Abstract
Growth of petal cells is a basis for expansion and morphogenesis (outward bending) of petals during opening of carnation flowers (Dianthus caryophyllus L.). Petal growth progressed through elongation in the early stage, expansion with outward bending in the middle stage, and expansion of the whole area in the late stage of flower opening. In the present study, four cDNAs encoding xyloglucan endotransglucosylase/hydrolase (XTH) (DcXTH1–DcXTH4) and three cDNAs encoding expansin (DcEXPA1–DcEXPA3) were cloned from petals of opening carnation flowers and characterized. Real-time reverse transcription-PCR analyses showed that transcript levels of XTH and expansin genes accumulated differently in floral and vegetative tissues of carnation plants with opening flowers, indicating regulated expression of these genes. DcXTH2 and DcXTH3 transcripts were detected in large quantities in petals as compared with other tissues. DcEXPA1 and DcEXPA2 transcripts were markedly accumulated in petals of opening flowers. The action of XTH in growing petal tissues was confirmed by in situ staining of xyloglucan endotransglucosylase (XET) activity using a rhodamine-labelled xyloglucan nonasaccharide as a substrate. Based on the present findings, it is suggested that two XTH genes (DcXTH2 and DcXTH3) and two expansin genes (DcEXPA1 and DcEXPA2) are associated with petal growth and development during carnation flower opening.
Keywords: Carnation, expansin, flower opening, gene expression, in situ XET staining, petal cell growth, xyloglucan endotransglycosylase/hydrolase (XTH)
Introduction
The vase life of cut ornamental flowers consists of the period from flower opening to senescence. It is necessary to slow down both processes in order to prolong the display time of the flowers. Many studies have been carried out on the mechanism of flower opening (van Doorn and van Meeteren, 2003; Yamada et al., 2009; Harada et al., 2010), and on senescence to develop technologies for retarding senescence (Hunter et al., 2004; Hoeberichts et al., 2007; van Doorn and Woltering, 2008). The molecular mechanism of flower opening has been under investigation using cut carnation flowers as a model ornamental.
Flower opening involves elongation, expansion, and outward bending of petals, which result from the enlargement of petal cells (Koning, 1984; Kenis et al., 1985; Evans and Reid, 1988). Sugar accumulation in petal cells reduces the petal water potential and promotes water influx into the petal cells, resulting in cell enlargement (Ho and Nichols, 1977; Evans and Reid, 1988; Ichimura et al., 2003). The influx of water relaxes the cell wall strength, resulting in cell enlargement. The plant cell wall is a strong fibrillar network arranged to give each cell a solid shape. Cell wall extensibility may be a growth-limiting factor for petal expansion (Yamada et al., 2009). Several studies have addressed the cell wall changes during flower opening (de Vetten and Huber, 1990; Yamada et al., 2009).
Two kinds of apoplastic enzymes are considered to be involved in cell wall loosening in plants (Buchanan et al., 2000). One of them, xyloglucan endotransglycosylase/hydrolase (XTH), catalyses a transglycosylation of xyloglucan, in which one chain of xyloglucan is cleaved and reattached to the non-reducing end of another xyloglucan chain (Fry et al., 1992; Nishitani and Tominaga, 1992; Vissenberg et al., 2003). Such a mechanism allows cellulosic microfibrils to undergo a transient slippage. XTH may also act in the realignment of the xyloglucan chain in different strata during cell enlargement and in the assembly of the wall when newly synthesized xyloglucans are incorporated. Expansins catalyse breakage of hydrogen bonds between cellulose and the cross-linking glucans, leading to cell wall extension without any detectable hydrolytic or transglucotic events (McQueen-Mason et al., 1992; Cosgrove, 2000). The coordinate action of XTH with expansins facilitates loosening of the cross-linking glycans that tether microfibrils, causing separation of the microfibrils, driven by the osmotic pressure of the cell, and finally cell enlargement (Buchanan et al., 2000).
O'Donoghue et al. (2002) showed that XTH activity is associated with flower opening in Sandersonia. Breeze et al. (2004) showed that an XTH gene was up-regulated in developing petals of opening Alstroemeria flowers. Laitinen et al. (2007) showed that Gerbera hybrida had several XTH genes and that their expression pattern during development of ray flowers differed; some genes were up-regulated in the early stage and down-regulated in the late stage of petal development, and others were affected in the opposite way.
The function of α-expansin (PhEXPA1) was characterized in Petunia hybrida petals by reducing its protein levels with a transgenic antisense approach (Zenoni et al., 2004). This study showed alterations in cell wall morphology and composition, suggesting that expansins play a role in the assembly of the cell wall by affecting either cellulose synthesis or deposition. In Mirabilis jalapa plants, α- and β-expansins were identified in opening and senescing flowers (Gookin et al., 2003). In general, α-expansin gene expression was high during maximal elongation of the floral tube and β-expansin was preferentially expressed during the early developmental stage of petunia flowers. More recently, it has been shown that rose plants have four XTH genes (RhXTH1–RhXTH4) and three α-expansin genes (RhEXPA1–RhEXPA3), and RhXTH1 and RhEXPA1 are mainly involved in expansion growth of rose petals (Takahashi et al., 2007; Yamada et al., 2009).
In the present study, the full-length cDNAs of four XTH genes (DcXTH1–DcXTH4) and three expansion genes (DcEXPA1–DcEXPA3) were cloned from carnation (Dianthus caryophyllus L.), and their expression profiles during growth and outward bending of petals during flower opening were examined. In addition, XTH was detected and localized in growing petals by activity staining of xyloglucan endotransglucosylase (XET) using a xyloglucan nonasaccharide labelled with a specific fluorescent dye as a substrate.
Materials and methods
Plant materials and sample preparation
Cut flowers of carnation (D. caryophyllus L. cv. Light Pink Barbara) that belongs to the spray category of carnation flower were harvested when the first flower out of 5–6 flower buds was nearly open at the nursery of a commercial grower in Miyagi prefecture. The flowers were transported dry to the laboratory at the Kyoto Prefectural Institute of Agricultural Biotechnology in Kyoto prefecture on the next day after harvest. They were put in plastic containers with their cut end in water under continuous light from white fluorescent lamps (14 μmol m−2 s−1) at 23 °C. The flower opening process was separated into the following six stages: stage 1, petals just emerged from buds; stage 2, petals elongated vertically; stage 3, petal clusters expanded; stage 4, outer petals start to bend outwards; stage 5, outer petals are bent outwards; stage 6, fully open flower with outer petals at right angles to the stem (Harada et al., 2010). Photographs of flowers at the respective opening stages and detached petals at stages 1 and 5 were shown previously (Fig. 1 in Harada et al., 2010). The 10 outermost petals were detached from five flowers at each stage, mixed (making a sample of 50 petals), and stored at –80 °C for extraction of RNA. Different tissues, i.e. leaves, stem, calyx, ovary, and style (actually style plus stigma), were detached from carnation plants with fully open flowers and stored as above.
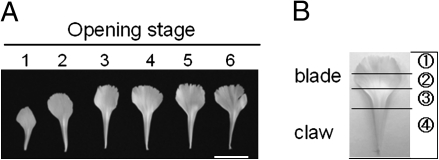
Fig. 1.
Profiles of petals detached from flowers at the respective opening stages (A) and separation of a petal into four different parts (B). Scale bar=2 cm.
In carnation the stigma is along the surface of the style (rather than a distinct structure as in other flowers). Therefore, the term ‘style’ was used to describe the stigma plus style herein.
RNA extraction
Total RNA was isolated by the cetyltrimetylammonim bromide (CTAB) method from petal tissues (Murray and Thompson, 1980; Chang et al., 1993) and precipitated with LiCl (Nakamura et al., 1991). Later, total RNA was isolated by the phenol–chloroform method as described by Harada et al. (2005). RNA was also extracted from other tissues of opening carnation flowers.
PCR cloning of cDNAs for XTHs and expansins
To obtain full-length composite cDNAs for XTH and expansins, a nearly identical strategy was used in which two or three partial-length cDNAs were obtained by ordinary RT-PCR, 5′-RACE (rapid amplification of cDNA ends; Frohman et al., 1988), and 3′-RACE techniques. The cDNAs were combined to make composite cDNAs. Then, to confirm the complete nucleotide sequences, full-length cDNAs were amplified with primers derived from both ends of the composite cDNAs and total RNAs obtained from opening carnation petals. The upstream and downstream primer pairs for PCR to obtain partial length cDNAs and full-length cDNAs, and the sizes (in base pairs) of amplificates and composite cDNAs are summarized in Supplemenatry Table S1, and their nucleotide sequences in Supplementary Table S2 available at JXB online. Detailed procedures for PCR cloning are given in Supplementary Table S3 with DcXTH1and DcXTH2 cDNAs as examples.
Nucleotide sequences were edited and analysed by GENETYX-WIN, and homology search was by the BLASTX program on the DDBJ website. Signal peptide sequences were predicted by PSORT. A homology search was conducted with the BLAST program.
Real-time PCR
Real-time RT-PCR for the quantification of transcripts for DcXTH1–DcXTH4 and DcEXPA1–DcEXPA3 was conducted using the LightCycler FastStart DNA Master SYBR Green I Kit and the LightCycler Instrument (Roche, Basel, Switzerland). cDNA was synthesized using ReverTra Ace (TOYOBO, Osaka, Japan) according to the manufacturer's instructions. Primer sets for the respective XTH and expansin cDNAs as well as carnation actin (DcACT1, acccession no. AY0073415) mRNA (Waki et al., 2001) are summarized in Supplementary Table S4 at JXB online. The sizes of amplificates ranged from 88 bp to 256 bp. PCR conditions were initial heating for 10 min at 95 °C followed by 40 cycles of 5 s at 95 °C, 5 s at 53 °C, and 4–11 s at 72 °C, in which the extension time was varied depending on the length of amplificates (1 s per 25mer). The absolute transcript level was calculated using a dilution series of a target sequence on LightCycler Software Ver. 3.5. DcACT1 was almost constant in transcript level throughout all stages and tissues, and was used to normalize the transcript level. Three RNA preparations per stage, obtained from three independent petal samples at each stage as described above, were used for analyses, and data were shown by the mean ±SE of the three preparations for each stage. Statistical analyses were carried out by Tukey's multiple range test using an online statistical analysis program MEPHAS (http://www.gen-info.osaka-u.ac.jp/testdocs/tomocom/).
In situ detection of XET activity
In situ detection of XTH in petal tissues was conducted by co-localization of XET activity and its donor xyloglucan polymer labelled with fluorescent dye [sulphorhodamine (SR)] at its reducing end according to Vissenburg et al. (2000). The reaction mixture was prepared to contain 6.5 mM XLLG-SR/2.5 mM MES-NaOH (pH 5.5), whereas the control mixture contained GGGG-SR instead of XLLG-SR. XLLG-SR and GGGG-SR are SR derivatives of xyloglucan nonasaccharide and cellotetraose, respectively. Square segments (5×5 mm2) were detached from three different parts (the claw, the boundary of the claw and blade, and the blade) of carnation petals using a razor blade, and immersed in the reaction or control mixtures (one segment per 300 μl of the mixture), and left under reduced pressure for 3 h at room temperature in the dark. Then the segments were washed in ethanol/formic acid/water (15:1:4 v/v/v) for 10 min to remove any remaining unreacted XLLG-SR or GGGG-SR, and further incubated overnight in 5% formic acid to remove apoplastic, non-wall-bound xyloglucan-SR. The tissue segments were rinsed in 25 mM MES-NaOH (pH 5.5), embedded in 3% (w/v) caboxymethylcellulose (CMC) gel, and frozen in cold hexane (–75 °C). Cryostat sections of 100 μm thickness were prepared using the Cryo-Film Transfer Kit (Fintec Co. Ltd, Tokyo, Japan) (Kawamoto, 2003). The film sections were put on slide glasses, and CMC in the section was removed by adding a water drop and then removing it. The sections were then mounted with glycerin under a cover glass. The sections were inspected under a fluorescence microscope (BX51, Olympus, Tokyo, Japan) using green (540 nm) excitation light and by IM Image Manager software (Leica, Wetzlar, Germany) and Aquacosmos software (Hamamatsu Photonics, Hamamatsu, Japan).
Results
Carnation flower opening and petal growth
The stages of flower opening were as reported by Harada et al. (2010). Stages 1 and 2 were regarded as the elongation period and stages 3–6 as the opening period. Notably, the growing petals started to bend outward at the boundary between the claw and blade at stage 4, and this bending proceeded with flower opening at stage 5. Figure 1 shows front views of petals detached from the outermost whorl of flowers at the respective opening stages and dissection of a petal into four parts, which were used for analyses in the experiments shown in Figs 4 and 5.
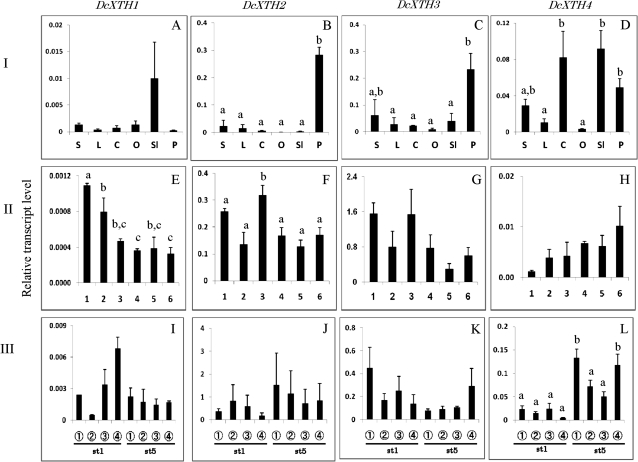
Fig. 4.
Comparison of transcript levels of DcXTH1–DcXTH4 genes among different tissues (I: A–D), and their changes in whole petals at each stage (II: E–H) and in different parts of petals (III: I–L) during flower opening. The six flower opening stages were defined as: stage 1, petals just emerged from buds; stage 2, petals elongated vertically; stage 3, petal clusters expanded; stage 4, outer petals start to bend outward; stage 5, outer petals are bent outward; stage 6, fully open flower with outer petals at right angles to the stem. Relative transcript levels are calculated by real-time RT-PCR with DcACT1 as a standard. Data are means ±SE of three independent samples, each prepared by mixing 10 outermost petals detached from five separate flowers at the respective stages. Significant difference (P <0.05) detected by Tukey's multiple range test is shown by different letters above the bars, only where there was a significant difference among determinations in each panel. S, stem; L, leaf; C, calyx; O, ovary; Sl, style+stigma; P, petal.
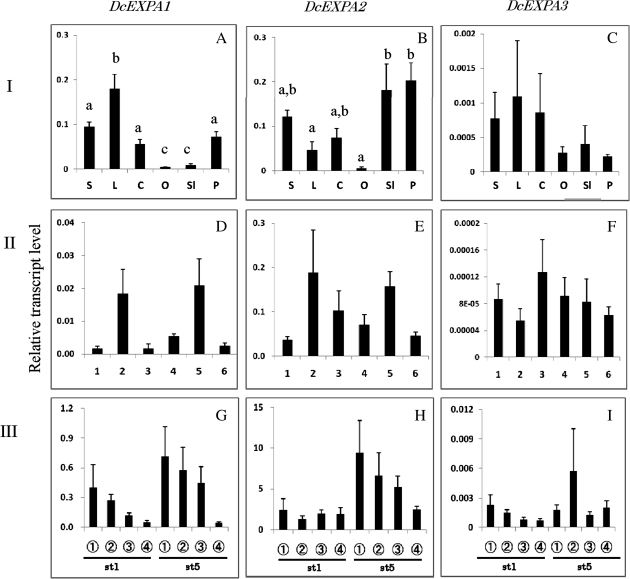
Fig. 5.
Comparison of transcript levels of DcEXPA1–DcEXPA3 genes among different tissues (I: A–C), and their changes in whole petals at each stage (II: D–F) and in different parts of petals (III: G–I) during flower opening. The procedures are the same as described in the legend to Fig. 4.
Cloning and structural analyses of DcXTH1–DcXTH4 and DcEXPA1–DcEXPA3
cDNAs encoding four putative XTHs and three putative expansins were isolated from opening petals of ‘Light Pink Barbara’ carnation using a combination of RT-PCR, 5′-RACE, and 3′-RACE techniques. After the composite cDNAs were obtained, the nucleotide sequences for the open reading frame (ORF) of all cDNAs, except one, were confirmed by cloning of the complete (full-length) cDNAs by PCR and sequencing. One XTH cDNA, designated DcXTH1 hereafter, was cloned first, and its composite cDNA was deposited, without further cloning of complete cDNA, in the NCBI database in 2006 with accession no. DQ449624. The other three XTH cDNAs were designated DcXTH2, DcXTH3, and DcXTH4, and deposited in the DDBJ with accession nos AB543809, AB542069, and AB542070, respectively. The three expansin cDNAs were designated DcEXPA1, DcEXPA2, and DcEXPA3, and deposited in the DDBJ with accession nos AB542071, AB542072, and AB542073, respectively.
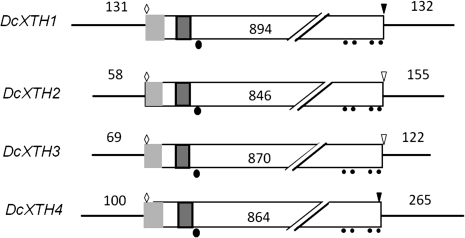
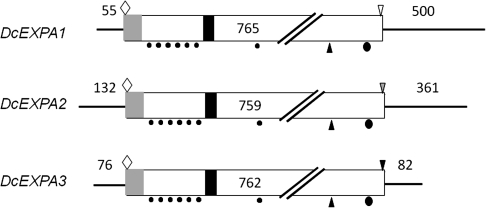
The structures of four XTH cDNAs are summarized in Fig. 2, which shows 5′-flanking sequences, ORFs, and 3′-flanking sequences. The cDNAs were 1157 bp long for DcXTH1, 1059 bp long for DcXTH2, 1061 bp long for DcXTH3, and 1229 bp long for DcXTH4. The predicted proteins consisted of 297 amino acids for DcXTH1, 281 amino acids for DcXTH2, 289 amino acids for DcXTH3, and 287 amino acids for DcXTH4, corresponding to the calculated molecular mass of 34.8, 31.7, 32.5, and 33.0 kDa, respectively. All of the four predicted proteins have a potential signal peptide sequence (e.g. amino acids 1–56 in DcXTH1), an active site motif (DFEFLG; e.g. amino acids 108–116 in DcXTH1), an N-glycosylation site (e.g. amino acid 107 in DcXTH1), and four cysteine residues (e.g. amino acids 231, 241, 277, and 290 in DcXTH1), probably responsible for the formation of intermolecular disulphide bonds. DcXTH1 has a stop codon TGA in common with DcXTH4, whereas both DcXTH2 and DcXTH3 have the stop codon TAG.
Fig. 2.
Structures of four XTH cDNAs cloned from petals of opening carnation flowers. Light shaded rectangle, signal peptide; dark shaded rectangle, active site motif; filled circles, cysteine residues; filled ellipsoid, N-glycosylation site; open diamond, ATG start codon; inverted open triangle, TGA stop codon; light shaded inverted triangle, TAG stop codon.
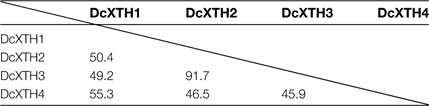
Table 1 shows the homology of the amino acid sequences of four predicted XTH proteins. DcXTH2 and DcXTH3 have a high homology of 91.7%, whereas the homology between other XTH proteins was 45.9–55.3%. The phylogenetic tree was constructed from the deduced amino acids sequences of four XTH proteins and 33 members of the Arabidopsis XTH family by the Clustral W program (Supplementary Fig. S1 at JXB online). DcXTH2 and DcXTH3 were clustered into group 1 of Arabidopsis XTH, whereas DcXTH1 and DcXTH4 clustered into group 2. XTHs belonging to group 1 and group 2 are thought to have xyloglucan endotransglucosylase activity as well as xyoglucan hydrolase activity (Rose et al., 2002). DcXTH2 and DcXTH3 proteins had high homology in amino acid sequences to BvXTH-1 (accession no. AY315428; Dimmer et al., 2004) of sugar beet (Beta vulgaris); that is, 76.2% and 78.2%, respectively. In contrast, DcXTH1 and DcXTH4 showed low homology to BvXTH-1; that is, 52.5% and 50.0%, respectively.
Table 1.
Homology (%) in amino acid sequences among predicted DcXTH1, DcXTH2, DcXTH3, and DcXTH4 proteins
 |
The structures of three expansin cDNAs are summarized in Fig. 3. The cDNAs were 1320 bp long for DcEXPA1, 1252 bp long for DcEXPA2, and 920 bp long for DcEXPA3. The predicted proteins consisted of 254 amino acids for DcEXPA1, 252 amino acids for DcEXPA2, and 253 amino acids for DcEXPA3, corresponding to the calculated molecular mass of 25.0, 26.8, and 27.8 kDa, respectively. Three predicted proteins have a potential signal peptide sequence (e.g. amino acids 1–22 in DcEXPA1), an HFD amino acid sequence (e.g. amino acids 129–131 in DcEXPA1), and conserved amino acids; that is, cysteine residues (e.g. amino acids 53, 81, 89, 96, 109, 123, and 157 in DcEXPA1), a valine residue (e.g. amino acid 225 in DcEXPA1), and a tryptophan residue (e.g. amino acid 248 in DcXTH1).
Fig. 3.
Structures of three expansin cDNAs cloned from petals of opening carnation flowers. Light shaded rectangle, signal peptide; dark shaded rectangle, active site motif; filled circles, cysteine residues; filled ellipsoid, tryptophan residue; open diamond, ATG start codon; inverted open triangle, TGA stop codon; dark shaded inverted triangle, TAG stop codon; light shaded inverted triangle, TAA stop codon.
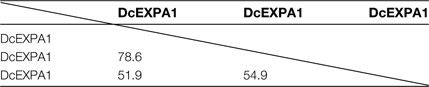
Table 2 shows the homology of amino acid sequences among three predicted expansin proteins. The homology between DcEXPA1 and DcEXPA2 was 78.6%, whereas that between DcEXPA1 and DcEXPA3 was 51.9% and that between DcEXPA2 and DcEXPA3 was 54.9%. The phylogenetic tree was constructed from the deduced amino acid sequences of three expansins and the expansins of Arabidopsis, rose, four o'clock, and petunia by the Clustral W program (Supplementary Fig. S2 at JXB online). DcEXPA1 and DcEXPA2 were clustered into the same clade, whereas DcEXPA3 clustered into the other clade.
Table 2.
Homology (%) in amino acid sequences among predicted DcEXPA1, DcEXPA2, and DcEXPA3 proteins
 |
Comparison of transcript levels of genes DcXTH1–DcXTH4 among different tissues, and their changes in whole petals and different parts of petals during flower opening
Figure 4A–D compares the relative transcript levels of DcXTH1–DcXTH4 in different tissues of carnation plants with opening flowers. Roughly speaking, transcripts of all the genes accumulated in flower tissues [petals, ovary, style (actually style plus stigma), and calyx] as opposed to vegetative tissues (stems and leaves). DcXTH2 and DcXTH3 transcripts were preferentially detected in large amounts in petals. DcXTH4 transcript was present in greater amounts in style, calyces, and petals than in stems and leaves. DcXTH1 transcript was detected, although not in significant amounts, in the style.
Figure 4E–H shows changes in transcript levels of DcXTH genes in whole petals at each opening stage. Transcripts of all the genes were already accumulated at flower opening stage 1 (bud), and they were detected until the flower opening stage 6 (fully open flower). Changes in transcript levels during flower opening were not significant for DcXTH3 and DcXTH4, and, even when significant, the differences were small for DcXTH1 and DcXTH2 (Fig. 4E, F).
Figure 4I–L shows transcript levels of DcXTH genes in four longitudinally successive parts of petals, from the tip of petals to the base of the claw, of flowers at stage 1 and stage 5. Transcripts of four DcXTH genes were detected in all parts of petals of both stages. However, transcript levels did not differ significantly irrespective of the flower opening stages and petal parts for all the genes, except for DcEXT4 in which significant but small differences were seen.
Comparison of transcript levels of genes DcEXPA1–DcEXPA3 among different tissues, and their changes in whole petals and different parts of petals during flower opening
Figure 5A–C shows the transcript levels of DcEXPA1, DcEXPA2, and DcEXPA3 in different tissues of carnation plants. Transcripts of all the genes were accumulated in green tissues (stem, leaf, and calyx). Among the three genes, DcEXPA2 transcripts were notably accumulated in floral tissues (petal and style).
Figure 5D–F shows the changes in transcript levels of three DcEXPA genes in whole petals at each flower opening stage, and Fig. 5G–I those in different parts of petals at opening stages 1 and 5. Transcripts of the three genes were detected in whole petals during flower opening stages, and all parts of petals at stages 1 and 5. However, no significant differences were found in transcripts levels of all the genes among flower opening stages and petal parts.
Detection of XEH activities in petal tissues
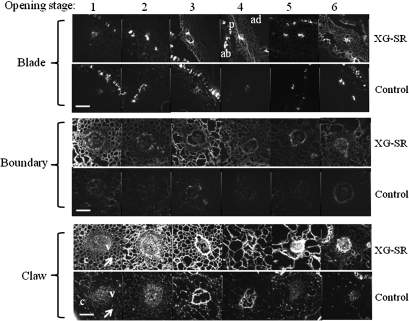
XET activity and xyloglucan were simultaneously localized in situ in carnation petal tissues according to fluorescence from wall-bound xyloglucan conjugated with XLLG-SR by the action of XET (Vissenberg et al., 2000). Preliminary experiments showed bright orange fluorescence emitted from the vascular bundles and the epidermis, whereas the parenchyma cell walls displayed a fainter staining. Therefore, fluorescence from the vascular bundles and surrounding parenchyma cells was examined in the claw, the boundary of the claw and blade, and the mid-blade of opening petals (Fig. 6). In the transverse sections of the claw, fluorescence emission was already high at stage 1, and increased more at stages 2 and 3, then declined thereafter. At the boundary of the claw and blade, fluorescence emission appeared to be bimodal, high at stage 1 and at stages 3–4. Fluorescence emission from the mid-blade was dense at stages 3, 4, and 6.
Fig. 6.
In situ detection of XET activity in carnation petal tissues by detecting fluorescence from wall-bound xyloglucan conjugated with XLLG-SR by the action of XET. Cryostat sections of 100 μm thick prepared by the thin film reinforcement method were used for observation with a fluorescence microscope. The control reaction mixture contained GGGG-SR instead of XLLG-SR. Arrows in the plate of the claw at opening stage 1 show the difference in fluorescence between the XLLG-SR-treated and the GGGG-SR-treated control tissues. Flower opening stages are similar to those described in the legend to Fig. 4. ab, abaxial epidermis; ad, adaxial epidermis; c, cell wall of parenchyma cells; p, parenchyma; v, vascular bundle. Scale bars=100 μm.
Discussion
Flower opening in carnation is attained by petal expansion and morphogenesis, mainly outward bending of growing petals, for which petal cell growth is essential. Petal cell growth consists of successive events, in which cell wall extension is an important event. In some flowers, as described in the Intoduction, XTH and expansin have been suggested to be probably responsible for this process. In the present study, cloning, characterization, and expression analyses of genes encoding XTHs and expansins were conducted with petals of opening carnation flowers.
DcXTHs
Four XTH cDNAs were cloned from carnation and designated DcXTH1, DcXTH2, DcXTH3, and DcXTH4. The four XTH proteins predicted from the cDNAs had molecular masses of 31.7–34.6 kDa, and had common structural features, which were conserved among XTH proteins from other plant sources—a signal peptide sequence, an active amino acid sequence, an N-glycosylation site, and four cysteine residues in the C-terminal region. Among the four carnation XTH proteins, the homology of amino acid sequences between DcXTH2 and DcXTH3 was 91.7%. DcXTH2 and DcXTH3 proteins had homology of 76.2% and 78.2%, respectively, to sugar beet BvXTH-1 protein (Dimmer et al., 2004). The homology was markedly high when compared with those with XTH proteins of different plant sources in the databases. This suggests a close relationship between carnation and sugar beet in terms of XTH proteins, which is in agreement with the fact that carnation (D. caryophyllus) and sugar beet (B. vulgaris) belong to the order Caryophyllales.
The XET activity was detected in all parts of growing carnation petals at all stages of flower opening. However, the activity tended to be high in the claw and boundary portions as compared with the blade.
The weak enzyme activity in the blade might be caused by decreased density of the fluorescent source because of parenchyma cell growth. This in situ observation of XET activity indicated the presence of XHT enzyme activity in growing petals of carnation, although the enzyme activity may differ depending on the portions and growing stage of the petals.
DcXTH2 and DcXTH3 transcripts were abundant in petals compared with other floral and vegetative (green) tissues (Fig. 4B, C), suggesting that they are major genes participating in the cell growth of growing carnation petals. In most cases, there were no significant differences in the transcript levels of DcXTH1–DcXTH4 genes among flower opening stages and petal parts at the flower opening stages 1 and 5. Even when differences were significant, they were small (double or threefold difference) (Figs 4E, F, L), which casts doubt on the biological relevance of the observed differences.
DcEXPA genes
Three full-length cDNAs for expansin genes, DcEXPA1, DcEXPA2, and DcEXPA3, were cloned from opening carnation flowers. The proteins predicted from cDNAs had molecular masses of 25.0–27.8 kDa, and structural features which were conserved among expansins of other plant sources; that is, a potential signal peptide sequence, an active amino acid sequence, seven cysteine residues in the N-terminal region, and specific valine and tryptophan residues. Among the three carnation expansins, the homology of amino acid sequences between DcEXPA1 and DcEXPA2 was high (78.6%).
With all the three expansin genes, transcripts were detected in petals and styles (actually style plus stigma) as well as leaves, stems, and calyces of carnation plants. However, it was noted that DcEXPA2 transcripts accumulated abundantly in petals and styles, and to a lesser extent DcEXPA1 transcripts accumulated in petals. These findings suggest the involvement of DcEXPA2 and DcEXPA1 gene expression in petal cell growth during carnation flower opening. Again, no significant differences were found in the transcript levels of DcEXPA1–DcEXPA3 genes among flower opening stages and petal parts at the flower opening stages 1 and 5.
Petal growth and morphogenesis
Taken together, the present findings suggested the expression of different XTH and expansin genes in growing carnation petals, in accordance with previous reports that showed the involvement of these genes in flower opening of other ornamentals (O'Donoghue et al., 2002; Gookin et al., 2003; Breeze et al., 2004; Zenoni et al., 2004; Laitinen et al., 2007; Takahashi et al., 2007; Yamada et al., 2009). Two XTH genes (DcXTH2 and DcXTH4) and two expansin genes (DcEXPA1 and DcEXPA2), out of the four and three genes, respectively, identified so far seemed to be involved mainly in petal growth and development during carnation flower opening.
There are two distinct events in growing petals of opening carnation flowers: growth (i.e. elongation and expansion) of the petals themselves and outward bending of the petals. At stages 1 and 2 of carnation flower opening, the elongation period defined by Harada et al. (2010), petals elongate mainly in the claw with moderate expansion of the blade. Then petals expand markedly in the blade at stages 3–6, the opening period (Harada et al., 2010). Meanwhile, the expanding petals reflexed at stages 4 and 5.
It is possible that XTHs and expansins function cooperatively or independently in these processes of petal growth and development. The interaction, if any, of the XTH and expansin genes and their roles in petal growth and morphogenesis remain to be elucidated.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Phylogenetic tree of four carnation XTH proteins and 33 members of the Arabidopsis XTH family. Amino acid sequences and accession numbers of 33 members of the Arabidopsis XTH family were obtained from Rose et al. (2002).
Figure S2. Phylogenetic tree of expansin proteins from carnation and other plant sources. Amino acid sequences of expansins of given plant sources, i.e. Arabidopsis thaliana, Petunia hybrida, Rosa hybrida, and Mirabilis jalapa were taken from Li et al. (2002), Zenoni et al. (2004), Yamada et al. (2009), and Gookin et al. (2003).
Table S1. Cloning of composite and complete cDNAs of XTH and expansins from petals of opening carnation flowers by PCR, 5'-RACE, and 3'-RACE techniques.
Table S2. Upstream and downstream primer sets used for cDNA cloning.
Table S3. Detailed procedures for PCR cloning for DcXTH1 and DcXTH2 cDNAs.
Table S4. Upstream (top) and downstream (bottom) primer sets used for real-time PCR.
Supplementary Material
Acknowledgments
This study was supported financially by a Grant-in-Aid (19380024 to S.S.) for Scientific Research from the Japan Society for the Promotion of Science.
References
- Breeze E, Wagstaff C, Harrison E, Bramke I, Rogers H, Stead A, Thomas B, Buchanan-Wollaston V. Gene expression patterns to define stages of post-harvest senescence in Alstroemeria petals. Plant Biotechnology Journal. 2004;2:155–168. doi: 10.1111/j.1467-7652.2004.00059.x. [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Gruissem W, Jones RL. Biochemistry and molecular biology of plants. Rockville, MD: American Society of Plant Physiologists; 2000. pp. 95–96. [Google Scholar]
- Chang S, Puryer J, Cairney J. A simple and efficient method for isolation of RNA from pine trees. Plant Molecular Biology Reports. 1993;11:113–116. [Google Scholar]
- Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- de Vetten NC, Huber DJ. Cell wall changes during the expansion and senescence of carnation (Dianthus caryophyllus) petals. Physiologia Plantarum. 1990;78:47–454. [Google Scholar]
- Dimmer E, Roden L, Cai D, Kingsnorth C, Mutasa-Gottgens E. Transgenic analysis of sugar beet xyloglucan endotransglucosylase/hydrolase Bv-XTH1 and Bv-XTH2 promoters reveal overlapping tissue-specific and wound-inducible expression profiles. Plant Biotechnology Journal. 2004;2:127–139. doi: 10.1046/j.1467-7652.2004.00056.x. [DOI] [PubMed] [Google Scholar]
- Evans RY, Reid MS. Changes in carbohydrates and osmotic potential during rhythmic expansion of rose petals. Journal of the American Society of Horticultural Science. 1988;113:884–888. [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proceedings of the National Academy of Sciences, USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochemical Journal. 1992;15:821–828. doi: 10.1042/bj2820821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gookin TE, Hunter DA, Reid MS. Temporal analysis of alpha and beta-expansin expression during floral opening and senescence. Plant Science. 2003;164:769–781. [Google Scholar]
- Harada T, Satoh S, Yoshioka T, Ishizawa K. Expression of sucrose synthase genes involved in enhanced elongation of pondweed (Potamogeton distinctus) turions under anoxia. Annals of Botany. 2005;96:683–692. doi: 10.1093/aob/mci220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Torii Y, Morita S, Masumura T, Satoh S. Differential expression of genes identified by suppression subtractive hybridization in petals of opening carnation flowers. Journal of Experimental Botany. 2010;61:2345–2354. doi: 10.1093/jxb/erq064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeberichts FA, van Doorn WG, Vorst O, Hall RD, van Wardragen MF. Sucrose prevents up-regulation of senescence-associated genes in carnation petals. Journal of Experimental Botany. 2007;58:2873–2885. doi: 10.1093/jxb/erm076. [DOI] [PubMed] [Google Scholar]
- Hunter DA, Lange NE, Reid SM. Physiology of flower senescence. In: Nooden LD, editor. Plant cell death processes. San Diego, CA: Academic Press; 2004. pp. 307–318. [Google Scholar]
- Ho LC, Nichols R. Translocation of 14C-sucrose in relation to changes in carbohydrate content in rose corollas cut at different stages of development. Annals of Botany. 1977;41:227–242. [Google Scholar]
- Ichimura K, Kawabata Y, Kishimoto M, Goto R, Yamada K. Shortage of soluble carbohydrates is largely responsible for short vase life of cut ‘Sonia’ rose flowers. Journal of the Japanese Society of Horticultural Science. 2003;72:292–298. [Google Scholar]
- Kawamoto T. Use of a new adhesive film for the preparation of multi-purpose fresh-frozen sections from hard tissues, whole animals and plants. Archives of Histology and Cytology. 2003;66:123–143. doi: 10.1679/aohc.66.123. [DOI] [PubMed] [Google Scholar]
- Kenis JD, Silvente ST, Trippi VS. Nitrogen metabolite and senescence-associated change during growth of carnation flowers. Physiologia Planarum. 1985;65:455–459. [Google Scholar]
- Koning RE. The role of plant hormones in the growth of the corolla of Gaillardia grandiflora (Asteraceae) ray flowers. American Journal of Botany. 1984;71:1–8. [Google Scholar]
- Laitinen RAE, Pollanen E, Teeri TH, Paula Elomaa, Kotilainen M. Trascriptional analysis of petal organogenesis in Gerbera hybrida. Planta. 2007;226:347–360. doi: 10.1007/s00425-007-0486-2. [DOI] [PubMed] [Google Scholar]
- Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, Mc-Queen-Mason S. Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiology. 2002;128:854–864. doi: 10.1104/pp.010658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell wall expansion in plants. The Plant Cell. 1992;4:1425–1433. doi: 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Ohto M, Yoshida N, Nakamura K. Sucrose-induced accumulation of α-amylase occurs concomitant with the accumulation of starch and sporamin in leaf-petiole cuttings of sweet potato. Plant Physiology. 1991;96:902–909. doi: 10.1104/pp.96.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani K, Tominaga R. Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. Journal of Biological Chemistry. 1992;267:21058–21064. [PubMed] [Google Scholar]
- O'Donoghue EM, Somerfield SD, Heyes JA. Organization of cell walls in Sandersonia aurantiaca floral tissues. Journal of Experimental Botany. 2002;53:513–523. doi: 10.1093/jexbot/53.368.513. [DOI] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K. The XTH family enzyme involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant and Cell Physiology. 2002;43:1421–1435. doi: 10.1093/pcp/pcf171. [DOI] [PubMed] [Google Scholar]
- Takahashi R, Fujitani C, Yamaki S, Yamada K. Analysis of the cell wall loosening proteins during rose flower opening. Acta Horticulturae. 2007;755:483–488. [Google Scholar]
- van Doorn WG, van Meeteren U. Flower opening and closure: a review. Journal of Experimental Botany. 2003;54:1801–1812. doi: 10.1093/jxb/erg213. [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Woltering EJ. Physiology and molecular biology of petal senescence. Journal of Experimental Botany. 2008;59:453–480. doi: 10.1093/jxb/erm356. [DOI] [PubMed] [Google Scholar]
- Vissenberg K, Martinez-Vilchez IM, Verbelen J-P, Miller JC, Fry SC. In vivo colocalization of xyloglucan endotransglycosylase activity and its donor substrate in the elongation zone of Arabidopsis roots. The Plant Cell. 2000;12:1229–1237. doi: 10.1105/tpc.12.7.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Van Sandt V, Fry SC, Verbelen J- P. Xyloglucan endotransglucosylase action is high in the root elongation zone and in the trichoblasts of all vascular plants from Selaginella to Zea mays. Journal of Experimental Botany. 2003;54:335–344. doi: 10.1093/jxb/erg024. [DOI] [PubMed] [Google Scholar]
- Waki K, Shibuya K, Yoshioka T, Hashiba T, Satoh S. Cloning of a cDNA encoding EIN3-like protein (DC-EIL1) and decrease in its mRNA level during senescence in carnation flower tissues. Journal of Experimental Botany. 2001;52:377–379. [PubMed] [Google Scholar]
- Yamada K, Takahashi R, Fujitani C, Mishima K, Yoshida M, Joyce DC, Yamaki S. Cell wall extensibility and effect of cell-wall-loosening proteins during rose flower opening. Journal of the Japanese Society of Horticultural Science. 2009;78:242–251. [Google Scholar]
- Zenoni S, Real L, Tornielli GB, et al. Downregulation of the Petunia hybrid α-expansin gene PhEXP1 reduces the amount of crystalline cellulose in cell walls and leads to phenotypic changes in petal limbs. The Plant Cell. 2004;16:295–308. doi: 10.1105/tpc.018705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.