Abstract
Rapamycin induced apoptosis in sarcoma cells is inhibited by IGF-1 through a signaling pathway independent of Ras-Erk-1/2 and Akt (Thimmaiah et al. Cancer Res. 63:364, 2003). IGF-1 induces Bad phosphorylation (S112, S136, S155) in a pathway involving PI3K and PKC (μ, ε, or θ) resulting in sequestering Bad from mitochondria and subsequently interacting with 14-3-3γ in the cytosol. Gene knockdown of Bad, Bid, Akt1, Akt2, PKC-μ, PKC-ε or PKC-θ was achieved by transient transfection using small interfering RNAs. Results indicate IGF-1 signaling to Bad requires activation of PI3K, PKC (μ, θ, ε) but not mTOR, Ras/Erk-1/2, PKA, or p90RSK. Wortmannin blocked the phosphorylation of PKC-μ (S744/748) suggesting that PI3K is required for the activation of PKCs. PKCs phosphorylate Bad under in vitro conditions and the association of phospho-Bad with PKC-μ or PKC-ε, as demonstrated by immunoprecipitation, indicated direct involvement of PKCs in Bad phosphorylation. To confirm these results, cells overexpressing pEGFP-N1, wt-Bad, or Bad with a single site mutated (S112A; S136A; S155A), two sites mutated (S112A/136A; S112A/155A; S136A/155A) or the triple mutant (TM) were tested. IGF-1 protected completely against rapamycin induced apoptosis in cells overexpressing wt-Bad, and mutants having either one or two sites of phosphorylation mutated. Knockdown of Bid using siRNA showed that Bid is not required for rapamycin induced cell death. Collectively, this data suggest that IGF-1 induced phosphorylation of Bad at multiple sites via a pathway involving PI3K and PKCs is important for protecting sarcoma cells from rapamycin induced apoptosis.
Keywords: IGF-1, Bad, PKC, rapamycin, apoptosis
Introduction
The BH3-only protein, Bad, is unique, since its functions are tightly regulated by serine phosphorylation (1, 2). In the hypophosphorylated form, Bad interacts with either Bcl-2 or Bcl-XL to neutralize their anti-apoptotic functions, and this neutralization is believed to account for its proapoptotic functions. Inactive Bad is highly phosphorylated by survival signals and binds to 14-3-3 scaffold proteins and thus cannot interact with Bcl-2 or Bcl-XL (2, 3). Five phosphorylation sites have been reported for Bad. Phosphorylation at Ser112 and Ser136 is involved in 14-3-3 binding (4). Published results reveal that phosphorylation of Ser136 is accomplished predominantly by Akt or p70S6 kinase (5, 6), whereas mitochondrially localized protein kinase A, Rsk, and Pak1 have all been shown to phosphorylate Ser112 (7) (8–10). Ser170 is another site that is phosphorylated in cytokine-dependent cell survival (11). Recent reports indicate that Ser128 is phosphorylated by cdc2 during induction of apoptosis in cerebellar granular neurons. Phosphorylation of Ser128 has also been implicated in dissociation of Bad from 14-3-3 (12).
Recently, it has been shown that phosphorylation of Bad at Thr-201 by JNK1 promotes glycolysis through activation of phosphofructokinase-1 (13). There is growing information in the literature that the BH3-only protein plays an essential role in cytokine deprivation induced apoptosis in mast cells (5). BH3-only members may initiate apoptosis by directly binding to the essential cell-death mediators Bax and Bak. Alternatively, they can act by engaging their pro-survival Bcl-2-like relatives (14). In one study, phosphorylation at Ser136 and association with 14-3-3 was found to be essential for its growth-promoting effect (15) whereas in the other study only association with Bcl-XL was shown to be required, independent of the phosphorylation state of Bad (16).
Mammalian isoforms of 14-3-3 that bind and modify the functions of a wide variety of critical signaling molecules. One function of 14-3-3 is to promote cell survival, as inhibition of apoptosis results from the binding of many 14-3-3 ligands, including Bad, ASK1, and forkhead transcription factors (17). 14-3-3 forms a very stable complex with phosphorylated Bad and plays a significant role in the regulation of Bad function.
Rapamycin, a selective inhibitor of mTORC1 signaling causes G1 phase accumulation, and under growth factor deficient conditions p53-independent apoptosis. We have shown that rapamycin-induced apoptosis is prevented by exogenous IGF-1 through a signaling pathway independent of Ras-Erk-1/2 and Akt (18), and that combining rapamycin with an antibody that blocks ligand binding to IGF-1R is synergistic against most sarcoma xenograft tumor models (19). Our initial observation that overexpression of Bcl-2 significantly protects cells from rapamycin-induced apoptosis stimulated us to extend both pharmacological and genetic studies to explore whether phosphorylation of Bad is involved in IGF-1 mediated rescue.
Materials and Methods
Inhibitors
Rapamycin, wortmannin, LY 294002, PD 98059, calphostin-C, chelerythrine chloride, KT 5720, forskolin, and phorbol 12-myristate 13-acetate (PMA) were dissolved in DMSO before being added to culture medium (final concentration 0.1%).
Cell lines and growth conditions
The human cell lines Rh1 and Rh30 have been described (20) and were grown in antibiotic-free RPMI-1640 medium supplemented with 10% fetal bovine serum and 2 mM l-glutamine at 37 °C in an atmosphere of 5% CO2. For serum-free experiments, cells were cultured in modified MN2E medium as previously described (20).
ApoAlert assay
We used the ApoAlert™ Annexin V-FITC Apoptosis kit (Clontech) as previously described (20).
Western blot analysis
Immunoblotting methods were as previously described (20) with minor modifications. The secondary antibody was either horseradish peroxidase–conjugated goat anti–rabbit IgG or horseradish peroxidase–conjugated goat anti–mouse IgG antibody. Immunoreactive protein was visualized by using Renaissance chemiluminescence reagent.
PKC kinase assay
Rh1 cells grown in MN2E were exposed to 0.1 % DMSO or calphostin-C (1.2 µM) for 2 h and then stimulated with IGF-1 (10 ng/ml) or PMA (1µM) for 30 min. We then used the PKC assay kit (Upstate Biotechnology) according to the manufacturer's instructions to analyze the amount of activated PKC [α, βII and γ, and ε, θ, λ, μ, δ and ζ]. The amount of incorporated radioactivity into the substrate was determined by scintillation counting. To calculate the actual PKC activity, background radioactivity associated with control IgG antibody immunoprecipitated samples was subtracted. The kinase assay was repeated three times.
Plasmids and transfection
The control vector pEGFP-N1 and the test plasmids, pEGFP-Bad (wild-type), or the same construct with a single mutation (Ser 112Ala; Ser 136Ala; Ser155Ala), double mutations (Ser112Ala/136Ala; Ser 112Ala/155Ala; Ser 136Ala/155Ala) and pEGFP-Bad (TM) (all three serine sites mutated to alanine) used for this study have been described previously (21). The cells were transfected using the FuGENE6 according to the manufacturer's instructions. Stable expression of single- or double-mutants of Bad was confirmed by western blot analysis using phospho-specific antibodies (data not shown).
siRNA experiments
Cells growing in McCoy’s 5A medium with 10% FBS were transfected with si-RNA control, siBad, siBid, siPKC-μ, siPKC-ε, siPKC-θ, siAkt1 or siAkt2 using lipofectamine 2000. The cells were then incubated at 37°C for 72h and the levels of protein expression of Bad, Bid, PKC-μ, PKC-ε, PKC-θ, Akt1, or Akt2 was analyzed by Western blot using their respective antibodies.
Binding of Bad to 14-3-3
Rh1, Rh1/GFP-Bad (wt) or Rh1/GFP-Bad (TM) cells cultured in MN2E medium were stimulated with IGF-1 or PMA for 30 min and lysed in 500 µl of M-PER buffer as described previously and immunoprecipitated using protein A/G PLUS agarose beads. Immunoprecipitates of Bad were immunoblotted with the antibodies developed against anti-14-3-3 (σ, γ, β, ε, θ, η, and ζ).
Results
Effect of growth factors on the phosphorylation of Bad
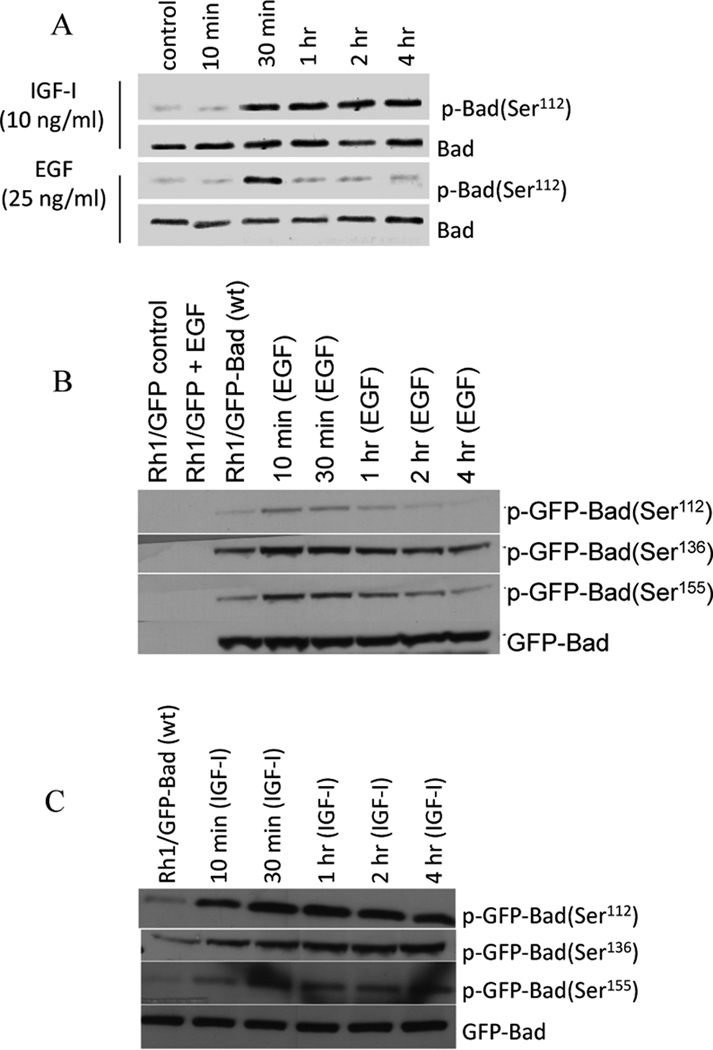
Because phosphorylation is critical for Bad inactivation (1, 2, 22–26), we first investigated whether IGF-1 (10 ng/ml), insulin (250 ng/ml), EGF (25 ng/ml) or PDGF (25 ng/ml) results in phosphorylation of Bad in Rh1 cells. As shown (Fig.1A), phosphorylation of Bad reached a maximum in 30 min after Rh1 cells were stimulated with either IGF-1 or EGF. Phospho-Bad specific antibodies to Ser 136 and Ser 155 were not sensitive enough for detecting endogenous Bad phosphorylation, consequently we examined the effect of these factors in Rh1 cells engineered to overexpress Bad (Rh1/GFP-Bad). As shown in Fig.1B, EGF stimulation led to a transient phosphorylation of Bad at Ser 112 and Ser 155. In contrast, IGF-1 stimulation resulted in prolonged phosphorylation at each site (Fig.1C). Similar kinetics for Bad phosphorylation were observed in Rh30 cells stimulated with IGF-1 (data not shown). These results demonstrate that IGF-1, or EGF treatment results in different patterns of Bad phosphorylation at multiple sites.
Figure 1. IGF-1 induces sustained phosphorylation of Bad while EGF induces transient phosphorylation of Bad.
Cells grown in MN2E medium were stimulated with IGF-1 (10 ng/mL) or EGF (25ng/mL) for different times. Phospho-Bad (serines 112, 136, or 155) was detected using phospho-specific antibodies. Identical results were obtained in at least three independent experiments.
A. Rh1 cells;
B–C Rh1/GFP-Bad (wt) cells.
IGF-1 or EGF stimulation of Bad phosphorylation is via PI3K but not ERK1/2, mTOR, or PKA
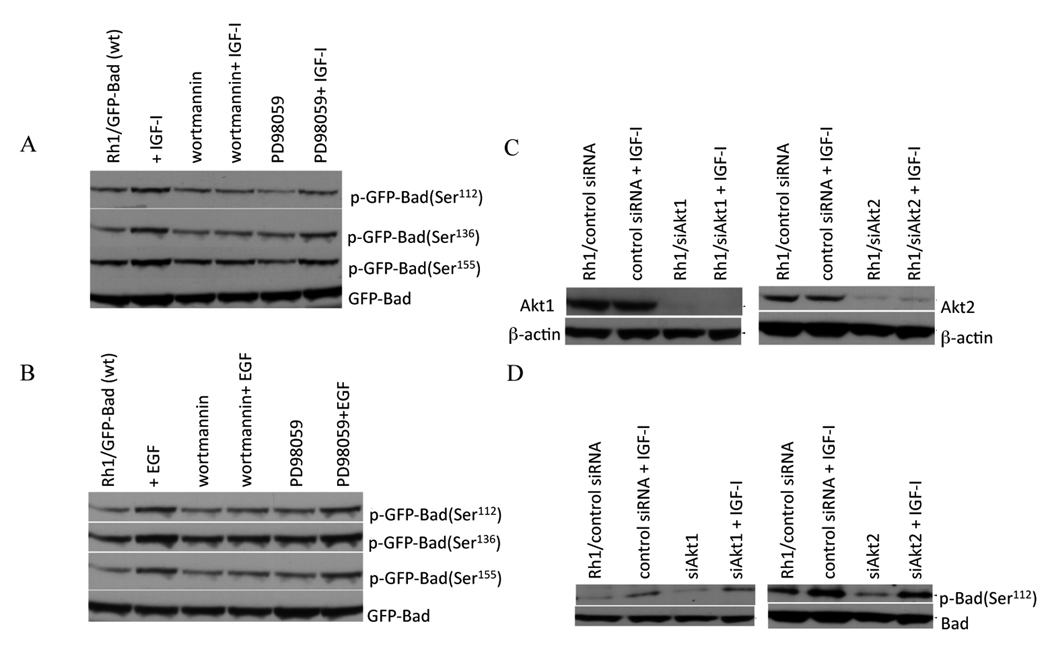
We (18), and others (27–31) have demonstrated that PI3K mediates IGF-1-mediated antiapoptotic signals. To determine whether PI3K or ERK-1/2 signaling was required for phosphorylation of each Bad site, Rh1/GFP-Bad cells were exposed to wortmannin (0.9 µM) or the MEK1/2 inhibitor PD98059 (30 µM) before stimulation with IGF-1 (Fig.2A) or EGF (Fig.2B). Wortmannin attenuated the IGF-1 or EGF-induced phosphorylation of Bad at each site whereas inhibition of MEK1/2 did not. PD98059 did inhibit the IGF-1 and EGF dependent activation of p90RSK as determined by a block in phosphorylation of Thr573 and Ser380 within the protein. The PKC inhibitor calphostin-C also failed to inhibit p90RSK phosphorylation in response to EGF or IGF-1 treatment, indicating that PKCs may not be required for the activation of p90RSK (Suppl. Fig.1A). To further determine if ERK-1/2 was important for IGF-1 dependent phosphorylation of Bad a dominant negative form of Ras (Ras N17) was expressed that completely inhibited Ras activation and ERK-1/2 phosphorylation induced by IGF-1 or EGF (18). Ras N17 failed to suppress phosphorylation of Bad (Ser112) after IGF-1 stimulation (Suppl. Fig.1B).
Figure 2. IGF-1 phosphorylates Bad at Ser112 via the PI3-kinase pathway independent of AKT.
A. Rh1/GFP-Bad cells were grown in MN2E, and treated with wortmannin, PD98059 for 2 h, and stimulated with IGF-1 for 30 min. Blots were probed with Bad phospho-specific antibodies.
B. Rh1/GFP-Bad cells were treated as in A but stimulated with EGF.
C. Rh1 cells were transfected with control siRNA or siRNA pools specific for Akt1 (left) or Akt2 (right). Transfected cells grown in MN2E medium were either stimulated with IGF-1, or not treated. Levels of Akt1 and Akt2 were determined by immunoblotting. Actin was used as control.
D. Rh1 cells were transfected and treated as in C. The levels of Bad (Ser112) were detected by immunoblotting. Results are representative of at least three independent experiments.
In addition, IGF-1-induced phosphorylation of Bad (Ser112) was not inhibited by rapamycin (Suppl. Fig.1C). Further, activation of PKA with forskolin, as measured by phosphorylation of CREB (inhibited by the PKA inhibitor KT5720), does not result in a phosphorylation of Bad (Suppl. Fig. 1D). This result excludes PKA in the IGF-1 mediated phosphorylation of Bad.
To further elucidate the role of Akt in Bad phosphorylation, Akt1 and Akt2 were silenced using siRNA specific for Akt1 or Akt2. siRNA greatly suppressed expression of Akt1 and Akt2 (Fig. 2C), however phosphorylation of Bad (Ser112) induced by IGF-1 was not abrogated (Fig.2D). Further, infection of Rh1 or Rh30 cells with ‘empty’ replication defective adenovirus (lacking the E1, and E3 regions) which results in massive activation of both the PI3K/Akt and MAPK pathways consistent with published data (32), failed to induce phosphorylation of Bad (Ser112) (Suppl. Fig. 2).
IGF-1 activates PKC (μ, θ, or ε) via PI3K
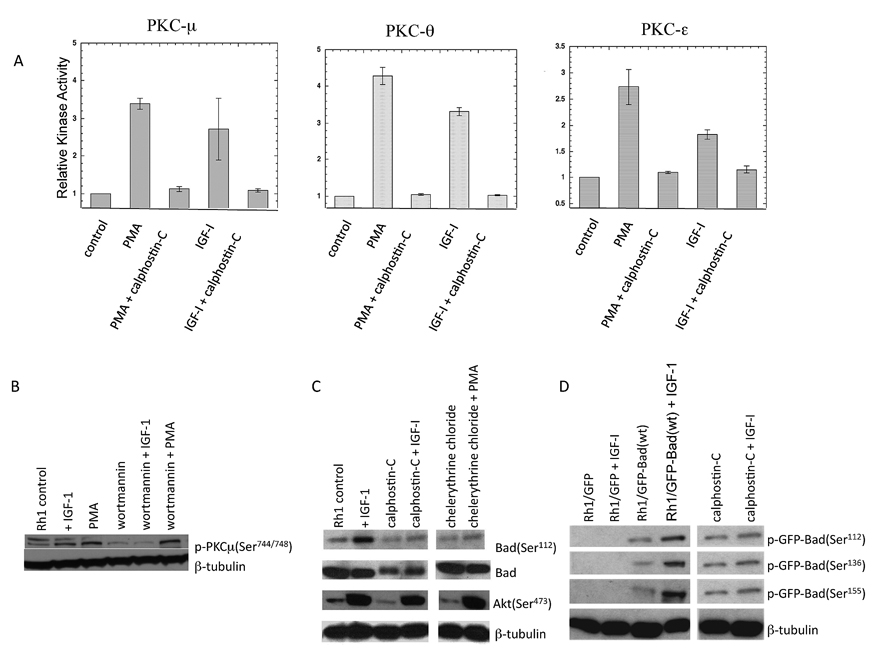
To determine the possible involvement of different isoforms of PKC in IGF-1 signaling, Rh1 cells were pre-incubated in MN2E medium with 0.1% DMSO or 1.2 µM of calphostin-C for 2 h before they were stimulated with IGF-1 or the PKC activator PMA (1 µM) for 30 min. Cell lysates were then immunoprecipitated with antibodies specific for PKC-α, PKC-βII, PKC-γ, PKC-δ, PKC-ε, PKC-θ, PKC-μ, PKC-λ, or PKC-ζ, and the resulting kinase activity of the immunocomplex was determined by measuring 32P phosphate transfer from ATP to peptide substrates. IGF-1 treatment resulted in a significant increase in the activity of only PKC-μ, PKC-θ and PKC-ε (Fig. 3A). PMA was used as a positive control for PKC activation. Further, as shown in Fig. 3A, activation of the three isoforms of PKC by IGF-1 or PMA was effectively blocked by calphostin-C.
Figure 3. PKCs-μ, θ, and ε are activated by IGF-1 and are inhibited by PKC and PI3K inhibitors.
A.Rh1 cells grown in MN2E were stimulated with IGF-1 or PMA for 30 min without or with calphostin-C for 2 h and immunoprecipitated using antibodies specific for PKC-μ, PKC-θ, PKC-ε. Immunocomplexes were then used to assay PKC activity. Activation of the three isoforms of PKC by IGF-1 (mean ± s.d.): PKC-μ (2.76 ± 0.81; n = 3); PKC-θ (3.31 ± 0.11; n = 3); PKC-ε (1.82 ± 0.10; n = 3).
B. Rh1 cells were grown under serum free conditions and treated as described above. The levels of phosphorylated PKC-μ (Ser 744/748) was determined in the presence and absence of PMA, wortmannin with or without IGF-1 stimulation.
C. Rh1 cells grown under serum free conditions were exposed to calphostin-C, or chelerythrine chloride for 2h and stimulated with IGF-1 or PMA for 30 min. After blotting phospho-specific antibodies directed against Bad Ser112 and Akt Ser473 were used to probe the blot. Total Bad and β-tubulin were used as controls.
D. Rh1/GFP, or Rh1/GFP-Bad (wt) cells grown in MN2E medium were incubated without or with calphostin-C for 2h and then stimulated with IGF-1 for 30 min. Cell lysates were prepared followed by SDS-PAGE and Western blot analysis. Phospho-specific antibodies detected the phosphorylated signals on GFP-Bad at Ser112, Ser136, and Ser155 in IGF-1 treated Rh1/GFP-Bad (wt) cells. The blot was stripped and reprobed with anti-β-tubulin antibody.
The results are representative of at least three independent experiments.
Wortmannin inhibited the IGF-1-induced phosphorylation of PKC-μ at Ser 744/748, but not PMA –induced phosphorylation (Fig.3B), indicating that PI3K plays an important role in IGF-1-mediated activation of PKC in Rh1 cells. To more directly assess the role of PKC in mediating Bad phosphorylation (10, 11, 33, 34), Rh1 cells grown in MN2E medium were exposed to potent inhibitors of PKC, calphostin-C (1.2 µM) or chelerythrine chloride (3.5 µM) for 2 h before stimulating with IGF-1 for 30 min (Fig.3C). Both calphostin-C and chelerythrine chloride suppressed IGF-1 stimulation of Bad (Ser112) phosphorylation, but failed to block phosphorylation of Akt. PMA, induced phosphorylation of Bad at Ser112 in Rh1 cells (10), is also blocked by treatment with chelerythrine chloride (Fig.3C). To test whether phosphorylation of Bad at other sites was PKC-dependent, Rh1/GFP-Bad cells grown in MN2E medium were exposed to calphostin-C for 2 h before stimulating with IGF-1 for 30 min. Immunoblotting revealed that calphostin-C blocked IGF-1 -induced Bad phosphorylation at each site (Fig.3D).
The simultaneous knockdown of genes for these three PKC-isoforms using siRNAs resulted in severe toxicity to cells. However, individual knockdown of PKC-μ, PKC-ε, or PKC-θ by means of siRNAs failed to inhibit the phosphorylation of Bad at Ser112 suggesting that the remaining PKC isoforms may be compensating for knockdown of an individual isoform (Suppl. Fig 3).
Direct interaction between PKC-μ or PKC-ε and Bad in Rh1 cells
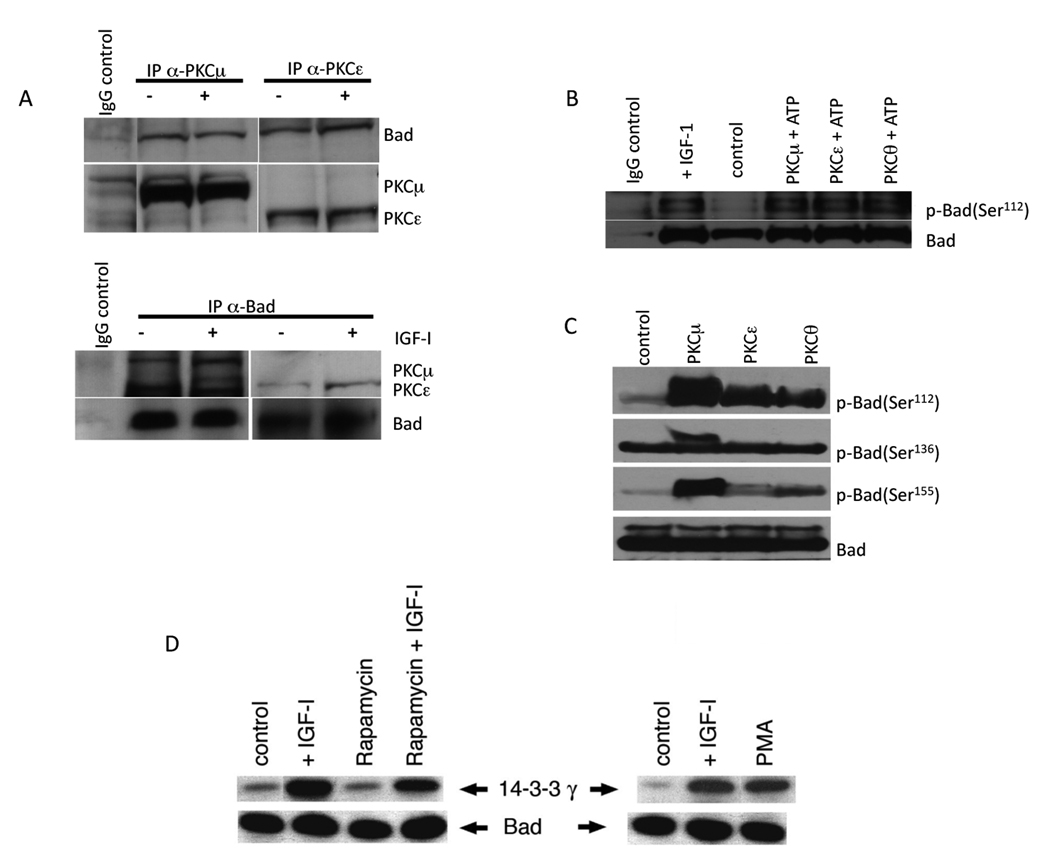
To test whether PKC can directly interact with Bad, Rh1 cells were stimulated with IGF-1 and cell lysates were immunoprecipitated with anti-PKC-μ, PKC-ε or anti-Bad antibodies and immunoblotted. The results demonstrate co-immunoprecipitation of PKC-μ or PKC-ε with Bad in Rh1 cells and provide evidence that there is a direct physical association (Fig.4A).
Figure 4. PKC-μ and -ε immunoprecipitate with Bad, and phosphorylate endogenous or recombinant Bad inducing association with 14-3-3γ.
A. Lysates from Rh1 cells were prepared after stimulation with IGF-1 (30 min) or after no stimulation and immunoprecipitated with antibodies specific for PKC-μ or PKC-ε or Bad. Immunoblots were probed with the anti-PKC or anti-Bad antibodies.
B. Endogenous Bad was immunoprecipitated from Rh1 cell lysates that had been stimulated with IGF-1 or not stimulated. Lysates were incubated with recombinant PKC isozymes without (control) or with addition of ATP. Proteins were separated by SDS-PAGE electrophoresis, and immunoblots were probed with antibodies against phospho-Bad (Ser112) or total Bad. IgG was used as a non-specific control.
C. Recombinant active PKC-μ, -ε, and -θ phosphorylate recombinant Bad. Recombinant Bad was incubated with active recombinant PKC isozymes in the absence (control) or presence of ATP. Proteins were separated by SDS-PAGE electrophoresis, and transferred to nitrocellulose membranes. The immunoblots were probed with antibodies against phospho-Bad (Ser112, Ser136, Ser155) or total Bad. The control reaction lacked ATP. The results were similar in at least three independent experiments.
D. IGF-1 stimulates the binding of Bad to 14-3-3γ under conditions where rapamycin induces apoptosis. Rh1 cells were grown under serum free conditions ± rapamycin, and ± IGF-1 for 5d (left panel) or stimulated with PMA or IGF-1 for 30 Min (right panel). Cell lysates were immunoprecipitated using anti-Bad antibody. The precipitated proteins were resolved by SDS-PAGE and immunoblotted using anti-14-3-3γ antibody. Results are representative of three independent experiments.
Active PKC (μ, θ, or ε) can directly phosphorylate endogenous Bad at Ser 112
To determine whether PKC can directly phosphorylate endogenous Bad, the lysates from serum starved Rh1 cells were immunoprecipitated with anti-Bad antibody and incubated with purified recombinant active PKC-μ, PKC-θ, or PKC-ε enzyme in a kinase buffer containing ATP (200 µM). The samples were resolved and immunoblotted for phospho-Bad at Ser 112 using phosphospecific antibody. IGF-1 stimulated phospho-Bad was used as a positive control. Results indicate that active PKC isoforms directly phosphorylate Bad at Ser 112 (Fig.4B). These findings suggest that PKC is a probable candidate for being the direct Bad kinase. Additional experiments were conducted in order to ascertain whether recombinant active PKC phosphorylates recombinant Bad. Recombinant Bad protein conjugated to agarose beads was incubated with purified active PKC-μ, PKC-θ, or PKC-ε for 30 min in an in vitro kinase assay. Phosphorylation of Bad was detected using phosphospecific antibodies to Ser 112, Ser136 or Ser155. As shown in Fig.4C, recombinant PKC-μ can directly phosphorylate recombinant Bad protein at all the three sites, whereas the activity of PKC-θ or -ε was weak against Ser155 and not detected against Ser 136.
IGF-1 stimulates the binding of Bad to 14-3-3γ under apoptotic conditions
We tested the interaction of Bad with seven different 14-3-3 isoforms (35). Immunoblotting with antibodies specific for particular isoforms of 14-3-3 (α, β, γ, θ, η, ζ, or ε) showed that both γ, and θ isoforms were expressed at high levels. We hypothesized that under apoptotic conditions induced by rapamycin treatment, Bad would not be associated with 14-3-3γ but that the protective effects of IGF-1 would stimulate binding. To test this hypothesis, Rh1 cells grown in MN2E medium were exposed to 0.1 % DMSO or rapamycin (100 ng/ml) in the absence or presence of IGF-1 continuously for 5 d. Lysates were prepared and Bad was immunoprecipitated using anti-Bad antibody, resolved by SDS-PAGE and immunoblotted using anti-14-3-3γ antibody. As shown in Fig. 4D, very little association of Bad with 14-3-3γ was seen in control cells or in the presence of rapamycin (apoptotic conditions). In contrast, treatment with IGF-1, in the absence or presence of rapamycin, resulted in a marked increase in Bad complexed with 14-3-3γ. These findings suggest a possible mechanism by which IGF-1 protects the cells from apoptosis induced by rapamycin through inducing binding of Bad to 14-3-3γ.
To determine whether PMA-induced phosphorylated Bad (Ser112) binds to 14-3-3 proteins, Rh1 cells cultured in MN2E medium were stimulated with IGF-1 or 1 µM PMA for 30 min, and samples processed as before. 14-3-3γ co-immunoprecipitated with Bad in cells stimulated with either IGF-1 or PMA but not from the non stimulated control cells (Fig.4D).
Bad phosphorylation at any site (Ser 112, Ser 136, Ser 155) facilitates IGF-1 rescue from rapamycin-induced apoptosis
To address the role of Bad phosphorylation in anti-apoptotic signaling, we tested the survival effect of IGF-1 in rapamycin treated Rh1 cells that were engineered to stably express pEGFP-N1 (vector control), wild type GFP-Bad, single and double phosphorylation site mutants, as well as the triple mutant (TM) containing amino acid substitutions Ser112Ala, Ser136Ala, and Ser155Ala. In preliminary experiments we determined that Rh1/GFP-Bad(wt), or Rh1/GFP-Bad(triple mutant) could be stably expressed in Rh1 cells (Suppl. Fig. 4A) and calphostin-C blocked IGF-1 induced phosphorylation of Bad at each site (Suppl. Fig.4B). As anticipated, no phosphorylation was detected in the Bad mutant where all three sites were mutated to alanine (Suppl. Fig. 4C). Overexpressed wild-type Bad bound 14-3-3γ whereas the triple mutant Bad failed to bind 14-3-3γ (Suppl. Fig. 4D).
The effect of rapamycin treatment on the viability of Rh1 cells expressing GFP-Bad (wt), or the Bad mutants was compared with the effect on the viability of an empty vector GFP expressing control cell line using the ApoAlert flow cytometric assay, and presented in Table 1 (cells expressing single, double or triple mutants of Bad). Expression of Bad wt and mutants was approximately similar (Suppl. Fig. 5). Consistent with our earlier findings, the apoptotic profile of Rh1 cells expressing pEGFP-N1 vector was almost identical with those of parental Rh1 cells with approximately 25% of the empty vector expressing cells positive both for annexin V-and propidium iodide staining. IGF-1 reduced this level to ~ 15 to 20%. Rapamycin treatment resulted in ~ 60% of control cells undergoing apoptosis and addition of IGF-1 resulted in almost complete protection from apoptosis. Similar results were obtained in cells expressing single or double Bad mutants, where IGF-1 completely protected against rapamycin-induced apoptosis. Expression of GFP-Bad (triple mutant) in Rh1 cells decreased the overall viability to a significant extent resulting in ~ 88% apoptosis under rapamycin-free culture conditions and IGF-1 could not reverse this effect completely (apoptotic population of 50%). This rescue probably reflects IGF-1 mediated abrogation of endogenous Bad function (~40% of cells rescued). Rapamycin treatment resulted in ~ 95% of cells being scored as apoptotic, and addition of IGF-1 resulted in the reduction of this population to ~76%, thus exhibiting only a marginal rescue effect. Taken together, our experiments with ectopic over-expression of phosphorylation deficient Bad mutants indicate that Bad phosphorylation at one of three serine residues is essential for the anti-apoptotic effects of IGF-1.
Table 1.
IGF-1 rescue from rapamycin-induced apoptosis in Rh1 cells expressing Bad single, double or triple phosphorylation mutants
| Cell line + Treatmenta | Viable cells (%) (Mean ± SD) |
Apoptotic cells (%) (Mean ± SD) |
|---|---|---|
| Rh1/GFP | 83.81 ± 6.53 | 15.91 ± 6.70 |
| Rh1/GFP + IGF-1 | 89.6 ± 5.54 | 10.07 ± 5.93 |
| Rh1/GFP + Rap | 51.53 ± 11.98 | 48.47 ± 12.42 |
| Rh1/GFP + Rap + IGF-1 | 86.89 ± 5.88 | 12.77 ± 5.99 |
| Rh1/GFP-Bad(wt) | 74.50 ± 2.29 | 25.50 ± 2.52 |
| Rh1/GFP –Bad(wt) + IGF-1 | 83.50 ± 4.27 | 16.50 ± 0.50 |
| Rh1/GFP-Bad(wt) + Rap | 40.00 ± 8.32 | 60.00 ± 9.40 |
| Rh1/GFP-Bad(wt) + Rap + IGF-1 | 79.50 ± 9.80 | 20.50 ± 1.32 |
| Rh1/GFP-Bad Ser 112Ala | 68.50 ± 9.90 | 31.50 ± 2.52 |
| Rh1/GFP Bad Ser 112Ala + IGF-1 | 84.00 ± 7.00 | 16.00 ± 2.55 |
| Rh1/GFP-Bad Ser 112Ala + Rap | 37.00 ± 2.65 | 63.00 ± 2.20 |
| Rh1/GFP-Bad Ser 112Ala + Rap + IGF-1 | 84.50 ± 2.29 | 15.50 ± 1.50 |
| Rh1/GFP-Bad Ser136Ala | 64.00 ± 8.72 | 36.00 ± 3.72 |
| Rh1/GFP-Bad Ser136Ala + IGF-1 | 85.00 ± 4.36 | 15.00 ± 2.61 |
| Rh1/GFP-Bad Ser136Ala + Rap | 38.00 ± 3.08 | 62.00 ± 1.74 |
| Rh1/GFP-Bad Ser136Ala + Rap + IGF-1 | 80.00 ± 6.21 | 20.00 ± 5.00 |
| Rh1/GFP-Bad Ser155Ala | 66.00 ± 5.29 | 34.00 ± 6.00 |
| Rh1/GFP-Bad Ser155Ala + IGF-1 | 80.00 ± 8.00 | 20.00 ± 5.00 |
| Rh1/GFP-Bad Ser155Ala + Rap | 38.00 ± 7.21 | 62.00 ± 4.80 |
| Rh1/GFP-Bad Ser155Ala + Rap + IGF-1 | 80.00 ± 7.00 | 20.00 ± 4.00 |
| Rh1/GFP-Bad Ser112Ala/136Ala | 67.50 ± 1.50 | 32.50 ± 1.50 |
| Rh1/GFP-Bad Ser112Ala/136Ala + IGF-1 | 85.00 ± 7.00 | 15.00 ± 2.00 |
| Rh1/GFP-Bad Ser112Ala/136Ala + Rap | 35.00 ± 3.00 | 65.00 ± 9.17 |
| Rh1/GFP-Bad Ser112Ala/136Ala + Rap + IGF-1 | 82.50 ± 3.50 | 17.50 ± 3.50 |
| Rh1/GFP-Bad Ser112Ala/155Ala | 57.00 ± 3.00 | 43.00 ± 2.87 |
| Rh1/GFP-Bad Ser112Ala/155Ala + IGF-1 | 84.50 ± 2.93 | 15.50 ± 1.77 |
| Rh1/GFP-Bad Ser112Ala/155Ala + Rap | 34.50 ± 1.50 | 65.50 ± 2.50 |
| Rh1/GFP-Bad Ser112Ala/155Ala + Rap + IGF-1 | 80.50 ± 2.50 | 19.50 ± 2.50 |
| Rh1/GFP-Bad Ser136Ala/155Ala | 53.50 ± 2.35 | 46.50 ± 2.35 |
| Rh1/GFP-Bad Ser136Ala/155Ala + IGF-1 | 85.00 ± 2.65 | 15.00 ± 2.65 |
| Rh1/GFP-Bad Ser136Ala/155Ala + Rap | 35.00 ± 3.00 | 65.00 ± 7.00 |
| Rh1/GFP-Bad Ser136Ala/155Ala + Rap + IGF-1 | 80.00 ± 8.50 | 20.00 ± 2.56 |
| Rh1/GFP-Bad (triple mutant) | 12.00 ± 4.00 | 88.00 ± 6.50 |
| Rh1/GFP-Bad (triple mutant) + IGF-1 | 50.00 ± 2.70 | 50.00 ± 3.20 |
| Rh1/GFP-Bad (triple mutant) + Rap | 5.00 ± 1.80 | 95.00 ± 5.60 |
| Rh1/GFP-Bad (triple mutant) + Rap + IGF-1 | 24.00 ± 3.70 | 76.00 ± 6.00 |
IGF-1, 10 ng/mL; rapamycin (Rap), 100 ng/ml, Annexin V-negative, propidium iodide-positive, < 1.5%.
Knockdown of Bad results in a partial increase of cytoprotection from rapamycin-induced apoptosis
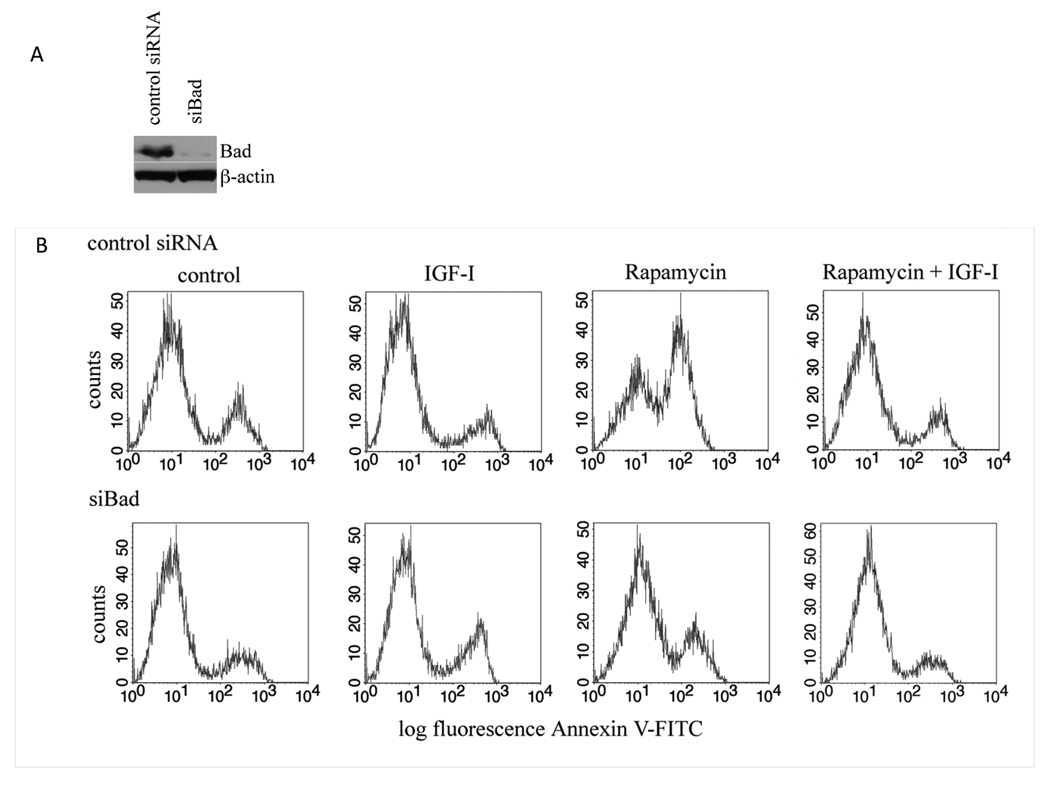
Our results indicate that IGF-1 can abrogate the proapoptotic activity of Bad by inducing its phosphorylation at multiple sites in sarcoma cells. To test this RNA interference was employed. Results show that siRNA can potently and specifically reduce Bad expression by more than 95%, whereas the control siRNA had no effect (Fig.5A). To determine whether Bad is responsible for rapamycin-induced apoptosis, we exposed Rh1/sicontrol RNA or Rh1/siBad cells to rapamycin for 4 days in the presence or absence of IGF-1, and determined the extent of apoptosis within each treatment population. Analysis of the results (Table-1, and Fig.5B) revealed that in Rh1/sicontrol cells rapamycin increased apoptosis by 31% whereas in Bad deficient cells rapamycin increase apoptosis by only 16%. In both conditions co-incubation with IGF-1 almost completely protected the cells. Thus, approximately 50% of the apoptosis induced by rapamycin appears to be mediated through Bad.
Figure 5. Downregulation of Bad protects cells from rapamycin-induced apoptosis.
A. Rh1 cells were transfected with siRNA pools against Bad or control RNAi. Cells were grown for anadditional 2 d, and levels of Bad determined by immunoblotting.
B. Rh1 cells transfected with control or Bad siRNA were grown in MN2E medium without treatment (control), with IGF-1, rapamycin or both reagents. Apoptosis was determined using the ApoAlert assay after 4 d treatment. Similar results were obtained in three separate experiments.
Discussion
The specific objectives of the present study were to determine whether Bad phosphorylation is required for IGF-1 rescue of Rh1 cells from rapamycin-induced apoptosis, and to define the IGF-1-Bad signaling pathway. We have dissected the signaling events leading to Bad phosphorylation at individual sites (Ser112, Ser136, and Ser155) and our results indicate that in response to IGF-1, or EGF, endogenous Bad is phosphorylated at Ser112 but not significantly at Ser136 or Ser155 in Rh1 and Rh30 cells, as evidenced by the failure to detect the phosphorylation sites of endogenous Bad at Ser136 and Ser155 by available phospho-specific antibodies, consistent with a previous report (36). In Rh1 cells, EGF induces only transient phosphorylation of Bad (Ser112), and does not afford protection from rapamycin-induced apoptosis, whereas IGF-1 stimulation leads to prolonged Bad (Ser112) phosphorylation and protection from apoptosis.
The observations that PI3K inhibitors blocked the IGF-1-induced phosphorylation of Bad demonstrate that PI3K mediates IGF-1 receptor-Bad signaling pathway, although phosphorylation of Akt(Ser473) is not necessary for Bad phosphorylation. Endogenous Akt3 expression in Rh1 cells is not detectable by Western blot analysis. Infection of cells with replication defective adenovirus resulted in a massive phosphorylation of Akt(Ser473) without phosphorylation of Bad. These data appear to confirm that Akt signaling is not involved in the Bad phosphorylation at Ser112 for this cell type.
IGF-1 stimulation of PKC-μ phosphorylation was inhibited by wortmannin placing PI3K upstream of PKCs in the IGF-1 signaling cascade. We focused on the potential role of PKCs in IGF-1 dependent phosphorylation of Bad. The PMA- or IGF-1-induced phosphorylation of Bad on Ser112 was drastically inhibited by calphostin-C or chelerythrine chloride, confirming that Bad phosphorylation at Ser112 is PKC-dependent. In vitro kinase assays using recombinant active PKC (μ, θ, ε) enzymes furnished direct evidence for the phosphorylation of both endogenous and recombinant Bad at serine 112, 136 or 155. We found that, of the three isoforms of PKC examined, only the recombinant active PKC-μ enzyme phosphorylates all three sites of recombinant Bad whereas PKC-θ or PKC-ε enzyme targets predominantly Ser 112 and to a lesser extent Ser155, and demonstrated no activity against Ser 136 on recombinant Bad. All three isoforms of recombinant active PKC enzymes phosphorylated endogenous Bad from Rh1 cells at Ser 112. We also observed a direct interaction between PKC-μ or PKC-ε and Bad as evidenced by immunoprecipitation, suggesting that PKC-μ or PKC-ε may be a physiological Bad kinases. Simultaneous silencing of PKC-μ, PKC-ε, and PKC-θ resulted in severe toxicity to Rh1 cells, but the individual knockdown of PKC-μ, PKC-ε, or PKC-θ by siRNA failed to inhibit the phosphorylation of Bad at Ser 112 suggesting that PKC-μ, PKC-ε, and PKC-θ isoforms may be able to compensate for one another.
Our findings that calphostin-C failed to block the IGF-1-induced phosphorylation of p90RSK but did block Bad phosphorylation, suggest that p90RSK is not involved in this PKC-mediated pathway. Similarly, pharmacologic and genetic evidence suggest that Erk-1/2, PKA, and mTOR are also not involved in IGF-1 dependent phosphorylation of Bad.
In Rh1 cells only 14-3-3γ was found to bind phosphorylated Bad following IGF-1 stimulation. Further, our observation that IGF-1 stimulates the interaction between 14-3-3γ and phospho-Bad under conditions where IGF-1 protects against rapamycin-induced apoptosis supports a possible role for this interaction in mediating the protective effects of IGF-1. To test the hypothesis that interaction of Bad with 14-3-3γ was required for IGF-1-mediated cell rescue, we used a genetic approach. Either wild type or phosphorylation defective GFP-Bad (TM) in which Ala residues were substituted for regulatory serines were expressed in Rh1 cells. Mutation of all the three serine residues in Bad to alanine abrogated its ability to be phosphorylated in response to IGF-1, and completely abolished its binding with 14-3-3γ.
The results of the apoptosis assays reveal Bad as a major player in rapamycin-induced cell death, and that IGF-1-mediated protection correlates well with phosphorylation and sequestration of Bad. In Rh1 cells stably expressing control vector, single or double mutants of Bad or Bad (wt) cells undergo apoptosis to an extent of ~ 25 to 50% in serum-free medium. However, addition of IGF-1 resulted in essentially complete protection. These results support the contention that Bad inactivation can be effected through phosphorylation of any one of the three regulatory serines. Expression of Bad (TM) offered the opportunity to determine the contribution played by Bad, as opposed to other proapoptotic molecules, in rapamycin-induced apoptosis. The viability of Bad (TM) cells was reduced to a significant extent and the apoptotic population increased to about 88% under serum-free conditions. In contrast to the phosphorylation-competent mutant of Bad, IGF-1 failed to protect Bad (TM) cells completely; IGF-1 reduced the apoptotic population from ~ 88 to 50%. This modest rescue probably is due to IGF-1 inducing phosphorylation and inactivation of endogenous Bad, but not altering the proapoptotic activity of the triple mutant. Rapamycin treatment increased the apoptotic population from ~88% to ~ 95%, and IGF-1 reduced this to ~ 76%, suggesting that it rescued the component contributed by the endogenous Bad, but not that contributed by the phosphorylation defective mutant. However, apoptosis contributed by the triple mutant makes analysis of the contribution of mutant and wild type Bad difficult.
To better understand the effect of rapamycin alone, Bad was silenced in Rh1 cells using siRNA. Rapamycin increased apoptosis by 31% in the siRNA control cells whereas it increased apoptosis by only 16% in siBad cells. These results suggest that Bad down regulation plays a significant role in protection from rapamycin-induced apoptosis. However, addition of IGF-1 to siBad cells resulted in almost complete protection.
This may indicate that a small fraction of endogenous Bad was still functional, or an alternative mechanism, other than Bad phosphorylation, contributes to partial protection from rapamycin-induced apoptosis. The apoptotic effect of rapamycin in Rh1 cells transfected with siRNA control, siBid with or without IGF-1 was also assessed. The results reveal that Bid (37) alone failed to play any role in the apoptosis caused by rapamycin (data not shown).
In summary, we suggest that the increase in Bad phosphorylation through a PI3K/PKC-mediated pathway by IGF-1 may represent an important pathway for the cytoprotective effect of this growth factor. Thus, IGF-1-induced phosphorylation of Bad by PKC and subsequent sequestration by14-3-3 proteins serves at least, in part, the mechanism by which IGF-1 protects Rh1 cells against rapamycin-induced apoptosis and offers a potential mechanism by which small molecule BH3 domain inhibitors or inhibitors of PKC may be valuable in changing the cellular response to rapamycin from cytostasis under normal physiological conditions where IGF-1 is present, to apoptosis
Supplementary Material
Acknowledgments
This work was supported by CA77776, CA96696, CA23099 and CA21765 (Cancer Center Support Grant) and by American, Lebaneses, Syrian Associated Charities (ALSAC).
The abbreviations used are
- Akt
Oncogene from AKR mouse thymoma
- ADB-II
assay dilution buffer-II
- IGF-1
Insulin-like growth factor I
- Erk-1/2
extracellular regulated kinase 1/2
- mTOR
mammalian target of rapamycin
- Rh
rhabdomyosarcoma
- PI3K
phosphoinositide 3’ kinase
- PKC
protein kinase C
- MAPK
mitogen-activated protein kinase
- PKA
cAMP-dependent protein kinase A
- BH
Bcl2 homology domain
- MEK
mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- CREB
cAMP-response element-binding protein
- GFP
green fluorescent protein
- EGF
epidermal growth factor
- PDGF
platelet-derived growth factor
- MN2E
modified N2E medium
- FACS
fluorescence-activated cell sorting
- DMSO
dimethylsulfoxide
References
- 1.Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 2.Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 3.Zha J, Harada H, Osipov K, Jockel J, Waksman G, Korsmeyer SJ. BH3 domain of BAD is required for heterodimerization with BCL-XL and pro-apoptotic activity. J Biol Chem. 1997;272:24101–24104. doi: 10.1074/jbc.272.39.24101. [DOI] [PubMed] [Google Scholar]
- 4.Alessi DR, Cuenda A, Cohen P, Dudley DT, Saltiel AR. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 5.Harada H, Andersen JS, Mann M, Terada N, Korsmeyer SJ. p70S6 kinase signals cell survival as well as growth, inactivating the pro-apoptotic molecule BAD. Proc Natl Acad Sci U S A. 2001;98:9666–9670. doi: 10.1073/pnas.171301998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datta SR, Dudek H, Tao X, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 7.Harada H, Becknell B, Wilm M, et al. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- 8.Schurmann A, Mooney AF, Sanders LC, et al. p21-activated kinase 1 phosphorylates the death agonist bad and protects cells from apoptosis. Mol Cell Biol. 2000;20:453–461. doi: 10.1128/mcb.20.2.453-461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimamura A, Ballif BA, Richards SA, Blenis J. Rsk1 mediates a MEK-MAP kinase cell survival signal. Curr Biol. 2000;10:127–135. doi: 10.1016/s0960-9822(00)00310-9. [DOI] [PubMed] [Google Scholar]
- 10.Tan Y, Ruan H, Demeter MR, Comb MJ. p90(RSK) blocks bad-mediated cell death via a protein kinase C-dependent pathway. J Biol Chem. 1999;274:34859–34867. doi: 10.1074/jbc.274.49.34859. [DOI] [PubMed] [Google Scholar]
- 11.Dramsi S, Scheid MP, Maiti A, et al. Identification of a novel phosphorylation site, Ser-170, as a regulator of bad pro-apoptotic activity. J Biol Chem. 2002;277:6399–6405. doi: 10.1074/jbc.M109990200. [DOI] [PubMed] [Google Scholar]
- 12.Konishi Y, Lehtinen M, Donovan N, Bonni A. Cdc2 phosphorylation of BAD links the cell cycle to the cell death machinery. Mol Cell. 2002;9:1005–1016. doi: 10.1016/s1097-2765(02)00524-5. [DOI] [PubMed] [Google Scholar]
- 13.Deng H, Yu F, Chen J, Zhao Y, Xiang J, Lin A. Phosphorylation of Bad at Thr-201 by JNK1 promotes glycolysis through activation of phosphofructokinase-1. J Biol Chem. 2008;283:20754–20760. doi: 10.1074/jbc.M800024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willis SN, Fletcher JI, Kaufmann T, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 15.Maslyar DJ, Aoki M, Vogt PK. The growth-promoting activity of the Bad protein in chicken embryo fibroblasts requires binding to protein 14-3-3. Oncogene. 2001;20:5087–5092. doi: 10.1038/sj.onc.1204662. [DOI] [PubMed] [Google Scholar]
- 16.Chattopadhyay A, Chiang CW, Yang E. BAD/BCL-[X(L)] heterodimerization leads to bypass of G0/G1 arrest. Oncogene. 2001;20:4507–4518. doi: 10.1038/sj.onc.1204584. [DOI] [PubMed] [Google Scholar]
- 17.Masters SC, Subramanian RR, Truong A, et al. Survival-promoting functions of 14-3-3 proteins. Biochem Soc Trans. 2002;30:360–365. doi: 10.1042/bst0300360. [DOI] [PubMed] [Google Scholar]
- 18.Thimmaiah KN, Easton J, Huang S, et al. Insulin-like growth factor I-mediated protection from rapamycin-induced apoptosis is independent of Ras-Erk1-Erk2 and phosphatidylinositol 3'-kinase-Akt signaling pathways. Cancer Res. 2003;63:364–374. [PubMed] [Google Scholar]
- 19.Kurmasheva RT, Dudkin L, Billups C, Debelenko LV, Morton CL, Houghton PJ. The insulin-like growth factor-1 receptor-targeting antibody, CP-751,871, suppresses tumor-derived VEGF and synergizes with rapamycin in models of childhood sarcoma. Cancer Res. 2009;69:7662–7671. doi: 10.1158/0008-5472.CAN-09-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hosoi H, Dilling MB, Shikata T, et al. Rapamycin causes poorly reversible inhibition of mTOR and induces p53-independent apoptosis in human rhabdomyosarcoma cells. Cancer Res. 1999;59:886–894. [PubMed] [Google Scholar]
- 21.Roberts ML, Virdee K, Sampson CP, Gordon I, Parone P, Tolkovsky AM. The combination of bcl-2 expression and NGF-deprivation facilitates the selective destruction of BAD protein in living sympathetic neurons. Mol Cell Neurosci. 2000;16:97–110. doi: 10.1006/mcne.2000.0867. [DOI] [PubMed] [Google Scholar]
- 22.Zhou XM, Liu Y, Payne G, Lutz RJ, Chittenden T. Growth factors inactivate the cell death promoter BAD by phosphorylation of its BH3 domain on Ser155. J Biol Chem. 2000;275:25046–25051. doi: 10.1074/jbc.M002526200. [DOI] [PubMed] [Google Scholar]
- 23.Virdee K, Parone PA, Tolkovsky AM. Phosphorylation of the pro-apoptotic protein BAD on serine 155, a novel site, contributes to cell survival. Curr Biol. 2000;10:R883. doi: 10.1016/s0960-9822(00)00843-5. [DOI] [PubMed] [Google Scholar]
- 24.Tan Y, Demeter MR, Ruan H, Comb MJ. BAD Ser-155 phosphorylation regulates BAD/Bcl-XL interaction and cell survival. J Biol Chem. 2000;275:25865–25869. doi: 10.1074/jbc.M004199200. [DOI] [PubMed] [Google Scholar]
- 25.Lizcano JM, Morrice N, Cohen P. Regulation of BAD by cAMP-dependent protein kinase is mediated via phosphorylation of a novel site, Ser155. Biochem J. 2000;349:547–557. doi: 10.1042/0264-6021:3490547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Datta SR, Katsov A, Hu L, et al. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell. 2000;6:41–51. [PubMed] [Google Scholar]
- 27.Yao R, Cooper GM. Growth factor-dependent survival of rodent fibroblasts requires phosphatidylinositol 3-kinase but is independent of pp70S6K activity. Oncogene. 1996;13:343–351. [PubMed] [Google Scholar]
- 28.Vemuri GS, McMorris FA. Oligodendrocytes and their precursors require phosphatidylinositol 3-kinase signaling for survival. Development. 1996;122:2529–2537. doi: 10.1242/dev.122.8.2529. [DOI] [PubMed] [Google Scholar]
- 29.Scheid MP, Lauener RW, Duronio V. Role of phosphatidylinositol 3-OH-kinase activity in the inhibition of apoptosis in haemopoietic cells: phosphatidylinositol 3-OH-kinase inhibitors reveal a difference in signalling between interleukin-3 and granulocyte-macrophage colony stimulating factor. Biochem J. 1995;312(Pt 1):159–162. doi: 10.1042/bj3120159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Datta K, Bellacosa A, Chan TO, Tsichlis PN. Akt is a direct target of the phosphatidylinositol 3-kinase. Activation by growth factors, v-src and v-Ha-ras, in Sf9 and mammalian cells. J Biol Chem. 1996;271:30835–30839. doi: 10.1074/jbc.271.48.30835. [DOI] [PubMed] [Google Scholar]
- 31.Dudek H, Datta SR, Franke TF, et al. Regulation of neuronal survival by the serinethreonine protein kinase Akt. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 32.O'Shea C, Klupsch K, Choi S, et al. Adenoviral proteins mimic nutrient/growth signals to activate the mTOR pathway for viral replication. Embo J. 2005;24:1211–1221. doi: 10.1038/sj.emboj.7600597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Z, Xin M, Deng X. Survival function of protein kinase C{iota} as a novel nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-activated bad kinase. J Biol Chem. 2005;280:16045–16052. doi: 10.1074/jbc.M413488200. [DOI] [PubMed] [Google Scholar]
- 34.Villalba M, Bushway P, Altman A. Protein kinase C-theta mediates a selective T cell survival signal via phosphorylation of BAD. J Immunol. 2001;166:5955–5963. doi: 10.4049/jimmunol.166.10.5955. [DOI] [PubMed] [Google Scholar]
- 35.Aitken A. 14-3-3 and its possible role in co-ordinating multiple signalling pathways. Trends Cell Biol. 1996;6:341–347. doi: 10.1016/0962-8924(96)10029-5. [DOI] [PubMed] [Google Scholar]
- 36.Wang SW, Denny TA, Steinbrecher UP, Duronio V. Phosphorylation of Bad is not essential for PKB-mediated survival signaling in hemopoietic cells. Apoptosis. 2005;10:341–348. doi: 10.1007/s10495-005-0808-4. [DOI] [PubMed] [Google Scholar]
- 37.Vogel A, Aslan JE, Willenbring H, et al. Sustained phosphorylation of Bid is a marker for resistance to Fas-induced apoptosis during chronic liver diseases. Gastroenterology. 2006;130:104–119. doi: 10.1053/j.gastro.2005.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.