Abstract
Grain development and its evolution in grasses remains poorly understood, despite cereals being our most important source of food. The grain, for which many grass species have been domesticated, is a single-seeded fruit with prominent and persistent endosperm. Brachypodium distachyon, a small wild grass, is being posited as a new model system for the temperate small grain cereals, but little is known about its endosperm development and how this compares with that of the domesticated cereals. A cellular and molecular map of domains within the developing Brachypodium endosperm is constructed. This provides the first detailed description of grain development in Brachypodium for the reference strain, Bd21, that will be useful for future genetic and comparative studies. Development of Brachypodium grains is compared with that of wheat. Notably, the aleurone is not regionally differentiated as in wheat, suggesting that the modified aleurone region may be a feature of only a subset of cereals. Also, the central endosperm and the nucellar epidermis contain unusually prominent cell walls that may act as a storage material. The composition of these cell walls is more closely related to those of barley and oats than to those of wheat. Therefore, although endosperm development is broadly similar to that of temperate small grain cereals, there are significant differences that may reflect its phylogenetic position between the Triticeae and rice.
Keywords: Brachypodium distachyon, development, endosperm, evolution, gene expression, grain structure
Introduction
Humans are largely dependent on domesticated cereals for the bulk of their calorie intake (http://faostat.fao.org). The use of grain as a source of food pre-dates cereal domestication by several tens of thousands of years, with wild grasses already providing a source of starch in the middle Stone Age (Mercader, 2009). Domestication has since improved the yield and productivity of grasses, significantly altering their physiology, development, and morphology in the process (Shewry, 2009). Rice and wheat occupy the top position in terms of human calorie intake from cereals but, while the genetic tractability of rice has revealed much about the molecular basis of the domestication process in tropical cereals, wheat is less accessible as an experimental system for the temperate cereals that lie within the Pooideae.
Brachypodium distachyon is the first member of the Pooideae subfamily to have its genome sequenced (Vogel et al., 2010) and so provides a valuable resource for studies on the more economically significant members of this family, such as wheat and barley (see, for example, Griffiths et al., 2006). Genome analyses efforts in the domesticated cereals, in terms of completed genome sequencing, have focused on rice, maize, and sorghum; but these are members of the Ehrhardtoideae and Panicoideae, respectively, so the Brachypodium genome and accompanying resources being developed should provide a more relevant model for temperate cereals (IRGSP, 2005; Paterson et al., 2009; Schnable et al., 2009; Vogel et al., 2010). Wheat and barley genome sequencing are underway, the latter due for completion in 2012 (Gill et al., 2004; Schulte et al., 2009). In addition, it should also provide insights into the effects of cultivation and domestication as this is the first wild grass to be sequenced.
The grain is a composite organ, composed of three genetically distinct tissues that include the two genetically distinct products of the fertilized female gametophyte and surrounding maternal tissues. The maternal tissues include the nucellus that surrounds the embryo sac and is itself surrounded by two protective layers—the inner and outer integuments. Outside the integuments is the carpel wall, which develops into the pericarp after fertilization. The products of the double fertilization event, the diploid embryo and the triploid endosperm, generally form the greater part of the mature grain and are protected by the maternal cell layers, the pericarp and, in most grasses, the floral organs, which become toughened as the grain ripens and can remain adherent during grain dispersal.
In cultivated cereals and most grasses, the endosperm is the largest compartment in the grain. Endosperm development progresses in the post-fertilization phase via the division of the central cell-derived triploid nucleus, whose descendents form a syncitial ring of nuclei around the central vacuole that divide and cellularize in a concerted sequence of anticlinal and periclinal cell divisions to eventually fill the central vacuole (Becraft, 2001; Olsen, 2001). The endosperm subsequently differentiates into functionally distinct subdomains, key among which are the aleurone and the central starchy endosperm, although the timing, identity, and relative sizes can vary depending on the species.
Most of the detailed developmental characterization of grain development has been carried out on domesticated species. For example, grain development in wheat and barley, particularly endosperm development, has been reasonably well studied (Olsen et al., 1992; Doan et al., 1996; Drea et al., 2005b; Wegel et al., 2005) and the developmental programmes are very similar, with key marker genes showing nearly identical expression patterns (Doan et al., 1996; Linnestad et al., 1998; Drea et al., 2005b). Other well-studied grains such as maize kernels and rice grains develop quite differently, with distinct tissue types, organization, and gene expression patterns (Brown et al., 1996; Opsahl-Ferstad et al., 1997; Gutierrez-Marcos et al., 2004; Sabelli and Larkins, 2009). However, since endosperm development in cereals is a distinctive process, a comparative view extending to all studied models both temperate and tropical is valuable. Specifically, the tractability of maize genetics has revealed that the cereal endosperm aleurone layers rely on input signals from the zygote, surrounding maternal tissues, and on positional information for proper differentiation (Costa et al., 2003; Gruis et al., 2006).
For this study, the events that occur during grain development in a wild grass, Brachypodium, at the cellular and molecular level were compared with those in wheat, one of the most closely related cultivated cereals whose development has been well characterized. While grain development is broadly similar, there are a number of significant differences in cellular differentiation and gene expression patterns that reflect significant developmental differences between Brachypodium and wheat.
Materials and methods
Plant growth conditions
The Bd-21 accession of Brachypodium distachyon and the Savanna cultivar of hexaploid wheat were used for a comparative analysis of grain development. Brachypodium plants were grown in a controlled-environment room under the following conditions: 22 h of light, 22 °C temperature, and 60% relative humidity. Wheat plants were grown as previously described (Drea et al., 2005b).
Grain development was monitored using a dissecting microscope and photographed for macro-morphological analyses. For comparison of tissue organization, fully developed green grains of Brachypodium and bread wheat were cut transversely at the mid point with a razor blade and observed under brightfield or UV fluorescence microscopy.
Tissue preparation for histology and RNA in situ hybridization (ISH)
For 4′,6-diamidino-2-phenylindole (DAPI) staining, caryopsis development was staged from the time of anthesis and individual florets were collected at 0, 2, 3, 4, 6, 8, 10, 15, and 20 days after anthesis (DAA). For RNA ISH experiments, caryopses were staged according to length. Caryopses were trimmed and fixed in formalin–acetic acid–alcohol (FAA; 3.7% formaldehyde, 5% acetic acid, 50% ethanol) and vacuum infiltrated for 15 min. After overnight fixation, samples were transferred to the Tissue Tek vacuum infiltration processor supplied by Bayer (Newbury, UK) for an automated dehydration/infiltration process as follows: 70% ethanol for 1 h at 35 °C; 80% ethanol for 1.5 h at 35 °C; 90% ethanol for 2 h at 35 °C; 100% ethanol for 1 h at 35 °C; 100% ethanol for 1.5 h at 35°C; 100% ethanol for 2 h at 35 °C; 100% xylene for 0.5 h at 35 °C; 100% xylene for 1.0 h at 35 °C; 100% xylene for 1.5 h at 35 °C; and molten paraffin wax (supplied by VWR International, Poole, UK) for 2 h at 60 °C. Samples were then transferred to the Tissue Tek embedding console (Bayer) for embedding in paraffin blocks.
Section preparation for ISH
Wax sections of 14 μm thickness were cut on a Leica Microtome (RM2125RT; Wetzlar, Germany) and organized sequentially on poly-lysine-coated slides (Grace Biolabs, supplied by Stratech Scientific, Soham, UK). After drying down at 42 °C overnight, the slides were checked by visual inspection using a dissecting microscope. Slides were de-waxed as previously described (Drea et al., 2005a) and used for staining or for RNA ISH.
Tissue staining
De-waxed sections of Brachypodium staged grains were stained with DAPI solution (Partec) for 20 min and viewed with a fluorescence microscope. Stained nuclei and some cell wall components produced a blue fluorescence with UV excitation (various cell wall components also fluoresce). Calcofluor (0.2%; a fluorescent brightener) was used to detect cell walls in staged grains.
Evans Blue vital staining
Staining was performed as described previously (Young and Gallie, 1999) with minor modifications. Briefly, thin sections were made by hand of mature grains that had been imbibed in distilled water overnight. Sections were immersed in a 0.1% Evans Blue solution for 4 min and then washed in several changes of distilled water for 3 h with agitation. Sections were immediately mounted in distilled water and photographed using dissecting and compound microscopes.
Blast analysis and sequence comparison
Wheat cDNA sequences were used in BLASTN searches against the Brachypodium genome 8× release (www.modelcrop.org) to identify potential markers with high nucleotide sequence similarity and single copy status. These results were used to design primers for RT-PCR and ISH (see Supplementary Table S1 available at JXB online).
RNA isolation and RT-PCR
Samples for three biological replicates were collected at each of the following stages: 3, 5, 8, 10, 15, and 20 DAA. Alternatively, grains were collected based on size. The husks were removed from each caryopsis before freezing in liquid nitrogen and storing at –70 °C. Cellular RNA was isolated using an RNeasy Plant Mini Kit (Qiagen). A 1 μg aliquot of total RNA was reverse-transcribed with an Omniscript RT Kit (Qiagen) using an oligo(dT) primer (Invitrogen). The final concentration of RNA and the cDNA content were assessed using a NanoDrop ND-1000 Spectrophotometer. PCR was performed for gene expression analyses using 0.5 μg of cDNA from each staged grain. Primers against selected Brachypodium genes (Table 1) were designed based on the Brachypodium draft genome (www.Brachypodium.org). For cDNA synthesis the poly(T) primer used was 5′GACTCGAGTCGACATCGA(T)17. Control primers were BdH4f and BdH4r: 5′ATGGATCCTCCAATCCAGAC3′ and TATTGTGTTGGACTCTGGTG3'; BdGAPDHf and BdGAPDHr (see Supplementary Table S1 at JXB online). The PCR products were loaded onto a 1% agarose gel containing ethidium bromide, and electrophoresis was performed. The gel was examined and photographed using a UV imager.
Table 1.
Thickness of the nucellar epidermis layer in mature grains and thickness of cell walls in relation to cell size (given as length of the cell in the longest axis) in endosperm of mature Brachypodium and wheat grains in transverse sections
| Brachypodium mature grain | Wheat mature grain | |
| Nucellar epidermis layer thickness (μm) | ||
| Adaxial site | 29.9±0.15 | 11±0.03 |
| Abaxial site | 16±0.15 | 14.9±0.15 |
| Lobes | 50.3±0.61 | 11.1±0.12 |
| Endosperm cells (μm) | ||
| Central endosperm—cell wall thickness | 4.4±0.15 | 2±0.06 |
| Central endosperm—cell length | 21.4±0.09 | 131.2±0.75 |
| Aleurone layer—cell length | 19.5±0.03 | 40.8±0.06 |
| Aleurone layer—cell wall thickness | 1.5±0.02 | 4.9±0.08 |
n = 5 and errors represent ±1 SD.
In situ hybridization
Hybridizations were carried out as previously described (Drea et al., 2005a) with minor modifications. Gene-specific regions were amplified by PCR from genomic DNA or cDNA and were either used directly to generate digoxigenin-labelled RNA probes (by using T7 RNA polymerase sites incorporated into the reverse primer) or first cloned into the pCR4 TOPO vector (Invitrogen) which has a T7 RNAP site flanking the inserted sequence.
Microscopy and image processing
For anatomical analyses and starch composition, 20 DAA grains from Brachypodium and 30 DAA grains from bread wheat were cut transversely across the widest part of the grain and sputter coated with gold. Samples were then analysed at 3 kV accelerating voltage in a Philips XL 30 FEG scanning electron microscope. Representative sections of Brachypodium grains were photographed with a Nikon E800 microscope (Tokyo, Japan) using a digital camera under bright-field conditions and a UV filter. Magnifications and camera settings remained unchanged for all images through all stages for Brachypodium sections.
Fourier transform-infrared (FT-IR) imaging
Transverse cell-wall-only thin cross-sections were prepared according the method described previously (Toole et al., 2007) with the sections being cut (1 μm thick) using an ultramicrotome rather than (50 μm thick) using a vibratome. FT-IR spectral images were then collected and each pixel within each image was classified and colour coded using the image analysis software ENVI 4.7 (Research Systems Inc., Boulder, CO, USA), according to a classification method that was developed to analyse barley and oat endosperm cell walls (G.A. Toole et al., unpublished results).
Results
Spike and floret structure in Brachypodium distachyon
Grass inflorescences are typically composed of several spikelets each containing one or more florets, the latter composed of the carpel, stamens, lodicules, palea, and lemma. Brachypodium has a simple spike-like racemose inflorescence and spikelets that often have awned lemmas. The inflorescence matures basipetally as the spikelets emerge, but within each spikelet grain maturation is acropetal (Khan and Stace, 1999). In Brachypodium, the terminal and lateral spikelets are indeterminate, with several diminished and underdeveloped florets at the apex. Spikelets are highly variable for the number of fertile florets that develop (4–20) and freely disarticulate below each floret at maturity. Each floret is subtended by two glumes. The spike of wheat resembles the spikelet of Brachypodium (Supplementary Fig. S1A at JXB online). The spikelet of wheat is a regular arrangement of five florets eventually producing up to four grains (the most distal floret remaining unfertilized) and subtended by two glumes. The florets of Bd21 observed here contained a single carpel and ovule, as in all grasses, but just two anthers. Generally grass florets, including those of all other members of the Brachypodium genus, contain three anthers (Hubbard, 1954). The anthers of B. distachyon are shorter than in perennial Brachypodium species and the flowers are cleistogamous (Khan and Stace, 1999).
Brachypodium is widely considered to be a model for temperate cereals (Draper et al., 2001; Opanowicz et al., 2008; Vogel et al., 2010) and its genome has recently been sequenced (Vogel et al., 2010), yet little is known about how grain structure and development compare with those cereals. As a first step, the overall shape and size of its grains was compared with that of related grasses and cereals (Fig.1). Brachypodium distachyon forms a caryopsis with an adherent pericarp (and also closely adhered lemma and palea), which is slender and ellipsoid and carries hairs at the apex. The Brachypodium grain is comparable in length with that of wheat, but contrasts starkly in terms of width and depth (Fig. 1A, and Supplementary Fig. S1B at JXB online). A comparison of grain dimensions in different grass species shows that the Brachypodium grain profile is narrow and flat with a width:depth ratio of 1.7 (Supplementary Fig. S1B), typical of grains of the closely related wild Elymus and Bromus genera with ratios of 1.43 and 1.76, respectively, and some wild wheats (data not shown). In contrast, typical hexaploid Triticum aestivum cultivars Soissons and Cadenza have more rounded profiles with an average width:depth ratio of 1.15.
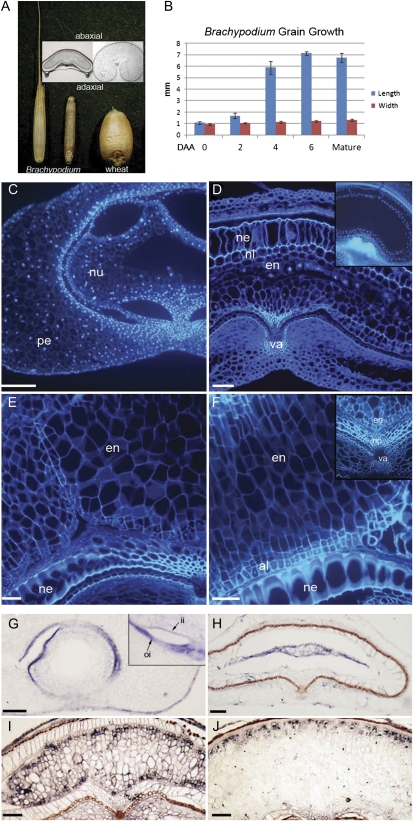
Fig. 1.
Development of the Brachypodium grain. (A) Brachypodium grain with and without husk compared with wheat grain. (B) Measurements of length and width of Brachypodium grain through development showing rapid elongation early in development; n = 6 and error bars represent ±1 SD. (C) DAPI-stained sections of Brachypodium grains at 3 DAA (days after anthesis); (D) 5 DAA; (E) 8 DAA; (F) 15 DAA; pe, pericarp; nu, nucellar tissue; ne, nucellar epidermis; nl, nucellar lysate; np, nucellar projection; en, endosperm; va, vascular tissue; al, aleurone. (G) Histone H4 mRNA ISH in grain sections at 3 DAA; (H) 5 DAA; (I) 8 DAA; (J) 15 DAA; ii, inner integument; oi, outer integument; all scale bars, 50 μm
Comparative cytological analysis of endosperm reveals significant differences in grain architecture between Brachypodium and wheat
To determine the cellular basis for these differences in grain profile, a cytological analysis of mature Brachypodium and wheat grains was performed, focusing on the endosperm which forms the major part of the grain in both species. In wheat, four major cell types constitute the endosperm, including transfer cells (modified aleurone), aleurone cells, starchy endosperm cells, and embryo-surrounding region cells.
In transverse section, the Brachypodium grain does not possess a distinctly indented hilum or ‘crease’ as is characteristic in wheat, but rather has a shallow concave indentation underlying the main vascular trace (Fig. 1A). The size of the nucellar projection region that extends from the vasculature towards the endosperm is vastly reduced in Brachypodium as compared with wheat, most notably later in development (Fig. 1F inset). Reflecting the flat shape of the caryopsis, the Brachypodium endosperm forms a crescent-shaped structure unlike the lobed structure that encloses the crease and nucellar projection in the rounder wheat grains. The region of the endosperm that corresponds to the modified aleurone in wheat forms a convex indentation in Brachypodium facing the nucellar projection region. Finally, the endosperm cavity is absent in Brachypodium (Fig. 1A).
To gain further insight into Brachypodium grain development, grains were staged using anthesis (0 DAA) as the reference point and their growth followed up to ∼15 DAA (Fig. 1C–F). The fertilized caryopsis (0 DAA) is ∼1 mm long and elongates rapidly between 2 and 6 DAA, and reaches its maximum length of 7–8 mm by 6 DAA (Fig. 1B). Using sectioned material to examine nuclear proliferation and lateral growth, four key stages of grain development were established (illustrated schematically in Fig. 6) corresponding to (i) syncytial endosperm and a circular central vacuole 0–3 DAA (Fig. 1C); (ii) an elongated, flat vacuole with a thin sliver of endosperm (at which time cellularization begins; 3–5 DAA) (Fig. 1D); (iii) cellularized endosperm with adaxial indentations (arrowed) flanking the nucellar projection 5–13 DAA (Fig. 1E); and (iv) fully filled endosperm with smaller peripheral aleurone cells discernible at 13–18 DAA (Fig. 1F). The entire process, from anthesis through to fully filled ripe grains, takes ∼24 d as opposed to an average of 35 d in wheat (depending on cultivar and growing conditions).
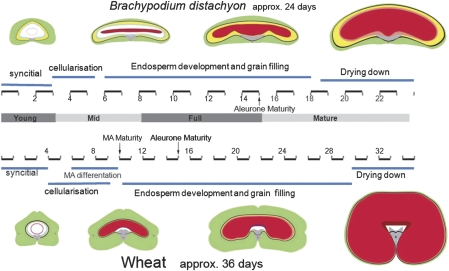
Fig. 6.
Schematic summarizing grain development. Brachypodium grain development compared with wheat focusing on the key stages in endosperm development—illustrations are not to scale and grain tissues are indicated by colour: pericarp, green; integuments; black; nucellar epidermis, yellow; nucellar lysate, blue; nucellar projection, grey; aleurone, pink; central endosperm, red; MA, modified aleurone. Numbers indicate DAA (days after anthesis). Stages indicated as young, mid, and full mature correspond to the material used in RT-PCR in Supplementary Fig. S2 at JXB online.
The different developmental stages were compared with corresponding stages in wheat grains (Fig. 1 and summarized in Fig. 6). Although the early post-fertilization stages are similar in both species, with endosperm nuclei proliferating and migrating around the periphery (Fig. 1C; Wegel et al., 2005), there are clear differences evident before cellularization. In wheat (Drea et al., 2005b) and barley (Olsen et al., 1992), nuclei form a highly regular arrangement around the periphery of the central vacuole that is seen in wheat just prior to cellularization. Cell division then re-orientates anticlinally in wheat and barley to produce internal layers of nuclei, but Brachypodium’s nuclear distribution appears more random at this point in development (5 DAA; Fig. 1D with wheat inset). The Brachypodium grain then becomes flattened dorso-ventrally and, by ∼8 DAA, the smaller (presumptive) aleurone cells form around the periphery of the endosperm and deep adaxial indentations become apparent in the endosperm flanking the nucellar projection (Fig. 1E). The nucellar tissue adjacent to the adaxial endosperm becomes compressed. At ∼15 DAA, small square cells around the edge of the central endosperm are clearly visible and appear uniformly distributed abaxially, adaxially, and laterally (Fig. 1F and inset). The nucellar projection is much reduced as compared with wheat (Drea et al., 2005b; Fig. 1F inset).
To map changes in cell cycle activity during grain development, a histone H4 transcript was used as a marker. Histone H4 is expressed in a DNA replication-dependent manner and is, therefore, a good indicator of cell cycle activity (Fobert et al., 1994) and can also provide an indication of cellularization and tissue differentiation (Drea et al., 2005b). At 3–5 DAA, there are high levels of histone H4 transcript in the outer pericarp layer and in the two integuments, inner and outer, as well as in the syncytial endosperm (Fig. 1G, H). In wheat grains at this stage, the histone transcript preferentially accumulates in the adaxial nucellar projection, whereas in Brachypodium the histone transcript is mainly present in syncytial endosperm (Drea et al., 2005b; Fig. 1H). The inner integument layer, marked naturally by the deposition of mucilage and a resulting brown colouration, is visible early in Brachypodium at a much earlier stage than in wheat grains. By 4 DAA, histone transcript is further restricted to the outer integument (Fig. 1G inset). The endosperm of Brachypodium at ∼5 DAA (Fig. 1H) is restricted to a slim sliver of material between robust nucellar layers in a flat grain. Histone signal is evenly distributed throughout the endosperm, indicating that the endosperm is not yet cellularized (Fig. 1H). After the endosperm has clearly cellularized (8 DAA), histone transcript is strongest along the periphery as in wheat, and is now distinctly patchy (Fig. 1I). This patchy expression pattern is due to asynchronized cell cycle progression in neighbouring cells combined with the tight S-phase specificity of histone gene expression (Fobert et al., 1994). Unlike in wheat grains at an equivalent developmental stage, however, there is no specific exclusion of transcript from the region presumed to equate to the modified aleurone overlying the crease (Fig. 1I). The disappearance of histone H4 transcript can indicate tissue differentiation (Drea et al., 2005b) and these differences in the timing between wheat and Brachypodium indicate that the aleurone region adjacent to the crease develops quite differently in the two species. Subsequently, as grain filling is completed, histone expression is preferentially localized to the abaxial endosperm as shown by a cross-section of a grain at ∼10 DAA (Fig. 1J). Taken together, these data indicate that there are distinct differences in the timing of differentiation in different regions of the Brachypodium grain and that these differ in a number of respects from wheat, particularly regarding the differentiation of the modified aleurone layer.
Molecular mapping of maternal and filial expression domains
The cytological analysis suggested that grain development was similar in many ways to that of wheat or barley, but that the timing and nature of cell type specification differed. To gain further insight into these differences, the molecular events associated with grain development were aligned using key marker genes (Supplementary Fig. S2 at JXB online) to map temporally and spatially domains of expression in the developing Brachypodium grain and relating these to the cytological events observed. Markers were selected based on tissue-specific markers in wheat (Drea et al., 2005b) and Brachypodium storage proteins (Laudencia-Chingcuanco and Vensel, 2008). A BLASTN survey of wheat endosperm- and cell type-specific genes against the Brachypodium genome revealed that there were generally lower levels of sequence similarity in the grouping expressed in the aleurone layers (Fig. 2A) as opposed to the groupings expressed in other tissues.
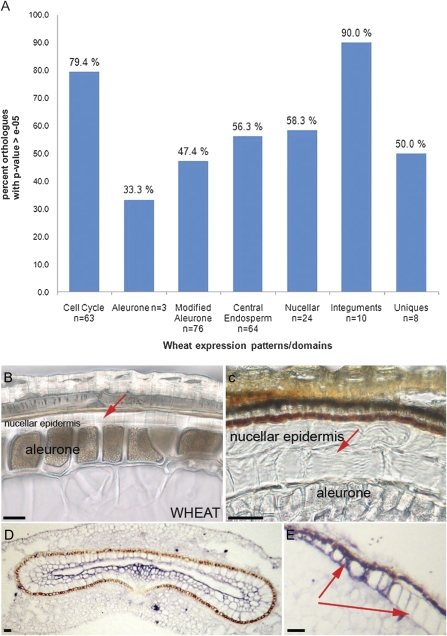
Fig. 2.
Analysis of nucellar tissue in Brachypodium grains. (A) Summary of BLASTN results for wheat sequences (showing tissue-specific gene expression patterns; Drea et al., 2005b) compared with the Brachypodium genome. (B) Brightfield micrograph of peripheral grain cell layers in mature wheat. (C) Brachypodium grains. Nucellar epidermis layers are indicated with red arrows and both nucellar epidermis and aleurone are labelled. (D) ISH of BdC13 showing expression in nucellar lysate of a 4 DAA (days after anthesis) cross-section; (E) BdC13 in the abaxial nucellar epidermis in a 15 DAA grain cross-section. Scale bars, 20 μm.
To select tissue-specific markers, Brachypodium genes were chosen that are most similar to known tissue-specific markers in wheat [i.e. those with high sequence similarity at the nucleotide level but which were not members of extensive gene families so as to maintain specificity (listed in Supplementary Fig. S2 at JXB online)]. For example, C13 endopeptidase or nucellain marks the nucellar lysate and nucellar epidermis—tissues that are degraded and compressed relatively early in wheat grain development (Linnestad et al., 1998; Drea et al., 2005b); pyruvate orthophosphate dikinase (PPDK) marks the single aleurone layer very specifically in wheat though its actual function is elusive given that it is involved in photosynthesis in green tissues converting phosphoenolpyruvate to pyruvate (Drea et al., 2005b; Hong-Gyu et al., 2005; Chastain et al., 2006); an α-galactosidase specifically marks the modified aleurone within the grain in wheat (Drea et al., 2005b). BdGLO1 and BdGLO2 were chosen because they have been shown to be the most abundant storage proteins in Brachypodium grain (Laudencia-Chingcuanco and Vensel, 2008) and provide useful markers for storage protein deposition in the endosperm during grain filling.
In all cases, except BdGLO2 and histone H4, a BLASTN query with the starting wheat nucleotide sequence resulted in a single highly significant (e <10−6) result at the nucleotide level for Brachypodium. More than one homologue for histone H4 and BdGLO2 was expected on the basis of previous analyses (Drea et al., 2005b; Laudencia-Chingcuanco and Vensel, 2008). The presence of only one copy of the PPDK is interesting given that maize, rice, and sorghum all have two PPDK genes, with maize and rice producing two proteins (chloroplast and cytosolic forms) from one of the genes (Imaizumi et al., 1997). The Brachypodium PPDK gene has a similar structure to the rice and maize gene, OsPPDKB and ppdkZm1, respectively, where a 5′ exon located distantly upstream provides a putative targeting peptide for chloroplast localization (Imaizumi et al., 1997).
All genes selected show expression in grains as judged by an RT-PCR survey of different tissues (Supplementary Fig. S2 at JXB online). Both BdGLO genes are expressed exclusively in the grain, specifically in the latter stages of grain development (10 DAA onwards) when the endosperm is fully cellularized. Some of the other selected genes are also expressed in non-grain tissues, and expression was detected in multiple tissues (Supplementary Fig. S2).
Maternal tissue organization in Brachypodium grains differs from that of wheat throughout development
The order and identity of tissue layers in the mature Brachypodium grain resemble those of wheat, but one very striking difference revealed by the cytological analysis is the persistence of the nucellar epidermis layer in Brachypodium. This reaches 50–60 μm at the thickest point in the lateral adaxial region (and in longitudinal sections is very pronounced at the distal or stigma end of the grain) and thinnest at the central abaxial point, where it is comparable with the thickness of the nucellar epidermis in wheat (Fig. 2B–E; Table 1). In wheat grain, the nucellar epidermis layer becomes compressed early in development and is very reduced even by 9 DAA. In Brachypodium, the inner and outer cell walls of the nucellar epidermis are thickened and minimally compressed on the sides of lobes. This suggests that these wall thickenings have a skeletal function, reinforcing the cell walls, and enhancing their resistance to deformation. In rice, the nucellar epidermis is also persistent, and these cells are specialized for transport of nutrients (Oparka and Gates, 1981; Ellis and Chaffey, 1987), being elongated in the principal direction of assimilate flow (i.e. circumferentially around the endosperm) and symplastically connected to adjacent cells by numerous plasmodesmata. Electron microscopy indicates that Brachypodium nucellar epidermal cells are also rich in plasmodesmata (data not shown).
ISH with BdC13 (endopeptidase/nucellain) showed enhanced expression in the nucellar lysate cell layers of young Brachypodium grains from 4 DAA (Fig. 2D). This is comparable with the expression pattern observed in wheat (Drea et al., 2005b). Expression was not detected in the nucellar epidermis except in later stages of development when transcript was present in the central abaxial region where the layer is at its narrowest point in the grain (Fig. 2E) and cells are thinnest (Table 1). The expression of this endopeptidase could correlate with specific degradation of cellular components and compression of the layer in this region.
Endosperm differentiation in Brachypodium and wheat
The outermost cell layer, or aleurone, is a functionally and structurally distinct but general feature of plant endosperm. The aleurone layer surrounds the central endosperm, except in the transfer cell region in temperate small grain cereals, and is composed of one (wheat) or more (barley) layers. The Brachypodium aleurone layer tends to be more irregular, being from one to three or more cells deep. It was noted that the Brachypodium aleurone layer appears to be structurally integrated with the central endosperm because it remains firmly attached to the endosperm during physical disruption of the grain (Fig. 3A). In wheat, the aleurone tends to adhere to the maternal tissues during disruption (Fig. 3B), a feature that is exploited during milling wheat grain and has the effect of separating the α-amylase activity in the aleurone from the starchy central endosperm.
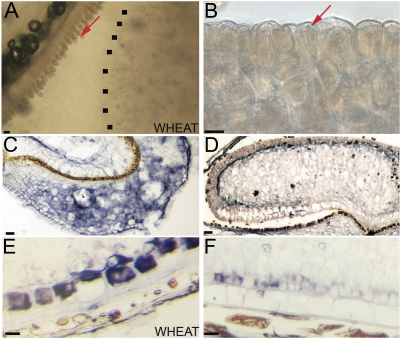
Fig. 3.
Analysis of aleurone tissue in Brachypodium and wheat grains. (A) Adherence of aleurone layer to the maternal tissues in wheat. (B) Adherence of aleurone layer to the endosperm in Brachypodium grains when grains are soaked in water and thin sections made with a sharp blade. The dotted line indicates the edge of the central endosperm. (C) ISH of BdPPDK showing expression in the pericarp of 4 DAA (days after anthesis) grains; (D) BdPPDK in the peripheral endosperm of 15 DAA grains. (E) Specific comparison of wheat PPDK with (F) BdPPDK in the aleurone layers. Scale bars, 20 μm.
The irregular nature and variable cell morphology of the aleurone layer make it difficult to determine the timing of its differentiation from the central endosperm using cytological examination. Therefore, to investigate the timing of aleurone at the molecular level, the temporal and spatial expression of presumptive aleurone-specific marker genes was examined. ISH using a BdPPDK probe suggests that the aleurone differentiates later or is less distinct in Brachypodium. BdPPDK transcript accumulated in the pericarp tissue at early stages (Fig. 3C) as was the case in wheat, but the expression in the peripheral endosperm was both weaker and occurred later during development than is the case in wheat (Fig. 3D–F). In wheat, expression is strong and coincides with differentiation of the peripheral endosperm, whereas in Brachypodium accumulation is only detected well after cellularization. In wheat the PPDK transcript accumulates specificially in the peripheral aleurone and is absent from the modified aleurone but, in Brachypodium, the distinction was less obvious.
The modified aleurone domain is absent in Brachypodium
To investigate further the possible absence of localized specialization within the aleurone layer, accumulation of an α-galactosidase transcript that was specifically expressed in the modified aleurone in wheat (Drea et al., 2005b) was examined. ISH with this probe did not produce any localized signal in the corresponding region in Brachypodium (data not shown), supporting the suggestion that the aleurone is not regionally differentiated.
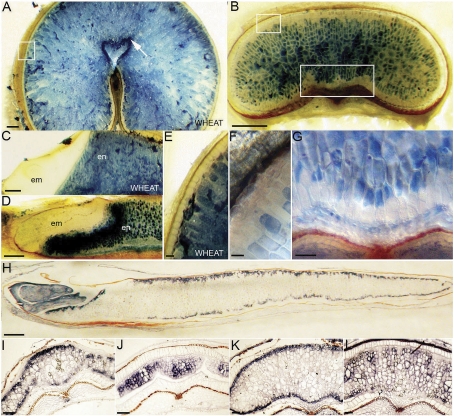
To confirm that modified aleurone is indeed absent, a vital dye, Evans Blue, was utilized to stain freshly cut sections of grains. Dead cells absorb and stain strongly with the blue dye (Young and Gallie, 1999). In the mature wheat grain, the only living tissues are the aleurone and embryo (Fig. 4A, C, E; Young and Gallie, 1999) so, while most of the aleurone of wheat excludes the dye and remains clear, the modified aleurone of the mature wheat grain stains strongly (Fig. 4A) because it is composed of dead cells. However, in the mature Brachypodium grain, while the aleurone layer and embryo exclude the dye as in wheat (Fig. 4B, D, F) so also does the region adjacent to the vascular bundle (Fig. 4B, G). This confirms that the Brachypodium endosperm is bounded by a continuous layer of living aleurone cells and does not possess a distinctive modified aleurone region. The distinction of peripheral and modified aleurone in other cereals such as wheat is made clear at the molecular level by the expression of genes in only one aleurone layer, such as PPDK. Since the expression pattern for this gene in Brachypodium grains did not definitively mark the aleurone as it does in wheat, another aleurone marker, BdGLO1, an orthologue of which is expressed in the aleurone of barley (Heck et al., 1993), was used. The BdGLO1 transcript is restricted to the aleurone layers and the embryo of the mature Brachypodium grain (Fig. 4H). A continuous peripheral pattern of expression extending into the adaxial region above the vasculature was detected from ∼7–8 DAA (Fig. 4I), confirming that the aleurone is not differentiated into discrete cell types as in wheat and barley. BdGLO1 transcript accumulation in the abaxial aleurone was found to extend into between one and four cell layers in an irregular manner, contrasting with a more discrete localization to a single cell layer in the adaxial layer (Fig. 4H).
Fig. 4.
Cellular and molecular domains within the Brachypodium endosperm. (A) Mature wheat and (B) Brachypodium grain cross-sections stained with Evans Blue. Boxed regions are shown in more detail in E, F, and G. Scale bars, 200 μm. (C) Wheat and (D) Brachypodium embryos are clear of staining. em, embryo; en; endosperm. Scale bars, 500 μm. (E) Wheat and (F) Brachypodium peripheral aleurone layers are clear of staining. Scale bars, 10 μm and 50 μm. (G) Brachypodium endosperm cells above the vascular/crease region are unstained. Scale bar, 50 μm. (H) BdGLO1expression in a mature grain long section. Scale bar, 500 μm. (I) BdGLO1expression at 7 DAA (days after anthesis) and (J) 20 DAA in grain cross-sections. (K) BdGLO2 expression at 7 DAA and (L) 20 DAA in grain cross-sections. Scale bars I–L, 50 μm.
In contrast to BdGLO1, BdGLO2 (a member of a family of genes related to the 12S globulins of oat) transcripts were localized to the central endosperm and excluded from the peripheral layers (Fig. 4J, L). In addition, BdGLO2 was not detected in the embryo (data not shown), in contrast to the strong expression of BdGLO1 (Fig. 4H). These detailed spatial and temporal expression patterns agree with the RT-PCR results indicating a slightly earlier onset of expression for BdGLO2 and indicate that specification of aleurone cell identity occurs after central endosperm proliferation has ceased (Supplementary Fig. S2 at JXB online).
Brachypodium central endosperm is composed of small thick-walled cells
Starch forms the major storage product of all the major cultivated cereals, accounting for up to 75% of the wheat grain (Bewley and Black, 1994). Brachypodium and wheat grains differ in the number and size of starch granules (Fig. 5A). While wheat endosperm cells were tightly packed with variable sized starch grains (Tomlinson et al., 2003), the equivalent cells in Brachypodium have few and small granules as judged by scanning electron microscopy (Fig. 5A). However, cell walls in the central endosperm of Brachypodium are very striking, reaching 4 μm (Fig. 5A). Measurements were taken from scanning elelctron micrographs of mature Brachypodium and wheat grains to quantify the differences in cell size and cell wall thicknesses (Fig. 5A; Table 1). Wheat endosperm cells are significantly larger than Brachypodium cells in both the aleurone and central endosperm, but the cell walls of Brachypodium endosperm cells are over twice as thick as wheat cells in the central endosperm. During germination, the cell walls of the central endosperm become attenuated to 2.38±0.74 μm and depleted of calcofluor-stained material at 6 d post-germination (Supplementary Fig. S3 at JXB online), suggesting mobilization of cell wall components during germination.
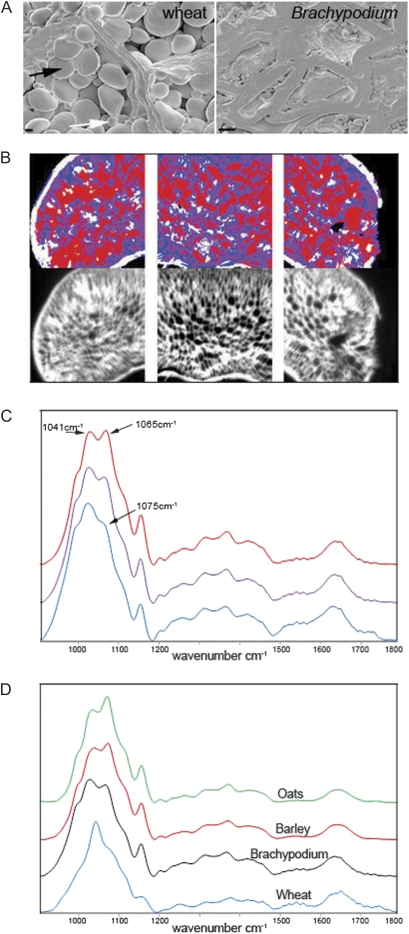
Fig. 5.
Composition of the mature Brachypodium grain. (A) Scanning electron micrographs of grain cross-sections showing packed starch grains in wheat in contrast to the small and few starch grains in Brachypodium and the very thick cell walls of its central endosperm. Large and small starch granules in wheat are indicated by black and white arrows, respectively. Scale bars, 5 μm. (B) FT-IR spectroscopic images above the visible images for a thin cell-wall-only cross-section of a Brachypodium grain. Blue pixels indicate spectra with a shoulder at 1075 cm−1, purple pixels a peak at 1065 cm−1, and red a peak at 1065 cm−1 greater in absorbance than the peak at 1041 cm−1. (C) The average spectra for each colour-coded region. (D) Average spectra for the endosperm cell walls of wheat, Brachypodium, barley, and oats.
As a first step towards determining the composition of Brachypodium cell walls and comparing these with other grains, FT-IR spectroscopy was carried out on the endosperm cell wall network and the spectra compared with those of other small grain cereals. FT-IR spectroscopic imaging reveals the composition and spatial distribution of cell wall components in situ, by coupling imaging techniques with infrared spectroscopic analysis on transverse de-starched tissue sections (Toole et al., 2007). The resultant spectral images were colour-coded based on β-glucan content (Fig. 5B, C). A peak at 1065 cm−1 is indicative of the β-glucan content (characteristically high in barley endosperm cell walls); in contrast, a shoulder at 1075 cm−1 indicates arabinoxylan (AX; typical of wheat). Arbitrary limits were set for each colour classification, with pixels where the spectra were more similar to that for AX (i.e. a shoulder at 1075 cm−1) being coloured blue (low β-glucan), those where the spectra showed a peak at 1065 cm−1 rather than a shoulder being coloured purple (medium β-glucan), and those with a β-glucan peak at 1065 cm−1, being higher than the main carbohydrate peak at 1041 cm−1, being coloured red (high β-glucan). The average spectra for each classification colour are shown in Fig. 5C and provide an indication of the relative abundance of the cell wall components. Thus the increased levels of blue and purple pixels indicate that the outer aleurone and subaleurone regions of the endosperm were higher in AX and lower in β-glucan as compared with the central region of the grain (which have increased red pixels). Figure 5D also shows the average spectra for similar thin sections of the endosperm cell walls of wheat (Toole et al., 2007), barley, and oats (G. A. Toole et al., unpublished results), and shows that Brachypodium endosperm cell walls were similar in general profile to barley and oats but with a lower β-glucan content. The additional shoulder at 980 cm−1 is most probably due to other insoluble cell wall hemicelluloses, and was found to be higher in Brachypodium than in the other cereals (Fig. 5D).
Discussion
Although Brachypodium grain development is broadly similar to that of wheat and the other temperate small grain cereals, there are a number of distinct differences. Some features such as the prominent and persistent nucellar epidermis may reflect its phylogenetic position as intermediate between rice and the Triticeae (Kellogg, 2001). However, other traits such as thick-walled endosperm cells and the lack of a modified aleurone may be distinctive for wild grasses.
Cell walls are a major component of the Brachypodium endosperm
Members of the Triticeae are generally rich in starch in the form of bimodal granules (Tomlinson et al., 2003), but Brachypodium endosperm cells tend to have smaller even-sized granules. The central endosperm cells tend to be much smaller than the equivalent cells in wheat with relatively thicker cell walls. The cell walls have a similar compositional profile to those of barley and oats in that they have more β-glucan than AXs but their overall composition is unique amongst the small grain cereals examined. Analysis of seedling leaf cell walls in Brachypodium also revealed profiles distinctive from those of wheat and barley. This was illustrated most dramatically by increased expression of BdCSL (cellulose synthase-like) genes (involved in β-D-glucan biosynthesis) and distinctive proportions of the constituents of hydroxycinnamic acids (Christensen et al., 2009).
Cell wall-associated materials may act as a storage product, and the germinating grain seems to re-mobilize these materials associated with the later stages of germination. A significant decrease in calcofluor staining was observed in the cell walls of the central endosperm between 3 d and 6 d post-germination (Supplementary Fig. S3B, D at JXB online) while the cell walls of the nucellar epidermis and aleurone remained relatively intact in comparison. Cell walls act in this capacity in a variety of seeds, including legumes and rice (McCleary and Matheson, 1976; Akiyama et al., 1998), while in other seeds they remain largely intact except for pores generated to aid mobilization of starch and protein reserves (Palmer, 1991). FT-IR analysis of the endosperm cell walls of Brachypodium showed a pronounced shoulder at 980 cm−1 that suggests the presence of insoluble hemicelluloses. Indeed, coffee beans are extremely hard because of hemicelluloses present in thick cell walls of the endosperm (Sutherland et al., 2004), and an alternative possible function, not mutually exclusive with storage, is to act as a physical barrier in seed defence. The overall high protein content relative to starch (Larre et al., 2010) and the predominance of glutelin-type storage proteins positions Brachypodium grain composition closer to that of oats than to that of wheat (Larre et al., 2010).
Hard grains (due to the thick cell walls) may have adaptive significance in relation to insect predation. Ants are a major grain predator for Brachypodium in some regions, and some ant species feed almost exclusively on grass grains. While a correlation with a particular grain composition is not clearly defined, in some studies granivorous ants generally favour elongated rather than round grains and grains that have obvious awns or hairs projecting (Pulliam and Brand, 1975). Some grains have mechanical properties, such as hardness, to protect against predation while positively influencing dispersal (Oliveras et al., 2008), and it seems likely that cell walls may contribute to this.
Differences in grain organization may reflect different grain-filling mechanisms
The generally accepted conduit for maternal nutritional supplies early in wheat endosperm development (Wang et al., 1994a, b, c) appears to be modified in Brachypodium. First, the nucellar projection is very much reduced throughout development. Secondly, the inner integument layer that forms the seed coat (a waterproof barrier) differentiates very early in the grain (4 DAA). Finally, the modified aleurone layer is absent as judged by several independent tests. This suggests that the modified aleurone layer is a distinctive feature of the Triticeae and can be addressed in a wider comparative analysis extending to neighbouring tribes. The modified aleurone layer is implicated as a major transfer tissue in wheat (Wang et al., 1994c). However, an alternative transport route may exist as the nucellar epidermis is persistent, similar to rice grains. Rice has two pathways involved in the transport of nutrients to the developing caryopsis: one via a pathway analogous to the nucellar projection pathway and the other via the nucellar epidermis (Krishnan and Dayanandan, 2003). Brachypodium grain conformation may be more similar to rice in this respect than to wheat, barley, or maize, which have obvious modified aleurone transfer layers adjacent to the maternal vascular crease. Rice has a vascular system extending the length of the grain, whereas the vascular tissue supplying the maize grain terminates at the junction of the funiculus and ovule (as in Arabidopsis). Maize, in turn, has a pronounced transfer cell layer within the endosperm, i.e. the basal endosperm transfer layer (BETL). Rice lacks this basal differentiation within its endosperm tissue and may utilize an alternative nucellar epidermis-mediated transport route (Ellis and Chaffey, 1987). Throughout caryopsis development in rice, the nucellar epidermis is present as an intact layer, but during the later stage of grain filling, the nucellar epidermis along with integument and pericarp becomes compressed. This compression coincides with continued endosperm expansion and filling. The structural collapse of nucellar epidermis blocks the flow of assimilates to endosperm and consequently inhibits further grain filling via this pathway (Ellis and Chaffey, 1987). As in rice, the lack of a transfer cell layer in Brachypodium is coupled with the persistence of a functional nucellar epidermis whose cross-walls are rich in plasmodesmata. The nucellar epidermis in Brachypodium is well developed along the whole caryopsis, and is minimally compressed by expanding endosperm (Fig. 2D). Therefore, it could play a major part in assimilate transport into the caryopsis. Indeed, the thickened cell walls of the nucellar epidermis may contribute to alternative carbon storage (Supplementary Fig. S3 at JXB online). Alternatively they may provide additional defence against predation.
Aleurone differentiation and organization
These results indicate that the aleurone differentiates relatively late in Brachypodium, lacks regional specialization, and is structurally integral with the central endosperm. Moreover, the number of cell layers contributing to the aleurone varies both locally and regionally—abaxially the aleurone appears to be several cells deep in terms of BdGLO1 expression patterns and vital staining but thinner and more homogeneous on the adaxial side. The number of cell layers in the aleurone is characteristic of the cereal species, with wheat, rye, oats, maize, and sorghum having a single aleurone layer and rice and barley having three layers (Kent and Evers, 1994). These layers have a high protein and lipid content but low starch. Several genes are expressed in the aleurone, either specifically or strongly up-regulated, and serve as good markers to follow its early development and subsequent differentiation.
Brachypodium has potential as a rapid and cost-effective model for dissecting numerous aspects of grass biology, providing a small rapid cycling model whose biology has not been influenced, at least directly, by humans. This paper provides a comprehensive cellular and molecular description of grain development in the reference strain, Bd21, providing a reference map that will underpin future work on genetic control of Brachypodium grain development. In addition it was used as the basis for the first detailed comparison of grain development in Brachypodium and related cereals. This analysis of grain development indicates that, although somewhat more closely allied to the Triticeae, Brachypodium is intermediate in many respects between this group and rice, with some features that may be unique to wild and forage grasses. Thus the Brachypodium grain is not only a good model for many aspects of cereal biology but it will also be informative in understanding the evolution of diversity in grain structure across the grasses. A greater understanding of the significant differences between the grains of non-crop species to the nutritional storehouses of cultivated cereals—which have been subject to intensive breeding during domestication—should provide knowledge to ensure greater yields and food security (Sabelli and Larkins, 2009).
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Macro-characteristics of the Brachypodium grain.
Figure S2. Brachypodium genes used as molecular markers for in situ hybridization (ISH).
Figure S3. Degradation of endosperm cell walls in early stages of grain germination.
Supplementary Material
Acknowledgments
MO was funded by an EU Re-Integration Fellowship. JHD acknowledges Gatsby and Leverhulme Research Grants. PH is funded by a BBSRC studentship and from a Royal Society Research Grant and University of Leicester start-up grant to SD. We thank David Garvin, Kay Denyer, Philippe Vain, and Peter Shaw for useful discussions. We also thank Anika Tailor for her contribution to the project.
References
- Akiyama T, Kaku H, Shibuya N. A cell wall-bound beta-glucosidase from germinated rice: purification and properties. Phytochemistry. 1998;48:49–54. doi: 10.1016/s0031-9422(97)01099-6. [DOI] [PubMed] [Google Scholar]
- Becraft PW. Cell fate specification in the cereal endosperm. Seminars in Cell and Developmental Biology. 2001;12:387–394. doi: 10.1006/scdb.2001.0268. [DOI] [PubMed] [Google Scholar]
- Bewley JD, Black M. Seeds. Physiology of development and germination. New York: Plenum Press; 1994. [Google Scholar]
- Brown R, Lemmon B, Olsen O-A. Development of the endosperm in rice (Oryza sativa L.): cellularization. Journal of Plant Research. 1996;109:301–313. [Google Scholar]
- Chastain C, Heck J, Colquhoun T, Voge D, Gu X-Y. Posttranslational regulation of pyruvate, orthophosphate dikinase in developing rice (Oryza sativa) seeds. Planta. 2006;224:924–934. doi: 10.1007/s00425-006-0259-3. [DOI] [PubMed] [Google Scholar]
- Christensen U, Alonso-Simon A, Scheller HV, Willats WG, Harholt J. Characterization of the primary cell walls of seedlings of Brachypodium distachyon—a potential model plant for temperate grasses. Phytochemistry. 2009;71:62–69. doi: 10.1016/j.phytochem.2009.09.019. [DOI] [PubMed] [Google Scholar]
- Costa LM, Gutierrez-Marcos JF, Brutnell TP, Greenland AJ, Dickinson HG. The globby1-1 (glo1-1) mutation disrupts nuclear and cell division in the developing maize seed causing alterations in endosperm cell fate and tissue differentiation. Development. 2003;130:5009–5017. doi: 10.1242/dev.00692. [DOI] [PubMed] [Google Scholar]
- Doan DN, Linnestad C, Olsen OA. Isolation of molecular markers from the barley endosperm coenocyte and the surrounding nucellus cell layers. Plant Molecular Biology. 1996;31:877–886. doi: 10.1007/BF00019474. [DOI] [PubMed] [Google Scholar]
- Draper J, Mur LA, Jenkins G, Ghosh-Biswas GC, Bablak P, Hasterok R, Routledge AP. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiology. 2001;127:1539–1555. [PMC free article] [PubMed] [Google Scholar]
- Drea S, Corsar J, Crawford B, Shaw P, Dolan L, Doonan JH. A streamlined method for systematic, high resolution in situ analysis of mRNA distribution in plants. Plant Methods. 2005a;1:8. doi: 10.1186/1746-4811-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drea S, Leader DJ, Arnold BC, Shaw P, Dolan L, Doonan JH. Systematic spatial analysis of gene expression during wheat caryopsis development. The Plant Cell. 2005b;17:2172–2185. doi: 10.1105/tpc.105.034058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JR, Chaffey NJ. Structural differentiation of the nucellar epidermis in the caryopsis of rice (Oryza sativa) Annals of Botany. 1987;60:671–675. [Google Scholar]
- Fobert PR, Coen ES, Murphy GJ, Doonan JH. Patterns of cell division revealed by transcriptional regulation of genes during the cell cycle in plants. EMBO Journal. 1994;13:616–624. doi: 10.1002/j.1460-2075.1994.tb06299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill BS, Appels R, Botha-Oberholster AM, et al. A workshop report on wheat genome sequencing: International Genome Research on Wheat Consortium. Genetics. 2004;168:1087–1096. doi: 10.1534/genetics.104.034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S, Sharp R, Foote TN, Bertin I, Wanous M, Reader S, Colas I, Moore G. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature. 2006;439:749–752. doi: 10.1038/nature04434. [DOI] [PubMed] [Google Scholar]
- Gruis DF, Guo H, Selinger D, Tian Q, Olsen OA. Surface position, not signaling from surrounding maternal tissues, specifies aleurone epidermal cell fate in maize. Plant Physiology. 2006;141:898–909. doi: 10.1104/pp.106.080945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Costa LM, Biderre-Petit C, Khbaya B, O'Sullivan DM, Wormald M, Perez P, Dickinson HG. maternally expressed gene1 is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. The Plant Cell. 2004;16:1288–1301. doi: 10.1105/tpc.019778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck GR, Chamberlain AK, Ho TH. Barley embryo globulin 1 gene, Beg1: characterization of cDNA, chromosome mapping and regulation of expression. Molecular and General Genetics. 1993;239:209–218. doi: 10.1007/BF00281620. [DOI] [PubMed] [Google Scholar]
- Hong-Gyu K, Sunhee P, Makoto M, Gynheung A. White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C4 type pyruvate orthophosphate dikinase gene (OsPPDKB) The Plant Journal. 2005;42:901–911. doi: 10.1111/j.1365-313X.2005.02423.x. [DOI] [PubMed] [Google Scholar]
- Hubbard CE. Grasses. London: Pelican Books; 1954. [Google Scholar]
- Imaizumi N, Ku MSB, Ishihara K, Samejima M, Kaneko S, Matsuoka M. Characterization of the gene for pyruvate, orthophosphate dikinase from rice, a C3 plant, and a comparison of structure and expression between C3 and C4 genes for this protein. Plant Molecular Biology. 1997;34:701–716. doi: 10.1023/a:1005884515840. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Kellogg EA. Evolutionary history of the grasses. Plant Physiology. 2001;125:1198–1205. doi: 10.1104/pp.125.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent NL, Evers AD. Technology of cereals. Amsterdam: Elsevier Science Ltd; 1994. [Google Scholar]
- Khan MA, Stace CA. Breeding relationships in the genus Brachypodium (Poaceae: Pooideae) Nordic Journal of Botany. 1999;19:13. [Google Scholar]
- Krishnan S, Dayanandan P. Structural and histochemical studies on grain-filling in the caryopsis of rice (Oryza sativa L.) Journal of Bioscience. 2003;28:455–469. doi: 10.1007/BF02705120. [DOI] [PubMed] [Google Scholar]
- Larre C, Pennick B, Bouchet V, Lollier O, Tranquet S, Denery-Papini S, Guillon F, Rogniaux H. Brachypodium distachyon grain: identification and subcellular localization of storage proteins. Journal of Experimental Botany. 2010;61:1771–1782. doi: 10.1093/jxb/erq050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudencia-Chingcuanco DL, Vensel WH. Globulins are the main seed storage proteins in Brachypodium distachyon. Theoretical and Applied Genetics. 2008;117:555–563. doi: 10.1007/s00122-008-0799-y. [DOI] [PubMed] [Google Scholar]
- Linnestad C, Doan DNP, Brown RC, Lemmon BE, Meyer DJ, Jung R, Olsen O-A. Nucellain, a barley homolog of the dicot vacuolar-processing protease, is localized in nucellar cell walls. Plant Physiology. 1998;118:1169–1180. doi: 10.1104/pp.118.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary BV, Matheson NK. Galactomannan utilization in germinating legume seeds. Phytochemistry. 1976;15:43–47. [Google Scholar]
- Mercader J. Mozambican grass seed consumption during the Middle Stone Age. Science. 2009;326:1680–1683. doi: 10.1126/science.1173966. [DOI] [PubMed] [Google Scholar]
- Oliveras J, Gómez C, Bas J, Espadaler X. Mechanical defence in seeds to avoid predation by a granivorous ant. Naturwissenschaften. 2008;95:501–506. doi: 10.1007/s00114-008-0349-0. [DOI] [PubMed] [Google Scholar]
- Olsen OA. ENDOSPERM DEVELOPMENT: cellularization and cell fate specification. Annual Review Plant Physiology and Plant Molecular Biology. 2001;52:233–267. doi: 10.1146/annurev.arplant.52.1.233. [DOI] [PubMed] [Google Scholar]
- Olsen OA, Potter RH, Kalla R. Histo-differentiation and molecular biology of developing cereal endosperm. Seed Science Research. 1992;2:117–131. [Google Scholar]
- Opanowicz M, Vain P, Draper J, Parker D, Doonan JH. Brachypodium distachyon: making hay with a wild grass. Trends Plant Science. 2008;13:172–177. doi: 10.1016/j.tplants.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Gates P. Transport of assimilates in the developing caryopsis of rice (Oryza sativa L.) Planta. 1981;152:388–396. doi: 10.1007/BF00385354. [DOI] [PubMed] [Google Scholar]
- Opsahl-Ferstad HG, Le Deunff E, Dumas C, Rogowsky PM. ZmEsr, a novel endosperm-specific gene expressed in a restricted region around the maize embryo. The Plant Journal. 1997;12:235–246. doi: 10.1046/j.1365-313x.1997.12010235.x. [DOI] [PubMed] [Google Scholar]
- Palmer GH. Enzymatic degradation of the endosperm cell walls of germinated sorghum. World Journal of Microbiology and Biotechnology. 1991;7:17–21. doi: 10.1007/BF02310913. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- Pulliam HR, Brand MR. The production and utilization of seeds in plains grassland of Southeastern Arizona. Ecology. 1975;56:1158–1166. [Google Scholar]
- Sabelli PA, Larkins BA. The development of endosperm in grasses. Plant Physiology. 2009;149:14–26. doi: 10.1104/pp.108.129437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Schulte D, Close TJ, Graner A, et al. The international barley sequencing consortium—at the threshold of efficient access to the barley genome. Plant Physiology. 2009;149:142–147. doi: 10.1104/pp.108.128967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry PR. Wheat. Journal of Experimental Botany. 2009;60:1537–1553. doi: 10.1093/jxb/erp058. [DOI] [PubMed] [Google Scholar]
- Sutherland PW, Hallett IC, MacRae E, Fischer M, Redgwell RJ. Cytochemistry and immunolocalisation of polysaccharides and proteoglycans in the endosperm of green Arabica coffee beans. Protoplasma. 2004;223:203–211. doi: 10.1007/s00709-004-0036-8. [DOI] [PubMed] [Google Scholar]
- Tomlinson K, Denyer K, Callow JA. Starch synthesis in cereal grains. In: Callow JA, editor. Advances in botanical research. London: Academic Press; 2003. pp. 1–61. [Google Scholar]
- Toole GA, Wilson RH, Parker ML, Wellner NK, Wheeler TR, Shewry PR, Mills EN. The effect of environment on endosperm cell-wall development in Triticum aestivum during grain filling: an infrared spectroscopic imaging study. Planta. 2007;225:1393–1403. doi: 10.1007/s00425-006-0448-0. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Garvin DF, Mockler TC, et al. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- Wang H-L, Offler CE, Patrick JW, Ugalde TD. The cellular pathway of photosynthate transfer in the developing wheat grain. I. Delineation of a potential transfer pathway using fluorescent dyes. Plant, Cell and Environment. 1994a;17:257–266. [Google Scholar]
- Wang H-L, Offler CE, Patrick JW. The cellular pathway of photosynthate transfer in the developing grain. II. A structural analysis and histochemical studies of the pathway from the crease phloem to the endosperm cavity. Plant, Cell and Environment. 1994b;18:373–388. [Google Scholar]
- Wang H-L, Patrick JW, Offler CE, Wang X-D. The cellular pathway of photosynthate transfer in the developing grain. III. A structural analysis and physiological studies of the pathway from the endosperm cavity to the starchy endosperm. Plant, Cell and Environment. 1994c;18:389–407. [Google Scholar]
- Wegel E, Pilling E, Calder G, Drea S, Doonan J, Dolan L, Shaw P. Three-dimensional modelling of wheat endosperm development. New Phytologist. 2005;168:253–262. doi: 10.1111/j.1469-8137.2005.01503.x. [DOI] [PubMed] [Google Scholar]
- Young TE, Gallie DR. Analysis of programmed cell death in wheat endosperm reveals differences in endosperm development between cereals. Plant Molecular Biology. 1999;39:915–926. doi: 10.1023/a:1006134027834. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.