Abstract
The three proline transporters of Arabidopsis thaliana (AtProTs) transport the compatible solutes proline and glycine betaine and the stress-induced compound γ-aminobutyric acid when expressed in heterologous systems. The aim of the present study was to show transport and physiological relevance of these three AtProTs in planta. Using single, double, and triple knockout mutants and AtProT-overexpressing lines, proline content, growth on proline, transport of radiolabelled betaine, and expression of AtProT genes and enzymes of proline metabolism were analysed. AtProT2 was shown to facilitate uptake of L- and D-proline as well as [14C]glycine betaine in planta, indicating a role in the import of compatible solutes into the root. Toxic concentrations of L- and D-proline resulted in a drastic growth retardation of AtProT-overexpressing plants, demonstrating the need for a precise regulation of proline uptake and/or distribution. Furthermore evidence is provided that AtProT genes are highly expressed in tissues with elevated proline content—that is, pollen and leaf epidermis.
Keywords: Arabidopsis, compatible solute, epidermis, pollen, proline, transport
Introduction
Organic solutes that accumulate at high concentrations in the cytoplasm in response to abiotic stress without interfering with primary metabolism are classified as ‘compatible’ solutes. The best known compatible solutes in plants are proline, glycine betaine, sugars, and polyols (Yancey, 2005; Verbruggen and Hermans, 2008). How exactly compatible solutes fulfil their protective role during stress is still a matter of debate; it appears that there exists a range of mechanisms such as supporting osmotic adjustment, protection of cellular structures, and regulation of cellular redox potential (Hare et al., 1998). High levels of compatible solutes are found not only under stress conditions, but also in plant organs that undergo dehydration as part of their development, such as pollen and seeds (Krogaard and Andersen, 1983; Chiang and Dandekar, 1995; Mondal et al., 1998; Schwacke et al., 1999; Schmidt et al., 2007). The key enzyme of proline biosynthesis, Δ1-pyrroline-5-carboxylate synthetase (P5CS), is encoded by two differentially regulated genes in Arabidopsis thaliana (Strizhov et al., 1997). AtP5CS1 is required for proline accumulation following osmotic stress, whereas AtP5CS2 is associated with embryo development (Székely et al., 2008).
The presence of compatible solutes in the phloem sap indicates that long-distance transport might be important for metabolism and/or stress tolerance (Girousse et al., 1996; Mäkelä et al., 1996). Likewise, uptake of compatible solutes from the soil can improve the plant's resistance to adverse environmental conditions (Räsänen et al., 2004). Although transporters for polyols (Klepek et al., 2005), glycine betaine, and proline (Rentsch et al., 1996; Schwacke et al., 1999) have been identified, their physiological role is poorly understood.
Amino acid transporters mediating the transport of proline have been identified in different gene families (Rentsch et al., 2007; Lehmann et al., 2010). The family of proline transporters (ProTs), with three members in Arabidopsis, transports proline, but no other proteinogenic amino acids (Rentsch et al., 1996). Although the ProTs were originally described as proline-selective transporters, later studies showed that in contrast to other amino acid permeases, ProTs from Arabidopsis, sugar beet, the mangrove Avicennia marina, and tomato, as well as barley ProT2 also transport glycine betaine (Rentsch et al., 1996; Schwacke et al., 1999; Waditee et al., 2002; Grallath et al., 2005; Yamada et al., 2009; Fujiwara et al., 2010). The tomato ProT1, barley ProT2, and the three Arabidopsis ProTs also recognize the stress-related compound γ-aminobutyric acid (GABA), though the affinity of the AtProTs and HvProT2 for GABA is lower than for proline or glycine betaine (Breitkreuz et al., 1999; Grallath et al., 2005; Fujiwara et al., 2010). The plasma membrane localization of AtProTs (Grallath et al., 2005), HvProT2 (Fujiwara et al., 2010), and sugar beet ProT (BvBet/ProT1; Yamada et al., 2009) suggests a function as a cellular uptake system in proline- and glycine betaine-accumulating species.
Although the intracellular localization, substrate selectivity, and affinity of the three Arabidopsis ProTs are similar, differences in expression indicate different roles in planta (Grallath et al., 2005). AtProT1:GUS staining was detected in the phloem of all organs analysed, suggesting a role in long-distance transport of proline (Grallath et al., 2005). AtProT2:GUS staining is found in the epidermis and cortex in roots and is detectable in leaves only after wounding. AtProT3 expression is restricted to the epidermis in leaves (Grallath et al., 2005). The expression of ProT family members is often associated with increased levels of proline. For example, tomato ProT1 transcripts are found exclusively in pollen that has high concentrations of proline (Schwacke et al., 1999). Transcript levels of Arabidopsis ProT2 and of the three mangrove ProT homologues increase in response to salt stress, as do the concentrations of proline and glycine betaine, respectively (Chiang and Dandekar, 1995; Rentsch et al., 1996; Hibino et al., 2001; Waditee et al., 2002). Salt stress also increases mRNA levels of HvProT in barley roots (Ueda et al., 2001). Despite these correlative changes, a role for ProTs in the translocation of proline or glycine betaine has been demonstrated only recently. Transgenic Arabidopsis lines overexpressing HvProT showed reduced shoot biomass and decreased proline accumulation (Ueda et al., 2008). Conversely, root cap-specific expression of HvProT resulted in increased accumulation of proline in the root tip and enhanced root elongation (Ueda et al., 2008).
To reveal the contribution of individual Arabidopsis ProTs to the distribution of proline within the plant, the focus of the present study was on organs and growth conditions known to be associated with AtProT expression. Using atprot knockout plants and overexpressing lines, it is demonstrated that AtProT1 and AtProT2 mediate proline transport in planta. It is shown that proline is unevenly distributed between the lower leaf epidermis and the remainder of the leaf, and that proline is the major free amino acid in mature Arabidopsis pollen, correlating with high expression of AtProT3 and AtProT1, respectively.
Materials and methods
Plant growth and transformation
Arabidopsis thaliana L. ecotype Columbia (Col-0) or Wassilewskija (Ws) and mutant lines were cultivated in a growth chamber under a photoperiod of 16 h of light (100–150 μmol m−2 s−1) and 8 h of darkness. Plants were grown in soil with a day/night temperature of 22 °C/18 °C at 65%/60% relative humidity.
The following Arabidopsis T-DNA insertion lines were used in this work: atprot1-1 (Salk_018050, ecotype Columbia), atprot2-1 (4B14, Feldmann line, ecotype Wassilewskija), atprot2-2 (CSJ1230, Wisconsin, ecotype Wassilewskija), atprot2-3 (Salk_067508, ecotype Columbia), and atprot3-2 (Salk_083340, ecotype Columbia), (Wisconsin lines: Arabidopsis Knockout Facility of the University of Wisconsin, Krysan et al., 1999; Salk lines: Alonso et al., 2003; Feldmann lines: PCR screening of mutants obtained from the ABRC stock center, Columbus, OH, USA, according to Krysan et al., 1996). The insertion sites were verified by sequencing flanking regions which were amplified by PCR using T-DNA left border primers (for Salk lines, 5′-GCGTGGACCGCTTGCTGCAACT-3′; for Feldmann line, 5′-GATGCACTCGAAATCAGCCAATTTTAGAC-3′; for Wisconsin line, 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′) and gene-specific primers (for atprot1-1, 5′-GGCAACAGTGAGGCAACCAGT-3′ and 5′-CATAGCTTTTGCATAGCATTC-3′; for atprot2-1, 5′-GCAGTTGAACAATTCGATCTCGAAGTCCC-3′ and 5′-GAAGCAAACATTGAGCCAATGCCATAGC-3′; for atprot2-2, 5′-GTGTGAAAGCTTAAGTGTTGAAGAACTTG-3′ and 5′-ACCCTAGTTTTCGCTATTAGGTCAAGACT-3′; for atprot2-3, 5′-TAAACAGTGCCTATGTGTTG-3′ and 5′-AGATCGATGACACTGACCTGT-3′; for atprot3-2, 5′-ACAATAACCATTTGGAGAGG-3′ and 5′-GAAAACTAGTGTAGCGGC-3′). Lines were back-crossed twice with the wild-type and selfed to isolate homozygous lines. Double and triple knockout lines were generated by crossing homozygous single atprot mutants (Col-0 background).
Plants were transformed by Agrobacterium tumefaciens- (GV3101 pMP90) mediated gene transfer using the floral dip method (Clough and Bent, 1998).
For axenic culture, surface-sterilized Arabidopsis seeds were vernalized at 4 °C in the dark for 2 d. Plants were cultivated for 10 d in square plates (12 cm) with 70 ml of AM medium per plate [2.2 g l−1 MS salts (Murashige and Skoog, 1962), 1% sucrose]. The medium contained 0.05% MES (pH 5.7) and was solidified with 0.7% agar.
DNA and RNA work
For overexpression in Arabidopsis, the AtProT1-cDNA and AtProT2-cDNA including 5'- and 3'-untranslated region sequences were isolated from pFL61 (Minet et al., 1992; Rentsch et al., 1996) and introduced into the SmaI and XbaI sites of pBinAR (Bevan, 1984).
Extraction of total RNA and RNA gel blot analysis were performed according to a phenol–SDS extraction method and a formaldehyde–formamide protocol, respectively (Ausubel et al., 1994). The fragments used for detection were labelled with [α-32P]dCTP using the Megaprime Kit (Amersham). For detection of AtProT1 expression, a 464 bp sequence was amplified from the AtProT1 cDNA using the primers 5′-GAGAATTCAGGCTCTAATGGTAAGAC-3′ and 5′-CATATGAGTACATGGACACA-3′. AtProT2 and AtP5CS expression was detected using the entire cDNAs for the labelling procedure. The fragment amplified from the AtP5CS1-cDNA might detect AtP5CS1 and AtP5CS2 mRNA due to high homology of the transcripts. For detection of AtPDH (proline dehydrogenase) a 1600 bp fragment of the AtPDH1 cDNA was amplified using a gene-specific primer 5′-CCCAACCTCTGATCTCC-3′ and a vector primer, possibly also detecting AtPDH2 transcripts.
For expression analysis of AtProT3, reverse transcription was performed using the RETROscript Kit (Ambion) with 2 μg of total RNA. The primers 5′-ACAATAACCATTTGGAGAGG-3′ and 5′-AAATCCAACTAAGAATAAATACG-3′ were used to amplify a full-length transcript of AtProT3 by reverse transcription-PCR (RT-PCR).
Amino acid analysis
The content of free proline in the leaf was determined as described by Bates et al. (1973). For HPLC analysis, free amino acids from mature and germinated pollen were extracted as described by Bieleski and Turner (1966) and analysed by ARC (Analytical Research and Services, University of Bern) according to Bidlingmeyer et al. (1984).
Preparation of Arabidopsis leaf epidermis
Arabidopsis plants were grown under long-day conditions for 3–4 weeks. The salt treatment was performed 24 h prior to the experiment by adding NaCl solution to the plants until a concentration of ∼200 mM NaCl was reached in the pot. After the leaf margin had been removed using a razor blade, the leaf was placed between two pieces of double-sided adhesive tape attached to a strip of plastic wrap. Tissues were separated by detaching the pieces of tape and immediately frozen in liquid nitrogen. A sample was composed of 10 leaves harvested from 10 individual plants [resulting in fresh weights of 10–20 mg (epidermis) and 60–100 mg (rest of the leaf)].
[14C]Glycine betaine uptake
Sixteen seedlings per genotype (64 seedlings in total) were grown for 10 d in square plates (12 cm) containing 70 ml of AM medium (0.6% agar). Per plate, 16 agar plugs (7 mm) were removed with a cut plastic pipette. To reach a final concentration of 500 μM glycine betaine, each of the holes was filled with 150 μl of labelling solution {14.6 mM glycine betaine, 1.3 μM [14C]glycine betaine (2 TBq mol−1) in liquid AM}. After 24 h the radioactivity was equally distributed in the plate (not shown). At each time point (6, 24, and 48 h) 10 seedlings per genotype were removed and washed thoroughly at 4 °C with AM containing 500 μM glycine betaine. For scintillation counting, seedlings were solubilized overnight using 500 μl of SOLUENE 350 (Perkin Elmer®). After addition of 4 ml of scintillation cocktail ULTIMA GOLD™ XR (Perkin Elmer®) the samples were counted using a Beckman LS6500 scintillation counter.
Pollen harvest and in vitro pollen germination
Mature pollen was harvested in bulk from several trays of flowering plants onto a nylon mesh (mesh size 15 μm), using a vacuum cleaner as described by Johnson-Brousseau and McCormick (2004). For germination, the pollen was transferred to an agar medium (pH 6.0) composed as described in Fan et al. (2001), but without myo-inositol. The pollen was transferred to the medium (5.5 cm plates) either by dipping the nylon mesh onto the agar surface or by placing the mesh on the medium (in the case of subsequent RNA extraction) and germinated in a humid chamber at 28 °C for 6 h. Due to this experimental set-up, the fresh weight of germinated pollen could not be determined. The extraction yielded similar amounts of amino acids per g pollen fresh weight from Col-0 and atprot1-1 plants. The relative amount of an amino acid was determined as the mean of three samples, each extracted from several nylon meshes with pollen.
Results
Characterization of atprot T-DNA insertion and 35S:AtProT-overexpressing lines
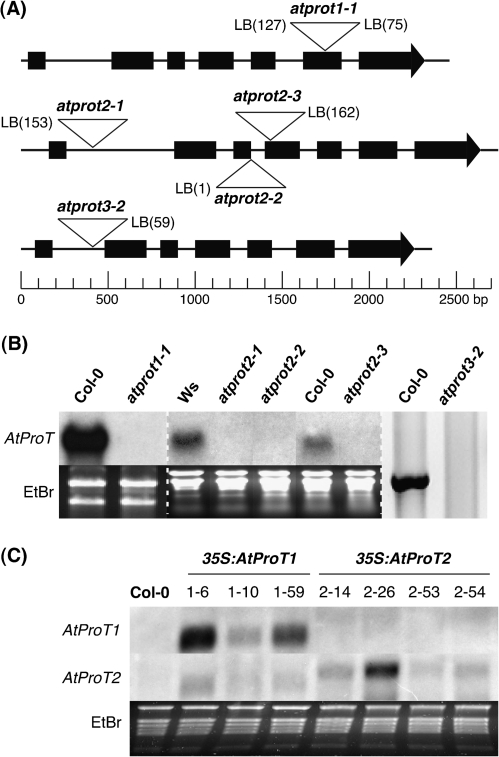
An overview of the T-DNA insertion sites is given in Fig. 1A. AtProT transcript levels were determined in tissues where high expression of the respective gene had been shown (Rentsch et al., 1996; Grallath et al., 2005; see below). RNA gel blot analysis showed that in the atprot1-1 mutant (Fig. 1B) and the three atprot2 lines (atprot2-1, atprot2-2, and atprot2-3, Fig. 1B), the respective transcripts cannot be detected and thus the insertion lines can be considered as null mutants. In the atprot3-2 mutant, no full-length transcript of AtProT3 could be amplified by RT-PCR (Fig. 1B). A fragment of ∼1000 bp downstream of the insertion site that contains an open reading frame was transcribed (data not shown), but expression of the respective 3' end fragment did not complement the proline transport-deficient Saccharomyces cerevisiae mutant strain 22574d, suggesting that the truncated AtProT3-2 transcript did not produce a functional protein in plants (data not shown; Jauniaux et al., 1987). Double and triple insertion lines were generated using the lines atprot1-1, atprot2-3, and atprot3-2. Additionally, lines overexpressing AtProT1 or AtProT2 under control of the constitutive 35S promoter were generated. Two lines of each construct were selected for further analyses, one line showing moderately (35S:AtProT1-59, 35S:AtProT2-14) and one line showing strongly (35S:AtProT1-6, 35S:AtProT2-26) increased AtProT mRNA levels in the T3 generation (Fig. 1C).
Fig. 1.
Molecular characterization of atprot T-DNA insertion mutants and AtProT-overexpressing lines. (A) Schematic representation of the exon–intron structure of AtProT1, AtProT2, and AtProT3 including the T-DNA integration sites. Genomic sequences were drawn to scale. Black boxes represent exons. The distance of the insertion site from the exon–intron border was verified by sequencing and is indicated as nucleotides in parentheses. (B) Expression of AtProT genes in several T-DNA insertion lines examined by northern blot (AtProT1 and AtProT2) or RT-PCR (AtProT3). RNA was isolated from pollen (AtProT1), salt-stressed seedlings (AtProT2), or leaves (AtProT3). A dashed line separates individual gels. The lanes of the RNA gel blot analysis of AtProT2 expression were regrouped for clarity. (C) RNA gel blot analysis of 35S:AtProT1 and 35S:AtProT2 lines (leaf tissue). Autoradiographic pictures in B and C have been adjusted in brightness levels.
When grown in soil or axenically, none of these single, double, and triple knockout mutants and overexpression lines showed phenotypical differences (e.g. shoot size and development, root length, flowering time). Also, the proline content of shoots and roots of plants grown in axenic culture and of rosette leaves of plants grown in soil did not differ between wild-type, knockouts, and overexpressing lines (not shown).
AtProT2 mediates uptake of glycine betaine from the rhizosphere
Heterologous expression of the AtProTs in S. cerevisiae demonstrated that the affinity for glycine betaine is higher than for L-proline (Grallath et al., 2005). Glycine betaine does not seem to be metabolized in Arabidopsis and no transporter other than the AtProTs has been reported to mediate transport of glycine betaine. As AtProT2 expression was primarily found in the cortex and epidermis of the roots (Grallath et al., 2005), a role for AtProT2 in the uptake of proline or glycine betaine into roots was suggested.
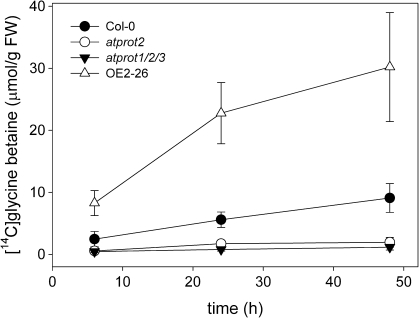
The uptake of [14C]glycine betaine in seedlings of wild-type plants, atprot2-3 mutant, triple knockout (atprot1-1 atprot2-3 atprot3-2), and 35S:AtProT2-26 lines showed that uptake was time dependent and highest in the overexpressing line (Fig. 2). Wild-type seedlings accumulated more [14C]glycine betaine than the atprot2 or the triple knockout mutant. No significant difference was observed between the latter. The overexpressing line accumulated three times as much [14C]glycine betaine as the wild type, which in turn imported 4.5 times the amount detected in atprot2 knockout plants, indicating that AtProT2 is the main uptake system for glycine betaine in roots.
Fig. 2.
Uptake of [14C]glycine betaine. Time-dependent uptake of 500 μM [14C]glycine betaine in Col-0, atprot2-3, triple knockout (atprot1-1 atprot2-3 atprot3-2), and 35S:AtProT2-26 (OE) seedlings. Values represent the mean of 10 separately measured seedlings ±SD. Comparable results were obtained in several biologically independent experiments. The differences between the wild type and all three mutant lines are statistically significant (one-way ANOVA, Scheffé test).
AtProT1 and AtProT2 transport L-proline in planta
Some D-amino acids such as D-serine or D-alanine have been shown to be detrimental for plant growth and development (Erikson et al., 2004). Transport studies using heterologous expression systems demonstrated that AtProT2 transports D- and L-proline at similar rates (Breitkreuz et al., 1999; CG and DR, unpublished).
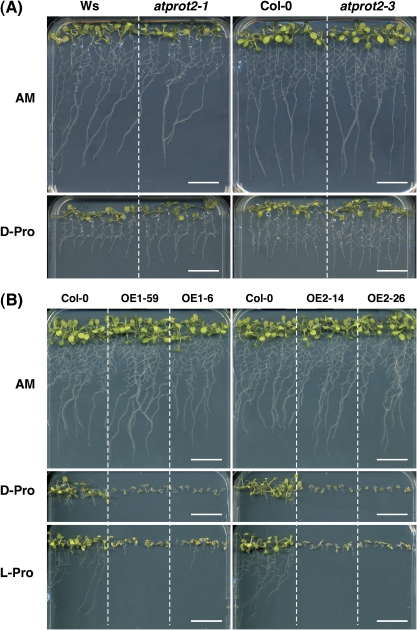
It was found that D-proline inhibited Arabidopsis root growth in a dose-dependent manner and differences in D-proline transport between wild-type and mutant plants were assessed. Two independent atprot2 mutants and the respective wild-type plants were germinated on AM medium and transferred to AM medium supplemented with 8 mM D-proline. D-Proline inhibited root growth of atprot2 seedlings to a lesser extent than growth of wild-type roots (Fig. 3A), indicating that the loss of AtProT2 activity decreased the net uptake of D-proline. Growth of the triple knockout line resembled that of the atprot2 mutant (not shown).
Fig. 3.
Growth on D-proline and high concentrations of D-proline. (A) atprot2-1 and atprot2-3 seedlings and the respective wild-type plants were germinated on AM medium and transferred to AM or medium supplemented with 8 mM D-proline for 5 d. Scale bar 2 cm. (B) Growth of Col-0 and 35S:AtProT (OE) seedlings was compared 10 d after germination on AM medium, AM supplemented with 4 mM D-proline and AM containing 50 mM L-proline. Scale bar=2 cm. (This figure is available in colour at JXB online.)
Similarly, plants overexpressing AtProT1 or AtProT2 were grown on AM medium supplemented with 4 mM D-proline and compared with the wild type (Fig. 3B). All genotypes showed a strong phenotypic response to D-proline. Shoot growth and root elongation were reduced in wild-type plants, while development of all tested 35S:AtProT lines was arrested shortly after unfolding of cotyledons (Fig. 3B).
Previous studies have demonstrated that high concentrations of exogenous L-proline also impair Arabidopsis growth (Hellmann et al., 2000; Mani et al., 2002). When plants were grown on AM medium containing 50 mM L-proline, shoot and root development was moderately inhibited in wild-type seedlings, whereas chlorosis and early cessation of growth was observed in 35S:AtProT plants (Fig. 3B). Taken together these results demonstrate that AtProT1 and AtProT2 mediate proline transport in planta.
High proline level in Arabidopsis leaf epidermis
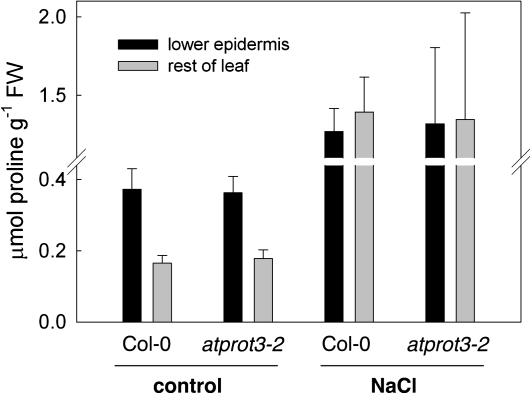
Zúñiga et al. (1989) demonstrated that in barley seedlings, proline distributes unevenly between leaf epidermis and mesophyll in water-stressed but not in control plants, whereas the concentration of glycine betaine is elevated in the epidermis of both stressed and unstressed barley leaves. As AtProT3 is expressed in the lower epidermis but not in the mesophyll of leaves (Grallath et al., 2005), proline distribution between the lower epidermis and the remaining leaf tissues was determined under control and under salt stress conditions, but found to be comparable in the wild type and the atprot3-2 mutant (Fig. 4). Interestingly, the concentration of proline in the lower epidermis was 1.5–2 times higher than the average concentration in the remaining leaf tissues (Fig. 4). Upon salt treatment, proline concentrations increased in particular in the remaining leaf tissues, so that the difference in concentrations between the lower epidermis and remaining leaf tissues was no longer observed (Fig. 4).
Fig. 4.
Proline distribution within the leaf. The proline content in the lower epidermis and in the rest of the leaf was determined in wild-type and atprot3-2 plants by ninhydrin assay. Plants were grown on soil for 3–4 weeks and watered normally or treated with 200 mM NaCl for 24 h prior to the analysis. Values represent the mean of six samples ±SD.
Altered glutamate and arginine levels in germinating pollen of the atprot1 mutant
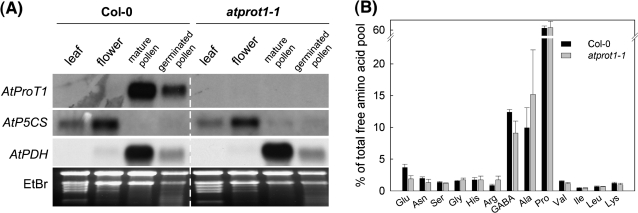
Pollen has repeatedly been reported to contain a high amount of free proline (Krogaard and Andersen, 1983; Mondal et al., 1998; Schwacke et al., 1999). Consistently, proline was found to be the most abundant amino acid in mature and germinated Arabidopsis pollen, accounting for 60–65% of the free amino acid pool (Supplementary Fig. S1 available at JXB online; Fig. 5B). Previously, expression of AtProT1 had been demonstrated in roots, leaves, and flowers (Rentsch et al., 1996). The present analysis shows that the AtProT1 transcript level is particularly high in pollen compared with leaf and flower tissue (Fig. 5A).
Fig. 5.
Molecular and biochemical analysis of the atprot1-1 mutant. (A) Expression of AtProT1, AtP5CS, and AtPDH in different organs of Col-0 and atprot1-1 plants examined by RNA gel blot analysis. The dashed line separates the half of the gel loaded with wild-type RNA from that loaded with RNA from atprot1-1 plants, which was mirrored for clarity. The autoradiographic pictures have been adjusted in brightness levels. (B) Free amino acid composition in germinated pollen of Col-0 and atprot1-1 plants. The relative amount of an amino acid is given as the mean of three composite samples ±SD.
Of the mRNA species present in mature pollen, many appear to be translated into protein only upon the onset of pollen germination (Mascarenhas, 1993). Two independent pollen transcriptome analyses detected an increase in AtProT1 transcript abundance during late stages of pollen maturation, suggesting that AtProT1 functions in post-pollination processes (Honys and Twell, 2004; Bock et al., 2006). Based on these observations, expression of AtProT1 was compared between mature and in vitro germinated pollen and found to decrease during pollen germination (Fig. 5A). Mature pollen of Col-0 and atprot1-1 plants does not differ in amino acid composition and content (Supplementary Fig. S1 at JXB online). In contrast, germinating pollen of atprot1-1 plants displayed minor differences regarding the free amino acids—that is, a lower and higher percentage of glutamate and arginine, respectively (Fig. 5B). The high level of GABA in germinating pollen of both genotypes might result from stress during in vitro germination and subsequent harvest (Shelp et al., 1999). Despite the altered concentrations of glutamate and arginine in atprot1-1 mutant pollen, in vitro germination assays and an in vivo transmission analysis did not reveal differences in germination rate or a significantly biased distribution of the AtProT1 alleles in the offspring (not shown).
Furthermore, transcript levels of AtP5CS (Δ1-pyrroline-5-carboxylate synthetase), the enzyme catalysing the first step in proline biosynthesis, and of AtPDH (proline dehydrogenase), the enzyme catalysing the oxidation of proline to pyrroline-5-carboxylate (P5C), were analysed by RNA gel blot in the atprot1-1 knockout (Fig. 5A). AtP5CS transcripts were abundant in leaf and flower tissue but could hardly be detected in pollen, in both Col-0 and atprot1-1 plants. In contrast, expression of AtPDH was high in pollen and decreased during pollen germination (Fig. 5A); however, lower AtPDH mRNA levels might also be caused by repression of PDH expression by sucrose present in the germination medium (Hanson et al., 2008). Similar results were obtained from several independent batches of plants, suggesting that there are no major differences in expression of AtP5CS and AtPDH between wild-type and atprot1-1 plants.
Discussion
AtProTs mediate proline and glycine betaine transport in planta
Evidence is provided that in planta, AtProTs transport D- and L-proline or glycine betaine, which were previously identified as substrates of AtProTs using heterologous expression systems (Breitkreuz et al., 1999; Grallath et al., 2005). First, the contribution of AtProT1 and AtProT2 to proline transport in planta is demonstrated by treatment with toxic concentrations of L-proline as well as D-proline, which are the cause of severe growth defects in 35S:AtProT plants. Secondly, after 6, 24, and 48 h seedlings of AtProT2-overexpressing line accumulated more [14C]glycine betaine than the wild type, which in turn contained more radiolabelled substrate than the atprot2 and the triple knockout plants. As the atprot2 mutant accumulates <25% of the [14C]glycine betaine imported by the wild type, AtProT2 seems to be the main glycine betaine uptake system in roots. AtProT2 expression is found in the epidermis and cortex of roots (Grallath et al., 2005), tissues which are involved in the uptake and radial transport of substances from the root medium. Because AtProT2 expression is induced under salt stress (Rentsch et al., 1996), the acquisition of compatible solutes from the rhizosphere might improve growth of Arabidopsis under water stress, as observed for seedlings of acacia (Räsänen et al., 2004), and is a common strategy found among bacteria (Sleator and Hill, 2002).
The proline distribution within the leaf changes under salt stress
The differential distribution of solutes between leaf tissues such as epidermis, mesophyll, and vascular bundles has been demonstrated in several plant species (Fricke et al., 1994; Karley et al., 2000). Here, it was shown that in Arabidopsis, the proline level in the lower leaf epidermis is higher than in the rest of the leaf (Fig. 4). Compared with mesophyll cells, the water status of cells in the lower epidermis is constantly challenged by stomatal and cuticular transpiration. Furthermore, a high vacuolar osmotic pressure in epidermal cells requires a high solute content in the cytosol, thus the concentration of compatible solutes is expected to be elevated.
Under salt stress, this differential accumulation was no longer observed, suggesting that the stress-related increase of proline is stronger in the mesophyll or in the vasculature than in the epidermis. In potato mesophyll cells, the cellular increase in proline in response to osmotic stress is primarily due to accumulation of proline in the chloroplast stroma (Büssis and Heineke, 1998). An AtP5CS1–green fluorescent protein (GFP) fusion protein has been reported to re-localize from the cytosol into the chloroplasts when Arabidopsis mesophyll cells are subjected to hyperosmotic stress (Székely et al., 2008). Therefore, less accumulation of proline in the epidermis under stress may also reflect the general scarcity of chloroplasts in this tissue. However, the highest increase in proline in water-stressed barley leaves was detected in the vasculature and the epidermis, whereas the proline level in mesophyll protoplasts remained rather constant (Zúñiga et al., 1989), demonstrating species-dependent differences in the distribution of compatible solutes under stress. To understand the physiological relevance of this phenomenon, it would be important to dissect the contribution of proline synthesis and transport, for instance by means of plants further impaired in the expression of AtP5CS or AtLHT1, an amino acid and proline transporter that is also expressed in the epidermis of Arabidopsis (Hirner et al., 2006).
Loss of AtProT1 activity changes amino acid composition in germinating but not in mature pollen
The accumulation of proline in Arabidopsis pollen argues for a role in stabilizing cellular structures during dehydration, high and low temperatures, or as a metabolic precursor and source of energy, as was proposed for other plants (Stanley and Linskens, 1974; Zhang and Croes, 1983; Mutters et al., 1989; Lansac et al., 1996; Schwacke et al., 1999). Székely et al. (2008) detected AtP5CS–GFP in Arabidopsis pollen, suggesting that biosynthesis contributes to proline accumulation. However, the low abundance of P5CS transcripts in pollen of Arabidopsis and tomato observed in other studies supports the idea that proline accumulation is attributed to import processes, though post-transcriptional regulation cannot be excluded (Fig. 5A; Fujita et al., 1998; Schwacke et al., 1999). Accumulation of proline which is independent of changes of P5CS expression has also been reported for tissues other than pollen—that is, in developing grapevine berries—again pointing to the contribution of proline transport or regulation at the post-transcriptional level (Stines et al., 1999).
The present data show that AtP5CS transcripts are present in flowers, but absent from pollen (Fig. 5A), suggesting that the proline generated in other parts of the flower may be imported into pollen. As pollen is symplasmically isolated, membrane transport is essential in sustaining its development during maturation and germination. It cannot be predicted whether transcripts that accumulate late in pollen development, such as AtProT1 mRNA, are translated in mature pollen, germinating pollen, or both (Mascarenhas, 1990). Likewise, the concomitant accumulation of AtPDH mRNA and high amounts of proline in mature pollen (Fig. 5A, Supplementary Fig. S1 at JXB online) suggests that AtPDH transcript abundance does not reflect PDH activity. Proline degradation has been detected in germinating pollen of Petunia (Zhang and Croes, 1983); therefore, translation of PDH might only start with the onset of pollen germination.
Germinated wild-type pollen differs from germinating atprot1 pollen particularly in those amino acids that are closely linked with proline metabolism (glutamate and arginine). Glutamate is the metabolic precursor of GABA and proline; it also represents the end-product of the degradation of proline and arginine. Thus it appears that during pollen germination the transport activity of AtProT1 affects amino acids other than proline, an effect that needs to be further investigated.
How do AtProTs contribute to proline transport in the plant?
None of the atprot T-DNA insertion lines (single, double, or triple knockouts) and overexpression lines analysed revealed differences in proline content, though the present results do not exclude changes at the subcellular or tissue level. Either transport of proline through AtProTs is not involved in the regulation of proline levels in planta, or plants can compensate for the lack or increase of AtProT activity—that is, other amino acid transporters and/or the regulation of proline metabolism might compensate for alterations in proline distribution. The findings of Ueda et al. (2008) suggest that metabolism responds to changes in proline transport. Overexpression of HvProT in Arabidopsis led to an increase in AtPDH expression and activity, parallel to a decrease in proline content of leaves (Ueda et al., 2008). The authors conjectured that enhanced accumulation of proline in leaves of 35S:HvProT plants might induce elevated AtPDH activity. Though altered expression of proline-metabolizing enzymes was not observed in the present mutant lines, altered proline transport may feed back directly on the metabolic pathway or alter flow through the pathway, keeping proline levels unchanged.
A functional overlap between AtProTs and transporters from other families is probably one of the main reasons why atprot mutants do not show a strong phenotype. The amino acid permease AtAAP2, expressed in major veins of leaves and stems (Hirner et al., 1998), or AtAAP3 in the root phloem (Okumoto et al., 2004) might counterbalance the altered AtProT1 activity in phloem tissue. In pollen, the lack of AtProT1 may be compensated by carriers with comparable expression patterns, for example by AtLHT5 or AtLHT7 (Foster et al., 2008; Hruz et al., 2008). Similarly, loss of AtProT2 function could be mitigated by the activity of AtLHT1 or AtAAP1, which mediate amino acid uptake into the root epidermis (Hirner et al., 2006; Lee et al., 2007; Svennerstam et al., 2007). Interestingly, AtLHT1 expression was also described in the leaf epidermis (Hirner et al., 2006), suggesting partial functional redundancy with AtProT3. To improve our understanding of their physiological significance, future analyses could benefit from studying plants impaired in the function of multiple transporters that show overlapping expression patterns and substrate selectivity.
Supplementary data
Supplementary data are available at JXB online.
Figure S1 Free amino acid composition of mature Col-0 and atprot1-1 pollen. The relative amount of an amino acid is given as the mean of four composite samples ±SD. Per gram pollen fresh weight, the extraction yielded between 0.75 mmol and 0.95 mmol amino acids.
Supplementary Material
Acknowledgments
This work was supported by grants from the Swiss National Science Foundation SNF (3100-064918 and SNF 3100A0-107507). We wish to thank Elisabeth Kuslys and Marianne Suter Grotemeyer for their technical assistance, Rebecca Alder and Christopher Ball for taking care of the plants, and Willi Tanner for designing the pollen harvesting system. We thank Mirco Hecht for testing the activity of the truncated AtProT3 protein. We are also grateful to Mechthild Tegeder (Washington State University, Pullman) for helpful comments on the manuscript.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current protocols in molecular biology. New York: Wiley; 1994. [Google Scholar]
- Bates LS, Waldren RP, Teare ID. Rapid determination of free proline for water-stress studies. Plant and Soil. 1973;39:205–207. [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Research. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidlingmeyer B, Cohen S, Tarvin T. Rapid analysis of amino acids using pre-column derivatization. Journal of Chromatography. 1984;336:93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- Bieleski RL, Turner NA. Separation and estimation of amino acids in crude plant extracts by thin-layer electrophoresis and chromatography. Analytical Biochemistry. 1966;17:278–293. doi: 10.1016/0003-2697(66)90206-5. [DOI] [PubMed] [Google Scholar]
- Bock KW, Honys D, Ward JM, Padmanaban S, Nawrocki EP, Hirschi KD, Twell D, Sze H. Integrating membrane transport with male gametophyte development and function through transcriptomics. Plant Physiology. 2006;140:1151–1168. doi: 10.1104/pp.105.074708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreuz KE, Shelp BJ, Fischer WN, Schwacke R, Rentsch D. Identification and characterization of GABA, proline and quaternary ammonium compound transporters from Arabidopsis thaliana. FEBS Letters. 1999;450:280–284. doi: 10.1016/s0014-5793(99)00516-5. [DOI] [PubMed] [Google Scholar]
- Büssis D, Heineke D. Acclimation of potato plants to polyethylene glycol-induced water deficit II. Contents and subcellular distribution of organic solutes. Journal of Experimental Botany. 1998;49:1361–1370. [Google Scholar]
- Chiang H, Dandekar AM. Regulation of proline accumulation in Arabidopsis thaliana (L.) Heynh during development and in response to desiccation. Plant, Cell and Environment. 1995;18:1280–1290. [Google Scholar]
- Clough S, Bent A. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Erikson O, Hertzberg M, Näsholm T. A conditional marker gene allowing both positive and negative selection in plants. Nature Biotechnology. 2004;22:455–458. doi: 10.1038/nbt946. [DOI] [PubMed] [Google Scholar]
- Fan LM, Wang YF, Wang H, Wu WH. In vitro Arabidopsis pollen germination and characterization of the inward potassium currents in Arabidopsis pollen grain protoplasts. Journal of Experimental Botany. 2001;52:1603–1614. [PubMed] [Google Scholar]
- Foster J, Lee YH, Tegeder M. Distinct expression of members of the LHT amino acid transporter family in flowers indicates specific roles in plant reproduction. Sexual Plant Reproduction. 2008;21:143–152. [Google Scholar]
- Fricke W, Leigh RA, Deri TA. Concentrations of inorganic and organic solutes in extracts from individual epidermal, mesophyll and bundle-sheath cells of barley leaves. Planta. 1994;192:310–316. [Google Scholar]
- Fujita T, Maggio A, García-Ríos M, Bressan RA, Csonka LN. Comparative analysis of the regulation of expression and structures of two evolutionarily divergent genes for Δ1-pyrroline-5-carboxylate synthetase from tomato. Plant Physiology. 1998;118:661–674. doi: 10.1104/pp.118.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Mitsuya S, Miyake H, Hattori T, Takabe T. Characterization of a novel glycinebetaine/proline transporter gene expressed in the mestome sheath and lateral root cap cells in barley. Planta. 2010;232:133–143. doi: 10.1007/s00425-010-1155-4. [DOI] [PubMed] [Google Scholar]
- Girousse C, Bournoville R, Bonnemain JL. Water deficit-induced changes in concentrations in proline and some other amino acids in the phloem sap of alfalfa. Plant Physiology. 1996;111:109–113. doi: 10.1104/pp.111.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallath S, Weimar T, Meyer A, Gumy C, Suter-Grotemeyer M, Neuhaus JM, Rentsch D. The AtProT family. Compatible solute transporters with similar substrate specificity but differential expression patterns. Plant Physiology. 2005;137:117–126. doi: 10.1104/pp.104.055079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J, Hanssen M, Wiese A, Hendriks MMWB, Smeekens S. The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of Asparagine Synthetase1 and Proline Dehydrogenase2. The Plant Journal. 2008;53:935–949. doi: 10.1111/j.1365-313X.2007.03385.x. [DOI] [PubMed] [Google Scholar]
- Hare PD, Cress WA, Van Staden J. Dissecting the roles of osmolyte accumulation during stress. Plant, Cell and Environment. 1998;21:535–553. [Google Scholar]
- Hellmann H, Funck D, Rentsch D, Frommer WB. Hypersensitivity of an Arabidopsis sugar signaling mutant toward exogenous proline application. Plant Physiology. 2000;123:779–789. doi: 10.1104/pp.123.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino T, Meng YL, Kawamitsu Y, et al. Molecular cloning and functional characterization of two kinds of betaine-aldehyde dehydrogenase in betaine-accumulating mangrove Avicennia marina (Forsk.) Vierh. Plant Molecular Biology. 2001;45:353–363. doi: 10.1023/a:1006497113323. [DOI] [PubMed] [Google Scholar]
- Hirner A, Ladwig F, Stransky H, Okumoto S, Keinath M, Harms A, Frommer WB, Koch W. Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. The Plant Cell. 2006;18:1931–1946. doi: 10.1105/tpc.106.041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirner B, Fischer WN, Rentsch D, Kwart M, Frommer WB. Developmental control of H+/amino acid permease gene expression during seed development of Arabidopsis. The Plant Journal. 1998;14:535–544. doi: 10.1046/j.1365-313x.1998.00151.x. [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biology. 2004;5:R85. doi: 10.1186/gb-2004-5-11-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertel L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauniaux JC, Vandenbol M, Vissers S, Broman K, Grenson M. Nitrogen catabolite regulation of proline permease in Saccharomyces cerevisiae. European Journal of Biochemistry. 1987;164:601–606. doi: 10.1111/j.1432-1033.1987.tb11169.x. [DOI] [PubMed] [Google Scholar]
- Johnson-Brousseau SA, McCormick S. A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. The Plant Journal. 2004;39:761–775. doi: 10.1111/j.1365-313X.2004.02147.x. [DOI] [PubMed] [Google Scholar]
- Karley AJ, Leigh RA, Sanders D. Where do all the ions go? The cellular basis of differential ion accumulation in leaf cells. Trends in Plant Science. 2000;5:465–470. doi: 10.1016/s1360-1385(00)01758-1. [DOI] [PubMed] [Google Scholar]
- Klepek Y, Geiger D, Stadler R, Klebl F, Landouar-Arsivaud L, Lemoine R, Hedrich R, Sauer N. Arabidopsis Polyol Transporter5, a new member of the monosaccharide transporter-like superfamily, mediates H+-symport of numerous substrates, including myo-inositol, glycerol, and ribose. The Plant Cell. 2005;17:204–218. doi: 10.1105/tpc.104.026641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogaard H, Andersen AS. Free amino acids of Nicotiana alata anthers during development in vivo. Physiologia Plantarum. 1983;57:527–531. [Google Scholar]
- Krysan PJ, Young JC, Sussman MR. T-DNA as an insertional mutagen in Arabidopsis. The Plant Cell. 1999;11:2283–2290. doi: 10.1105/tpc.11.12.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Tax F, Sussman MR. Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proceedings of the National Academy of Sciences USA. 1996;93:8145–8150. doi: 10.1073/pnas.93.15.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansac AR, Sullivan CY, Johnson BE. Accumulation of free proline in sorghum (Sorghum bicolor) pollen. Canadian Journal of Botany. 1996;74:40–45. [Google Scholar]
- Lee YH, Foster J, Chen J, Voll LM, Weber APM, Tegeder M. AAP1 transports uncharged amino acids into roots of Arabidopsis. The Plant Journal. 2007;50:305–319. doi: 10.1111/j.1365-313X.2007.03045.x. [DOI] [PubMed] [Google Scholar]
- Lehmann S, Funck D, Szabados L, Rentsch D. Proline metabolism and transport in plant development. Amino Acids. 2010;39:949–962. doi: 10.1007/s00726-010-0525-3. [DOI] [PubMed] [Google Scholar]
- Mäkelä P, Peltonen-Sainio P, Jokinen K, Pehu E, Setälä H, Hinkkanen R, Somersalo S. Uptake and translocation of foliar-applied glycinebetaine in crop plants. Plant Science. 1996;121:221–230. [Google Scholar]
- Mani S, Van de Cotte B, Van Montagu M, Verbruggen N. Altered levels of proline dehydrogenase cause hypersensitivity to proline and its analogs in Arabidopsis. Plant Physiology. 2002;128:73–83. [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas JP. Gene activity during pollen development. Annual Review of Plant Physiology and Plant Molecular Biology. 1990;41:317–338. [Google Scholar]
- Mascarenhas JP. Molecular mechanisms of pollen tube growth and differentiation. The Plant Cell. 1993;5:1303–1314. doi: 10.1105/tpc.5.10.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F. Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. The Plant Journal. 1992;2:417–422. doi: 10.1111/j.1365-313x.1992.00417.x. [DOI] [PubMed] [Google Scholar]
- Mondal AK, Parui S, Mandal S. Analysis of the free amino acid content in the pollen of nine Asteraceae species of known allergenic activity. Annals of Agricultural and Environmental Medicine. 1998;5:17–20. [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Mutters RG, Ferreira LGR, Hall AE. Proline content of the anthers and pollen of heat-tolerant and heat-sensitive cowpea subjected to different temperatures. Crop Science. 1989;29:1497–1500. [Google Scholar]
- Okumoto S, Koch W, Tegeder M, Fischer WN, Biehl A, Leister D, Stierhof YD, Frommer WB. Root phloem-specific expression of the plasma membrane amino acid proton co-transporter AAP3. Journal of Experimental Botany. 2004;55:2155–2168. doi: 10.1093/jxb/erh233. [DOI] [PubMed] [Google Scholar]
- Räsänen LA, Saijets S, Jokinen K, Lindström K. Evaluation of the roles of two compatible solutes, glycine betaine and trehalose, for the Acacia senegal–Sinorhizobium symbiosis exposed to drought stress. Plant and Soil. 2004;260:237–251. [Google Scholar]
- Rentsch D, Hirner B, Schmelzer E, Frommer WB. Salt stress-induced proline transporters and salt stress-repressed broad specificity amino acid permeases identified by suppression of a yeast amino acid permease-targeting mutant. The Plant Cell. 1996;8:1437–1446. doi: 10.1105/tpc.8.8.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D, Schmidt S, Tegeder M. Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Letters. 2007;581:2281–2289. doi: 10.1016/j.febslet.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Schmidt R, Stransky H, Koch W. The amino acid permease AAP8 is important for early seed development in Arabidopsis thaliana. Planta. 2007;226:805–813. doi: 10.1007/s00425-007-0527-x. [DOI] [PubMed] [Google Scholar]
- Schwacke R, Grallath S, Breitkreuz KE, Stransky E, Stransky H, Frommer WB, Rentsch D. LeProT1, a transporter for proline, glycine betaine, and γ-amino butyric acid in tomato pollen. The Plant Cell. 1999;11:377–392. doi: 10.1105/tpc.11.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelp BJ, Bown AW, McLean MD. Metabolism and functions of γ-aminobutyric acid. Trends in Plant Science. 1999;4:446–452. doi: 10.1016/s1360-1385(99)01486-7. [DOI] [PubMed] [Google Scholar]
- Sleator RD, Hill C. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiology Reviews. 2002;26:49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x. [DOI] [PubMed] [Google Scholar]
- Stanley RG, Linskens HF. Berlin: Springer Verlag; 1974. Pollen. Biology biochemistry management. [Google Scholar]
- Stines AP, Naylor DJ, Høj PB, van Heeswijck R. Proline accumulation in developing grapevine fruit occurs independently of changes in the levels of Δ1-pyrroline-5-carboxylate synthetase mRNA or protein. Plant Physiology. 1999;120:923–931. doi: 10.1104/pp.120.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strizhov N, Ábrahám E, Ökrész L, Blickling S, Zilberstein A, Schell J, Koncz C, Szabados L. Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. The Plant Journal. 1997;12:557–569. doi: 10.1046/j.1365-313x.1997.00557.x. [DOI] [PubMed] [Google Scholar]
- Svennerstam H, Ganeteg U, Bellini C, Näsholm T. Comprehensive screening of Arabidopsis mutants suggests the lysine histidine transporter 1 to be involved in plant uptake of amino acids. Plant Physiology. 2007;143:1853–1860. doi: 10.1104/pp.106.092205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Székely G, Ábrahám E, Csépő Á, et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. The Plant Journal. 2008;53:11–28. doi: 10.1111/j.1365-313X.2007.03318.x. [DOI] [PubMed] [Google Scholar]
- Ueda A, Shi W, Sanmiya K, Shono M, Takabe T. Functional analysis of salt-inducible proline transporter of barley roots. Plant and Cell Physiology. 2001;42:1282–1289. doi: 10.1093/pcp/pce166. [DOI] [PubMed] [Google Scholar]
- Ueda A, Shi W, Shimada T, Miyake H, Takabe T. Altered expression of barley proline transporter causes different growth responses in Arabidopsis. Planta. 2008;227:277–286. doi: 10.1007/s00425-007-0615-y. [DOI] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C. Proline accumulation in plants: a review. Amino Acids. 2008;35:753–759. doi: 10.1007/s00726-008-0061-6. [DOI] [PubMed] [Google Scholar]
- Waditee R, Hibino T, Tanaka Y, et al. Functional characterization of betaine/proline transporters in betaine-accumulating mangrove. Journal of Biological Chemistry. 2002;277:18373–18382. doi: 10.1074/jbc.M112012200. [DOI] [PubMed] [Google Scholar]
- Yamada N, Promden W, Yamane K, Tamagake H, Hibino T, Tanaka Y, Takabe T. Preferential accumulation of betaine uncoupled to choline monooxygenase in young leaves of sugar beet—Importance of long-distance translocation of betaine under normal and salt-stressed conditions. Journal of Plant Physiology. 2009;166:2058–2070. doi: 10.1016/j.jplph.2009.06.016. [DOI] [PubMed] [Google Scholar]
- Yancey P. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. Journal of Experimental Biology. 2005;208:2819–2830. doi: 10.1242/jeb.01730. [DOI] [PubMed] [Google Scholar]
- Zhang HQ, Croes AF. Proline metabolism in pollen: degradation of proline during germination and early tube growth. Planta. 1983;159:46–49. doi: 10.1007/BF00998813. [DOI] [PubMed] [Google Scholar]
- Zúñiga G, Argandoña VH, Corcuera LJ. Distribution of glycine-betaine and proline in water stressed and unstressed barley leaves. Phytochemistry. 1989;28:419–420. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.