: row 1, column 2). After the SAM completed the vegetative to inflorescence transition, the SAM initiated flowers (Fig. 1A), which are subtended by cryptic bracts (Long and Barton, 2000). To determine the interplay between STM and FT or FD, the inflorescence phenotypes of single and double mutant combinations were characterized. In these analyses, loss of function alleles of ft-2 or fd-3 was combined with a weak allele of stm, stm-10, to determine the role of these gene products in floral specification.
Fig. 1.
Interplay between STM and FT/FD is crucial for inflorescence development. (A) Wild-type, (B) ft, (C) stm-10, (D–F) ft stm-10, (G) fd-3, and (H–J) fd stm-10 inflorescences. The Class I phenotype for (D) ft stm-10 and (H) fd stm-10 initiated inflorescences that were morphologically similar to stm-10; however, these shoots initiate approximately 2-fold more cauline leaves than stm-10 before terminating with flowers (inset: Table 2). The (E) ft stm-10 and (I) fd-3 stm-10 non-flowering Class II phenotype produced inflorescences that initiated cauline leaves indefinitely. In the Class III inflorescence shoots, (F) ft-2 stm-10 and (J) fd-3 stm-10 terminated without producing a single flower generating an ‘umbrella’ phenotype. Close up of the Class II non-flowering producing (K) ft-2 stm-10 and (L) fd-3 stm-10 apices. Bar=0.5 mm. (M–N) Scanning Electron Microscopy images of fd-3 stm-10 umbrella like apices. Arrows point at stipules. (M) Bar=1 mm and (N) Bar=0.25 mm. (O) Histological longitudinal cross section through an fd-3 stm-10 umbrella-like apex. Bar=0.3 mm. (This figure is available in colour at JXB online.)
Table 1.
Floral specification
| 1 | 2 | 3 | 4 | |
| Genotype | Ns | CLs | FLs | %FL |
| 1. Wild type | 42.1 (2.5) | 3.2 (0.47) | 40.5 (3.6) | 96% |
| 2. ft-2 | 44.1 (4.1) | 9.9 (1.2) | 39.8 (3.1) | 90% |
| 3. fd-3 | 42.7 (3.6) | 7.0 (0.8) | 40.1 (4.2) | 93% |
| 4. stm-10 | 9.4 (2.4) | 6.9 (1.8) | 2.3 (1.2) | 24% |
| 5. ft-2 stm-10 | 17.7 (7.2) | 15.5 (6.9) | 2.1 (1.5) | 11% |
| 6. fd-3 stm-10 | 22.5 (9.4) | 20.5 (9.6) | 1.9 (1.3) | 8% |
| 7. 35S:FT | 8.1 (1.7) | 1.3 (0.44) | 6.6 (2.1) | 81% |
| 8. 35S:FT stm-10 | 9.7 (4.3) | 4.4 (1.0) | 5.1 (4.1) | 52% |
| 9. 35S:FD | 32.3 (2.7) | 2.6 (0.49) | 29.5 (3.2) | 91% |
| 10. 35S:FD stm-10 | 10.4 (2.4) | 7.2 (1.7) | 3.1 (2.7) | 29% |
The average number of nodes (Ns), cauline leaves (CLs), and flowers (FLs) was determined for each genotype. Standard deviation was determined and displayed in parentheses. In our analysis, Ns, CLs, and FLs were quantified in the Class I ft-2 stm-10 and fd-3 stm-10. The percentage of flowering was determined by dividing the average number of flowers by the average number organs produced by the inflorescence shoot. *Note: Student's t test was performed (P <0.0001).
In ft-2 and fd-3 mutants, the SAM initiated 3.1-fold and 2.2-fold more coflorescence meristems subtended by cauline leaves than wild-type plants, respectively, indicating that the FT–FD complex plays a role in the specification of floral meristems (Fig. 1B, G; Table 1: rows 2 and 3, column 2) (Ruiz-García et al., 1997; Abe et al., 2005; Wigge et al., 2005). Overall, the inflorescences of ft-2 and fd-3 produced similar numbers of flowers as the wild type (Table 1: rows 1–3, column 3). In stm-10, an inflorescence shoot typically produced 4–11 (average=6.9) cauline leaves and 0–4 (average=2.3) floral nodes before shoot growth ceased with the formation of a terminal flower (Fig. 1C, inset; Table 1: row 4, columns 2 and 3). The shoots of stm-10 also displayed an internode patterning defect (data not shown). In stm-10 plants, the terminal growth habit of the primary reproductive shoots resulted in the outgrowth of secondary coflorescences shoots, which also produced terminal flowers. This pattern of growth, arrest, and the initiation of higher order coflorescences, repeated with each successive shoot resulting in a bushy phenotype (data not shown).
Genetic studies showed that three classes of inflorescence phenotypes were produced when ft-2 and fd-3 were combined with stm-10 (Fig. 1D–F and H–J, respectively; Table 2: columns 1–3). The first phenotypic class had an overall morphology similar to stm-10 inflorescences; however, this class of ft-2 stm-10 and fd-3 stm-10 reproductive shoots produced, on average, 2.2-fold and 2.9-fold more cauline leaves than the stm-10 plants, respectively, before the formation of the terminal flower (Fig. 1D, H; Table 2: column 1). The second inflorescence phenotypic class displayed a non-flower-producing phenotype in which these inflorescences initiated cauline leaves for 90–200 d before the plants senesced without producing a single flower (Fig. 1E, I, K, L; Table 2: column 3). These inflorescences produced up to 100 cauline leaves with shoots growing up to 78 cm in length (data not shown). Internode patterning defects observed in ft-2 stm-10 and fd-3 stm-10 were probably due to the decrease in STM function, since the inflorescences of stm-10 often produced shortened internodes. Taken together, the fact that ft-2 stm-10 and fd-3 stm-10 produced significantly more cauline leaves before the SAM was converted into a terminal flower showed that the combined functions of STM and FT/FD are crucial for the specification of flower meristems.
Table 2.
Penetrance of the classes of inflorescence phenotypes produced in ft stm-10 and fd stm-10
| 1 | 2 | 3 | |
| Phenotypes | stm-10-like | Umbrella | Non-flower producing |
| 1. ft stm-10 | 72% | 10% | 18% |
| 2. fd stm-10 | 46% | 23% | 31% |
In the third phenotypic class of inflorescences produced by ft-2 stm-10 and fd-3 stm-10, the SAM terminated with a compact cluster of cauline leaves, resembling an umbrella (Fig. 1F, J; Table 2: column 2). The umbrella-like inflorescence shoots initiated 8–18 (average=12.7) cauline leaves before growth terminated, without producing a single flower. Examination of the shoot apices of these plants showed that mature leaves directly emanated from the centre of the inflorescence apex (Fig. 1M). Scanning electron microscopy (SEM) demonstrated that the umbrella apex of fd-3 stm-10 lacked a meristem (Fig. 1N) (note: stipules were detected at the base of the cauline leaves). Histological examination of these umbrella apices showed that meristems were not evident in shoot apex of fd-3 stm-10 (Fig. 1O). Similar results were obtained with ft-2 stm-10 umbrella-like shoots (data not shown). These results indicate that FT and FD function with STM to maintain meristem integrity during inflorescence development.
Specification of coflorescence meristems requires STM and FT–FD
Since FT plays a role in coflorescence specification (Ruiz-García et al., 1997; Kanrar et al., 2008), the role of STM and FD/FT in the formation of coflorescence meristems was investigated. Coflorescences develop in the axils of cauline leaves in wild-type, ft-2, and fd-3 (Fig. 2A, B, C, respectively). In stm-10 plants, coflorescences developed in the axils of cauline leaves 55% of the time (Fig. 2D, H; Table 3: row 4, column 6). However, 45% of the cauline leaves produced by stm-10 inflorescence were devoid of coflorescence development and referred to as the solitary cauline leaf phenotype (Fig. 2E, I; Table 3: row 4, columns 1 and 5). Histological analyses of stm-10 inflorescences showed that coflorescence meristems were not specified in all cauline leaf axils (Fig. 2L). During the later stages of growth, coflorescence meristems developed into reproduction shoots (Fig. 2M). After termination of the primary stm-10 shoot, AMs failed to develop in the axils of the solitary cauline leaves (Fig. 2O).
Fig. 2.
Coflorescence meristem specification. Coflorescences develop in the axils of cauline leaves in (A) wild-type, (B) ft-2, and (C) fd-3 plants. In stm-10, (D) coflorescence shoots develop in the axils of some cauline leaves, (E) while the remaining leaves displayed an empty axil phenotype, which were devoid of coflorescence development (arrows). (F) ft-2 stm-10 and (G) fd-3 stm-10 inflorescence stems displayed an increased number of empty leaf axils. (H) Scanning electron microscopy image of a coflorescence shoot that developed in the axil of a cauline leaf in stm-10 (arrowhead points to coflorescence shoot). Scanning electron image of empty leaf axils in (I) stm-10, (J) ft-2 stm-10, and (K) fd-3 stm-10. (L) Longitudinal cross-section through an stm-10 inflorescence apex (arrow points to empty leaf axil and arrow-head points to a coflorescence meristem). (M) Transverse section through an stm-10 cauline leaf with a young coflorescence that developed in the axil. (O) After termination of shoot development, coflorescence meristems failed to develop in the axils of solitary cauline leaves in stm-10. (P) Longitudinal section through an fd-3 stm-10 non-flowering apex. Arrows point at the empty cauline leaf axils. (H, I) Bar=0.5 mm. (J, K) Bar=0.25 mm. (L–P) Bar=0.5 μm. cl, cauline leaf; s, stem. (This figure is available in colour at JXB online.)
Table 3.
Production of the ‘solitary’ cauline leaves displayed on the main shoot
| 1 | 2 | 3 | 4 | 5 | 6 | |
| Genotype | SCLs | Range | SD | n | %SCL | %CL |
| 1. Wild type | 0 | 0 | 0 | 51 | 0% | 100% |
| 2. ft-2 | 0 | 0 | 0 | 51 | 0% | 100% |
| 3. fd-3 | 0 | 0 | 0 | 51 | 0% | 100% |
| 4. stm-10 | 3.1 | 1–7 | 1.4 | 51 | 45% | 55% |
| 5. ft-2 stm-10 | 11.5 | 3–32 | 6.73 | 51 | 74% | 26% |
| 6. fd-3 stm-10 | 16.6 | 5–44 | 9.1 | 51 | 81% | 19% |
| 7. 35S:FT | 0 | 0 | 0 | 51 | 0% | 100% |
| 8. 35S:FT stm-10 | 0.69 | 0–2 | 0.54 | 51 | 16% | 84% |
| 9. 35S:FD | 0 | 0 | 0 | 51 | 0% | 100% |
| 10. 35S:FD stm-10 | 1.5 | 0–3 | 0.87 | 51 | 21% | 79% |
The average number of solitary cauline leaves (SCLs) produced by the inflorescences was determined for each genotype. The standard deviation (SD) was determined and the number (n) of shoots examined is also displayed in the table. The % of SCLs produced was calculated by dividing the averages number of SCLs by the average number of total cauline leaves with and without a coflorescence shoot in its axil (Table 2). The percentage of cauline leaves containing a coflorescence shoot in the leaf axil is also displayed (%CLs). In our analysis, SCLs were quantified in Class I shoots of ft-2 stm-10 and fd-3 stm-10. Student's t test was performed (P <0.0001).
Inflorescence shoots of ft-2 stm-10 and fd-3 stm-10 showed a marked decrease in the development of coflorescences such that 74% and 81% of the inflorescences initiated solitary cauline leaves, respectively (Fig. 2F, G; Table 3: rows 5 and 6, columns 1 and 5). SEM analysis of the ft-2 stm-10 and fd-3 stm-10 showed that coflorescence meristems were not detected in the axils of the solitary cauline leaves (Fig. 2J, K). Further, histological analysis showed that AM formation was not apparent during the early stages of cauline leaf development in fd-3 stm-10 (Fig. 2P, arrows). Similar results were obtained with ft-2 stm-10 (data not shown). The substantial decrease in the specification of coflorescence meristems in ft-2 stm-10 and fd-3 stm-10 resulted in plants that were less bushy than stm-10 (data not shown). Taken together, these results showed that the combined functions of STM and FT or FD are crucial for the specification of coflorescence meristems.
Ectopic FT partially restores reproductive meristems and structures in stm-10 plants
Previous studies indicate that FT activity is partially required for coflorescence specification (Ruiz-García et al., 1997; Kanrar et al., 2008). However, it has not been demonstrated that FT can promote the formation of AMs during inflorescence development. To determine if ectopic FT can induce AM formation, the development of coflorescence shoots in 35S:FT stm-10 plants was examined. Results showed that ectopic expression of FT in stm-10 increased the specification of coflorescence shoots in the axils of cauline leaves from 55% to 84% (Fig. 3C, F; Table 3: row 8, columns 1, 5, and 6). Likewise, 35S:FD stm-10 displayed a 24% increase in coflorescence specification compared to stm-10 (Table 3; row 10, columns 1, 5, and 6). The restoration of coflorescence specification was also apparent in high order coflorescence shoots of 35S:FT stm-10 (Fig. 3G) and 35S:FD stm-10 plants (data not shown). The increase in coflorescence specification in the reproductive shoots of 35S:FT stm-10 and 35S:FD stm-10 resulted in plants that were extremely bushy compared to stm-10 (data not shown). Taken together, these results showed that both FT and FD function is not only required for coflorescence formation there but that these floral integrators also promote the specification of AMs. Moreover, increased levels of FT and FD partially compensate for STM, when the function of this homeodomain protein is reduced during reproductive development.
Fig. 3.
Ectopic FT partially rescues the reproductive defects displayed in stm-10. (A) 35S:FT plant. Inflorescence shoots of (B) stm-10 and (C) 35S:FT stm-10. (D–F) Close up of cauline leaf on the inflorescence shoot of (D) 35S:FT, (E) stm-10, and (F) 35S:FT stm-10. (G) 35S:FT stm-10 secondary coflorescence shoot initiated multiple cauline leaves and axillary shoots. (E) Arrows point to the empty leaf axils in the stm-10 inflorescence. (H) Terminal flower in (H) 35S:FT and (I) stm-10. (J) Close up of an inflorescence shoot initiating mutiple flowers in 35S:FT stm-10. (K) 35S:FT silique. Arrow-head points at the silique. (L, M) Carpel-like organs developed in an stm-10 flower. Bar=0.5 mm. (N) At a low penetrance, siliques develop in 35S:FT stm-10 plants. Arrow-heads point to the siliques. (This figure is available in colour at JXB online.)
In Arabidopsis, the inflorescences of 35S:FT plants transition rapidly to flower production, initiating fewer coflorescence shoots subtended by cauline leaves than wild-type plants (Fig. 3A; Table 1: row 7, column 2). 35S:FT inflorescences produced 5–8 (average=6.6) floral nodes before the SAM was transformed into a floral meristem (Fig. 3H; Table 1: row 7, column 3). If STM is a crucial component, which functions with FT and FD to specify flowers, then an increase in the levels of FT and/or FD in stm-10 may augment the floral specification potential in these shoots. Results showed that, 35S:FT stm-10 initiated flowers earlier in inflorescence development than stm-10 (Table 1: row 8, column 2). Moreover, 35S:FT stm-10 plants, on average, produced twice as many flowers as stm-10 (Fig. 3J, N; Table 1: row 8, column 3). Unlike FT, ectopic expression of FD in stm-10 had little effect on the timing of flower specification (Table 1: row 10, column 2). Taken together, these results indicate that increased levels of FT can partially restore meristem activity and floral specification potential when STM levels are limiting.
STM function is required for carpel formation and development (Clark et al., 1996; Endrizzi et al., 1996; Scofield et al., 2007; Yu et al., 2009). Carpels specified during flower patterning developed into the fruits or siliques in wild-type plants (data not shown) and 35S:FT (Fig. 3K). Unlike wild-type and 35S:FT flowers, stm-10 displayed a marked reduction in the specification of carpels (Fig. 3L, M) (Clark et al., 1996; Endrizzi et al., 1996; Scofield et al., 2007; Yu et al., 2009). Interestingly, 4% of the 35S:FT stm-10 plants initiated fused carpels (Fig. 3N), which produced 3–5 seeds. When germinated, these seeds gave rise to plants with the 35SFT stm-10 phenotype (data not shown). Taken together, these experiments show that increased levels of FT can partially compensate for a reduction in STM activity during carpel development.
Expression of flower meristem identity genes in fd-3 stm-10 non-flower producing shoots
During inflorescence development, flowers are specified on the flanks of the SAM by the activity of flower meristem identity genes (Liu et al., 2009). To determine if STM acts with FT/FD to specify flower meristem identity, the expression patterns of AP1 and LFY were examined in wild-type, fd-3, stm-10, and the non-flower producing fd-3 stm-10 inflorescence apices. In situ hybridization was performed in fd-3 stm-10, since the non-flowering phenotype was less penetrant in ft-2 stm-10 (Table 2, column 3).
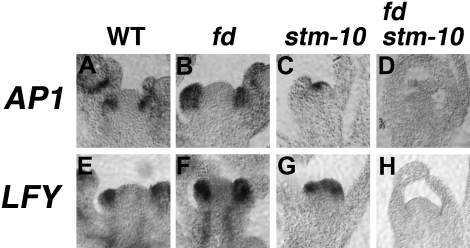
In the wild type, AP1 transcripts accumulate in floral meristems and eventually become restricted to whorls 1 and 2 during the later stages of flower development (Mandel et al., 1992) (Fig. 4A). In fd-3, the onset of AP1 expression in AMs is delayed because flower meristems are converted to coflorescence meristems during the early stages of inflorescence development (Wigge et al., 2005). However, once fd-3 shoots complete the transition from coflorescence to flower production, the expression pattern for AP1 was similar to the wild type (Wigge et al., 2005) (Fig. 4B). In stm-10, AP1 transcripts were detected in the SAM, possibly during the formation of the terminal flower (Fig. 4C). AP1 transcripts were also detected in developing sepal and petal primordia (data not shown). Consistent with the non-flower producing phenotype, AP1 was not detected in the fd-3 stm-10 inflorescence apices (Fig. 4D). Because LFY controls AP1 expression in parallel with FT–FD, the expression patterns of LFY in fd-3 stm-10 were examined. In wild-type and fd-3, LFY transcripts were visualized in cells on the flanks of the SAM and in developing flower meristems (Weigel et al., 1992) (Fig. 4E, F). LFY transcripts were also detected in the terminal flower meristems of stm-10 (Fig. 4G). However, in fd-3 stm-10, LFY expression was dramatically reduced in the shoot apices of these non-flower-producing inflorescences (Fig. 4H). Taken together, the in situ hybridization results show that the combined functions of STM and FT–FD are crucial for activating LFY and AP1.
Fig. 4.
Expression analysis of flower meristem identity. Localization of AP1 transcripts in (A) wild-type, (B) fd-3, (C) stm-10, and (D) fd-3 stm-10 reproductive apices. LFY mRNA was localized in (E) wild-type, (F) fd-3, (G) stm-10, and (H) fd-3 stm-10 Class II inflorescence shoot tips. Bar=50 μm.
MADS-box floral integrator genes are expressed in the SAM of fd-3 stm-10 non-flower-producing shoots
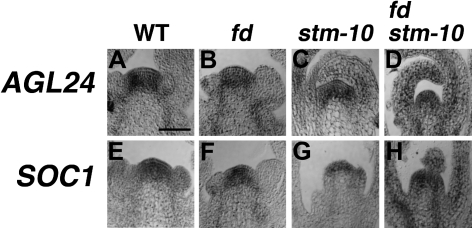
The floral integrator genes SOC1 and AGL24 are expressed in the inflorescence meristem and function together to activate LFY during reproductive development (Lee et al., 2008; Liu et al., 2008). Because LFY is not expressed in the non-flower-producing shoots of fd-3 stm-10, the expression pattern of AGL24 and SOC1 was examined during inflorescence development. In the wild type, AGL24 and SOC1 transcripts were detected in the inflorescence meristem (Borner et al., 2000; Samach et al., 2000; Yu et al., 2002; Michaels et al., 2003) (Fig. 5A, E, respectively). During inflorescence development, AGL24 and SOC1 mRNAs localized to the SAM in fd-3 and stm-10 reproductive shoots (Fig. 5B, C, F, G). Interestingly, in the non-flower-producing inflorescence shoots of fd-3 stm-10, both AGL24 and SOC1 transcripts were visualized in the SAM (Fig. 5D, H, respectively). Thus, in the absence of FD, SOC1 and AGL24 partially depend on the function of STM in order to activate LFY.
Fig. 5.
Expression of floral integrator genes. Expression patterns for AGL24 were determined in (A) wild-type, (B) fd-3, (C) stm-10, and (D) fd-3 stm-10 Class II inflorescence apices. Localization of SOC1 mRNA in (E) wild-type, (F) fd-3, (G) stm-10, and (H) fd-3 stm-10 reproductive shoot apices. Bar=50 μm.
Discussion
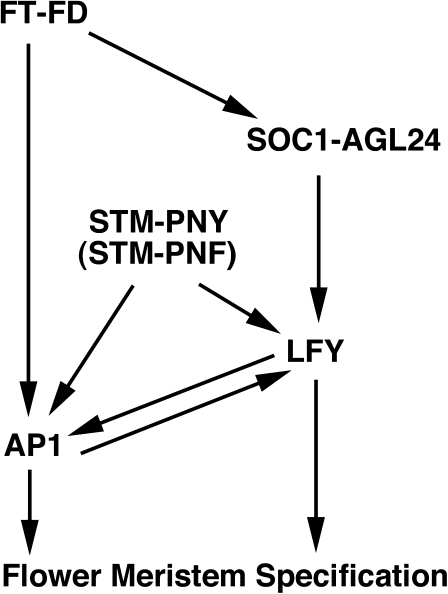
The floral transition is a pivotal phase change event, which establishes reproductive growth patterns that are often distinct from vegetative modes of development (Benlloch et al., 2007). It is proposed that, in response to flowering time signals, floral integrators must somehow act with the proteins that control meristem maintenance and function in order to establish and maintain inflorescence patterns of growth. FT is the universal florigen signal that promotes flowering and regulates meristem activity (Kobayashi and Weigel, 2007; Turck et al., 2008; Zeevaart, 2008; Shalit et al., 2009). In the SAM, FT associates with FD and together these proteins mediate the floral transition as well as flower meristem specification (Kobayashi and Weigel, 2007; Turck et al., 2008; Zeevaart, 2008). At the molecular level, how FT regulates meristem activity is not well understood. STM and related KNOX proteins regulate meristem maintenance and reproductive pattern events during shoot maturation (Hake et al., 2004; Scofield and Murray, 2006; Hay and Tsiantis, 2009). In this paper, a reduction in FT or FD enhances the floral and coflorescence specification phenotypes displayed in stm-10. At the same time, ectopic FT or FD restores coflorescence specification in stm-10 plants. An increase in the levels of FT augments the floral specification potential of stm-10 inflorescence meristems. Thus, STM, FT, and FD play a fundamental role in the specification of axillary meristems during reproductive development. It is proposed that STM–PNY and STM–PNF complexes act with FT and FD to promote the formation of coflorescence meristems as well as specify flower meristem identity (Figure 6).
Fig. 6.
A model for flower specification during Arabidopsis inflorescence development. It is proposed that STM-PNY/PNF dimers and the floral integrator complexes FT–FD and AGL24-SOC1 function to specify flower meristem identity. In this model, STM-PNY/PNF function with AGL24-SOC1 to regulate LFY. The late flower meristem identity gene AP1 requires the combined functions of FT–FD, STM-PNY/PNF, and LFY-UFO complexes as well as SPLs.
Studies in tomato indicate that the homologue of FT called SINGLE FLOWER TRUSS (SFT) acts as a general plant growth regulator, which functions to promote meristem determinacy (Shalit et al., 2009). The inhibitor of SFT, SELF-PRUNING (SP), which is a homologue of TFL1, acts to control the terminal growth effect of SFT in shoot meristems (Shalit et al., 2009). Previous studies show that increased levels of FT convert the indeterminate inflorescence meristem into a determinate meristem with floral identity in Arabidopsis (Kardailsky et al., 1999; Kobayashi and Weigel, 2007). In tomato, F1 progeny derived from crossing sft mutants with different tomato varities, which are homozygous for SFT produce significantly more fruit than the parental lines (Krieger et al., 2010). The heterozygous effect of SFT produces shoots that display a decrease in meristem determinacy, indicating that the levels of SFT is crucial for shoot architecture and productivity. In this study, ectopic FT promotes the formation of coflorescence and floral meristems as well as carpels during inflorescence and flower development, respectively, in stm-10 plants. The fact that ectopic FT partially suppresses some of the reproductive phenotypes of STM indicates that these factors act to regulate inflorescence and floral development.
Networks controlling flower meristem identity
In Arabidopsis, flower meristem identity is specified on the flanks of the SAM by the activity of LFY. The promoter of LFY integrates floral inductive cues mediated by long-day photoperiod and gibberellin (Blazquez and Weigel, 2000). SOC1 and AGL24 encode MADS-box proteins that are induced by multiple flowering time pathways in the SAM (Borner et al., 2000; Samach et al., 2000; Yu et al., 2002; Michaels et al., 2003; Moon et al., 2003). Recent studies indicate that the SOC1–AGL24 complex positively regulates LFY transcription in response to floral inductive cues (Yu et al., 2002; Moon et al., 2003; Lee et al., 2008). Studies in this paper show that flower meristem specification is reduced and often completely impaired in stm-10 fd-3 and stm-10 ft-2 plants. Transcripts for AGL24 and SOC1 localize to the SAM in the stm-10 fd-3 non-flower producing inflorescence shoots. However, LFY is not expressed in these fd-3 stm-10 inflorescence shoots. Therefore, the SOC1–AGL24 complexes are partially dependent upon STM for the activation of LFY. In Fig. 6, it is proposed that SOC1–AGL24 complexes require STM–PNY/STM–PNF for the activation of LFY.
In yeast, specific mating cell types are specified by the co-operative interaction between the MADS-box protein minichromosome maintenance protein 1 (MCM1) and the homeodomain proteins Mating type-a (MATa) or MATalpha (Johnson, 1995). In plants, BELL1 (BEL1), the founding member of the BLH class of transcription factors, associates with the AGAMOUS and SEPALATA3 MADS-box dimer, possibly forming a trimeric complex, which acts to specify integument cell identity during ovule development (Brambilla et al., 2007). Therefore, it may be possible that PNY/PNF–STM complexes directly associate with AGL24–SOC1 dimers and/or tetramers to specify flower meristem identity by activating LFY.
LFY functions to specify flower meristem identity by activating the late flower meristem identity genes, including AP1 (Parcy et al., 1998; Wagner et al., 1999). LFY interacts with the F-box protein UNUSUAL FLORAL ORGANS (UFO), which is an orthologue of the Petunia DOUBLE TOP (DOT) protein (Hepworth et al., 2006; Chae et al., 2008; Souer et al., 2008). Recent studies showed that UFO and DOT function in specifying flower meristem identity (Hepworth et al., 2006; Souer et al., 2008). Taken together, the LFY–UFO complex directly regulates AP1 in a pathway parallel with FT–FD (Fig. 6) (Ruiz-García et al., 1997; Abe et al., 2005; Wigge et al., 2005). Previous studies showed that FT–FD requires PNY and PNF for flower formation and the activation of AP1 (Kanrar et al., 2008). Because AP1 expression is not detected in the non-flower producing fd-3 stm-10 shoots, it is proposed that STM–PNY/PNF functions with the FT–FD and LFY–UFO complexes co-operatively to regulate AP1 during the later stages of flower meristem specification (Fig. 6).
Acknowledgments
We would like to thank Dr Patricia Springer to critical reading of this manuscript. We also thank Dr Xuemei Chen and Dr Detlef Weigel for the pKY89 and pDW124 plasmids used to synthesize the AP1 and LFY in situ probes, respectively. We are grateful to Dr David Carter, the Academic Coordinator for the Microscopy Core Facility, for help with the confocal microscopy and image analysis. Lastly, the authors thank the two anonymous reviewers for helpful comments on this manuscript. This work was funded by NSF, grant number IOB-0615774. N Ung was supported by NIH, grant number GM062756.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- Bao X, Franks RG, Levin JZ, Liu Z. Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. The Plant Cell. 2004;16:1478–1489. doi: 10.1105/tpc.021147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nature Genetics. 2008;40:1136–1141. doi: 10.1038/ng.189. [DOI] [PubMed] [Google Scholar]
- Barton MK. Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Developmental Biology. 2009;341:95–113. doi: 10.1016/j.ydbio.2009.11.029. [DOI] [PubMed] [Google Scholar]
- Barton MK, Poethig RS. Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development. 1993;119:823–831. [Google Scholar]
- Benlloch R, Berbel A, Serrano-Mislata A, Madueno F. Floral initiation and inflorescence architecture: a comparative view. Annals of Botany. 2007;100:659–676. doi: 10.1093/aob/mcm146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T, Leyser O. Something on the side: axillary meristems and plant development. Plant Molecular Biology. 2006;60:843–854. doi: 10.1007/s11103-005-2763-4. [DOI] [PubMed] [Google Scholar]
- Bhatt AM, Etchells JP, Canales C, Lagodienko A, Dickinson H. VAAMANA: a BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene. 2004;328:103–111. doi: 10.1016/j.gene.2003.12.033. [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Weigel D. Integration of floral inductive signals in Arabidopsis. Nature. 2000;404:889–892. doi: 10.1038/35009125. [DOI] [PubMed] [Google Scholar]
- Bleckmann A, Simon R. Interdomain signaling in stem cell maintenance of plant shoot meristems. Molecules and Cells. 2009;27:615–620. doi: 10.1007/s10059-009-0094-z. [DOI] [PubMed] [Google Scholar]
- Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S. A MADS domain gene involved in the transition to flowering in Arabidopsis. The Plant Journal. 2000;24:591–599. doi: 10.1046/j.1365-313x.2000.00906.x. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development. 1993;119:721–743. [Google Scholar]
- Brambilla V, Battaglia R, Colombo M, Masiero S, Bencivenga S, Kater MM, Colombo L. Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. The Plant Cell. 2007;19:2544–2556. doi: 10.1105/tpc.107.051797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME, Groover AT, Fontana JR, Martienssen RA. Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development. 2003;130:3941–3950. doi: 10.1242/dev.00620. [DOI] [PubMed] [Google Scholar]
- Chae E, Tan QK, Hill TA, Irish VF. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development. 2008;135:1235–1245. doi: 10.1242/dev.015842. [DOI] [PubMed] [Google Scholar]
- Champagne C, Sinha N. Compound leaves: equal to the sum of their parts? Development. 2004;131:4401–4412. doi: 10.1242/dev.01338. [DOI] [PubMed] [Google Scholar]
- Chuck G, Muszynski M, Kellogg E, Hake S, Schmidt RJ. The control of spikelet meristem identity by the branched silkless1 gene in maize. Science. 2002;298:1238–1241. doi: 10.1126/science.1076920. [DOI] [PubMed] [Google Scholar]
- Clark SE, Jacobsen SE, Levin JZ, Meyerowitz EM. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development. 1996;122:1567–1575. doi: 10.1242/dev.122.5.1567. [DOI] [PubMed] [Google Scholar]
- Cole M, Nolte C, Werr W. Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Research. 2006;34:1281–1292. doi: 10.1093/nar/gkl016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodsworth S. A diverse and intricate signalling network regulates stem cell fate in the shoot apical meristem. Developmental Biology. 2009;336:1–9. doi: 10.1016/j.ydbio.2009.09.031. [DOI] [PubMed] [Google Scholar]
- Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. The Plant Journal. 1996;10:967–979. doi: 10.1046/j.1365-313x.1996.10060967.x. [DOI] [PubMed] [Google Scholar]
- Eriksson S, Bohlenius H, Moritz T, Nilsson O. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. The Plant Cell. 2006;18:2172–2181. doi: 10.1105/tpc.106.042317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S, Smith HM, Holtan H, Magnani E, Mele G, Ramirez J. The role of knox genes in plant development. Annual Review of Cell and Developmental Biology. 2004;20:125–151. doi: 10.1146/annurev.cellbio.20.031803.093824. [DOI] [PubMed] [Google Scholar]
- Hamant O, Pautot V. Plant development: a TALE story. Comptes Rendus Biologies. 2010;333:371–381. doi: 10.1016/j.crvi.2010.01.015. ? [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nature Genetics. 2006;38:942–947. doi: 10.1038/ng1835. [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. A KNOX family TALE. Current Opinion in Plant Biology. 2009;12:593–598. doi: 10.1016/j.pbi.2009.06.006. [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Klenz JE, Haughn GW. UFO in the Arabidopsis apex is required for floral-meristem identity and bract suppression. Planta. 2006;223:769–778. doi: 10.1007/s00425-005-0138-3. [DOI] [PubMed] [Google Scholar]
- Jackson D. In situ hybridisation in plants. In: Bowles DJ, Gurr SJ, McPherson M, editors. Molecular plant pathology: a practical approach. Oxford: Oxford University Press; 1991. pp. 163–174. [Google Scholar]
- Johnson AD. Molecular mechanisms of cell-type determination in budding yeast. Current Opinion in Genetics and Development. 1995;5:552–558. doi: 10.1016/0959-437x(95)80022-0. [DOI] [PubMed] [Google Scholar]
- Kanrar S, Bhattacharya M, Arthur B, Courtier J, Smith HM. Regulatory networks that function to specify flower meristems require the function of homeobox genes PENNYWISE and POUND-FOOLISH in Arabidopsis. The Plant Journal. 2008;54:924–937. doi: 10.1111/j.1365-313X.2008.03458.x. [DOI] [PubMed] [Google Scholar]
- Kanrar S, Onguka O, Smith HM. Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta. 2006;224:1163–1173. doi: 10.1007/s00425-006-0298-9. [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Laudencia-Chingcuanco D, Smith LG, Hake S. Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development. 1997;124:3045–3054. doi: 10.1242/dev.124.16.3045. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Weigel D. Move on up, it's time for change: mobile signals controlling photoperiod-dependent flowering. Genes and Development. 2007;21:2371–2384. doi: 10.1101/gad.1589007. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Vanderveen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Molecular and General Genetics. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Krieger U, Lippman ZB, Zamir D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nature Genetics. 2010;42:459–463. doi: 10.1038/ng.550. [DOI] [PubMed] [Google Scholar]
- Lee J, Oh M, Park H, Lee I. SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. The Plant Journal. 2008;55:832–843. doi: 10.1111/j.1365-313X.2008.03552.x. [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Gustafson-Brown C, Pinyopich A, Ditta GS, Yanofsky MF. Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. The Plant Cell. 1999;11:1007–1018. doi: 10.1105/tpc.11.6.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development. 2008;135:1481–1491. doi: 10.1242/dev.020255. [DOI] [PubMed] [Google Scholar]
- Liu C, Thong Z, Yu H. Coming into bloom: the specification of floral meristems. Development. 2009;136:3379–3391. doi: 10.1242/dev.033076. [DOI] [PubMed] [Google Scholar]
- Long J, Barton MK. Initiation of axillary and floral meristems in Arabidopsis. Developmental Biology. 2000;218:341–353. doi: 10.1006/dbio.1999.9572. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. The Plant Journal. 2003;33:867–874. doi: 10.1046/j.1365-313x.2003.01671.x. [DOI] [PubMed] [Google Scholar]
- Moon J, Lee H, Kim M, Lee I. Analysis of flowering pathway integrators in Arabidopsis. Plant and Cell Physiology. 2005;46:292–299. doi: 10.1093/pcp/pci024. [DOI] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. The Plant Journal. 2003;35:613–623. doi: 10.1046/j.1365-313x.2003.01833.x. [DOI] [PubMed] [Google Scholar]
- Nilsson O, Lee I, Blaazquez MA, Weigel D. Flowering-time genes modulate the response to LEAFY activity. Genetics. 1998;150:403–410. doi: 10.1093/genetics/150.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D. A genetic framework for floral patterning. Nature. 1998;395:561–566. doi: 10.1038/26903. [DOI] [PubMed] [Google Scholar]
- Pnueli L, Gutfinger T, Hareven D, Ben-Naim O, Ron N, Adir N, Lifschitz E. Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. The Plant Cell. 2001;13:2687–2702. doi: 10.1105/tpc.010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig RS. Phase change and the regulation of developmental timing in plants. Science. 2003;301:334–336. doi: 10.1126/science.1085328. [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E. Evolution and development of inflorescence architectures. Science. 2007;316:1452–1456. doi: 10.1126/science.1140429. [DOI] [PubMed] [Google Scholar]
- Roeder AH, Ferrandiz C, Yanofsky MF. The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Current Biology. 2003;13:1630–1635. doi: 10.1016/j.cub.2003.08.027. [DOI] [PubMed] [Google Scholar]
- Ruiz-García L, Madueño F, Wilkinson M, Haughn G, Salinas J, Martínez-Zapater JM. Different roles of flowering-time genes in the activation of floral initiation genes in Arabidopsis. The Plant Cell. 1997;9:1921–1934. doi: 10.1105/tpc.9.11.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutjens B, Bao D, van Eck-Stouten E, Brand M, Smeekens S, Proveniers M. Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. The Plant Journal. 2009;58:641–654. doi: 10.1111/j.1365-313X.2009.03809.x. [DOI] [PubMed] [Google Scholar]
- Sablowski R. Flowering and determinacy in Arabidopsis. Journal of Experimental Botany. 2007;58:899–907. doi: 10.1093/jxb/erm002. [DOI] [PubMed] [Google Scholar]
- Saddic LA, Huvermann B, Bezhani S, Su Y, Winter CM, Kwon CS, Collum RP, Wagner D. The LEAFY target LMI1 is a meristem identity regulator and acts together with LEAFY to regulate expression of CAULIFLOWER. Development. 2006;133:1673–1682. doi: 10.1242/dev.02331. [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288:1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- Schultz EA, Haughn GW. Genetic analysis of the floral initiation process (FLIP) in Arabidopsis. Development. 1993;119:745–765. [Google Scholar]
- Scofield S, Dewitte W, Murray JA. The KNOX gene SHOOT MERISTEMLESS is required for the development of reproductive meristematic tissues in Arabidopsis. The Plant Journal. 2007;50:767–781. doi: 10.1111/j.1365-313X.2007.03095.x. [DOI] [PubMed] [Google Scholar]
- Scofield S, Murray JA. KNOX gene function in plant stem cell niches. Plant Molecular Biology. 2006;60:929–946. doi: 10.1007/s11103-005-4478-y. [DOI] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Krober S, Amasino RA, Coupland G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes and Development. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalit A, Rozman A, Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y, Lifschitz E. The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proceedings of the National Academy of Sciences, USA. 2009;106:8392–8397. doi: 10.1073/pnas.0810810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HM, Campbell BC, Hake S. Competence to respond to floral inductive signals requires the homeobox genes PENNYWISE and POUND-FOOLISH. Current Biology. 2004;14:812–817. doi: 10.1016/j.cub.2004.04.032. [DOI] [PubMed] [Google Scholar]
- Smith HM, Hake S. The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. The Plant Cell. 2003;15:1717–1727. doi: 10.1105/tpc.012856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer E, Rebocho AB, Bliek M, Kusters E, de Bruin RA, Koes R. Patterning of inflorescences and flowers by the F-box protein DOUBLE TOP and the LEAFY homolog ABERRANT LEAF AND FLOWER of petunia. The Plant Cell. 2008;20:2033–2048. doi: 10.1105/tpc.108.060871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano S, Niihama M, Smith HM, Tasaka M, Aida M. gorgon a novel missense mutation in the SHOOT MERISTEMLESS gene, impairs shoot meristem homeostasis in Arabidopsis. Plant and Cell Physiology. 2010;51:621–634. doi: 10.1093/pcp/pcq028. [DOI] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annual Review of Plant Biology. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Reiser L, Hake S. Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene knotted1. Development. 2000;127:3161–3172. doi: 10.1242/dev.127.14.3161. [DOI] [PubMed] [Google Scholar]
- Wagner D, Sablowski RWM, Meyerowitz EM. Transcriptional activation of APETALA1 by LEAFY. Science. 1999;285:582–584. doi: 10.1126/science.285.5427.582. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- William DA, Su Y, Smith MR, Lu M, Baldwin DA, Wagner D. Genomic identification of direct target genes of LEAFY. Proceedings of the National Academy of Sciences, USA. 2004;101:1775–1780. doi: 10.1073/pnas.0307842100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiology. 2005;139:770–778. doi: 10.1104/pp.105.066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Xu Y, Tan EL, Kumar PP. AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proceedings of the National Academy of Sciences, USA. 2002;99:16336–16341. doi: 10.1073/pnas.212624599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Patibanda V, Smith HM. A novel role of BELL1-like homeobox genes, PENNYWISE and POUND- FOOLISH, in floral patterning. Planta. 2009;229:693–707. doi: 10.1007/s00425-008-0867-1. [DOI] [PubMed] [Google Scholar]
- Zeevaart JAD. Leaf-produced floral signals. Current Opinion in Plant Biology. 2008;11:541–547. doi: 10.1016/j.pbi.2008.06.009. [DOI] [PubMed] [Google Scholar]