Abstract
In plants, cell proliferation and polarized cell differentiation along the adaxial–abaxial axis in the primordium is critical for leaf morphogenesis, while the temporal–spatial relationships between these two processes remain largely unexplored. Here, it is reported that microRNA396 (miR396)-targeted Arabidopsis growth-regulating factors (AtGRFs) are required for leaf adaxial–abaxial polarity in Arabidopsis. Reduction of the expression of AtGRF genes by transgenic miR396 overexpression in leaf polarity mutants asymmetric leaves1 (as1) and as2 resulted in plants with enhanced leaf adaxial–abaxial defects, as a consequence of reduced cell proliferation. Moreover, transgenic miR396 overexpression markedly decreased the cell division activity and the expression of cell cycle-related genes, but resulted in an increased percentage of leaf cells with a higher ploidy level, indicating that miR396 negatively regulates cell proliferation by controlling entry into the mitotic cell cycle. miR396 is mainly expressed in the leaf cells arrested for cell division, coinciding with its roles in cell cycle regulation. These results together suggest that cell division activity mediated by miR396-targeted AtGRFs is important for polarized cell differentiation along the adaxial–abaxial axis during leaf morphogenesis in Arabidopsis.
Keywords: Arabidopsis, AtGRFs, cell differentiation, cell division, leaf polarity, miR396
Introduction
The development of multicellular organisms relies on the generation of an appropriate number of cells and timely acquisition of specialized cell functions. In flowering plants, leaves are determinate organs, whose development is mediated by the temporal–spatial regulation of cell proliferation and differentiation (Gutierrez, 2005; Ramirez-Parra et al., 2005; Fleming, 2006). Leaf morphogenesis is conceptually divided into three processes: primordia initiation, establishment of polarity, and leaf expansion (Sinha, 1999). After initiation from the peripheral zone of shoot apical meristem (SAM), cells in leaf primordia rapidly divide and are subsequently specified to establish asymmetric growth along the proximo-distal, medio-lateral, and adaxial–abaxial axes (Hudson, 2000; Bowman et al., 2002).
The establishment of the adaxial–abaxial axis is most critical for leaf morphogenesis (Sussex, 1954; Waites and Hudson, 1995; Bowman et al., 2002). In recent years, several families of transcription factor genes have been identified as the key determinants of adaxial or abaxial cell fate. Members of the KANADI (KAN) and YABBY (YAB) families and two AUXIN RESPONSE FACTOR genes (ARF3/ETT and ARF4) play important roles in determination of abaxial cell fate (Eshed et al., 2004; Pekker et al., 2005). In contrast, three genes in the class III homeodomain-leucine zipper (HD-ZIP III) family, namely PHB, PHV, and REV, are expressed in the leaf adaxial domain and redundantly promote adaxial leaf fate (McConnell et al., 2001; Emery et al., 2003). Transcripts of HD-ZIP III genes are the targets of microRNA165 and 166 (miR165/166) (Rhoades et al., 2002; Tang et al., 2003), whereas ARF3/4 are targeted by a trans-acting small interfering RNA (ta-siRNA), termed tasiR-ARF (Allen et al., 2005; Fahlgren et al., 2006). Moreover, the Myb-domain protein ASYMMETRIC LEAVES 1 (AS1) and the LOB domain protein AS2 function together to promote leaf adaxial identity by repressing the abaxial-expressed KAN genes and miR165/166 (Byrne et al., 2000; Semiarti et al., 2001; Iwakawa et al., 2002; Kumaran et al., 2002; Xu et al., 2002, 2003; Lin et al., 2003; Li et al., 2005; Wu et al., 2008), while KAN genes antagonize the roles of HD-ZIP III genes (Emery et al., 2003) and directly repress AS2 expression by binding to its promoter (Wu et al., 2008). In addition, the 26S proteasome and ribosome proteins were recently implicated to play important roles in promoting the establishment of leaf polarity at the protein regulation level (Huang et al., 2006; Pinon et al., 2008; Yao et al., 2008).
In contrast to the adaxial–abaxial axis, only a few genes were known to be involved in patterning of the proximo-distal (leaf length direction) and medio-lateral (leaf width direction) axes of leaves (Tsukaya, 2006). Growth along the leaf length is governed by ROTUNDIFOLIA3 (ROT3), which regulates polarized cell expansion (Kim et al., 1998), and ROT4, which inhibits cell proliferation to regulate specifically cell number in the leaf length direction (Narita et al., 2004). Leaf expansion in the width direction is affected by ANGUSTIFOLIA (AN) and AN3. AN regulates cell expansion in the width direction probably through controlling the cortical microtubule arrangement in leaf cells (Folkers et al., 2002; Kim et al., 2002). AN3, which encodes a putative coactivator for the Arabidopsis growth-regulating factor (AtGRF) family of transcription factors (J. H. Kim et al., 2003), also called AtGIF1 (GRF-interacting factor1), predominantly controls cell number in the leaf width direction by modulating cell proliferation activity (Horiguchi and Tsukaya, 2003; Kim and Kende, 2004; Horiguchi et al., 2005). The Arabidopsis AtGRF family consists of nine members, while AtGIF1 has another two homologues, AtGIF2 and AtGIF3. Both AtGRF and AtGIF genes appear to perform redundant functions to promote and/or maintain cell proliferation activity in leaves (J. H. Kim et al., 2003; Kim and Kende, 2004; Horiguchi et al., 2005; Lee et al., 2009), whereas at least seven AtGRF genes (AtGRF1–4 and AtGRF7–9) are targeted by miR396, which is encoded by two loci, MIR396a and MIR396b, and attenuate cell proliferation activity by repression of expression of the targeted AtGRF genes (Jones-Rhoades and Bartel, 2004; Liu et al., 2009; Rodriguez et al., 2010).
Although a number of factors have been found to specify leaf polarity, the temporal–spatial relationships of cell proliferation and differentiation during leaf morphogenesis remain largely unexplored. Recently, a novel gene, AS1/2 ENHANCER7 (AE7), which is required for both leaf adaxial–abaxial polarity formation and normal cell proliferation, was characterized. It was also found that the previously characterized 26S proteasome mutant ae3-1 (Huang et al., 2006) and ribosome mutant ae5-1 (Yao et al., 2008) exhibited not only leaf adaxial–abaxial polarity but also cell proliferation defects, and it was therefore proposed that normal cell proliferation may be essential for establishment of leaf polarity (Yuan et al., 2010). However, because the 26S proteasome and ribosome are widely recognized as essential machinery for many biological processes, and AE7 encodes a member of a functionally unknown protein family conserved in eukarytoes, whether this proposal is a general mechanism and how the functions of these proteins in cell proliferation specifically regulate leaf polarity need further investigation.
In this work, it is reported that cell proliferation mediated by miR396-targeted AtGRFs is required for leaf adaxial–abaxial polarity formation during leaf morphogenesis. It is further shown that miR396 negatively regulates cell proliferation in leaves by controlling the entry into the mitotic cell cycle, coincident with its expression in leaf cells arrested for cell division. Because the cells unable to enter into the mitotic cell cycle often undergo enlargement and expansion to start differentiation (Gutierrez, 2005, 2009; De Veylder et al., 2007; Nieuwland et al., 2009), these data together with previous results strongly suggest that active cell division in the primordium is important for leaf adaxial–abaxial polarity formation, and highlight that miR396-targeted AtGRFs may mediate coordination processes of cell division coupled with cell differentiation during leaf morphogenesis.
Materials and methods
Plant materials and growth conditions
The as1-1, as2-1, and T-DNA insertion mutant atgif1/an3 (SALK_150407) were obtained from the ABRC. All the plant materials used in this study are in the Columbia-0 (Col-0) background. Plant growth was according to previous conditions (Chen et al., 2000).
Plasmid construction and plant transformation
For transgenic miR396 overexpression, the 316 bp MIR396A and MIR396B precursor fragments were PCR-amplified with genomic DNA from wild-type Col plants. The DNA fragments were cloned into a modified pCAMBIA2300 vector, behind the duplicated cauliflower mosaic virus 35S promoter, to generate the constructs 35S:miR396a and 35S:miR396b. For generation of miR396-resistant versions of AtGRF9, the miR396 target site in AtGRF9 was altered by introducing synonymous mutations using an overlap-PCR method. The modified AtGRF9 gene was then cloned into a modified pCAMBIA1300 vector, behind the duplicated 35S promoter. For promoter–GUS (β-glucuronidase) constructs, the 3125 bp and 3132 bp 5′ upstream sequences of MIR396A and MIR396B were PCR-amplified and cloned into the entry vector pENTR/D-TOPO. The resulting DNA fragments were then recombined into the GATEWAY destination vector pMDC162A (Curtis and Grossniklaus, 2003), to generate p396a:GUS and p396b:GUS constructs. All constructs were verified by nucleotide sequencing for inserts and introduced into the Agrobacterium strain GV3101. The transgenic plants were generated by the floral dip method (Clough and Bent, 1998). The sequences of primers used for PCR are listed in Supplementary Table S1 available at JXB online.
Real-time RT-PCR (qRT-PCR)
Total RNAs were extracted from leaves of 13-day-old seedlings using TRIzol reagents (Invitrogen), and reverse transcription was performed with 2 μg of total RNA using a kit (Fermentas, Vilnius, Lithuania). Real-time PCR was performed according to previous methods (Li et al., 2005). Primer sequences are listed in Supplementary Table S1 at JXB online.
In situ hybridization and GUS staining
In situ hybridizations were performed as previously described (Kim et al., 1998) using 13-day-old seedlings. Detection of miR396 by in situ hybridization was performed using the complementary locked nucleic acid (LNA)-modified DNA probe, 5′-AgTTcAAgAAaGCtGGaA-3′, where the lowercase letters represent LNAs. Other probe preparations and GUS staining were according to previous methods (Li et al., 2005). The sequences of primers used for probe preparation are listed in Supplementary Table S1 at JXB online.
Microscopy
Scanning electron microscopy (SEM) and preparation of thin-section specimens were according to previous methods (Chen et al., 2000). For differential interference contrast microscopy (DICM) observation, leaves were first fixed in FAA (formalin:acetic acid:70% ethanol, 1:1:8) overnight and then cleared in a chloral solution (chloral hydrate:glycerol:water, 8:2:1) (Tsuge et al., 1996). The determined unit area for cell counting was located in leaves taken at 50% above the base and half way between the midrib and the leaf margin. The unit area was first photographed and then the number of palisade cells within the area was counted.
Flow cytometric analysis
A 0.1 g aliquot of 8-day-old leaves was harvested from 13-day-old seedlings and freshly chopped with a razor blade in a Petri dish containing 2 ml of nuclear isolation and stain buffer (NIM-DAPI 10, Beckman Coulter, CA, USA). The chopped leaves were then filtered through a 40 μm mesh and the isolated nuclei were analysed with a Quanta™ SC flow cytometer (Beckman Coulter). Results from two biological replicates, each with duplicated samples, were analysed.
Results
miR396-targeted AtGRF transcription factors are required for establishment of leaf polarity
To explore how cell proliferation defects affect establishment of leaf polarity, the transgenic plants overexpressing MIR396a and MIR396b under the control of the cauliflower mosaic virus 35S promoter (35S:miR396a and 35S:miR396b, respectively) were first characterized. Compared with the wild type (Supplementary Fig. S1A at JXB online), transgenic 35S:miR396a (Supplementary Fig. S1B) and 35S:miR396b (Supplementary Fig. S1C) plants displayed narrow and slightly small rosette leaves, resembling atgif1/an3 mutants (Horiguchi et al., 2005) (Supplementary Fig. S1D). Furthermore, similarly to atgif1/an3 (Supplementary Fig. S1H), the size of the palisade mesophyll cells in 35S:miR396a (Supplementary Fig. S1F) and 35S:miR396b (Supplementary Fig. S1G) plants was clearly bigger than those of the wild-type (Supplementary Fig. S1E), while the cell numbers in those plants are significantly reduced (Supplementary Fig. S1I). qRT-PCR analysis revealed that the transcription levels of the nine AtGRF genes including seven predicated AtGRF gene targets in 35S:miR396a and 35S:miR396b plants were all decreased (Supplementary Fig. S1J), indicating that miR396 negatively regulates the cell proliferation in leaves by repressing the expression of AtGRF genes, consistent with reported results (Liu et al., 2009; Rodriguez et al., 2010).
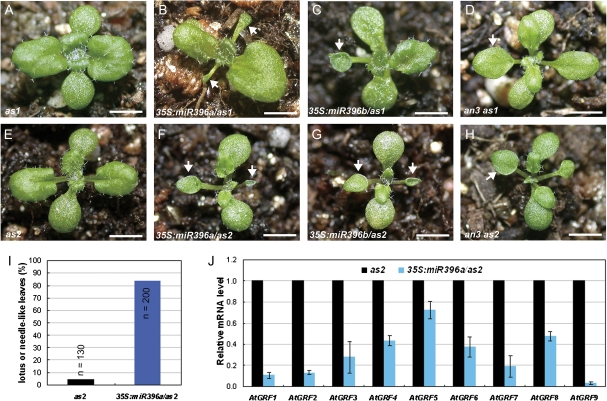
The roles of miR396-targeted AtGRFs in establishment of leaf polarity were then investigated by introducing the 35S:miR396a and 35S:miR396b constructs into the leaf polarity mutants as1-1 (Fig. 1A) and as2-1 (Fig. 1E), which exhibit only weak leaf adaxial–abaxial polarity defects (Byrne et al., 2000; Semiarti et al., 2001; Iwakawa et al., 2002; Kumaran et al., 2002; Xu et al., 2002, 2003; Lin et al., 2003). In contrast, the 35S:miR396/as1-1 (Fig. 1B, C) and 35S:miR396/as2-1 (Fig. 1F, G) transgenic plants produced stronger leaf polarity defects; the first two rosette leaves typically appeared lotus like and, in some extreme cases, needle like. In the F3 progeny of three independent homozygous lines, >80% of 35S:miR396a/as2 plants produced lotus-like or needle-like leaves in the first pair of true leaves, while <5% of as2 plants generated lotus-like leaves (Fig. 1I), indicating that miR396-AtGRFs and AS1/2 pathways synergistically regulate leaf adaxial–abaxial formation.
Fig. 1.
miR396-targeted AtGRF transcription factors are required for establishment of leaf polarity. (A–H) Morphological observations of the as1 (A), 35S:miR396a/as1 (B), 35S:miR396b/as1 (C), an3 as1 (D), as2 (E), 35S:miR396a/as2 (F), 35S:miR396b/as2 (G), and an3 as2 (H) plants. Arrowheads in B–D and F–H indicate the lotus- or needle-like leaves. (I) Lotus- or needle-like leaves were present more frequently in 35S:miR396a/as2 plants than in the as2 mutant. The first pair of rosette leaves was analysed for the presence of lotus- or needle-like leaves. n, numbers of plants analysed; (J) qRT-PCR to analyse the transcription levels of AtGRF gens in 14-day-old seedlings of as2 and 35S:miR396a/as2 plants. Bars=1 cm in A–H.
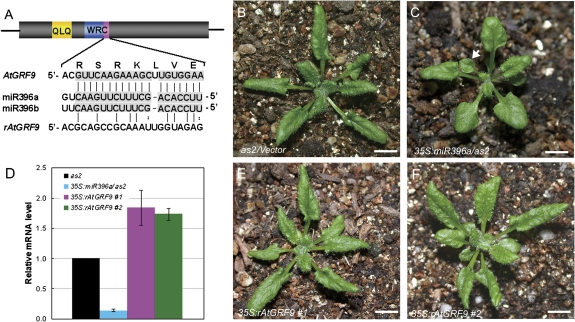
The transcription levels of AtGRF genes were analysed in 35S:miR396a/as2 transgenic plants by qRT-PCR. Similar to 35S:miR396/Col transgenic plants, the expression levels of AtGRF genes were decreased in 35S:miR396a/as2 (Fig. 1J), suggesting that the leaf polarity defects in 35S:miR396a/as2 plants were due to the decreased expression of AtGRF genes. To support these results, the double mutants an3 as1 and an3 as2 were also generated. The phenotype of the an3 as1 (Fig. 1D) and an3 as2 (Fig. 1H) double mutants largely resembled that of 35S:miR396/as1 and 35S:miR396/as2 plants, respectively. To confirm this further, the construct 35S:rAtGRF9, which carries the miR396-resistant version of AtGRF9 (rAtGRF9), was made and introduced into 35S:miR396a/as2 plants. The miR396 target sequence in AtGRF genes is located at the end of the region encoding the conserved WRC domain (Fig. 2A). Disruption of the miR396-binding sites in AtGRF9 could result in the accumulation of their stabilized transcripts in transgenic plants. By qRT-PCR, it was found that the AtGRF9 mRNA in the double transgenic plants was increased to levels higher than in the wild type, in contrast to the much lower accumulation in 35S:miR396a/as2 plants (Fig. 2D). Accordingly, it was observed that the leaf polarity defects (as indicated by the appearance of lotus-like leaves, Fig. 2C) of 35S:miR396/as2-1 could be rescued by the introduced rAtGRF9 gene (Fig. 2E, F) but not by an empty vector (Fig. 2B). Taken together, these results suggested that the AtGRF–AtGIF complexes are required for establishment of leaf polarity, and the reduction of AtGRF gene expression by transgenic miR396 overexpression resulted in the leaf polarity defects in 35S:miR396a/as2 plants.
Fig. 2.
Introduction of the miR396-resistant AtGRF9 gene into a 35S:miR396a/as2 plant partially rescued the leaf polarity defects. (A) Diagram of the miR396 target sites of the wild type and a modified version of AtGRF9. The conserved QLQ motif in AtGRFs is indicated in yellow, and the WRC motif is shown in blue, with the miR396 target region marked in purple. (B, C) Morphological observation of the 27-day-old seedlings of as2 (B) and 35S:miR396a/as2 (C). The arrowhead in C indicates the lotus-like leaf. (D) qRT-PCR analysis of the AtGRF9 transcript levels in as2, 35S:miR396a/as2, and 35S:miR396a/as2 expressing the miR396-resistant AtGRF9 gene. (E, F) Morphology of the 27-day-old seedlings of 35S:miR396a/as2 expressing the miR396-resistant AtGRF9 gene driven by the 35S promoter. #1, line1; #2, line2. Bars=1 cm in B, C, E, and F.
Enhanced defects of abaxial–adaxial polarity in the leaves of 35S:miR396a/as2 plants
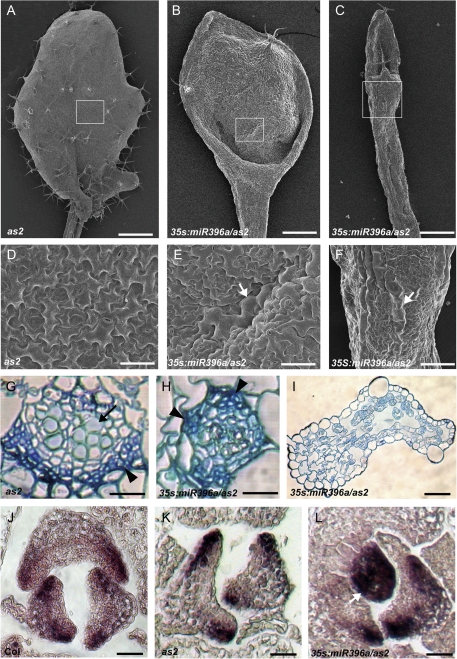
To better understand the roles of miR396 and AtGRF genes in leaf patterning, the leaf phenotype of 35S:miR396a/as2 transgenic plants was analysed in more detail by SEM. The adaxial epidermis of both wild-type and as2 leaves was characterized by an undulating surface comprising uniformly sized pavement cells (Fig. 3A, D), while smaller pavement cells interspersed with some long and narrow cells were noted in the abaxial epidermis (data not shown). The epidermal cell patterns of 35S:miR396 plants were normal (data not shown). However, the adaxial surface of the lotus-like leaves of 35S:miR396a/as2 transgenic plants comprised a mosaic of both adaxial and abaxial epidermal cells, characterized by the presence of abaxialized long and narrow cells in the adaxial surface (Fig. 3B, E). The extremely narrow and needle-like leaves of 35S:miR396a/as2 plants exhibited long and narrow abaxial epidermal cells (Fig. 3C, F), suggesting that this leaf is abaxialized.
Fig. 3.
Enhanced adaxial–abaxial defects of leaf polarity by transgenic miR396 overexpression in the as2 mutant. (A–C) Comparison of the first rosette leaf of as2 (A) with the lotus-like (B) and needle-like (C) leaves of 35S:miR396a/as2. (D–F) Epidermal cells on the adaxial surface of as2 (D), 35S:miR396a/as2 lotus-like (E) and needle-like (F) leaves. Arrowheads in E and F indicate the long and narrow abaxialized-featured epidermal cells. (G–I) Transverse sections through the blade–petiole junction region showed vascular patterns of leaves in as2 (G), 35S:miR396a/as2 lotus-like (H) and needle-like (I) leaves. Arrows and arrowheads in G and H show xylem and phloem, respectively. (J–L) In situ hybridization to analyse the FIL transcripts in Col (J), as2 (K), and 35S:miR396a/as2 (L) seedlings. The arrowhead in L indicates the leaf primordium uniformly expressing FIL. Bars=1 mm in A, 500 μm in B and C, 50 μm in D and G–I, and 100 μm in E, F, and J–L.
The vascular pattern of 35S:miR396a/as2 plants was further analysed by transverse sectioning through the blade–petiole conjugation region. In the wild-type or as2 leaves, vascular bundles in this region showed a pattern whereby xylem develops on the adaxial pole and phloem is located on the abaxial pole (Fig. 3G). In the 35S:miR396 transgenic plants, the vascular pattern in the expanded rosette leaves did not show obvious changes (data not shown). In contrast, the lotus-like leaves of 35S:miR396a/as2 plants exhibited a phloem-surrounding-xylem structure (Fig. 3H), while the needle-like leaves of 35S:miR396a/as2 plants exhibited no apparent vascular bundle but numerous intercellular spaces (Fig. 3I).
To obtain molecular evidence that miR396-targeted AtGRFs are required for establishment of leaf polarity, the expression pattern of a YABBY family gene, FIL/YAB1, was examined by in situ hybridization. FIL is usually expressed on the leaf abaxial domain of both the wild type (Fig. 3J) and the as2 mutant (Fig. 3K). In contrast, FIL was expressed throughout the entire primordium of the needle-like leaves of 35S:miR396a/as2 plants and seemed to accumulate to higher levels in the other leaves (Fig. 3L). These results together indicated that miR396-targeted AtGRFs are required for leaf patterning and genetically regulate the expression of leaf polarity-controlling genes.
miR396 negatively regulates cell proliferation by controlling entry into the mitotic cell cycle
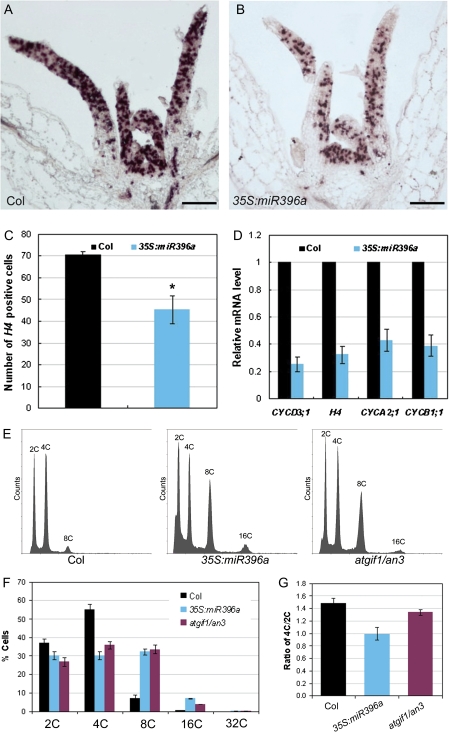
In Arabidopsis, the proliferating cells in lateral organ primordium usually divide in the mitotic cell cycle, which follows the common scheme of G1, S, G2, and M phases (Inze and De Veylder, 2006; Gutierrez, 2009), to multiplify and promote organ growth and development. To gain insights into how the roles of miR396 and AtGRFs in cell proliferation affect establishment of leaf polarity, the expression pattern of one of the cell cycle marker genes, histone H4, was analysed by in situ hybridization. Histone H4 is predominantly expressed in the proliferating cells in the DNA replication-related S phase of the cell cycle and thus might mark the cells undergoing mitotic cell division (Reichheld et al., 1995, 1998). Compared with the wild type (Fig. 4A), the number of leaf cells expressing histone H4 was dramatically reduced in the 35S:miR396a plants (Fig. 4B, C). In addition to histone H4, the expression of several other cell cycle-related marker genes was also analysed by qRT-PCR. Cyclin D3;1 (CycD3;1) is mainly expressed at proliferating cells in the G1 phase and drives the G1/S transition together with cyclin-dependent kinase A (CDKA) (Dewitte et al., 2003; Menges et al., 2006). CYCA2;1 is involved in S/G2 transition, and CYCB1;1 in G2/M transition (Doerner et al., 1996; Shaul et al., 1996). The expression of all the tested marker genes was reduced in 35S:miR396a plants with a 2- to 4-fold change (Fig. 4D), indicating that the defective cell proliferation in leaves of 35S:miR396 plants could be due to the reduced cell division activity. The expression of cell cycle-related genes was also examined in as2 and 35S:miR396a/as2 plants. Similarly, the number of cells expressing histone H4 was reduced (Supplementary Fig. S2B, C at JXB online), and the mRNA levels of cell cycle marker genes were reduced even more (5- to 15-fold change) in 35S:miR396a/as2 plants (Fig. S2D) as compared with those in the as2 mutant (Fig. S2A, D), indicating that the cell division activity mediated by miR396-targeted AtGRFs is important for establishment of leaf adaxial–abaxial polarity.
Fig. 4.
miR396 negatively regulates cell proliferation by controlling entry into the mitotic cell cycle. (A and B) In situ hybridization analysis of histone H4 expression in shoot apices of 13-day-old wild-type (A) and 35S:miR396a (B) seedlings. (C) The number of histone H4-positive cells in leaves of the 13-day-old wild-type and 35S:miR396a seedlings. The asterisk indicates a statistically significant difference by t-test (P <0.05). (D) Expression of cell cycle-related genes in 13-day-old wild-type and 35S:miR396a seedlings. (E–G) Ploidy level distribution analysis. The ∼8-day-old leaves from 13-day-old seedling of the wild type, 35S:miR396a, and atgif1/an3 were used to examine the cell nuclear ploidy levels (E). The percentages of cells with different nuclear ploidy levels (F) and the ratio of 4C/2C cells (G) were calculated. Bars=100 μm in A and B. (This figure is available in colour at JXB online.)
To dissect further the phase of the cell cycle affected by transgenic miR396 overexpression, a flow cytometric analysis was performed with nuclei from leaves of ∼8 d old to examine the nuclear ploidy level. The atgif1/an3 leaves were analysed as a control. The percentages of 8C and 16C cells in 35S:miR396 and atgif1/an3 plants were significantly higher than those in the wild-type plants, whereas the percentages of 2C and 4C cells were reduced accordingly (Fig. 4E, F). Because it has been proposed that at this time point leaf cells have the capacity to divide and 2C and 4C cells could represent the G1 and G2 cells, respectively (Takahashi et al., 2008), the ratio of 4C/2C cells in 8-day-old leaves was compared further. As shown in Fig. 4G, the ratio of 4C/2C cells was slightly decreased in 35S:miR396 and atgif1/an3 leaves compared with those of the wild type.
It is known that in response to a variety of physiological signals or developmental cues, proliferating cells can exit from the mitotic cell cycle to an alternative cycle, the endoreplication cycle (endocycle), for expansion/differentiation, whereby cells replicate the full genome to increase the ploidy level (from 2C to 2nC) but fail to undergo neclear division to produce progeny cells (De Veylder et al., 2007; Gutierrez, 2009). The switch to the endocycle also requires that, at least, CYCD3;1 expression is turned off (Dewitte and Murray, 2003). Because both the number of cells undergoing mitotic cell division and the cell cycle marker genes including CYCD3;1 were reduced, but the percentage of leaf cells with higher ploidy levels was increased in 35S:miR396 plants, these results indicate that miR396 appears to play important roles in controlling the entry into the mitotic cell cycle.
miR396 is mainly expressed in leaf cells arrested for cell division
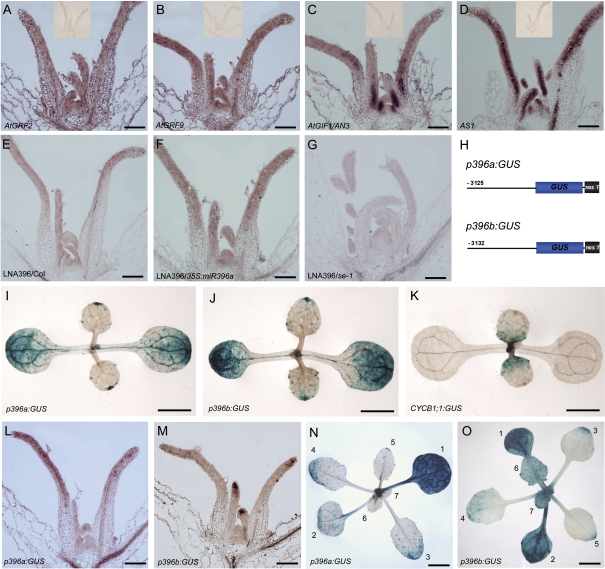
To analyse the temporal–spatial regulation of cell division by miR396–AtGRF modules, the expression patterns of AtGRF2, AtGRF9, and AtGIF1/AN3 were first examined by in situ hybridization. AtGRF2 and AtGRF9 were expressed in the SAM and young leaf primordium (Fig. 5A, B). Along the adaxial–abaxial axis, they were expressed uniformly throughout the young leaf primordia, similar to the pattern of AS1 (Fig. 5D), which is believed to be expressed in the whole primordium (Byrne et al., 2000). Interestingly, the transcripts of AtGIF1/AN3 appeared to be highly accumulated in the proximal part of leaf primordia (Fig. 5C), in which cells are rapidly proliferating, consistent with the previous results obtained by promoter–GUS assay (Horiguchi et al., 2005).
Fig. 5.
miR396 is mainly expressed in the cells arrested for cell division. (A–D) In situ hybridization analysis of AtGRF2, AtGRF9, AN3, and AS1 expression in shoot apices. The insets show no signals detected by the sense probes of the corresponding genes. (E–G) The antisense LNA probe detected miR396 in the shoot apex of wild-type (E), 35S:miR396a (F), and se-1 (G) seedlings. (H) Diagram of the structures of p396a:GUS and p396b:GUS constructs. (I–K) GUS staining to analyse the GUS distribution patterns in the 9-day-old p396a:GUS (I), p396b:GUS (J), and CYCB1;1:GUS (K) seedlings. (L and M) In situ hybridization to analyse the GUS distribution pattern in the p396a:GUS (L) and p396b:GUS (M) seedlings, respectively. (N and O) GUS staining to analyse the GUS distribution pattern in the 24-day-old seedlings of p396a:GUS (N) and p396b:GUS (O). The numbers 1–7 indicate the number of leaves. Bars=100 μm in A–G, L, and M, 0.5 cm in I–K, N, and O.
The expression pattern of miR396 was further investigated using an antisense LNA-modified DNA probe, which could detect the mature products of both MIR396a and MIR396b. As shown in Fig. 5E, the LNA signals were detected in the SAM and leaf primordia, and appeared to have a higher level in the distal part of primoridia, but no apparent polar distribution along the adaxial–abaxial axis was observed. The specificity of the hybridization signal was confirmed using the seedlings of 35S:miR396a transgenic plants (Fig. 5F; high miR396 level) and se-1 (Fig. 5G; low miR396 level), in which the LNA probe detected strong signals or almost no signals, respectively. Due to the relatively low accumulation of miR396 revealed by LNA in situ hybridization, the functional domain of the promoters of MIR396A and MIR396B in the leaves was therefore characterized to better understand the expression pattern of miR396. The ∼3.2 kb 5' upstream regions of MIR396A and MIR396B (counting from the first nucleotide of the mature miR396 sequences) were cloned to represent their respectiveputative promoters. The fragments were fused with the GUS reporter gene to generate the p396a:GUS and p396b:GUS constructs (Fig. 5H), which were then introduced into wild-type plants. By examining the GUS staining signals, it was found that dynamic but similar patterns for both constructs were detected from the cotyledon, shoot apex, and rosette leaves (Fig. 5I, J, L–O). In situ hybridization with a GUS-specific probe revealed that the GUS signals were distributed in both the adaxial and abaxial domains (Fig. 5L, M), suggesting that miR396 may not have a polar distribution along the adaxial–abaxial axis in leaves. In contrast, the GUS staining signals appeared to be dynamically distributed along the proximal–distal axis of leaves and varied during leaf development. In the cotyledon, the GUS signals were very strong (Fig. 5I–J), in contrast to almost no signals of CYCB1;1:GUS (Colon-Carmona et al., 1999) in the cotyledon. In young leaves, the GUS signals were strong in the tip and distal parts, opposite to the expression pattern of CYCB1;1:GUS. However, when the leaves gradually become mature, the GUS signals progressively accumulate from the distal to the proximal part of developing leaves, with strong activity in the whole mature leaves (Fig. 5N, O). It should be pointed out that the expression patterns of miR396 revealed by promoter–GUS assay do not completely coincide with those by LNA in situ hybridization analysis. This may be due to the non-cell-autonomous movement of mature miR396 to adjacent cells from the original biosynthetic cells. Because the leaf primordia emerging from the SAM first undergo proliferative cell division, and subsequently the progression of cell division arrest in leaves moves from the distal leaf tip to the proximal leaf part in Arabidopsis (Donnelly et al., 1999), the results together indicate that miR396 is mainly expressed in the leaf cells arrested for the cell cycle in the distal part of either developing leaves or mature leaves.
Effects of miR396 misexpression on the establishment of leaf adaxial–abaxial polarity
To investigate how the temporal–spatial regulation of cell division by miR396 affects the establishment of leaf polarity, the effects of miR396 expression driven by tissue-specific promoters in different shoot apex domains of the as2 mutant on establishment of leaf polarity was analysed. The promoter of SHOOT MERISTEMLESS (STM) was used for expression in the SAM (J. Y. Kim et al., 2003), those of AS1 (Byrne et al., 2000) and AN3 (Horiguchi et al., 2005) (Fig. 5C) for young leaf primordia, that of REV for the adaxial domain of young leaf primordia and the central domain of SAM (Emery et al., 2003; Otsuga et al., 2001; Prigge et al., 2005), and that of FIL for the abaxial domain of young leaf primordia (Siegfried et al., 1999; Watanabe and Okada, 2003). All the constructs were introduced into the as2 mutant background and >20 independent transgenic plants were analysed for each construct. As shown in Fig. 6, it was found that the tissue-specific reduction of cell division in leaf primordia by pAS1:miR396a (Fig. 6B) and pAN3:miR396a expression (Fig. 6C) in the as2 mutant (Fig. 6A) showed enhanced leaf polarity defects in transgenic plants, as indicated by the presence of lotus- or needle-like leaves, indicating that cell division in the rapidly proliferating primordia is more important for establishment of leaf polarity. Typically, the leaf polarity defects in the pAN3:miR396a/as2 transgenic plants were stronger than those of pAS1:miR396a/as2 plants, which may reflect the consequence of their different expression domain in leaf primordia. Transgenic miR396 expression driven by the promoter of REV, FIL, and STM in as2 mutants failed to enhance the polarity defects of as2, except for the slightly small and dwarf plant architecture (Fig. 6D–F), suggesting that either the activities of these three promoters were not strong enough or reduction of cell division in the specific adaxial/abaxial domain of young leaf primordia and the SAM had minor effects on the establishment of leaf polarity.
Fig. 6.
Effects on establishment of leaf polarity of miR396 misexpression in the as2 mutant. (A–F) Morphology of the 16-day-old seedlings of as2 (A) and as2 expressing pAS1:miR396a (B), pAN3:miR396a (C), pREV:miR396a (D), pFIL:miR396a (E), and pSTM:miR396a (F). Bars=1 cm in A–F. (This figure is available in colour at JXB online.)
Discussion
Previously, computational prediction implied that at least seven AtGRF genes contain the target sites of miR396 (Jones-Rhoades and Bartel, 2004), and recent transgenic experiments (Liu et al., 2009; Rodriguez et al., 2010) and the characterization of 35S:miR396a and 35S:miR396b transgenic plants in this study (Supplementary Fig. S1 at JXB online) all demonstrated that miR396 negatively regulates cell proliferation by repressing AtGRF gene expression in Arabidopsis. To date, miR396 and its cognate GRF targets have been found to be present in >30 plant species from gymnosperms to monocots and eudicots (Axtell and Bartel, 2005; Sunkar and Jagadeeswaran, 2008), indicating that miR396–GRF regulatory modules have an ancient origin and may be important for flowering plant evolution and development.
Cell division, expansion, and differentiation are pivotal processes necessary for organogenesis. A major developmental question is how cells acquire specific fates in conjunction with cell division and integration into developmental processes. During leaf morphogenesis, cell division and differentiation along the adaxial–abaxial axis result in the two leaf faces being distinct in both structure and biological function (McConnell et al., 2001). In this study, the cell division activity mediated by miR396-targeted AtGRFs was found to be required for leaf adaxial–abaxial polarity and trichome formation, revealing tight coordination of cell division and differentiation during leaf morphogenesis. It was observed that transgenic miR396 overexpression in as1/2 mutants resulted in plants with enhanced leaf adaxial–abaxial polarity, which was correlated with the decreased cell division activity accompanying the decreased expression of AtGRF genes (Supplementary Fig. S2 at JXB online). The recent characterization of ae7 and the previously reported 26S proteasome mutant ae3-1 and ribosome mutant ae5-1 showed that these three mutations affected both leaf adaxial–abaxial polarity and cell proliferation, and it was therefore proposed that normal cell proliferation may be essential for the establishment of leaf polarity (Yuan et al., 2010. Studies on the roles of miR396–AtGRF modules in the establishment of leaf polarity provide further evidence and extend this proposal, suggesting that normal cell division is coupled with polarized cell proliferation and is important for the establishment of leaf polarity.
It is known that the switch from cell proliferation to differentiation often coincides with the switch from the mitotic cell cycle to the endocycle during leaf development (Boudolf et al., 2004). Although not always, the endoreplication is frequently correlated with an increase in cell size. In leaves, the existence of the compensation syndrome, in which a decrease in cell number triggers an increase in mature cell size, also suggests the tight coordination of cell proliferation and expansion/differentiation (Tsukaya, 2006, 2003). Similarly to atgif/an3, the reduction of cell numbers in 35S:miR396 leaves was accompanied by the enlargement of cell size (Supplementary Fig. S1 at JXB online) and an increased percentage of 8C and 16C cells, suggesting that miR396 has functions in promoting the entry of cells into the endocycle for expansion/differentiation. Cell differentiation in leaf primordia along the adaxial–abaxial axis (McConnell et al., 2001) usually occurs in a sequential fashion for an ultimately functional leaf, and the abaxial spongy cells stop proliferating and start to differentiate earlier than the tightly arranged adaxial palisade cells (Yuan et al., 2010). Therefore, it is possible that cell proliferation defects caused by transgenic miR396 overexpression in the as1/2 mutant background may result in cell divison stopping earlier, and thereby early differentiation of abaxial cells and insufficient cells for differentiation into the leaf adaxial domain, which in turn might lead to the premature establishment of abaxial cell fate accompanied by ectopic expression of abaxial-specific genes such as FIL in the leaf primordia, because the adaxial and abaxial domains specified by the polarity-related genes are mutually antagonized.
The negative roles of miR396 in cell proliferation appear to be due to the reduced cell division activity. First, the numbers of dividing cells in the leaf primordia of 35S:miR396 were decreased, as revealed by the decreased number of cells expressing histone H4. Secondly, the expression of cell cycle-related genes including CYCD3;1, histone H4, CYCA2;1, and CYCB1;1 was found to be reduced in 35S:miR396 plants. Finally, the percentages of 8C and 16C cells in 35S:miR396a and atgif/an3 leaves were significantly increased. All these results indicate that miR396 negatively regulates cell proliferation by controlling entry into the mitotic cell cycle. This is also consistent with the observation that the leaves of 35S:miR396 plants displayed a reduced cell number but enlarged cell size (Supplementary Fig. S1 at JXB online), because the cells unable to enter into the mitotic cell cycle could undergo endoreplication to increase the ploidy level and cell size in leaves. Thus, miR396 may have functions to prevent mitotic cell division and thereby promote cell differentiation during leaf development.
Analysis of the expression pattern of miR396 further supported the roles of miR396 in controlling entry into the mitotic cell cycle. By in situ hybridization and promoter–GUS assays, miR396 was found to accumulate mainly in the distal part of developing leaves, progressively from the distal to the proximal part, whereas it appears to accumulate throughout almost the whole cotyledon and mature leaves in cells which have stopped dividing, coinciding with its roles in controlling entry into the mitotic cell cycle. In addition, the putative promoters of both MIR396A and MIR396B exhibited high activities in the leaf trichomes (Supplementary Fig. S3 at JXB online). Leaf trichomes are large single cells originating from the epidermis, in which trichomes are the first epidermal cells that begin to differentiate from the developing leaf primordia (Hulskamp et al., 1994; Larkin et al., 1996). After trichome fate commitment the cells stop dividing and switch to the endocycle, resulting in a DNA content of 32C on average in the mature trichome (Hulskamp et al., 1994; Hulskamp, 2004; Schellmann and Hulskamp, 2005). Thus, the mature miR396 is presumably biosynthesized in the single trichome cells to inhibit their further division, consistent with the role of miR396 in controlling entry into the mitotic cell cycle.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. miR396 negatively regulates cell proliferation by repressing the expression of AtGRF genes in the leaves.
Figure S2. In situ hybridization and qRT-PCR analyses of cell cycle-related genes in as2 and 35S:miR396a/as2 transgenic plants.
Figure S3. Expression of p396a:GUS and p396b:GUS in leaf trichomes.
Table S1. The sequences of the primers used in this study.
Supplementary Material
Acknowledgments
We thank the ABRC for the seeds of SALK_150407, and X. Gao for SEM. This research was supported by grants from the Chief Scientist Program of Shanghai Institutes for Biological Sciences, the Chinese National Scientific Foundation (30630041/30721061/30800601), and the Chinese Academy of Sciences (KSCX2-YW-N-016/057, 2008KIP205).
References
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Bartel DP. Antiquity of microRNAs and their targets in land plants. The Plant Cell. 2005;17:1658–1673. doi: 10.1105/tpc.105.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Vlieghe K, Beemster GT, Magyar Z, Torres Acosta JA, Maes S, Van Der Schueren E, Inze D, De Veylder L. The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. The Plant Cell. 2004;16:2683–2692. doi: 10.1105/tpc.104.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Eshed Y, Baum SF. Establishment of polarity in angiosperm lateral organs. Trends in Genetics. 2002;18:134–141. doi: 10.1016/s0168-9525(01)02601-4. [DOI] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature. 2000;408:967–971. doi: 10.1038/35050091. [DOI] [PubMed] [Google Scholar]
- Chen C, Wang S, Huang H. LEUNIG has multiple functions in gynoecium development in Arabidopsis. Genesis. 2000;26:42–54. doi: 10.1002/(sici)1526-968x(200001)26:1<42::aid-gene7>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Colon-Carmona A, You R, Haimovitch-Gal T, Doerner P. Spatio-temporal analysis of mitotic activity with a labile cyclin–GUS fusion protein. The Plant Journal. 1999;20:503–508. doi: 10.1046/j.1365-313x.1999.00620.x. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inze D. The ins and outs of the plant cell cycle. Nature Reviews Molecular Cell Biology. 2007;8:655–665. doi: 10.1038/nrm2227. [DOI] [PubMed] [Google Scholar]
- Dewitte W, Murray JA. The plant cell cycle. Annual Review of Plant Biology. 2003;54:235–264. doi: 10.1146/annurev.arplant.54.031902.134836. [DOI] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JM, Jacqmard A, Kilby NJ, Murray JA. Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. The Plant Cell. 2003;15:79–92. doi: 10.1105/tpc.004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerner P, Jorgensen JE, You R, Steppuhn J, Lamb C. Control of root growth and development by cyclin expression. Nature. 1996;380:520–523. doi: 10.1038/380520a0. [DOI] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Developmental Biology. 1999;215:407–419. doi: 10.1006/dbio.1999.9443. [DOI] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Current Biology. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Izhaki A, Baum SF, Floyd SK, Bowman JL. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development. 2004;131:2997–3006. doi: 10.1242/dev.01186. [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Current Biology. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- Fleming AJ. The co-ordination of cell division, differentiation and morphogenesis in the shoot apical meristem: a perspective. Journal of Experimental Botany. 2006;57:25–32. doi: 10.1093/jxb/eri268. [DOI] [PubMed] [Google Scholar]
- Folkers U, Kirik V, Schobinger U, et al. The cell morphogenesis gene ANGUSTIFOLIA encodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO Journal. 2002;21:1280–1288. doi: 10.1093/emboj/21.6.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C. Coupling cell proliferation and development in plants. Nature Cell Biology. 2005;7:535–541. doi: 10.1038/ncb0605-535. [DOI] [PubMed] [Google Scholar]
- Gutierrez C. The Arabidopsis book. Rockville, MD: American Society of Plant Biologists; 2009. The Arabidopsiscell division cycle. http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. The Plant Journal. 2005;43:68–78. doi: 10.1111/j.1365-313X.2005.02429.x. [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Tsukaya H. Characterization of a narrow leaf mutant of Arabidopsis, angustifolia3, suggests the presence of a mechanism controlling cell division orientation in the leaf-width direction. Plant and Cell Physiology. 2003;44:S107–S107. [Google Scholar]
- Huang W, Pi L, Liang W, Xu B, Wang H, Cai R, Huang H. The proteolytic function of the Arabidopsis 26S proteasome is required for specifying leaf adaxial identity. The Plant Cell. 2006;18:2479–2492. doi: 10.1105/tpc.106.045013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson A. Development of symmetry in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 2000;51:349–370. doi: 10.1146/annurev.arplant.51.1.349. [DOI] [PubMed] [Google Scholar]
- Hulskamp M, Misera S, Jurgens G. Genetic dissection of trichome cell-development in Arabidopsis. Cell. 1994;76:555–566. doi: 10.1016/0092-8674(94)90118-x. [DOI] [PubMed] [Google Scholar]
- Hulskamp M. Plant trichomes: a model for cell differentiation. Nature Reviews Molecular Cell Biology. 2004;5:471–480. doi: 10.1038/nrm1404. [DOI] [PubMed] [Google Scholar]
- Inze D, De Veylder L. Cell cycle regulation in plant development. Annual Review of Genetics. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- Iwakawa H, Ueno Y, Semiarti E, et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant and Cell Physiology. 2002;43:467–478. doi: 10.1093/pcp/pcf077. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Kim GT, Shoda K, Tsuge T, Cho KH, Uchimiya H, Yokoyama R, Nishitani K, Tsukaya H. The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO Journal. 2002;21:1267–1279. doi: 10.1093/emboj/21.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Uchimiya H. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes and Development. 1998;12:2381–2391. doi: 10.1101/gad.12.15.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Choi D, Kende H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. The Plant Journal. 2003;36:94–104. doi: 10.1046/j.1365-313x.2003.01862.x. [DOI] [PubMed] [Google Scholar]
- Kim JH, Kende H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2004;101:13374–13379. doi: 10.1073/pnas.0405450101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Yuan Z, Jackson D. Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development. 2003;130:4351–4362. doi: 10.1242/dev.00618. [DOI] [PubMed] [Google Scholar]
- Kumaran MK, Bowman JL, Sundaresan V. YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. The Plant Cell. 2002;14:2761–2770. doi: 10.1105/tpc.004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin JC, Young N, Prigge M, Marks MD. The control of trichome spacing and number in Arabidopsis. Development. 1996;122:997–1005. doi: 10.1242/dev.122.3.997. [DOI] [PubMed] [Google Scholar]
- Lee BH, Ko JH, Lee S, Lee Y, Pak JH, Kim JH. The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiology. 2009;151:655–668. doi: 10.1104/pp.109.141838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xu L, Wang H, Yuan Z, Cao X, Yang Z, Zhang D, Xu Y, Huang H. The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. The Plant Cell. 2005;17:2157–2171. doi: 10.1105/tpc.105.033449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Shuai B, Springer PS. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial–abaxial patterning. The Plant Cell. 2003;15:2241–2252. doi: 10.1105/tpc.014969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Song Y, Chen Z, Yu D. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiologia Plantarum. 2009;136:223–236. doi: 10.1111/j.1399-3054.2009.01229.x. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- Menges M, Samland AK, Planchais S, Murray JA. The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. The Plant Cell. 2006;18:893–906. doi: 10.1105/tpc.105.039636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita NN, Moore S, Horiguchi G, Kubo M, Demura T, Fukuda H, Goodrich J, Tsukaya H. Overexpression of a novel small peptide ROTUNDIFOLIA4 decreases cell proliferation and alters leaf shape in Arabidopsis thaliana. The Plant Journal. 2004;38:699–713. doi: 10.1111/j.1365-313X.2004.02078.x. [DOI] [PubMed] [Google Scholar]
- Nieuwland J, Scofield S, Murray JAH. Control of division and differentiation of plant stem cells and their derivatives. Seminars in Cell and Developmental Biology. 2009;20:1134–1142. doi: 10.1016/j.semcdb.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. REVOLUTA regulates meristem initiation at lateral positions. The Plant Journal. 2001;25:223–236. doi: 10.1046/j.1365-313x.2001.00959.x. [DOI] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. The Plant Cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon V, Etchells JP, Rossignol P, Collier SA, Arroyo JM, Martienssen RA, Byrne ME. Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development. 2008;135:1315–1324. doi: 10.1242/dev.016469. [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. The Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Parra E, Desvoyes B, Gutierrez C. Balance between cell division and differentiation during plant development. International Journal of Developmental Biology. 2005;49:467–477. doi: 10.1387/ijdb.052001er. [DOI] [PubMed] [Google Scholar]
- Reichheld JP, Gigot C, Chaubet-Gigot N. Multilevel regulation of histone gene expression during the cell cycle in tobacco cells. Nucleic Acids Research. 1998;26:3255–3262. doi: 10.1093/nar/26.13.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld JP, Sonobe S, Clement B, Chaubet N, Gigot C. Cell cycle-regulated histone gene expression in synchronized plant cells. The Plant Journal. 1995;7:245–252. [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development. 2010;137:103–112. doi: 10.1242/dev.043067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S, Hulskamp M. Epidermal differentiation: trichomes in Arabidopsis as a model system. International Journal of Developmental Biology. 2005;49:579–584. doi: 10.1387/ijdb.051983ss. [DOI] [PubMed] [Google Scholar]
- Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development. 2001;128:1771–1783. doi: 10.1242/dev.128.10.1771. [DOI] [PubMed] [Google Scholar]
- Shaul O, Mironov V, Burssens S, Van Montagu M, Inze D. Two Arabidopsis cyclin promoters mediate distinctive transcriptional oscillation in synchronized tobacco BY-2 cells. Proceedings of the National Academy of Sciences, USA. 1996;93:4868–4872. doi: 10.1073/pnas.93.10.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development. 1999;126:4117–4128. doi: 10.1242/dev.126.18.4117. [DOI] [PubMed] [Google Scholar]
- Sinha N. Leaf development in angiosperms. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:419–446. doi: 10.1146/annurev.arplant.50.1.419. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Jagadeeswaran G. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biology. 2008;8:37. doi: 10.1186/1471-2229-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussex IM. Experiments on the cause of dorsoventrality in leaves. Nature. 1954;174:351–352. [Google Scholar]
- Takahashi N, Lammens T, Boudolf V, Maes S, Yoshizumi T, De Jaeger G, Witters E, Inze D, De Veylder L. The DNA replication checkpoint aids survival of plants deficient in the novel replisome factor ETG1. EMBO Journal. 2008;27:1840–1851. doi: 10.1038/emboj.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes and Development. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge T, Tsukaya H, Uchimiya H. Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development. 1996;122:1589–1600. doi: 10.1242/dev.122.5.1589. [DOI] [PubMed] [Google Scholar]
- Tsukaya H. Organ shape and size: a lesson from studies of leaf morphogenesis. Current Opinion in Plant Biology. 2003;6:57–62. doi: 10.1016/s1369526602000055. [DOI] [PubMed] [Google Scholar]
- Tsukaya H. Mechanism of leaf-shape determination. Annual Review of Plant Biology. 2006;57:477–496. doi: 10.1146/annurev.arplant.57.032905.105320. [DOI] [PubMed] [Google Scholar]
- Waites R, Hudson A. phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus. Development. 1995;121:2143–2154. [Google Scholar]
- Watanabe K, Okada K. Two discrete cis elements control the abaxial side-specific expression of the FILAMENTOUS FLOWER gene in Arabidopsis. The Plant Cell. 2003;15:2592–2602. doi: 10.1105/tpc.015214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Lin WC, Huang T, Poethig RS, Springer PS, Kerstetter RA. KANADI1 regulates adaxial–abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proceedings of the National Academy of Sciences, USA. 2008;105:16392–16397. doi: 10.1073/pnas.0803997105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Xu Y, Dong A, Sun Y, Pi L, Xu Y, Huang H. Novel as1 and as2 defects in leaf adaxial–abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development. 2003;130:4097–4107. doi: 10.1242/dev.00622. [DOI] [PubMed] [Google Scholar]
- Xu Y, Sun Y, Liang W, Huang H. The Arabidopsis AS2 gene encoding a predicted leucine-zipper protein is required for the leaf polarity formation. Acta Botanica Sinica. 2002;44:1194–1202. [Google Scholar]
- Yao Y, Ling Q, Wang H, Huang H. Ribosomal proteins promote leaf adaxial identity. Development. 2008;135:1325–1334. doi: 10.1242/dev.017913. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Luo D, Li G, Yao X, Wang H, Zeng M, Huang H, Cui X. The Plant Journal. 2010. Characterization of the AE7 gene in Arabidopsis suggests that normal cell proliferation is essential for leaf polarity establishment. (in press) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.