Abstract
In maize, water stress at flowering causes loss of kernel set and productivity. While changes in the levels of sugars and abscisic acid (ABA) are thought to play a role in this stress response, the mechanistic basis and genes involved are not known. A candidate gene approach was used with association mapping to identify loci involved in accumulation of carbohydrates and ABA metabolites during stress. A panel of single nucleotide polymorphisms (SNPs) in genes from these metabolic pathways and in genes for reproductive development and stress response was used to genotype 350 tropical and subtropical maize inbred lines that were well watered or water stressed at flowering. Pre-pollination ears, silks, and leaves were analysed for sugars, starch, proline, ABA, ABA-glucose ester, and phaseic acid. ABA and sugar levels in silks and ears were negatively correlated with their growth. Association mapping with 1229 SNPs in 540 candidate genes identified an SNP in the maize homologue of the Arabidopsis MADS-box gene, PISTILLATA, which was significantly associated with phaseic acid in ears of well-watered plants, and an SNP in pyruvate dehydrogenase kinase, a key regulator of carbon flux into respiration, that was associated with silk sugar concentration. An SNP in an aldehyde oxidase gene was significantly associated with ABA levels in silks of water-stressed plants. Given the short range over which decay of linkage disequilibrium occurs in maize, the results indicate that allelic variation in these genes affects ABA and carbohydrate metabolism in floral tissues during drought.
Keywords: ASI, abscisic acid, association mapping, drought, flower set, kernel set
Introduction
Drought causes serious losses in crop productivity, and future climate change is predicted to exacerbate its frequency and severity due to altered rainfall patterns and higher temperatures. In maize (Zea mays L.), development of the female inflorescence and its floral parts is vulnerable to delay or arrest by water deficit and other abiotic stresses, leading to substantial losses in grain production (reviewed in Boyer and Westgate, 2004). Studies which have compared the response of maize to drought at various stages of development have identified drought at flowering and early kernel development as the most damaging to grain yield (Grant et al., 1989). Similar vulnerabilities to stress at flowering and early seed formation are found in many other species, including rice (Oryza sativa L.) (Kato, 2008), wheat (Triticum aestivum L.) (Ghiglione et al., 2008), barley (Hordeum vulgare L.) (Arisnabarreta and Miralles, 2006), soybean (Glycine max L.) (Ball et al., 2000), and chickpea (Cicer arietinum L.) (Leport et al., 2006). When populations of diverse maize genotypes are screened for their response to drought at flowering, the stress affects development of male floral organs to a lesser extent than female floral organs, and the delay in silking (growth of the elongated stigma) creates a substantial anthesis–silking interval (ASI) in affected genotypes (Bolanos and Edmeades, 1996). In segregating germplasm, the ASI often has a strong negative correlation with yield and has been used successfully as a selection criterion in tropical maize breeding programmes (Barker et al., 2005; Monneveux et al., 2006). Selection for tolerance to water stress (WS) at flowering has revealed that genetic differences in ability of the female floral parts (the ear and silk) to maintain growth during stress episodes is associated with improved yield. Grain yield under drought is correlated with improved partitioning of growth toward the ear and silks relative to growth of tassels and other plant organs within a plant (Bolanos et al., 1992; Monneveux et al., 2006). However, the mechanistic basis and genes involved in the ASI trait are not known.
Several lines of evidence have suggested that the maintenance of silk and ear development during water deficit, and achievement of high seed set, relates to maintenance of sugar supply to these organs (Zinselmeier et al., 1999). Consistent with carbohydrate transport into floral organs playing an important role, water deficit has been shown to delay expression of invertases in floral and kernel tissues, where they are thought to enhance carbohydrate transport (Zinselmeier et al., 1995). Water deficit decreases glucose levels in pedicels of ovaries (McLaughlin and Boyer, 2004a, b; Setter and Parra, 2010), and it decreases Ivr2, a soluble invertase, in floral parts at the pre- and post-pollination phases (Andersen et al., 2002; Qin et al., 2004). In leaves, water deficit increases Ivr2 expression, which, by hydrolysing sucrose into its two hexose moieties, can enhance the contribution sugars make to leaf osmotic potential (Kim et al., 2000). WS also alters expression of some starch pathway enzymes in floral tissues (Zinselmeier et al., 2002) and depletes starch reserves in floral parts (Zinselmeier et al., 1999), suggesting that carbohydrate deprivation plays a role in initiating flower and kernel abortion.
The stress hormone abscisic acid (ABA) may also be involved in limiting kernel set during water deficit. WS increases the ABA concentration in reproductive tissues, which correlates with decreased kernel set (reviewed in Setter 2006). Exogenous application of ABA at the early phase of kernel development decreases kernel growth and cell division at the early phase of development when kernel set/abortion decisions are made. There is also evidence that signalling interactions between sugars and ABA may play a role in regulating development in kernels during stress, as has been found in other plant systems (Dekkers et al., 2008).
To date, mutant screens have not identified genes associated with maize kernel set in water deficit conditions. Several mutants that affect inflorescence development have been found (reviewed in Vollbrecht and Schmidt, 2009). These include ramosa3, which results in an abnormal ear with lengthened and indeterminate floral meristems. This gene encodes trehalose-6-phosphate phosphatase (Satoh-Nagasawa et al., 2006), an enzyme that has been associated with abiotic stress response and with signalling of carbohydrate status (Y. Zhang et al., 2009). Genes have also been cloned for several viviparous mutants, which encode enzymes in the ABA synthesis pathway or are involved in ABA signalling (Cao et al., 2007; Suzuki et al., 2008). However, while these mutants represent intriguing candidates as potential determinants of stress responses of floral and kernel organs, they have extreme phenotypic effects. Our interest with respect to genetic improvement of crops is in allelic variation that spans a less extreme range, because quantitative traits such as yield under drought are unlikely to be optimal if an important gene function is severely altered or entirely knocked out. Thus, to identify genes with allelic variation relevant to crop improvement, it is valuable to investigate crop breeding stock.
To identify regions of the maize genome that affect morphological, metabolic, and enzymatic traits related to drought response, quantitative trait locus (QTL) studies have been performed within populations constructed of crosses between parents representing two or more contrasting lines. Ribaut et al. (2004) identified several QTLs for the ASI and associated morphology in a biparental recombinant inbred line (RIL) population, and, using the same population, Welcker et al. (2007) found that some of these QTLs co-localized with those for leaf growth traits in water deficit. There has also been progress in identifying QTLs for carbohydrate and ABA accumulation during stress. Pelleschi et al. (2006) used a biparental RIL population and a 9 d water deficit at flowering to identify several QTLs for leaf carbohydrate contents, enzyme activities for sugar and starch pathways, and ABA; Capelle et al. (2010) identified QTLs for kernel moisture and ABA content in embryos and endosperms at late embryogenesis and kernel desiccation; and Tuberosa and Salvi (2007) identified QTLs for ABA accumulation in leaves that relate to root morphology in response to drought. However, QTL studies with biparental populations only sample a small proportion of the total allelic diversity expected to be present in diverse germplasm. Furthermore, to locate the causal gene underpinning a significant QTL requires a lengthy and involved process of fine mapping. In the current study, the approach was to use association mapping of candidate genes in diverse germplasm (Yu and Buckler, 2006; Myles et al., 2009). Association mapping in a population of diverse lines allows sampling of greater genetic diversity. It also provides higher mapping resolution because more recombination events have occurred during historical diversification than during production of a biparental mapping population. A diverse panel of 350 maize genotypes was created from the collection of breeding stocks for tropical and subtropical maize at the International Maize and Wheat Improvement Center (CIMMYT), which is expected to capture substantial molecular and functional diversity.
Given the evidence that drought response of maize ear and silk development involves altered fluxes of carbohydrate and ABA pathways, these metabolic pathways were targeted for investigation. The candidate gene approach was used to identify single nucleotide polymorphisms (SNPs) in genes from metabolic pathways and in regulatory systems that control reproductive development and drought tolerance. These SNPs were used to genotype the association mapping panel, which was also phenotyped for traits involving the accumulation of carbohydrate and ABA metabolites. The candidate gene approach targets genes with known functions related to the traits of interest to increase the likelihood of finding meaningful trait associations. The association mapping panel contains lines from breeding programmes for tropical and subtropical maize, so the allelic variation in it is more likely to be useful to crop improvement programmes. The objectives of this study were to use association mapping of a diverse panel of maize inbred lines and candidate genes from putative drought-related metabolic pathways to identify genes which significantly affect carbohydrate and ABA accumulation and have roles in stress tolerance.
Materials and methods
Plant material and growth conditions
Maize inbred breeding lines representing a wide diversity of tropical and subtropical sources were chosen for the association mapping of genes affecting metabolite traits (Supplementary Table S1 available at JXB online). Lines were selected from inbreds released by CIMMYT (CMLs, http://apps.cimmyt.org/english/wps/obtain_seed/cimmytCMLS.htm), national programmes, and CIMMYT experimental lines. Plant growth and watering treatment was conducted at CIMMYT's experimental station in Tlaltizapan, Mexico as described by Ribaut et al. (1996) during the dry (winter) seasons of 2004/05 (designated TL05A) and 2005/06 (designated TL06A). In TL05A, 460 lines were included; in TL06A, some of the earliest and latest lines were removed from the analysis such that 400 lines were used (Supplementary Table S1). Given that SNP data were obtained on a subset of 350 lines (described below), and phenotypic data on these lines were used for association analysis, all data reported here are for this subset of lines. Lines were classified into three maturity groups (early=1, medium=2, late=3) based on preliminary trials where male and female flowering data were collected. Sowing dates for each group were staggered to achieve approximate flowering synchrony among groups, and to ensure that irrigation could be withdrawn on a single date and all lines would experience stress at the flowering stage of development. Group 3 was sown first, and sowing of group 2 and group 1 was delayed by 4 d and 8 d, respectively. This achieved approximate synchrony in male flowering, as the average number of days from sowing to anthesis in groups 1, 2, and 3 was 90, 94, and 98, respectively, in TL05A, and 89, 95, and 99 in TL06A. For each group of inbreds, a well-watered (WW; only in TL06A) and severe drought stress (WS) trial was laid out in an alpha (0,1) lattice design, with two replications. Plots consisted of 2.5 m rows, 0.75 m apart, with 0.2 m in-row spacing. The WW trials were irrigated about every 2 weeks by furrow irrigation. The WS trials were also irrigated every 2 weeks until 20 d before anthesis, when water was withdrawn according to a previously established protocol (Banziger et al., 2001).
Tissue sampling
At 2 and 4 weeks after withholding irrigation, corresponding to ∼1 week before and ∼1 week after anthesis, respectively, five leaf disks of 2 cm diameter were sampled from upper leaves of each plot. Samples were immediately submersed in 5 ml of ice-chilled 80% (v/v) methanol. Primary ears were bagged before silk emergence to prevent their pollination until after sampling. At 0 d and 7 d after anthesis, unpollinated silk and ear tissues were sampled from five plants per plot. The apical 2 cm of ears (ear tips) and a segment of silks attached to the ear tip and extending 2 cm acropetal from the ear tip were dissected out and immediately submersed in 25 ml of ice-chilled 80% (v/v) methanol in the field. Within 4 h following sampling, sealed tubes of leaf, silk, and ear samples were transferred to –18 °C storage. In TL05A, 1.8 ml aliquots of 80% methanol extracts from leaf, silk, and ear tissue were dried under vacuum; in TL06A, 0.2 ml aliquots of 80% methanol extracts were dried at 45 °C with forced air ventilation; and the samples were used for sugar and ABA metabolite assay. Residual solids were dried and weighed, and an aliquot was ground to a fine powder for assay of starch as described below.
ABA and metabolites
ABA and its metabolites, ABA-glucose ester (ABA-GE) and phaseic acid (PA), were separated using solid phase extraction, modified from the method of Setter et al. (2001). C18 columns in 96-well format (model: DSC-18, 25 mg packing material, Supelco, Belleionte, PA, USA) were initially wetted with 400 μl of 95% (v/v) ethanol:water per well, followed by equilibration to initial conditions with 600 μl of 30% (v/v) acidified methanol solution (30% methanol, 69% distilled water, 1% glacial acetic acid). Extracts were reconstituted in 100 μl of 30% acidified methanol solution, and 8 μg of bromecresol green was added as a chromatograph tracer. Extracts were loaded and solvents were drawn through the columns under slight vacuum to provide flow rates of ∼50 μl min−1 or less. ABA-GE, PA, and hydrophilic compounds were eluted with 280 μl of 30% acidified methanol solution; ABA was eluted with 200 μl of 65% methanol plus 1% glacial acetic acid. Absorbance of bromecresol green was determined for each fraction; these values were used to correct for ABA mis-elution into hydrophilic fractions, which averaged <5%. Fractions were dried at 35 °C with forced air ventilation and stored at –18 °C. ABA, ABA-GE, and PA were determined by enzyme-linked immunosorbant assay (ELISA). ABA ELISA was performed according to the procedure of Setter et al. (2001). A similar assay was used for PA, except that plates were pre-coated with a PA–bovine serum albumin conjugate (synthesized according to Gergs et al., 1993) and the primary antibody was monoclonal PA3-2-B3, kindly supplied by E. W. Weiler, Ruhr University, Bochum, Germany (Gergs et al., 1993). ABA-GE was hydrolysed to free ABA by incubating for 2 h at 60 °C in 2 M NH4OH, samples were dried, and the released ABA was assayed as before. Each sample was assayed at least twice for each compound and replicate assays were averaged.
Carbohydrate and proline assay
Glucose and sucrose were determined in aliquots of reconstituted 80% methanol extracts by a peroxidase/glucose oxidase (PGO) coupled enzyme method (Setter et al., 2001), with slight modification. Glucose content was determined using 200 μl of PGO reagent; when the reaction was complete, absorbance was measured at 490 nm using a plate reader (model 750, Cambridge Technology, Watertown, MA, USA). Sucrose was hydrolysed to glucose and fructose by adding invertase solution to the sample aliquots and analysing total glucose with PGO. For starch determination, a 12 mg aliquot of powdered tissue dry matter was exhaustively extracted/filtered with 80% methanol to remove free sugars, and gelatinized in H2O at 80 °C for 2 h. After cooling, starch was completely hydrolysed to glucose with α-amylase and amyloglucosidase (Setter et al., 2001), and assayed for glucose by PGO. Proline was assayed by the method of Bates et al. (1973), modified by substituting citric acid or acetic acid for phosphoric acid (to avoid sugar interference; Magne and Larher, 1992), and heating at 80 °C to permit use of 96-well plates. As an alternative to toluene solvent partitioning, Ruhemann's purple absorbance of amino acid reaction products was depleted by providing an extended aqueous incubation at room temperature.
Phenotypic data analysis
The model used for determining watering treatment effects included inbred genotype (G), treatment (T), and G×T interaction as sources of variation, and each trait was analysed using the linear model in R (version 2.7.1, R Foundation for Statistical Computing, http://www.r-project.org/). The model for combined analysis over years considered each treatment×year combination as an environment such that sources of variation were G, environment (E), and G×E. Broad sense heritability was calculated using the combined data as:
where σG2 is the genotypic variance, σGE2 is the genotype×environment variance, σe is the plot residual variance, and η and r are the number of environments and replicate plots, respectively (Cooper et al., 1996). Correlations were calculated using Pearson's statistic in the cor procedure of R.
Candidate gene SNPs
A panel of 1536 SNPs and flanking sequences from 582 genes were analysed from the chosen candidate genes, as described by Yan et al. (2009). In brief, PCR primers were developed for 1–3 maize contigs (http://magi.plantgenomics.iastate.edu/) per candidate gene (depending on the length of the gene) and were tested for amplification. In total, ∼10 000 SNPs were tested in initially selected candidate genes. From these SNPs, a panel of 1536 SNPs from 582 genes was designed using the Assay Design Tool™ (Illumina Inc., San Diego, CA, USA) (Yan et al., 2009). Supplementary Table S2 at JXB online lists the SNP panel used in this study. Marker context sequences (SNP and flanking nucleotide sequence) are available at http://www.panzea.org. The 1536-plex SNP genotyping assay (Illumina GoldenGate™, BeadArray™ platform, Illumina Inc.) was used to genotype the inbreds. Of these, after removal of those unsuccessfully called or monomorphic (Yan et al., 2009), 1229 SNPs in 540 genes gave usable information and were employed in the present analysis. For SNP 191, manual checking of data indicated a potential hybridization and SNP-call error. Accordingly, inbred samples were sequenced using primers spanning this SNP, and corrected SNP calls were used in the association analysis.
Association analysis
To correct for population stratification, data from the 1229 SNPs in 350 inbred lines were subjected to principal components analysis (PCA), and the top five axes, which explained 7.2% of the variation, were used to create a population structure matrix (P matrix) (Price et al., 2006; Yu et al., 2006; Z. Zhang et al., 2009). To account for relatedness among individuals, all the SNP data were used to generate a relative kinship matrix of similarity between each pair of lines in the association panel (the K matrix) using the program SPAGeDi vs 1.02 and the Loiselle algorithm (Hardy and Vekemans, 2002; Yan et al., 2009). The P matrix was fit as a fixed effect, and K matrices were incorporated as a covariate structure of random effect representing the total of polygene effects in a mixed linear model (MLM). Each SNP was fit as a fixed effect one at a time to test the association between the SNP and phenotype. The analysis was performed using the TASSEL software package (Version 2.1; http://www.maizegenetics.net). Variance components of polygene effects and the residual were estimated using a simplification of the likelihood equations employing the EMMA algorithm (Kang et al., 2008), implemented in TASSEL (Bradbury et al., 2007; Z. Zhang et al., 2009). To correct for multiple comparisons, a family-wise probability level, α ≤0.10 was chosen, and a Bonferroni-corrected threshold probability based on individual tests, β, was calculated as β ≤ 0.10/n, where n was the number of individual trait–SNP combinations tested.
Phylogenetic analysis
To determine relationships between members of gene families in maize and other species, phylogenetic trees were constructed using TreeView version 1.6.6 software (http://taxonomy.zoology.gla.ac.uk/rod/treeview. html) based on multiple alignment of translated amino acid sequences using ClustalX (version 2.0.12) with the Neighbor–Joining method and bootstrap analysis of 1000 replicates, and with the following settings: Matrix Gonnet, Gapopen 10.0, Gapext 0.2, Gapdist 4, Endgap –1.
Results
Analysis of environments and phenotypes
Irrigation was withdrawn in advance of flowering so that water deficit was imposed on unpollinated ears when growth and development of silks and ears was under way. Dry weights (DWs) of ear tips and silks provide an indication of the magnitude and timing of treatment effects. In 2006, the WS treatment decreased average ear tip DW by 11% at 0 days after anthesis (DAA), and 32% at 7 DAA (Table 1). WS decreased DWs of silks 4% at 0 DAA and 24% at 7 DAA. Thus at 0 DAA water deficit had just begun to inhibit ear and silk growth, and at 7 DAA stress effects were severe. Although in 2005 a WW irrigation trial was not conducted, changes in ear and silk DW in the 0–7 DAA interval suggested that the WS treatment in 2005 was a more mild stress than the treatment provided in 2006 (Table 1).
Table 1.
Averages, standard deviations, and broad sense heritabilities for traits measured on ears and silks in 2005–2006
| Organ | Trait | Sampling stage, DAA | Units | 2006 | 2005 | 2006–2005 | |||
| WS averagea | WW average | WS average | Phenotypic SD | Broad sense heritability | G×E | ||||
| Growth | |||||||||
| Ear | DW | 0 | g | 0.47*** | 0.53 | 0.38 | 0.16 | 0.62 | *** |
| Ear | DW | 7 | g | 0.56*** | 0.81 | 0.59 | 0.20 | 0.49 | *** |
| Silk | DW | 0 | g | 0.50 | 0.52 | 0.53 | 0.17 | 0.31 | *** |
| Silk | DW | 7 | g | 0.57*** | 0.76 | 0.74 | 0.22 | 0.43 | *** |
| Phenology | |||||||||
| Tassel | Anthesis | d from sowing | 90.9*** | 95.8 | 96.1 | 4.71 | 0.82 | *** | |
| Carbohydrates | |||||||||
| Ear | Glc | 0 | mmol g−1 DW | 0.37 | 0.38 | 0.86 | 0.17 | 0.30 | *** |
| Ear | Glc | 7 | mmol g−1 DW | 1.20*** | 1.08 | 1.11 | 0.26 | 0.41 | *** |
| Ear | Tot. sugar | 0 | mmol g−1 DW | 1.22*** | 1.02 | 1.59 | 0.30 | 0.25 | *** |
| Ear | Tot. sugar | 7 | mmol g−1 DW | 2.18*** | 1.59 | 1.64 | 0.38 | 0.31 | *** |
| Ear | Suc | 0 | mmol g−1 DW | 0.85*** | 0.64 | 0.72 | 0.20 | 0.41 | * |
| Ear | Suc | 7 | mmol g−1 DW | 0.98*** | 0.49 | 0.53 | 0.25 | 0.43 | ** |
| Ear | Frac. suc | 0 | 0.69*** | 0.62 | 0.45 | 0.08 | 0.62 | *** | |
| Ear | Frac. suc | 7 | 0.44*** | 0.30 | 0.31 | 0.10 | 0.52 | *** | |
| Ear | Starch | 0 | mmol g−1 DW | 1.79*** | 2.21 | 2.41 | 0.34 | 0.44 | *** |
| Ear | Starch | 7 | mmol g−1 DW | 1.61*** | 1.28 | 1.92 | 0.48 | 0.47 | *** |
| Silk | Glc | 0 | mmol g−1 DW | 1.19 | 1.16 | 1.41 | 0.25 | 0.22 | *** |
| Silk | Glc | 7 | mmol g−1 DW | 1.94* | 2.00 | 1.19 | 0.36 | 0.45 | *** |
| Silk | Tot. sugar | 0 | mmol g−1 DW | 2.03 | 2.08 | 1.83 | 0.43 | 0.22 | *** |
| Silk | Tot. sugar | 7 | mmol g−1 DW | 2.27*** | 2.57 | 1.51 | 0.44 | 0.47 | *** |
| Silk | Suc | 0 | mmol g−1 DW | 0.83*** | 0.92 | 0.40 | 0.21 | 0.13 | *** |
| Silk | Suc | 7 | mmol g−1 DW | 0.34*** | 0.55 | 0.32 | 0.17 | 0.20 | *** |
| Silk | Frac. suc | 0 | 0.40*** | 0.43 | 0.21 | 0.05 | 0.12 | *** | |
| Silk | Frac. suc | 7 | 0.14*** | 0.21 | 0.20 | 0.06 | 0.17 | *** | |
| Silk | Starch | 0 | mmol g−1 DW | 0.111** | 0.121 | 0.129 | 0.05 | 0.28 | *** |
| Silk | Starch | 7 | mmol g−1 DW | 0.051** | 0.055 | 0.097 | 0.03 | 0.65 | *** |
| ABA metabolites and proline | |||||||||
| Ear | ABA | 0 | pmol g−1 DW | 5384*** | 1337 | 3200 | 1608 | 0.43 | *** |
| Ear | ABA | 7 | pmol g−1 DW | 5155*** | 1344 | 3228 | 1611 | 0.31 | *** |
| Ear | ABA-GE | 0 | pmol g−1 DW | 445*** | 215 | 1073 | 362 | 0.26 | *** |
| Ear | ABA-GE | 7 | pmol g−1 DW | 885*** | 191 | 1108 | 486 | 0.41 | *** |
| Ear | PA | 0 | pmol g−1 DW | 605*** | 442 | 432 | 193 | 0.47 | *** |
| Ear | PA | 7 | pmol g−1 DW | 382*** | 199 | 409 | 196 | 0.13 | * |
| Silk | ABA | 0 | pmol g−1 DW | 8142*** | 3402 | 3607 | 2179 | 0.20 | *** |
| Silk | ABA | 7 | pmol g−1 DW | 6217*** | 2372 | 3276 | 2032 | 0.41 | *** |
| Silk | ABA-GE | 0 | pmol g−1 DW | 2147*** | 1161 | 1811 | 891 | 0.10 | ** |
| Silk | ABA-GE | 7 | pmol g−1 DW | 5127*** | 1119 | 1660 | 1662 | 0.21 | *** |
| Silk | PA | 0 | pmol g−1 DW | 399*** | 207 | 542 | 163 | 0.18 | *** |
| Silk | PA | 7 | pmol g−1 DW | 191*** | 152 | 267 | 102 | 0.43 | *** |
| Ear | Proline | 0 | μmol g−1 DW | 68*** | 45 | 42 | 18 | 0.47 | *** |
| Ear | proline | 7 | μmol g−1 DW | 65*** | 32 | 58 | 19 | 0.37 | *** |
| Silk | Proline | 0 | μmol g−1 DW | 147* | 142 | 42 | 22 | 0.11 | *** |
| Silk | Proline | 7 | μmol g−1 DW | 124*** | 105 | 33 | 27 | 0.34 | *** |
Phenotypic standard deviations and broad sense heritabillities represent composite estimates from ANOVAs on combined data sets of 2005 and 2006. Statistical significance of comparisons between water stress (WS) and well-watered (WW) treatments, and for genotype by treatment interaction (G×E) are indicated where applicable.
Statistical significance of means comparisons between WS and WW.
*, **, *** Significant at P ≤ 0.05, 0.01, 0.001, respectively.
ABA-GE, abscisic acid-glucose ester; DW, dry weight; Frac. suc, fraction of sugar as sucrose; Glc, glucose; PA, phaseic acid; Suc, sucrose; Tot. sugar, total sugar.
In 2006, WS increased the concentration of total sugars per gram of residual DW in ear tips (ear total sugar) at both 0 and 7 DAA (Table 1). Although increasing sugar concentration may be a mechanism for increasing stress tolerance by contributing osmotically active solutes, it could also represent an indirect effect of slowed development in response to WS. Slowed development and delayed cell wall synthesis may simply have delayed the dilution of sugar per g DW that occurs during development. Sugar composition provided an indication of tissue developmental stage. In the interval from 0 to 7 DAA, sugar composition in ears of WW controls shifted from predominantly sucrose at 0 DAA to predominantly glucose at 7 DAA. The glucose concentration served as an indicator of the total hexose pool, which typically contains approximately equal amounts of glucose and fructose (Setter et al., 2001). Consistent with slowed development, WS restrained the shift in sugar composition so that at 7 DAA WS ears still had 44% of total sugar as sucrose, whereas in WW controls it had decreased to 30%. In 2005, WS sugar levels in ears generally indicated a less severe effect than in 2006, as WS decreased ear sucrose at 7 DAA to 31% of total sugar. Starch concentration in ear tips in 2006 also declined more in WW controls during development from 0 to 7 DAA than in WS ears, but in silks WS treatment had a relatively small effect on sugar and starch concentration and composition throughout development.
In 2006, at both 0 and 7 DAA, the ear tip ABA concentration per g residual DW increased ∼4-fold in water-stressed plants compared with WW controls (Table 1). Given that ear growth was only slightly inhibited at 0 DAA (as described above), ABA accumulation was an early event in stress development. The earliness of the ABA response was also observed in the data for ABA metabolites in ears, silks, and leaves. Compared with WW controls, WS ear tips, silks, and leaves had about 2–3 times as much ABA-GE and PA at 0 DAA (Tables 1, 2). Thus, substantial increases in ABA and its metabolites were observed at an early stage (0 DAA) of stress, in all tissues.
Table 2.
Averages, standard deviations, and broad sense heritabilities for traits measured on leaves in 2005–2006
| Organ | Trait | Sampling stage, weeka | Units | 2006 | 2005 | 2006–2005 | |||
| WS averageb | WW average | WS average | Phenotypic SD | Broad sense heritability | G×E | ||||
| Leaf | SLW | 2 | mg cm−2 | 5.85*** | 5.62 | 6.28 | 0.57 | 0.57 | *** |
| Leaf | SLW | 4 | mg cm−2 | 5.95** | 6.05 | 6.44 | 0.55 | 0.46 | *** |
| Leaf | Glc | 2 | nmol cm−2 | 77*** | 90 | 133 | 46 | 0.48 | *** |
| Leaf | Glc | 4 | nmol cm−2 | 87*** | 112 | 129 | 53 | 0.53 | *** |
| Leaf | Tot. sugar | 2 | nmol cm−2 | 1056 | 1081 | 1421 | 51 | 0.40 | *** |
| Leaf | Tot. sugar | 4 | nmol cm−2 | 460*** | 681 | 1231 | 308 | 0.37 | *** |
| Leaf | Suc | 2 | nmol cm−2 | 977 | 989 | 1281 | 320 | 0.39 | *** |
| Leaf | Suc | 4 | nmol cm−2 | 370*** | 564 | 1096 | 271 | 0.34 | *** |
| Leaf | Frac. glc | 2 | 7.7*** | 8.9 | 8.7 | 2.7 | 0.62 | *** | |
| Leaf | Frac. glc | 4 | 18.6*** | 16.6 | 10.0 | 4.3 | 0.55 | *** | |
| Leaf | Proline | 2 | nmol cm−2 | 198*** | 164 | 98 | 30 | 0.25 | ** |
| Leaf | Proline | 4 | nmol cm−2 | 239*** | 137 | 120 | 39 | 0.04 | *** |
| Leaf | ABA | 2 | pmol cm−2 | 15.75*** | 5.13 | 5.36 | 4.52 | 0.28 | *** |
| Leaf | ABA | 4 | pmol cm−2 | 6.29*** | 2.53 | 11.57 | 3.74 | 0.11 | |
| Leaf | ABA-GE | 2 | pmol cm−2 | 9.48*** | 3.36 | 8.90 | 4.17 | 0.30 | *** |
| Leaf | ABA-GE | 4 | pmol cm−2 | 18.55*** | 14.82 | 18.02 | 8.68 | 0.34 | *** |
| Leaf | PA | 2 | pmol cm−2 | 13.51*** | 5.55 | 7.48 | 5.26 | 0.64 | *** |
| Leaf | PA | 4 | pmol cm−2 | 9.53*** | 5.29 | 17.57 | 7.18 | 0.12 | *** |
| Plant | Ear height | m | 0.734** | 0.716 | 0.700 | 0.15 | 0.70 | *** | |
| Plant | Plant height | m | 1.232 | 1.240 | 1.250 | 0.20 | 0.61 | *** |
Phenotypic standard deviations and broad sense heritabillities represent composite estimates from ANOVAs on combined data sets of 2005 and 2006. Statistical significance of comparisons between water stress (WS) and well-watered (WW) treatments, and for genotype by treatment interaction (G×E) are indicated where applicable.
Weeks after withholding irrigation.
Statistical significance of means comparisons between WS and WW.
*, **, *** Significant at P ≤ 0.05, 0.01, 0.001, respectively.
ABA-GE, abscisic acid-glucose ester; Frac. glc, fraction of sugar as glucose; Glc, glucose; PA, phaseic acid; SLW, specific leaf weight; Suc, sucrose; Tot. sugar, total sugar.
Heritability
To evaluate the consistency across environments of genotypic values for the measured traits, broad sense heritability (H2) was calculated based on data from the tested environments in 2006 and 2005 (Table 1). Heritability of days to anthesis was high (0.82), while H2 for the fraction of sugar as sucrose in ears was somewhat lower (0.62 and 0.52 at 0 DAA and 7 DAA, respectively). Heritability for DW of ears and silks at 7 DAA was 0.49 and 0.43, respectively. In general, intermediate H2 was found for most carbohydrate and ABA traits in ears and leaves, and in silks at 7 DAA when stress was well developed. Even though ABA synthesis was rather low in the WW environment and was increased by stress, heritability of ABA metabolite traits at 7 DAA was intermediate across both WW and WS environments, indicating that while G×E was statistically significant (Table 1), genotypes behaved consistently across WW and WS.
Phenotypic correlations
To evaluate further the relationships between traits, and the robustness of these relationships across environments, phenotypic correlations were calculated for traits measured in 2006 under WW and WS conditions. In the WS treatment, average ear DW for the 0 and 7 DAA harvests, an indicator of ear growth, was strongly negatively correlated with concentrations of sucrose (–0.61), ABA (–0.61), and ABA-GE (–0.64) (Table 3). However, the correlation between ear DW and ear starch was positive in WS (0.29) and not significant in WW, suggesting that the correlations were not due to dilution related to a genotype's growth rate. Similar negative correlations were found in the WW treatment, although for ABA and ABA-GE they were not as strong, as expected given that ABA synthesis is relatively low in WW conditions.
Table 3.
Correlation matrix for ear (E) traits in 2006; data from water stress plots (lower left) and well-watered plots (upper right) of 350 maize inbreds
| E_DW | E_Glc | E_Tsug | E_Suc | E_Fsuc | E_Str | E_Pro | E_ABA | E_ABA-GE | E_PA | S_DW | |
| E_DW | –0.36 | –0.71 | –0.65 | –0.48 | –0.04 | –0.03 | –0.36 | –0.46 | –0.43 | 0.63 | |
| E_Glc | –0.23 | 0.61 | 0.10 | –0.39 | –0.22 | 0.13 | 0.27 | 0.24 | 0.23 | –0.22 | |
| E_Tsug | –0.61 | 0.63 | 0.79 | 0.36 | –0.04 | 0.10 | 0.24 | 0.33 | 0.32 | –0.50 | |
| E_Suc | –0.61 | 0.08 | 0.80 | 0.80 | 0.15 | 0.01 | 0.13 | 0.27 | 0.26 | –0.50 | |
| E_Fsuc | –0.39 | –0.56 | 0.22 | 0.70 | 0.32 | –0.11 | 0.05 | 0.17 | 0.15 | –0.41 | |
| E_Str | 0.29 | –0.48 | –0.35 | –0.09 | 0.27 | –0.45 | –0.28 | –0.15 | –0.17 | 0.07 | |
| E_Pro | –0.33 | 0.33 | 0.34 | 0.19 | –0.09 | –0.52 | 0.20 | 0.09 | 0.09 | –0.17 | |
| E_ABA | –0.61 | 0.25 | 0.45 | 0.39 | 0.18 | –0.42 | 0.40 | 0.74 | 0.48 | –0.39 | |
| E_ABA-GE | –0.64 | 0.32 | 0.53 | 0.45 | 0.17 | –0.48 | 0.46 | 0.66 | 0.44 | –0.44 | |
| E_PA | –0.49 | 0.07 | 0.23 | 0.25 | 0.19 | –0.23 | 0.17 | 0.47 | 0.40 | –0.32 | |
| S_DW | 0.53 | –0.27 | –0.40 | –0.31 | –0.08 | 0.33 | –0.37 | –0.39 | –0.48 | –0.28 |
Shown are Pearson's correlation coefficients for average of plots sampled 0 d and 7 d after anthesis. Values < –0.18 or > +0.18 are significant at P ≤ 0.001. Values < –0.40 or >0.40 are in bold. Also shown is silk (S) dry weight.
ABA, abscisic acid; ABA-GE, ABA-glucose ester; DW, dry weight; Fsuc, fraction of sugar as sucrose; Glc, glucose; PA, phaseic acid; Pro, proline; Str, starch; Suc, sucrose; Tsug, total sugar.
Silk DW was also negatively correlated with sugar and ABA concentration in both WS and WW environments (Table 4). Silk and ear DWs had moderately strong correlations with each other; and for each metabolite, moderately strong correlations were found between ear and silk tissues as well (Tables 3, 4). This suggests that each of these floral tissues provides similar information on genotypic tendencies and response to environment.
Table 4.
Correlation matrix for silk (S) traits in 2006; data from water stress plots (lower left) and well-watered plots (upper right) of 350 maize inbreds
| S_DW | S_Glc | S_Tsug | S_Suc | S_Fsuc | S_Str | S_Pro | S_ABA | S_ABA-GE | S_PA | E_DW | |
| S_DW | –0.57 | –0.49 | –0.21 | 0.19 | –0.33 | –0.73 | –0.35 | –0.45 | –0.51 | 0.63 | |
| S_Glc | –0.53 | 0.90 | 0.48 | –0.13 | 0.15 | 0.54 | 0.15 | 0.29 | 0.31 | –0.23 | |
| S_Tsug | –0.58 | 0.89 | 0.80 | 0.27 | 0.20 | 0.53 | 0.12 | 0.23 | 0.33 | –0.18 | |
| S_Suc | –0.46 | 0.41 | 0.77 | 0.72 | 0.22 | 0.34 | 0.05 | 0.08 | 0.25 | –0.06 | |
| S_Fsuc | –0.16 | –0.09 | 0.29 | 0.72 | 0.16 | 0.00 | –0.05 | –0.13 | 0.01 | 0.11 | |
| S_Str | –0.20 | 0.12 | 0.18 | 0.19 | 0.19 | 0.24 | 0.06 | 0.06 | 0.14 | –0.3 | |
| S_Pro | –0.68 | 0.35 | 0.45 | 0.45 | 0.25 | 0.12 | 0.28 | 0.45 | 0.44 | –0.53 | |
| S_ABA | –0.50 | 0.25 | 0.34 | 0.36 | 0.30 | –0.02 | 0.51 | 0.61 | 0.51 | –0.18 | |
| S_ABA-GE | –0.40 | 0.14 | 0.16 | 0.16 | 0.14 | –0.10 | 0.43 | 0.45 | 0.44 | –0.32 | |
| S_PA | –0.52 | 0.33 | 0.41 | 0.40 | 0.19 | 0.06 | 0.38 | 0.45 | 0.23 | –0.31 | |
| E_DW | 0.53 | –0.25 | –0.18 | –0.04 | 0.10 | –0.08 | –0.44 | –0.26 | –0.28 | –0.22 |
Shown are Pearson's correlation coefficients for average of plots sampled 0 d and 7 d after anthesis. Values < – 0.18 or > +0.18 are significant at P ≤ 0.001. Values < –0.40 or >0.40 are in bold. Also shown is ear (E) dry weight.
ABA, abscisic acid; ABA-GE, ABA-glucose ester; DW, dry weight; Fsuc, fraction of sugar as sucrose; Glc, glucose; PA, phaseic acid; Pro, proline; Str, starch; Suc, sucrose; Tsug, total sugar.
In contrast to ears and silks, in WS leaves, ABA metabolites (ABA, ABA-GE, and PA) were weakly correlated with each other and with carbohydrates (Table 5). In WW leaves they were moderately well correlated. Further examination of correlations indicated that values remained modestly consistent from 2 to 4 weeks after withholding irrigation. For example, the correlation between values at 2 and 4 weeks was 0.60 for leaf sucrose and 0.45 for leaf ABA (Supplementary Table S3 at JXB online).
Table 5.
Correlation matrix for leaf (L) traits in 2006; data from water stress plots (lower left) and well-watered plots (upper right) of 350 maize inbreds
| L_Slw | L_Glc | L_Tsug | L_Suc | L_%glc | L_Pro | L_ABA | L_ABA-GE | L_PA | |
| L_Slw | 0.40 | 0.51 | 0.51 | –0.03 | 0.19 | 0.35 | 0.31 | 0.43 | |
| L_Glc | 0.12 | 0.71 | 0.62 | 0.62 | 0.00 | 0.26 | 0.34 | 0.15 | |
| L_Tsug | 0.23 | 0.67 | 0.98 | 0.05 | 0.14 | 0.24 | 0.26 | 0.21 | |
| L_Suc | 0.24 | 0.52 | 0.98 | –0.05 | 0.16 | 0.24 | 0.24 | 0.20 | |
| L_%glc | –0.02 | 0.68 | 0.10 | –0.05 | –0.19 | 0.12 | 0.26 | 0.02 | |
| L_Pro | 0.31 | –0.08 | 0.02 | 0.05 | –0.06 | 0.22 | 0.11 | 0.19 | |
| L_ABA | 0.15 | –0.13 | –0.26 | –0.26 | 0.08 | 0.10 | 0.51 | 0.53 | |
| L_ABA-GE | 0.08 | 0.06 | –0.09 | –0.12 | 0.18 | 0.23 | 0.30 | 0.41 | |
| L_PA | 0.28 | –0.07 | –0.14 | –0.13 | 0.06 | 0.20 | 0.21 | 0.27 |
Shown are Pearson's correlation coefficients for average of plots sampled 0 d and 7 d after anthesis. Values < –0.18 or > +0.18 are significant at P ≤ 0.001. Values < –0.40 or > 0.40 are in bold.
ABA, abscisic acid; ABA-GE, ABA-glucose ester; Fsuc, fraction of sugar as sucrose; Glc, glucose; PA, phaseic acid; Pro, proline; SLW, specific leaf weight; Str, starch; Suc, sucrose; Tsug, total sugar.
Association mapping
Candidate genes for ABA and carbohydrate metabolic pathways and for signal transduction of ABA and carbohydrate status were identified from a review of plant biology literature. Due to the importance of ear and silk development in drought response, and the likelihood that additional regulatory pathways would play roles in drought response, the current study joined forces with an ongoing SNP identification project which was focused on genes in a broader range of reproductive development, metabolic, and regulatory genes (project title: ‘Molecular and Functional Diversity in the Maize Genome’, http://www.nsf.gov, award number 0321467). Table 6 summarizes the functional classification of the 540 candidate genes in the resulting panel of 1229 SNPs. Included were ∼15 genes encoding enzymes in the carotenoid pathway leading to ABA synthesis, and ∼40 in signalling and transcription factors that are affected by ABA. In the category ‘stress tolerance’ were downstream targets of ABA signalling, such as members of the dehydrin gene family, as well as genes encoding heat shock factors, antioxidant-related enzymes, and enzymes for synthesis of compatible osmolytes. About 60 candidate genes encoding sugar and starch pathway enzymes were included. Given the emphasis in the current project on factors influencing ear and silk development in response to water deficit, genes having roles in regulation of reproductive development were of potential importance. About 22% of the genes were in this category, including genes associated with phytochrome, flowering time, and genes that regulate floral morphological development.
Table 6.
Functional classification of genes containing the SNPs utilized in the association tests
| Gene classification | No. of genes | Fraction of all genes | Subclass fraction |
| ABA and stress tolerance | 92 | 0.17 | |
| ABA synthesis | 0.19 | ||
| ABA regulatory pathways | 0.47 | ||
| Stress tolerance | 0.34 | ||
| Carbohydrate and sugar regulation(enzymes, transcription factors, signalling) | 60 | 0.11 | |
| Other metabolic genes (enzymes and transporters) | 81 | 0.15 | |
| Regulation of reproductive development (transcription factors, signalling) | 120 | 0.22 | |
| Other regulation (transcription factors, signalling) | 180 | 0.33 | |
| Other | 7 | 0.01 |
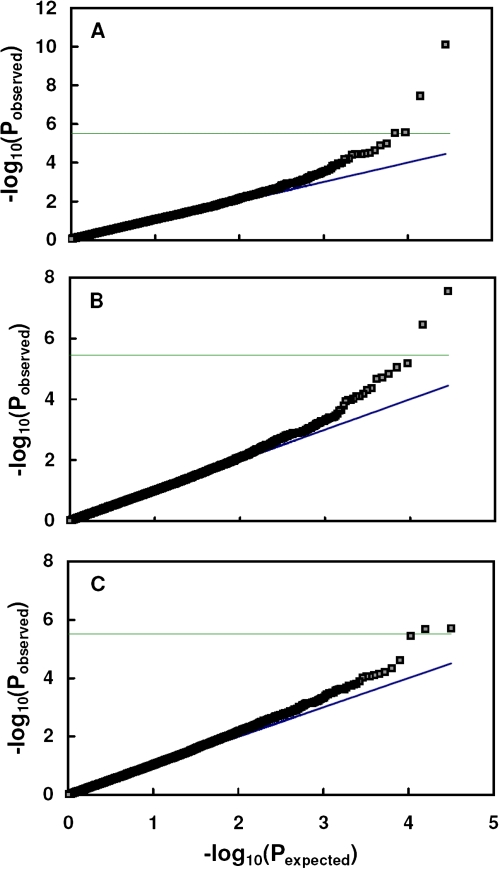
A mixed linear model (MLM) was used with the P and K matrices to perform association analysis between the metabolic traits and the 1229 SNPs. To control for population structure, which might be associated with genetic stratification due to adaptation among lines developed within a certain geographic region or diversifying selection, PCA was used to identify such stratification and correspondingly adjust data to remove these effects (Price et al., 2006). The PCA method has been shown to provide results comparable with alternative approaches such as the STRUCTURE algorithm (Zhao et al., 2007). Previous work with this panel of SNPs indicated that population structure for a broad panel of maize lines consists of four distinct subpopulations of temperate (stiff-stalk and non-stiff-stalk), tropical/subtropical, and mixed (Yan et al., 2009). However, within the panel of 350 tropical/subtropical lines in the current work, population structure was relatively modest, accounting for just 7.2% of variation. To test for association, the expected P-values were generated under the null hypothesis that there is no association between the traits and markers. The distribution under the null hypothesis does not depend on population structure or kinship, although the alternative hypothesis does. As listed in Tables 1 and 2, 58 growth and metabolite traits were included in each watering treatment and each year, where a trait is defined as a sampled characteristic measured in one tissue at one particular sampling date. Quantile–quantile (Q–Q) plots (Fig. 1), which enable visual identification of associations versus non-association, indicated that for each environment, only a few SNP–trait associations had P-values that deviated noticeably from the null hypothesis of no association (the 1:1 diagonal line). This agrees with the expectation that true SNP–trait associations are rare. The plots also show that the mixed model was quite effective in accounting for population structure and relative kinship, as indicated by close agreement for the preponderance of SNP–trait data points to the 1:1 line for Pobserved versus Pexpected, and the substantial deviation from this line for only a few data points. The horizontal line on each plot marks the threshold above which the error rate is ≤0.10 on a multiple-comparison basis (Bonferroni corrected); the SNP–trait associations which exceeded the threshold are assumed to be true associations. Given the large number of trait–SNP combinations tested, it was necessary to use a familywise error rate. The relatively conservative Bonferroni method of controlling the familywise error rate was used to increase the likelihood that claimed associations are true positives; the error rate was set at 10%, a commonly used value.
Fig. 1.
Quantile–quantile plot of test statistics for SNP–trait associations. (A) Traits evaluated in well-watered plots in 2006. (B) Traits evaluated in water-stressed plots in 2006. (C) Traits evaluated in water-stressed plots in 2005. The solid diagonal lines represent agreement between observed and expected probability distributions assuming null SNP–trait association. Squares represent ordered probabilities for each SNP–trait association analysed, with adjustment for population structure and kinship using MLM. The horizontal lines represent threshold probabilities for significant SNP–trait associations that deviate from the null distribution for multiple comparisons by P ≤0.10. (This figure is available in colour at JXB online.)
Significant SNP–trait associations
The SNP–trait associations which exceeded the threshold for multiple comparisons are shown in detail in Table 7, which also indicates the location of each significant SNP according to chromosome and bin number, and the gene in which each SNP is located. The SNP associations were all with metabolites in reproductive tissues (silk and ear), while none was in leaves. Relatively few associations were observed in the WS treatment in 2005, which is not surprising given the relatively later onset and less severe stress imposed in that year (Tables 1, 2). All the associations were with sugars, ABA, and the ABA catabolite PA; none was with starch or proline. A more extended list of the top 41 SNP–trait associations (beyond those listed in Table 7; with P ≤10−4, uncorrected for multiple comparisons) is shown in Supplementary Table S4 at JXB online.
Table 7.
SNP–trait associations which deviated from the null hypothesis for multiple testing with a Bonferroni-corrected threshold P ≤ 0.10
| SNP no. | SNP context sequence number | Chr. | Chr. bin | SNP position | SNP | MAF | N | Organ | Metabolite | Date | Treatment | Year | P | Gene harbouring SNP | Gene function |
| 186 | PZB01403.4 | 1 | 1.11 | 285274032 | A/G | 0.054 | 332 | Silk | ABA | 7 | WS | 2006 | 2.8×10−8 | GRMZM2G124260 | Aldehyde oxidase, ZmAO3 |
| 251 | PZB02017.1 | 2 | 2.03 | 20958246 | A/T | 0.085 | 342 | Ear | Suc | 7 | WS | 2005 | 1.9×10−6 | GRMZM2G173784 | Casein kinase II, regulatory subunit |
| 255 | PZA03635.1 | 2 | 2.03 | 21202350 | C/T | 0.085 | 342 | Ear | Suc | 7 | WS | 2005 | 2.1×10−6 | GRMZM2G021044 | SET domain-containing protein |
| 465 | PZD00027.3 | 3 | 3.05 | 169757661 | A/C | 0.090 | 345 | Ear | PA | 0 | WW | 2006 | 7.8×10−11 | GRMZM2G110153 | MADS-domain transcription factor |
| 465 | PZD00027.3 | 3 | 3.05 | 169757661 | A/C | 0.090 | 345 | Ear | PA | 7 | WW | 2006 | 3.6×10−8 | GRMZM2G110153 | MADS-domain transcription factor |
| 947 | PZA03368.1 | 7 | 7.05 | 162878175 | C/T | 0.074 | 350 | Silk | Tot sug | 7 | WS | 2006 | 3.5×10−7 | GRMZM2G064848 | Pyruvate dehydrogenase kinase |
| 1145 | PZA03573.1 | 9 | 9.07 | 150138200 | A/G | 0.120 | 326 | Silk | Tot sug | 7–0 | WW | 2006 | 2.7×10−6 | GRMZM2G092497 | DNA cytosine methyltransferase |
| 1198 | PZA03569.2 | 10 | 10.06 | 138760685 | T/G | 0.063 | 335 | Ear | PA | 7 | WW | 2006 | 3.0×10−6 | GRMZM2G125023 | Aquaporin, MIP |
SNP number (Yan et al. 2009); SNP context sequence (www.panzea.org); Chr., chromosome number; Chr. bin, bin number for maize chromosome; SNP position in base pairs numbered from the top of the respective chromosome (www.maizesequence.org Release 4a.53); MAF, minor allele fraction (fraction of lines containing the minor allele); N. number of maize genotypes for which trait data was usable for testing the SNP–trait association; P, probability that an SNP–trait association is due to random effects, uncorrected for multiple comparisons; maize identification number for gene harbouring the indicated SNP (www.maizesequence.org Release 4a.53).
SNPs 465, 1145, and 1198 were associated with traits measured in a WW environment. The two most significant of these associations (P ≤7.8×10−11 and 3.6×10−8, respectively) were for SNP 465, and both were for PA (sampled in ears at 0 and 7 DAA). In WS, SNP 465 was weakly associated with ABA in ears and silks at 7 DAA in 2006 (P ≤3.4×10−3 and P ≤2.7×10−3, respectively), below the threshold for significance corrected for multiple comparisons. Analysis with a fixed-effects model in GLM (Z. Zhang et al., 2009), indicated that marker R2, the proportion of total variation explained by the marker but not by the other terms in the model, was 0.14 and 0.11, respectively, for SNP 465 with ear PA at 0 and 7 DAA, respectively. SNP 465 is within the gene for the MADS-box transcription factor ZmMADS16, a maize homologue of the Arabidopsis (Arabidopsis thaliana L.) regulatory gene PISTILLATA (PI), which regulates development of floral parts (Arora et al., 2007; Supplementary Fig. S1 at JXB online).
In the WS treatment in 2006, two SNP–trait associations substantially exceeded the threshold for significance on a multiple-comparison basis: SNPs 186 (P ≤3.1×10−8) and 947 (P ≤3.1×10−7) (Table 7, Fig. 1B). SNP 947, which is located in the gene encoding a regulatory protein kinase for mitochondrial pyruvate dehydrogenase (ZmPDK2; Thelen et al., 1998), was associated with total sugar concentration in silks. The SNP is within the histidine kinase-like domain in exon 6 of this gene.
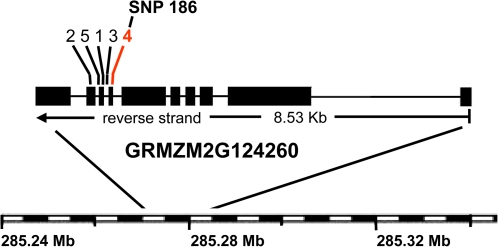
SNP 186 is within a gene annotated as aldehyde oxidase (AO) on chromosome 1, in bin 1.11, related to the Arabidopsis abscisic aldehyde oxidase, AAO3. Analysis with a fixed-effects model in GLM (Z. Zhang et al., 2009) indicated that marker R2 for this SNP was 0.10. Further analysis indicated that this SNP is in the N-terminal half of the gene, in the region of the active site for AO within exon 7 (Fig. 2). In addition, two other AOs were found in bin 1.11, located ∼90 kb and 140 kb downstream of the gene containing SNP 186. Each of these AOs contained a complete set of domains defining it as a member of the AO/xanthine dehydrogenase (XDH) gene family (Schwarz and Mendel, 2006): a molybdenum cofactor (Moco)-binding domain in the N-terminal half of the encoded protein, an FAD-binding domain in the C-terminal half, and iron–sulphur cluster-binding domains ([2Fe–2S]-binding) near the C-terminal end.
Fig. 2.
Gene structure of aldehyde oxidase ZmAO3 (GRMZM2G124260) on chromosome 1, bin 1.11. Gene location on the physical map of chromosome 1 is indicated in Mb. The locations of SNPs used in this study are shown, labelled with the most significant digit of context sequences PZB01403.1 to .5. Significant trait association was found for SNP 186 (in PZB01403.4). (This figure is available in colour at JXB online.)
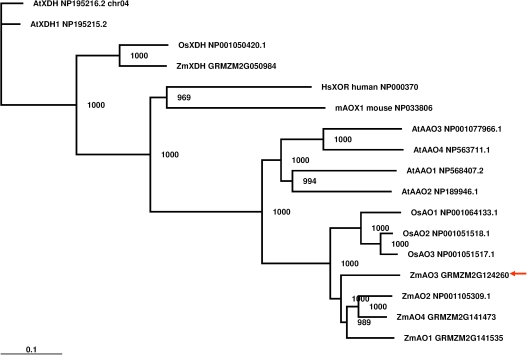
Phylogenetic analysis with partially characterized members of AO gene families in Arabidopsis (indicated with ‘At’ preceding the gene name) and uncharacterized members from rice (indicated with ‘Os’) showed that the gene containing SNP 186 is more closely related to the AO group (AtAAO1, AtAAO2, AtAAO3, AtAAO4, OsAO1, OsAO2, and OsAO3) than to the similar XDH group (AtXDH1 NP195215.2, AtXDH NP195216.2, and OsXDH NP001050420.1) (Fig. 3). An AO gene located 90 kb downstream from SNP 186 (GRMZM2G141535) and an AO on chromosome 5, bin 5.01, were previously reported and named ZmAO1 and ZmAO2, respectively (Sekimoto et al., 1998). The gene containing SNP 186 (GRMZM2G124260) was designated as ZmAO3, and the fourth AO gene 50 kb downstream of ZmAO1 (GRMZM2G141473) was designated as ZmAO4 (Fig. 3).
Fig. 3.
Phylogenetic tree of aldehyde oxidases. The arrow identifies gene in which significant association with the tissue ABA level was found. The tree was constructed using TreeView version 1.6.6 software with the Neighbor–Joining method based on ClustalX alignments of aldehyde oxidases (AOs) and xanthine dehydrogenases (XDHs) from maize (Zm prefix), rice (Os), Arabidopsis (At), human, and mouse, rooted with AtXDH NP195216.2 as the outgroup. Maize sequences are from the maize genome sequencing project (GRMZM2G prefix; http://www.maizesequence.org/index.html Release 4a.53 or 3b.50). The number at the node indicates the bootstrap value (1000 replicates). Bar=0.1 amino acid substitutions per site. (This figure is available in colour at JXB online.)
Although the plant AOs in this study were distinct from plant XDHs, the animal AOs (HsXOR and mAOX1) were intermediate between plant AOs and XDHs (Fig. 2; Supplementary Fig. S2 at JXB online). Among plant AOs, clusters formed according to species rather than AO subfamily. It was not possible to infer which of the maize AOs is most closely related to Arabidopsis AAO3, which has been shown to catalyse the last step of ABA synthesis (Seo et al., 2000a, b). In addition to the strong evidence for association with ABA accumulation in water-stressed silks at 7 DAA (Table 7), among the SNP–trait associations with P ≤0.001 (uncorrected for multiple comparisons), all four associations with SNP 186 were for traits involving ABA and ABA metabolites in silks and ears. This suggests that the basis of the association was abscisic aldehyde oxidase activity of this AO.
Discussion
It is widely recognized that maize is most sensitive to water deficit at the stage around flowering and early kernel development (Grant et al., 1989; Setter et al., 2001). Genetic selection for a shorter ASI has proven to be an effective method by which maize tolerance to drought stress can be improved (Barker et al., 2005; Monneveux et al., 2006; Ribaut et al., 2009). Variation in ASI occurs because ear and silk growth and development are drastically decreased during stress, but pollen development is less so, and genotypes differ in the extent of such decreases (Edmeades et al., 1993). Although the basis for maize vulnerability at flowering and the biology of shortened ASI leading to improved drought tolerance are not understood, evidence indicates that superior lines are better able to sustain ear and silk growth in water deficit environments and thereby partition scarce resources to reproductive rather than vegetative organs (Bolanos et al., 1992; Monneveux et al., 2006).
In the present study the objective was to impose water deficit to coincide with this vulnerable phase, and with sufficient severity to reveal genetic differences between lines. The WS environments in this study substantially decreased ear tip and silk growth from 0 to 7 DAA and genetic variation was sufficient that genotypic differences were significant (Table 1). Another indicator that the WS environments were satisfactory for phenotyping is that WS increased the ABA concentration in all tissues. Concentrations of ABA and ABA catabolites increased at 0 DAA, before large effects on ear and silk DW, and were sustained at 7 DAA (ears) or were attenuated somewhat (silk and leaf), as expected in plants subjected to long-term water stress (Wang et al., 2002).
In the germplasm panel studied, ear and silk DW were negatively correlated with concentrations of sucrose and ABA in ears and silks. ABA signalling up-regulates numerous genes with roles in stress tolerance, such as those encoding proteins that stabilize the macromolecular structure in desiccating cells and enzymes for processing reactive oxygen species (Bartels and Sunkar, 2005; Setter, 2006). However, the negative correlations observed here indicate that genotypes which accumulated higher concentrations of sugars and ABA tended to have lower rates of ear and silk growth. A possible basis for the negative correlation involving ABA is its inhibitory effects on meristematic tissues, analogous to those in buds and growing seed embryos, where it arrests outgrowth and holds tissues quiescent such that energy is conserved (Gubler et al., 2005). Indeed, previous studies of maize, wheat, and soybean seeds at early stages of development indicated that elevated ABA concentration inhibits development and decreases seed set (Liu et al., 2005; Setter, 2006).
It is possible that arrested development also underlies the negative correlation between growth and sugar concentration, as those genotypes which are more responsive to stress may become arrested at an early stage of development when cell walls are thin and a higher proportion of biomass is in sugars. Another indication of delayed development is that WS diminished the decline from 0 DAA to 7 DAA of the sucrose fraction in ears (Table 1). This is consistent with previous reports that water deficit delays or inhibits expression of invertase activity in pedicel and pericarp tissues of unpollinated maize ears, and hence a smaller fraction of sucrose is hydrolysed to hexoses (Andersen et al., 2002; McLaughlin and Boyer, 2004a, b). In contrast to sugars, ear starch concentration was positively correlated with ear DW, such that growth was less in ears with poorer ability to maintain carbohydrate supplies. This agrees with previous studies in which stress was associated with depletion of starch storage reserves from ear tissues and, when this occurs, it leads to loss of kernel set (Zinselmeier et al., 1999).
Association mapping
In the present investigation, a panel of maize inbreds was chosen to represent a diverse range of tropical and subtropical germplasm. Given the diverse background and population history of this panel, it can be expected that historical recombination has degraded linkage disequilibrium (LD) to only short distances. Previous work has indicated that LD extends <1 kb in maize landraces (Flint-Garcia et al., 2003) and <2 kb for diverse inbreds (Remington et al., 2001), although genomic regions differ in their rate of LD breakdown (Gore et al., 2009). Hence, a significant SNP–trait association can potentially provide gene-level resolution for gene(s) responsible for the observed variation in trait phenotypes. For example, gene-level resolution is demonstrated in a 3.5 kb region of the genome surrounding SNP 186 (ZmAO3). Analysis of LD decay in 27 diverse maize inbred lines using SNPs from this candidate gene study and the high density maize HapMap project (Gore et al., 2009) shows that LD rapidly approaches nominal levels within 800 bp (Supplementary Fig. S3 at JXB online). Furthermore, analysis of LD decay in the panel of 350 inbreds indicates rapid LD breakdown near SNP 186 (total distance, 487 bp; median r2, 0.04).
ABA and aldehyde oxidases
SNP 186 is within a gene that encodes an AO (EC 1.2.3.1), as defined by the conserved sequence among AOs, including binding domains for two non-identical [2Fe–2S] clusters and FAD cofactor, and a binding domain at the active site for Moco, which is essential for the enzymatic activity (Schwarz and Mendel, 2006). All AOs are quite similar in amino acid sequence and have the same domain structure as XDH (EC 1.17.1. 4). As has been found in other organisms, AOs in plants catalyse a wide range of reactions, including synthesis of ABA, indole acetic acid, and benzoic acid (Seo et al., 2000b; Ibdah et al., 2009). In Arabidopsis leaves and seeds, mutant analysis and tests of substrate specificity provide evidence that AtAAO3 encodes an AO that catalyses the oxidation of abscisic aldehyde to ABA (Seo et al., 2004), while analysis of AAO4 indicates that it catalyses benzoic acid synthesis in seeds (Ibdah et al., 2009).
Studies of human (Homo sapiens L.) and mouse (Mus musculus L.) AOs (HsXOR and mAOX1, respectively) have used site-directed mutagenesis to determine the roles of certain amino acids in the active site region to define substrate specificity (Yamaguchi et al., 2007; Schumann et al., 2009). These studies have indicated that alterations in the substrate-binding amino acids in the active site at positions 806 and 884 (mouse numbering) affect substrate specificity. In plants, the position equivalent to 806 is conserved as an alanine in all published AOs of maize, rice, and Arabidopsis, except AtAAO2 (Supplementary Fig. S2 at JXB online). In contrast, at position 884, and at the amino acid sequences surrounding this site, plant AOs are quite variable, offering the possibility that this region determines an AO's substrate specificity and reaction rate as defined by substrate affinity (Km) and catalytic turnover rate (kcat). SNP 186 maps to within 254 amino acids of the site equivalent to 884, and this SNP was the closest to this site among the five SNPs investigated here in this gene. Thus, allelic variation at amino acid 884 is closely linked to SNP 186 and provides a plausible molecular explanation for the SNP–trait associations observed in this study.
In Arabidopsis, AAO3 has been reported to be the main AO gene for ABA synthesis in leaves. The evidence includes AAO3 mutants (aao3), which have drastically lower leaf ABA levels and resultant phenotypic effects, and the demonstration that heterologous expression of AAO3 in yeast is capable of catalysing ABA synthesis from abscisic aldehyde (Seo et al., 2000a). More recent work has shown that AAO3 is expressed in a wide range of tissues and contributes to ABA synthesis in Arabidopsis seeds and siliques as well as leaves (Gonzalez-Guzman et al., 2004; Koiwai et al., 2004; Seo et al., 2004). However, even in null mutants, ABA accumulation and seed dormancy occur to some extent (Seo et al., 2004), indicating that one or more of the other AO genes also contribute to ABA synthesis in these tissues. Previous study of genes encoding enzymes in the ABA synthesis pathway have indicated that with respect to ABA accumulation during water deficit, the primary up-regulated genes are members of the 9-cis-epoxycarotenoid dioxygenase (NCED) gene family (Tan et al., 1997, 2003). However, studies have shown that when tissues are exposed to low water potentials, AAO3 expression in leaves and seeds is also up-regulated (Gonzalez-Guzman et al., 2004; Zdunek-Zastocka et al., 2004; Endo et al., 2008). In roots where carotenoid pathway flux is relatively low, the upstream enzyme phytoene synthase-3 is up-regulated in response to WS in parallel with up-regulation of NCED expression and accumulation of ABA (Li et al., 2008).
Pyruvate dehydrogenase kinase
SNP 947 was strongly associated with total sugar level in silks of WS plants at 7 d after pollination. This SNP was located at amino acid 301 within the ATPase/histidine kinase domain of the gene encoding pyruvate dehydrogenase kinase isoform 2 (PDK2; Thelen et al., 1998). Hence, its location is probably within LD distance of polymorphisms that affect the activity of this enzyme. PDK2 is a protein kinase that regulates mitochondrial pyruvate dehydrogenase complex (mtPDC), the key enzyme that determines entry of carbon metabolites into the respiratory pathway. In growing and metabolically active organs such as silks, the respiratory pathway represents a large sink for carbon, and flux through this pathway could affect sugar levels. In rice, transgenic plants expressing an RNAi of OsPDK1 have an altered rate of vegetative growth, and expression of OsPDK1 is regulated by gibberellin (Jan et al., 2006). Miernyk et al. (2007) showed that osmotic stress leads to phosphorylation of mtPDC, which would lower its activity and consumption of carbon in respiration. In the current study, silks of water-stressed plants had higher sugar concentrations (on average, across all genotypes; Table 1), which might reflect a greater share of carbon accumulated as sugars rather than used in respiratory metabolism.
Phaseic acid and the MADS-box gene
SNP 465 was among the strongest SNP–trait associations in the current work, associating with the level of PA in ears of WW plants, and weakly with ABA in ears and silks under WS. Although ABA synthesis and catabolism are elevated during WS, such that the concentrations of both ABA and PA are higher in WS than WW conditions (Table 1), ABA synthesis, transport, and catabolism occur in WW conditions as well (Wang et al., 2002). Since ABA is a growth inhibitor, and its accumulation in ear and silk tissue is negatively correlated with growth of these tissues in WW as well as WS conditions (Tables 3, 4), control of such metabolism may be part of the growth regulatory system in maize ovaries. Consistent with this interpretation, in WW tomato ovaries at the pre-pollination stage, ABA levels are relatively high, and upon pollination they decline as the levels of dihydrophaseic acid catabolite rise and expression of ABA 8′-hydroxylase genes is induced (Nitsch et al., 2009). SNPs within genes for ABA 8′-hydroxylase were not included in the current investigation. SNP 465 is located in a MADS-box transcription factor, ZmMADS16 (formerly called Zmm16), with close homology to rice MADS-box genes OsMADS2 and OsMADS4, and with Arabidopsis floral homeotic gene, PI (Münster et al., 2001; Arora et al., 2007; Supplementary Figure S1). Expression of ZmMADS16 is specific to young floral organs (Münster et al., 2001; Whipple et al., 2004). Maize ZmMADS16 is capable of binding to the corresponding Arabidopsis B-class partner proteins to form a complex that binds DNA, and rescues Arabidopsis PI mutants (Whipple et al., 2004). Thus, the association of SNP 465 with PA accumulation in maize ears (Table 7) may be revealing a link to a hormonal role in the floral development regulated by ZmMADS16.
The current study surveyed SNPs in a wide range of genes including many with putative involvement in drought response, carbohydrate and ABA metabolism, and reproductive organ development. Nevertheless, given that LD breakdown occurs within a short kilobase range from an SNP, the candidate gene approach in maize is limited to the choice of genes surveyed. For example, among ABA synthesis enzymes, SNPs in just one NCED gene were included, whereas the maize genome contains at least six NCED genes of which five are expressed in maize kernels (Capelle et al., 2010). In Arabidopsis, there are five NCED genes, comprising a family of genes with distinct specificities for expression in various tissues and in response to environment (Tan et al., 2003). In addition, the current study included SNPs in only certain sugar and starch metabolism genes. Even so, by including a wide diversity of maize genotypes representing material being employed in tropical maize breeding programmes, the allelic variation sampled has the potential of being useful in the future for maize improvement.
In summary, the current work used an association approach to identify SNPs that affect metabolite accumulation in developing floral tissues of maize in response to water stress. This work demonstrated the utility of the association approach to identify candidate genes that impact metabolic traits in populations of particular interest for the improvement of maize drought tolerance. Allelic variation in ZmAO3 affected the extent to which ABA and ABA catabolites accumulate in floral tissues during drought and an SNP in a MADS-box floral homeotic gene affected PA accumulation in ear tissue. Several SNP–trait associations involved carbohydrate levels, including one for mtPDK2, a key regulator of carbon flux into respiration. While the associations suggest causal links between SNP markers and traits, these findings provide the first step toward establishing such connections. These associations merit investigation to validate them further and to utilize the findings to improve maize response to stress. Given that this study is the first to use the SNP association approach with phenotypic measurements of metabolic and ABA traits in WS, it supports further work that exploits more extensive sets of high density markers that are currently under development.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Phylogenetic tree of MADS-box transcription factors.
Figure S2. Alignment of amino acid sequences for aldehyde oxidases and xanthine dehydrogenases in maize (Zm), rice (Os), and Arabidopsis (At).
Figure S3. Plot of squared correlations of allele frequencies (r2) against distance between polymorphic sites (SNPs) in the aldehyde oxidase ZmAO3 candidate gene.
Table S1. Maize inbreds used in the association analysis.
Table S2. Panel of SNPs in candidate genes.
Table S3. Correlation matrix for ear (E), silk (S), and leaf (L) traits in 2006; data from water stress plots (lower left) and well-watered plots (upper right) of 350 maize hybrids.
Table S4. SNP–trait associations with P <10−4, uncorrected for multiple comparisons.
Supplementary Material
Acknowledgments
We thank Anqi Xing (National Maize Improvement Center of China, China Agricultural University, 100193 Beijing, China) for re-sequencing in the region of ZmAO1, C. Sanchez, E. Huerta, L. Ma, G. Liu, and C. Chiang for technical assistance, and J. Crouch and J. Wilkinson for their critical review of the manuscript. This work was supported by a grant from the Generation Challenge Programme.
References
- Andersen MN, Asch F, Wu Y, Jensen CR, Naested H, Mogensen VO, Koch KE. Soluble invertase expression is an early target of drought stress during the critical, abortion-sensitive phase of young ovary development in maize. Plant Physiology. 2002;130:591–604. doi: 10.1104/pp.005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisnabarreta S, Miralles D-J. Floret development and grain setting in near isogenic two- and six-rowed barley lines (Hordeum vulgare L.) Field Crops Research. 2006;96:466–476. [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics. 2007;8:242. doi: 10.1186/1471-2164-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball RA, Purcell LC, Vories ED. Short-season soybean yield compensation in response to population and water regime. Crop Science. 2000;40:1070–1078. [Google Scholar]
- Banziger M, Edmeades GO, Beck D, Bellon M. Breeding for drought and nitrogen stress tolerance in maize. From theory to practice. Mexico, D.F: CIMMYT; 2001. [Google Scholar]
- Barker T, Campos H, Cooper M, Donlan D, Edmeades GO, Habben J, Schussler JR, Wright D, Zinselmeier C. Improving drought tolerance in maize. Plant Breeding Reviews. 2005;25:173–253. [Google Scholar]
- Bartels D, Sunkar R. Drought and salt tolerance in plants. Critical Reviews in Plant Sciences. 2005;24:23–58. [Google Scholar]
- Bates LS, Waldren RP, Teare ID. Rapid determination of free proline for water stress studies. Plant and Soil. 1973;39:205–207. [Google Scholar]
- Bolanos J, Edmeades GO. The importance of the anthesis–silking interval in breeding for drought tolerance in tropical maize. Field Crops Research. 1996;48:65–80. [Google Scholar]
- Bolanos J, Edmeades GO, Martinez L. Eight cycles of selection for drought tolerance in lowland tropical maize: III. Responses in drought-adaptive physiological and morphological traits. Field Crops Research. 1992;31:269–286. [Google Scholar]
- Boyer JS, Westgate ME. Grain yields with limited water. Journal of Experimental Botany. 2004;55:2385–2394. doi: 10.1093/jxb/erh219. [DOI] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Cao X, Costa LM, Biderre-Petit C, Kbhaya B, Dey N, Perez P, McCarty DR, Gutierrez-Marcos JF, Becraft PW. Abscisic acid and stress signals induce viviparous1 expression in seed and vegetative tissues of maize. Plant Physiology. 2007;143:720–731. doi: 10.1104/pp.106.091454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelle V, Remoue C, Moreau L, et al. QTLs and candidate genes for desiccation and abscisic acid content in maize kernels. BMC Plant Biology. 2010;10:2. doi: 10.1186/1471-2229-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M, DeLacy IH, Basford KE. Relationships among analytical methods used to analyse genotypic adaptation in multi-environment trials. In: Cooper M, Hammer GL, editors. Plant adaptation and crop improvement. Wallingford, UK: CAB International; 1996. pp. 193–224. [Google Scholar]
- Dekkers B, Schuurmans J, Smeekens S. Interaction between sugar and abscisic acid signalling during early seedling development in Arabidopsis. Plant Molecular Biology. 2008;67:151–167. doi: 10.1007/s11103-008-9308-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmeades GO, Bolanos J, Hernandez M, Bello S. Causes for silk delay in a lowland tropical maize population. Crop Science. 1993;33:1029–1035. [Google Scholar]
- Endo A, Sawada Y, Takahashi H, et al. Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiology. 2008;147:1984–1993. doi: 10.1104/pp.108.116632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint-Garcia SA, Thornsberry JMS, Buckler ES. Structure of linkage disequilibrium in plants. Annual Review of Plant Biology. 2003;54:357–374. doi: 10.1146/annurev.arplant.54.031902.134907. [DOI] [PubMed] [Google Scholar]
- Gergs U, Hagemann K, Zeevaart JAD, Weiler E-W. The determination of phaseic acid by monoclonal antibody-based enzyme immunoassay. Botanica Acta. 1993;106:404–410. [Google Scholar]
- Ghiglione HO, Gonzalez FG, Serrago R, Maldonado SB, Chilcott C, Cura JA, Miralles DJ, Zhu T, Casal JJ. Autophagy regulated by day length determines the number of fertile florets in wheat. The Plant Journal. 2008;55:1010–1024. doi: 10.1111/j.1365-313X.2008.03570.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Abia D, Salinas J, Serrano R, Rodriguez PL. Two new alleles of the abscisic aldehyde oxidase 3 gene reveal its role in abscisic acid biosynthesis in seeds. Plant Physiology. 2004;135:325–333. doi: 10.1104/pp.103.036590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore MA, Chia J-M, Elshire RJ, et al. A first-generation haplotype map of maize. Science. 2009;326:1115–1117. doi: 10.1126/science.1177837. [DOI] [PubMed] [Google Scholar]
- Grant RF, Jackson BS, Kiniry JR, Arkin GF. Water deficit timing effects on yield components in maize. Agronomy Journal. 1989;81:61–65. [Google Scholar]
- Gubler F, Millar AA, Jacobsen JV. Dormancy release, ABA and pre-harvest sprouting. Current Opinion in Plant Biology. 2005;8:183–187. doi: 10.1016/j.pbi.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes. 2002;2:618–620. [Google Scholar]
- Ibdah M, Chen Y-T, Wilkerson CG, Pichersky E. An aldehyde oxidase in developing seeds of Arabidopsis converts benzaldehyde to benzoic acid. Plant Physiology. 2009;150:416–423. doi: 10.1104/pp.109.135848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan A, Nakamura H, Handa H, Ichikawa H, Matsumoto H, Komatsu S. Gibberellin regulates mitochondrial pyruvate dehydrogenase activity in rice. Plant and Cell Physiology. 2006;47:244–253. doi: 10.1093/pcp/pci241. [DOI] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y. Preflowering abortion reduces spikelet number in upland rice (Oryza sativa L.) under water stress. Crop Science. 2008;48:2389–2395. [Google Scholar]
- Kim J-Y, Mahe A, Brangeon J, Prioul J-L. A maize vacuolar invertase, Ivr2, is induced by water stress. Organ/tissue specificity and diurnal modulation of expression. Plant Physiology. 2000;124:71–84. doi: 10.1104/pp.124.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwai H, Nakaminami K, Seo M, Mitsuhashi W, Toyomasu T, Koshiba T. Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiology. 2004;134:1697–1707. doi: 10.1104/pp.103.036970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leport L, Turner NC, Davies SL, Siddique KHM. Variation in pod production and abortion among chickpea cultivars under terminal drought. European Journal of Agronomy. 2006;24:236–246. [Google Scholar]
- Li F, Vallabhaneni R, Wurtzel ET. PSY3, a new member of the phytoene synthase gene family conserved in the poaceae and regulator of abiotic stress-induced root carotenogenesis. Plant Physiology. 2008;146:1333–1345. doi: 10.1104/pp.107.111120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Jensen CR, Andersen MN. A review of drought adaptation in crop plants: changes in vegetative and reproductive physiology induced by ABA-based chemical signals. Australian Journal of Agricultural Research. 2005;56:1245–1252. [Google Scholar]
- Magne C, Larher FR. High sugar content of extracts interferes with colorimetric determination of amino acids and free proline. Analytical Biochemistry. 1992;200:116–118. doi: 10.1016/0003-2697(92)90285-f. [DOI] [PubMed] [Google Scholar]
- McLaughlin JE, Boyer JS. Glucose localization in maize ovaries when kernel number decreases at low water potential and sucrose is fed to the stems. Annals of Botany. 2004a;94:75–86. doi: 10.1093/aob/mch123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin JE, Boyer JS. Sugar-responsive gene expression, invertase activity, and senescence in aborting maize ovaries at low water potentials. Annals of Botany. 2004b;94:675–689. doi: 10.1093/aob/mch193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk JA, Szurmak B, Tovar-Mendez A, Randall DD, Muszynska G. Is there a signal transduction pathway that links events at the plasma membrane to the phosphorylation state of the mitochondrial pyruvate dehydrogenase complex? Physiologia Plantarum. 2007;129:104–113. [Google Scholar]
- Monneveux P, Sanchez C, Beck D, Edmeades GO. Drought tolerance improvement in tropical maize source populations: evidence of progress. Crop Science. 2006;46:180–191. [Google Scholar]
- Münster T, Ursula Wingen L, Faigl W, Werth S, Saedler H, Theißen G. Characterization of three GLOBOSA-like MADS-box genes from maize: evidence for ancient paralogy in one class of floral homeotic B-function genes of grasses. Gene. 2001;262:1–13. doi: 10.1016/s0378-1119(00)00556-4. [DOI] [PubMed] [Google Scholar]
- Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE, Buckler ES. Association mapping: critical considerations shift from genotyping to experimental design. The Plant Cell. 2009;21:2194–2202. doi: 10.1105/tpc.109.068437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch LMC, Oplaat C, Feron R, Ma Q, Wolters-Arts M, Hedden P, Mariani C, Vriezen WH. Abscisic acid levels in tomato ovaries are regulated by LeNCED1 and SlCYP707A1. Planta. 2009;229:1335–1346. doi: 10.1007/s00425-009-0913-7. [DOI] [PubMed] [Google Scholar]
- Pelleschi S, Leonardi A, Rocher JP, Cornic G, de-Vienne D, Thevenot C, Prioul J-L. Analysis of the relationships between growth, photosynthesis and carbohydrate metabolism using quantitative trait loci (QTLs) in young maize plants subjected to water deprivation. Molecular Breeding. 2006;17:21–39. [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Qin L, Trouverie J, Chateau-Joubert S, Simond-Cote E, Thevenot C, Prioul J-L. Involvement of the Ivr2-invertase in the perianth during maize kernel development under water stress. Plant Science. 2004;166:371–379. [Google Scholar]
- Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proceedings of the National Academy of Sciences, USA. 2001;98:11479–11484. doi: 10.1073/pnas.201394398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribaut J-M, Betran J, Monneveux P, Setter T. Drought tolerance in maize. In: Bennetzen JL, Hake SC, editors. Handbook of maize: its biology. New York: Springer; 2009. pp. 311–344. [Google Scholar]
- Ribaut J-M, Hoisington D, Banziger M, Setter TL, Edmeades GO. Genetic dissection of drought tolerance in maize: a case study. In: Nguyen HT, Blum A, editors. Physiology and biotechnology integration for plant breeding. Vol. 15. New York: Marcel Dekker; 2004. pp. 571–609. [Google Scholar]
- Ribaut JM, Hoisington DA, Deutsch JA, Jiang C, González-de-León D. Identification of quantitative trait loci under drought conditions in tropical maize. 1. Flowering parameters and the anthesis–silking interval. Theoretical and Applied Genetics. 1996;92:905–914. doi: 10.1007/BF00221905. [DOI] [PubMed] [Google Scholar]
- Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D. A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature. 2006;441:227–230. doi: 10.1038/nature04725. [DOI] [PubMed] [Google Scholar]
- Schumann S, Terao M, Garattini E, Saggu M, Lendzian F, Hildebrandt P, Leimkuehler S. Site directed mutagenesis of amino acid residues at the active site of mouse aldehyde oxidase AOX1. PLoS One. 2009;4(10):e7433. doi: 10.1371/journal.pone.0005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G, Mendel RR. Molybdenum cofactor biosynthesis and molybdenum enzymes. Annual Review of Plant Biology. 2006;57:623–647. doi: 10.1146/annurev.arplant.57.032905.105437. [DOI] [PubMed] [Google Scholar]
- Sekimoto H, Seo M, Kawakami N, Komano T, Desloire S, Liotenberg S, Marion-Poll A, Caboche M, Kamiya Y, Koshiba T. Molecular cloning and characterization of aldehyde oxidases in Arabidopsis thaliana. Plant and Cell Physiology. 1998;39:433–442. doi: 10.1093/oxfordjournals.pcp.a029387. [DOI] [PubMed] [Google Scholar]
- Seo M, Aoki H, Koiwai H, Kamiya Y, Nambara E, Koshiba T. Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant and Cell Physiology. 2004;45:1694–1703. doi: 10.1093/pcp/pch198. [DOI] [PubMed] [Google Scholar]
- Seo M, Koiwai H, Akaba S, Komano T, Oritani T, Kamiya Y, Koshiba T. Abscisic aldehyde oxidase in leaves of Arabidopsis thaliana. The Plant Journal. 2000a;23:481–488. doi: 10.1046/j.1365-313x.2000.00812.x. [DOI] [PubMed] [Google Scholar]
- Seo M, Peeters AJM, Koiwai H, Oritani T, Marion-Poll A, Zeevaart JAD, Koornneef M, Kamiya Y, Koshiba T. The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proceedings of the National Academy of Sciences, USA. 2000b;97:12908–12913. doi: 10.1073/pnas.220426197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setter TL. The role of abscisic acid under water-limited conditions. In: Ribaut JM, editor. Drought adaptation in cereals. New York: Food Products Press–Haworth; 2006. pp. 505–530. [Google Scholar]
- Setter TL, Flannigan BA, Melkonian J. Loss of kernel set due to water deficit and shade in maize: carbohydrate supplies, abscisic acid, and cytokinins. Crop Science. 2001;41:1530–1540. [Google Scholar]
- Setter TL, Parra R. Relationship of carbohydrate and abscisic acid levels to kernel set in maize under postpollination water deficit. Crop Science. 2010;50:980–988. [Google Scholar]
- Suzuki M, Latshaw S, Sato Y, Settles AM, Koch KE, Hannah LC, Kojima M, Sakakibara H, McCarty DR. The maize Viviparous8 locus, encoding a putative ALTERED MERISTEM PROGRAM1-like peptidase, regulates abscisic acid accumulation and coordinates embryo and endosperm development. Plant Physiology. 2008;146:1193–1206. doi: 10.1104/pp.107.114108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. The Plant Journal. 2003;35:44–56. doi: 10.1046/j.1365-313x.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JAD, McCarty DR. Genetic control of abscisic acid biosynthesis in maize. Proceedings of the National Academy of Sciences, USA. 1997;94:12235–12240. doi: 10.1073/pnas.94.22.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen JJ, Muszynski MG, Miernyk JA, Randall DD. Molecular analysis of two pyruvate dehydrogenase kinases from maize. Journal of Biological Chemistry. 1998;273:26618–26623. doi: 10.1074/jbc.273.41.26618. [DOI] [PubMed] [Google Scholar]
- Tuberosa R, Salvi S. Scale and complexity in plant systems research: gene–plant–crop relations. Wageningen UR Frontis Series; 2007. From QTLs to genes controlling root traits in maize; pp. 15–24. [Google Scholar]
- Vollbrecht E, Schmidt RJ. Development of the inflorescences. In: Bennetzen JL, Hake SC, editors. Handbook of maize: its biology. New York: Springer; 2009. pp. 13–40. [Google Scholar]
- Wang Z, Mambelli S, Setter T-L. Abscisic acid catabolism in maize kernels in response to water deficit at early endosperm development. Annals of Botany. 2002;90:623–630. doi: 10.1093/aob/mcf239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker C, Boussuge B, Bencivenni C, Ribaut JM, Tardieu F. Are source and sink strengths genetically linked in maize plants subjected to water deficit? A QTL study of the responses of leaf growth and of anthesis–silking interval to water deficit. Journal of Experimental Botany. 2007;58:339–349. doi: 10.1093/jxb/erl227. [DOI] [PubMed] [Google Scholar]
- Whipple CJ, Ciceri P, Padilla CM, Ambrose BA, Bandong SL, Schmidt RJ. Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development. 2004;131:6083–6091. doi: 10.1242/dev.01523. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Matsumura T, Ichida K, Okamoto K, Nishino T. Human xanthine oxidase changes its substrate specificity to aldehyde oxidase type upon mutation of amino acid residues in the active site: roles of active site residues in binding and activation of purine substrate. Journal of Biochemistry. 2007;141:513–524. doi: 10.1093/jb/mvm053. [DOI] [PubMed] [Google Scholar]
- Yan J, Shah T, Warburton ML, Buckler ES, McMullen MD, Crouch J. Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS One. 2009;4:e8451. doi: 10.1371/journal.pone.0008451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Buckler ES. Genetic association mapping and genome organization of maize. Current Opinion in Biotechnology. 2006;17:155–160. doi: 10.1016/j.copbio.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- Zdunek-Zastocka E, Omarov RT, Koshiba T, Lips HS. Activity and protein level of AO isoforms in pea plants (Pisum sativum L.) during vegetative development and in response to stress conditions. Journal of Experimental Botany. 2004;55:1361–1369. doi: 10.1093/jxb/erh134. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RAC, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ. Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiology. 2009;149:1860–1871. doi: 10.1104/pp.108.133934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Buckler ES, Casstevens TM, Bradbury PJ. Software engineering the mixed model for genome-wide association studies on large samples. Briefings in Bioinformatics. 2009;10:664–675. doi: 10.1093/bib/bbp050. [DOI] [PubMed] [Google Scholar]
- Zhao K, Aranzana MJ, Kim S, Lister C, Shindo C, Tang C, Toomajian C, Zheng H, Dean C, Marjoram P, Nordborg M. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 2007;3:e4. doi: 10.1371/journal.pgen.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinselmeier C, Jeong BR, Boyer JS. Starch and the control of kernel number in maize at low water potentials. Plant Physiology. 1999;121:25–36. doi: 10.1104/pp.121.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinselmeier C, Sun Y, Helentjaris T, Beatty M, Yang S, Smith H, Habben J. The use of gene expression profiling to dissect the stress sensitivity of reproductive development in maize. Field Crops Research. 2002;75:111–121. [Google Scholar]
- Zinselmeier C, Westgate ME, Schussler JR, Jones RJ. Low water potential disrupts carbohydrate metabolism in maize (Zea mays L.) ovaries. Plant Physiology. 1995;107:385–391. doi: 10.1104/pp.107.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.