Introduction
Translational mobility is involved in every process in neurobiology—released neurotransmitter diffuses through the synaptic cleft in search of receptor targets, membrane receptors traffic to synaptic sites in the neuron, RNA and other cargo are transported to distal dendrites, and cell signaling is mediated by circuits of diffusing proteins. However, accurate methods for quantifying translational mobility have not been widespread due to their technical demands and complicated analysis. Over the past decade, this trend has begun to reverse as major advancements have been made in the application of quantitative optical techniques, specifically fluorescence recovery after photobleaching (FRAP or FPR), fluorescence correlation spectroscopy (FCS), raster image correlation spectroscopy (RICS), and single-particle tracking (SPT), that can quantitatively analyze molecular diffusion and concentration within living cells with high spatial and temporal resolution.
Here we present a review of these optical methods to measure translational mobility in three dimensions of fluorescent molecules in neurons and address the strengths, limitations, and potential artifacts of each of the methodologies. In addition, we discuss the use of two-photon excitation in conjunction with these techniques. Two-photon excitation provides inherent optical sectioning, a well defined focal volume, and benefits of increased viability and decreased photobleaching and phototoxicity. Focusing these highly sensitive techniques on the analysis of molecular mobility in neurons opens up a new realm of quantitative spatial and temporal information about the dynamics of molecules in the intracellular milieu.
Choice of fluorescent probes and delivery
All of these optical methods rely on the ability to tag the molecule of interest with a fluorescent label and then deliver them to the intracellular environment (Fig. 1). The choice of the fluorophores and method of delivery depends heavily on the technique being used. FRAP experiments require the fluorescent molecule to be sensitive to photobleaching to minimize the bleach intensity and duration, while FCS, RICS, and SPT experiments benefit from fluorophores optimized for high quantum yields and excellent photostability. The most important trait is that the fluorescent probes need to be free from nonspecific interactions with molecules or structures in the cell and should interfere as little as possible with the biological activity of the protein being studied.
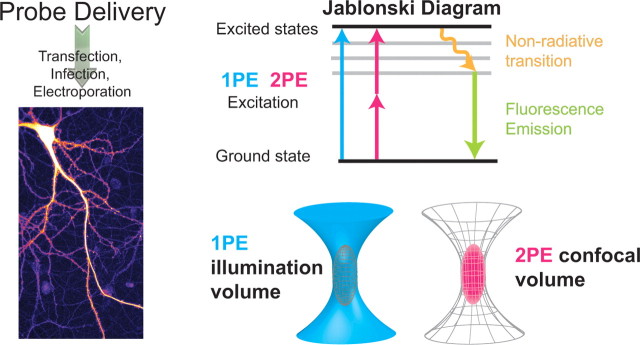
Figure 1.
Probe delivery and characteristics. Left, Probe delivery. The first step toward quantification of translational mobility is the introduction of the fluorescently tagged molecules to the cell of interest. Fluorescent probes that are conjugated with the molecule of interest or genetically encoded are delivered to the interior of the cell most commonly by transfection, microinjection, electroporation, viral infection, or some combination. Here, a dissociated hippocampal neuron was transfected using liposome-mediated gene transfer for high expression of GFP (the image is pseudocolored according to fluorescence intensity). The delivery method strongly depends on the optical technique and tagging method being used. Top right, The Jablonski diagram compares the difference between the electronic states of a fluorescent molecule excited by 1PE or 2PE. For 1PE, a higher-energy photon (shown in blue) is used to generate fluorescence, while for 2PE, two lower-energy photons (shown in magenta) are combined to excite the fluorophore from the ground state to an excited state. The fluorescence emission is used in each optical technique for quantifying the mobility of molecules through a specified volume. Bottom right, A comparison between the excitation (illumination) volume with 1PE or 2PE is shown. To achieve a confocal volume with 1PE (gray mesh), a pinhole placed in the detection path is introduced to spatially restrict the signal, while 2PE has an intrinsic spatial confinement by the square of the intensity profile of the laser (magenta ellipsoid).
A variety of different methods of fluorescently labeling molecules have successfully been implemented for these applications, such as in vitro labeling of molecules, quantum dot labeling, and genetically encoded fluorescent proteins. Small fluorescent organic tags, such as the Alexa dye series, have distinct advantages—small size (∼300–1000 Da), commercial availability with chemical reactivities to label either amines or cysteines, and a wide array of molecules with unique excitation and emission characteristics. Their use requires that the molecule to be investigated is produced, purified, labeled, and then delivered to living cells through microinjection, electroporation, or other chemical delivery methods (Craig, 1998). Note that this approach is not limited to proteins; any molecule with appropriate reactive sites can be labeled, including lipids, nucleic acids, glycolipids, etc. Quantum dots (Q Dots), water-soluble semiconductor nanoparticles, have also been coupled to biological molecules for optical applications, particularly for SPT experiments. Advantages of these molecules are their brightness, photostability, and large absorption cross sections, but their bulky, large size (∼10–40 nm; often larger than the molecule of interest) and blinking properties introduce complications that must be considered for optical tracking and mobility studies (Chan et al., 2002; Pinaud et al., 2010). Although delivering quantum dots to the cell is challenging due to their size, several methods have been developed, including delivery using endocytic mechanisms, lipid- and polymer-mediated delivery, and microinjection. An attractive alternative is genetically encoded tags, such as green fluorescent protein (GFP; ∼28 kDa) or one of its many colored or photoactivatable variants (for review, see Shaner et al., 2005). The gene for one of these fluorescent molecules is fused to the gene of the protein of interest, and the DNA can then be introduced into neurons using standard transfection techniques (e.g., lipid-mediated carriers, biolistics, or electroporation) or infected through the use of engineered virus particles. The type of delivery depends, in part, on the labeling method but also on the required concentrations of fluorescent molecules for the technique being used (range: FRAP in the high nanomolar to micromolar; FCS in the lower nanomolar; RICS from nanomolar to micromolar; SPT observes single molecules).
Additionally, advances have been made in the development of new methods that couple genetically encoded fusion proteins of interest that are not directly fluorescent but act as targets that can be covalently labeled with chemical fluorophores. Examples of such systems include the self-labeling proteins, SNAP-tags [22 kDa (Keppler et al., 2003)] and HaloTags [33 kDa (Los et al., 2008)], both of which rapidly, specifically, and irreversibly label fusion proteins under physiological conditions. Complementary protein-labeling systems have been developed that use phosphopantetheinyl transferases (PPTases) to perform the covalent attachment of small fluorescent molecules to genetically encoded short peptide tags (∼8 kDa), specifically PCP and ACP domains (Johnsson et al., 2005; Yin et al., 2006). However, these systems are currently restricted to use with cell-surface proteins. Titering of the fluorescent ligand permits some control over the level of fluorescently tagged molecule to be examined, which is a significant advantage.
FRAP
FRAP was introduced as a technique to measure the diffusion of fluorescently labeled molecules bound to the plasma membrane of living cells (Axelrod et al., 1976a). Since then, it has found widespread use in cell biology and the neurosciences [for a few examples, see Svoboda et al. (1996), Majewska et al. (2000), Bats et al. (2007), and Yoshii and Constantine-Paton (2007)]. In a FRAP experiment, the fluorescence of a labeled probe is initially monitored at low intensity for a short time (typically a few seconds) to establish a baseline fluorescence intensity; Fo (Fig. 2, FRAP panel). A brief, high-intensity light exposure is used to photobleach a portion of the fluorescently labeled molecules in the focal volume. The laser is rapidly attenuated, and the rate at which unbleached fluorophores diffuse into the focal volume from the surrounding environment is assessed. The resulting recovery curve has an amplitude proportional to the initial local bleaching of the fluorophore and recovery kinetics that depend on the diffusion of fluorescent molecules back into the observation volume. Slower recovery results in a slower calculated diffusion coefficient. FRAP provides another critical piece of information about the system—whether an immobile fraction exists. During the initial round of bleaching, the freely mobile and immobile pools are bleached simultaneously; however, if the immobile pool is significant, the recovery curve will not return to the initial baseline. An immobile fraction is revealed when the asymptotic value of the postbleach monitoring fluorescence (F(∞)) is less than the prebleach fluorescence (Fo) (Fig. 2, FRAP panel).
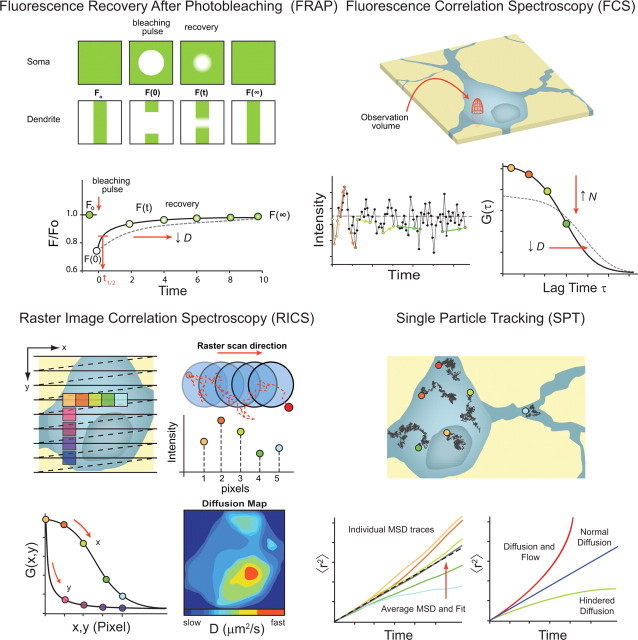
Figure 2.
Comparison of optical methods used for quantifying translational mobility in neurons. Top left, FRAP. Typically, a higher concentration of labeled probe is required for FRAP, since the technique depends on the irreversible photobleaching of a population of fluorophores and assessment of the rate of recovery back into the volume. In a FRAP experiment, a short, intense pulse of light is used to photobleach a portion of the fluorescently labeled molecules in the focal volume positioned in a region of interest, immediately after which the laser is attenuated and the rate at which new fluorophores diffuse back into the focal volume is measured. Using an appropriate model, the recovery curve is fit, and a diffusion coefficient is determined. Examples of bleaching in the soma or in a dendrite are shown (top rows). A model FRAP curve (shown below) indicates the different phases of an experiment: Fo is the baseline fluorescence intensity immediately before photobleaching, F(0) is the fluorescence intensity after photobleaching, and F(∞) is the fluorescence at the asymptote of the recovery (the color of the dots indicates the level of fluorescence being recovered). Data fitting extracts quantitative values from the recovery time course. Slower recovery (longer t1/2) represents a slower diffusion coefficient (see dashed curve). Top right, FCS. For FCS experiments, the excitation beam can be “parked” at any location in the cell. When molecules diffuse through the observation volume, florescence is emitted and the corresponding intensity fluctuations are measured over time (bottom left). The autocorrelation gives a measure of self-similarity after a certain time delay. Since the correlation of the position of any given molecule with respect to its original position decreases as the molecule diffuses, the average values of the products of points separated by shorter time intervals (orange colors points) are larger than those for longer times (green colored points). The fluctuations are then autocorrelated to produce the decay function (bottom right) that depends on the average number of molecules in the focal volume element (N) and diffusion (D). Decreasing values of D shift the autocorrelation curve to the right, while increasing N decreases the amplitude of G(τ) (see dashed curve). Bottom left, RICS. A diagram model of a raster scan on a preselected neuron is shown (top left). Horizontal lines represent the scan to collect data, and dashed lines represent the return and shift of the scanned volume between periods of data collection. The colored boxes are discrete parts of the scan used to calculate the spatial correlation of molecules moving within the sampled regions (bottom left). Molecules are captured for a longer period of time in the x dimension because of the rapid sampling in that axis, but it is less likely to find the same molecules in the spatial correlation of the y dimension because they have diffused away during the longer sampling interval between line scans. The focal volume, illustrated as blue circles, is oversampled to capture adequate information for calculating the molecules rate of movement (top right). A randomly diffusing particle is shown along with the scanned volume element (pixel), and the histogram below shows the fluorescence intensity that would be detected in each of the pixels. Fitted with the appropriate model, the number of molecules (N) and the diffusion (D) can be extracted for particular regions of interest (ROIs). By analyzing continuous ROIs through the image, a spatial map for the diffusion molecules of interest can be obtained (bottom right). In this example, diffusion is fairly homogenous through the cytoplasm but shows more rapid diffusion in the nucleus. Bottom right, SPT, accomplished with video microscopy, uses numerical algorithms to select single bright molecules on the images to “track” their position as the video elapses. This builds a map of many single trajectory paths in the same cell. Random walk traces are then converted to MSD versus time plots to analyze their transport properties (bottom left). At short timescales, single-particle traces follow nicely a linear relationship with time, but at longer times, the statistical nature of random walks allows them to deviate from one another. Thus, the standard deviation cannot be judged from a single curve but requires an average curve [see average of these 5 traces (gray line)) and its linear fit (dashed black line)]. The slope of this curve corresponds to the diffusion coefficient, and in this case the diffusion is constant. When the diffusion coefficient or the slope of MSD versus time is not constant, other types of mobility corresponding to different transport properties can be assumed, such as diffusion with flow or hindered diffusion (bottom right).
In the past, the most common application of FRAP has been for studying two-dimensional (2D) membrane systems. For this, the bleached area for FRAP experiments was confined to a 2D geometry by use of thin samples, such as a membrane, or by use of low numerical objectives to generate a cylindrical beam in a thick sample (Axelrod et al., 1976a,b; Seksek et al., 1997). FRAP was later extended to determine 3D diffusion of fluorophores in cells using one-photon excitation (1PE) photobleaching with a high NA lens and confocal detection (Blonk et al., 1993). This is problematic for the case of 1PE with a high NA objective due to the extended double cone of bleached fluorophore, which cannot be mathematically defined in a simple enough form to derive fitting functions for the determination of diffusion coefficients (Brown et al., 1999). Two-photon excitation (2PE) offers an ideal extension of conventional FRAP techniques where volume issues are minimized (Svoboda et al., 1996; Brown et al., 1999). Using 2PE with high numerical aperture objectives, fluorescence excitation and photobleaching can be confined to subfemtoliter volumes (see Fig. 1 for a comparison between 1PE and 2PE volumes), significantly increasing the potential for sampling different regions of the same cell, point by point. Therefore, data can be collected by “parking the beam” of a multiphoton microscope at different positions throughout the cell to create a 3D diffusion map [for technical details, please refer to Brown et al. (1999) and Zipfel and Webb (2001)]. The spatial pattern of photobleaching recovery has also been used to determine the mobility of molecules within confined geometries of organelles, such as the nucleus, endoplasmic reticulum, mitochondria, and Golgi complex, and within membranes (Sbalzarini et al., 2005; Tagawa et al., 2005; Mitra and Lippincott-Schwartz, 2010) and between organelles (Presley et al., 1997; Ward et al., 2001). This is particularly useful for neurons where geometric constraints and compartmentalization are critical but requires special consideration of complex shape and defined boundaries to avoid underestimated diffusion coefficients.
FRAP has transformed our understanding of molecular dynamics and cellular organization in living cells, but optimization of the experimental conditions and careful consideration of the analysis and theory are required for accurate measurements of translational mobility. One limitation is that relatively high intracellular concentrations of fluorescent probes are typically required to achieve a reliable signal over background for FRAP experiments. Introducing or expressing molecules at these concentrations may saturate endogenous binding partners and therefore shift the system away from its normal equilibrium, reporting a faster diffusion coefficient. Another concern with photobleaching techniques is that the brief high-intensity pulses required to rapidly photobleach the molecules might introduce artifacts. Severing of polymers or cross-linking of macromolecules within the focal volume is a concern, as are potential unwanted chemical reactions due to the generation of free radicals as a result of the photobleaching process. The fluorophores themselves can also enter different quantum states whose time course of recovery is unrelated to diffusion (Periasamy et al., 1996). High-intensity laser pulses from one- and two-photon excitation have also been found to induce photo-unbinding under certain conditions, which can also influence the interpretation of recovery curves (Akaaboune et al., 2002; Heinze et al., 2009). However, these experimental artifacts can be minimized or eliminated by careful monitoring of the bleaching depth and intensity in conjunction with appropriate controls [see Brown et al. (1999), Zipfel and Webb (2001), and Mavrakis et al. (2008) for technical details about FRAP].
A related and complementary technique, photoactivation, works by converting molecules to a fluorescent state by brief exposure to high-intensity irradiation and then following these molecules as they reequilibrate into the surrounding cellular environment (Lippincott-Schwartz et al., 2003). Photoactivation of caged compounds and proteins has been used for more than 20 years but has reemerged in popularity with the development of genetically encoded photoactivatable fluorescent proteins. Fluorescence photoactivation of these proteins is rapid and yields a stable signal over time, which allows them to be used for studying protein dynamics, such as diffusion and compartmental transport, both in cultured neurons and in vivo (Tsuriel et al., 2006; Sato et al., 2007; Blanpied et al., 2008). A particular benefit of this technique is the high signal-to-noise ratio, particularly in the earliest points of the photoactivation experiment, but on the other hand, the signal dissipates significantly as it moves throughout the neighboring regions.
FCS
FCS analyzes fluorescence intensity fluctuations of molecules diffusing through the illuminated volume (Fig. 2, FCS panel) rather than relying on photobleaching of fluorophores. Introduced almost 40 years ago (Magde et al., 1972; Ehrenberg and Rigler, 1974; Elson and Magde, 1974), FCS is exquisitely sensitive, provides fast temporal and high spatial resolution, and offers access to a multitude of measurement parameters in real time. Such advantages have been realized by applying FCS to measurements of translational mobility in neurons [for a few examples, see Terada et al. (2000), Gennerich and Schild (2002), Meissner and Häberlein (2003), and Stagi et al. (2005)]. FCS is based on the concept that at equilibrium, the number of fluorescent molecules fluctuates about a mean value due to individual molecules diffusing in and out of the observation volume. Data collection starts at an arbitrary time 0, and at increasing time intervals, the fluctuations in the measured fluorescence are compared to the average fluorescence in the form of an autocorrelation function, G(τ). The intensity autocorrelation is a statistical tool that compares intensities across different points in time. Given that the detected intensity is related to the dwell time of the molecule in the illumination volume, a decay function such as the one shown in Figure 2 (FCS panel) is expected. A typical curve is constructed with at least 10 s of data collection, which is repeated multiple times (5–10) to increase statistical reliability of the data.
The power of FCS measurements is that they can access a wide variety of parameters simultaneously. Any factor that alters the fluorescence fluctuations can be detected using such analysis. Photophysical events, such as blinking or triplet state, are detected in the microsecond and faster timescales, and these events occur while the same molecules remain resident in the illumination volume. The translational mobility of a molecular species can be derived from the same experimental autocorrelation curve where the timescales of the decay of G(τ) can be directly related to the diffusion coefficient (D). Molecules moving faster (smaller hydrodynamic radius) enter and exit the volume more quickly and therefore exhibit a faster correlation decay and faster D. In addition to the mobility, direct assessment of the concentration of molecule species is possible—the number of fluorophores (N) in the focal volume is inversely proportional to the amplitude of the autocorrelation function. All values are extracted from the experimental data by fitting to the appropriate physical model in which the free parameters correspond to the desired experimental quantities.
The observation of only a small number of fluctuating molecules in a small volume can be achieved using 1PE with confocal detection but can be accomplished more effectively with 2PE, which optically defines a subfemtoliter volume and permits the use of more efficient, wide-field detection (Berland et al., 1995; Schwille et al., 1999; Zipfel and Webb, 2001). The major advantages of 2PE include the following: minimal cumulative photobleaching of the fluorophores, high axial resolution of the focal volume with inherent optical sectioning, and background suppression of cellular autofluorescence. Most important is the benefit of increased cell viability, which becomes critical due to the longer measurement time [see Kim et al. (2007) for technical discussion of the implementation of FCS for intracellular measurements].
FCS requires two key features of the molecular system under investigation—low probe concentrations (in the nanomolar range) and mobility. Too many molecules in the focal volume result in a higher average fluorescence intensity but with smaller relative contributions from each molecule, which decreases the amplitude of the FCS curve. Artifacts due to the appearance of sparse aggregates diffusing through the focal volume can greatly distort the autocorrelation function, and subpopulations of different diffusive species are difficult to distinguish based on diffusion alone. To differentiate subpopulations of molecules by fitting routines, their diffusion coefficient must differ by at least 1.6-fold (Meseth et al., 1999), an important consideration to differentiate molecules that exist in multiple forms (e.g., monomer vs oligomer). The counts per molecule or molecular brightness is an alternative parameter that may be used to distinguish the formation of monomers versus oligomers. The second necessary property is that molecules must be mobile for FCS measurements to induce a decay of the autocorrelation function, and D of ∼500–0.01 μm2/s is accessible using this technique. The need for molecules to exit the focal volume means that the immobile fraction is not accessible to FCS. Longer diffusion times, due to the large size of the molecule or increased viscosity (as in membranes), require longer acquisition times to acquire the full decay of the autocorrelation function. In practice, photobleaching becomes problematic with slower diffusion rates, since the molecules remain in the focal volume for an extended period. The bleaching of molecules will distort both the determination of molecular concentration and diffusion times. However, corrective calculations for photobleaching, removal of unexpected fluorescent spikes, and background can be implemented in postcollection data analysis on raw data (Eggeling et al., 2001; Hess and Webb, 2002).
Super-resolution microscopy has recently brought a new dimension to FCS measurements. By decreasing the illumination volume, one can sample different regions of a cell at much greater resolution, and because the number of molecules within the volume is decreased, it is possible to make correlation measurements at higher concentrations of fluorescent molecules. One example of this approach is to combine the power of stimulated emission depletion (STED) nanoscopy (Hell and Wichmann, 1994) and FCS. STED relies on the overlap between a standard excitation laser and a second “depletion laser” that has an optically engineered point-spread function to deplete fluorescent molecules from the periphery of the focal volume. The maximum resolution attained to date is 6 nm, which is 25- to 50-fold better than the 250 nm resolution obtained with diffraction-limited microscopy techniques (Rittweger et al., 2009). An example of STED-FCS in live cells is the direct observation of dynamics of membrane lipids (Eggeling et al., 2009). Certainly, such advancements will find important applications in evaluating diffusion in small neuronal compartments such as dendrites and synaptic spines.
RICS
Image correlation spectroscopy (ICS) considers not a single fixed point in space, as with FRAP and FCS, but rather a field of view that can be large enough to include a significant area of the cell. This is important in assessing intracellular protein dynamics that have an associated spatial heterogeneity and spatial compartmentalization. Several methods using the main concept behind ICS have been implemented with a major goal of extracting dynamic information concerning molecular mobility when collecting and analyzing imaging data [for review, see Kolin and Wiseman (2007), Nohe and Petersen (2007), and Digman et al. (2009)]. Here we focus our attention on RICS, which can access a range of timescales relevant to intracellular diffusion. A number of successful applications of RICS have recently appeared and include studying diffusion from heterogeneous solutions (Gielen et al., 2009), examining the dynamics of calmodulin mobility (Sanabria et al., 2008), focal adhesion dynamics (Digman et al., 2008, 2009), and lipid diffusion (Gielen et al., 2009). In addition, RICS can be accomplished on standard confocal microscopes, with 1P or 2P excitation, making the equipment for data acquisition immediately available to many investigators.
As the acronym suggests, RICS uses the raster scan to generate confocal images of particular regions of interest (Fig. 2, RICS panel). Each point, or pixel, on the image registers the emitted fluorescence from the sample with the where and when information embedded on the register. RICS capitalizes on the natural scanning pattern of a confocal microscope where the pixels on each horizontal line are separated from each other on the microsecond timescale, lines are separated on the millisecond timescale, and full frames are separated on the second timescale. Fluctuation analysis can then be performed to extract the mobility of molecules within subregions of the scanned image.
A key factor for RICS is determination of an appropriate scan rate—the faster the molecules diffuse, the faster the needed scan rates. An average image size for RICS is 256 × 256 pixels, which means a scan rate (dictated by the pixel dwell time) of ∼20 μs/pixel would be sufficient to capture a broad range of diffusion coefficients relevant to cell biology (D of ∼100–0.001 μm2/s). Another important setting in RICS is an accurate “zoom” that will capture the mobility of the fluorescent molecules. A rule of thumb is that the chosen pixel size (e.g., 0.05 μm) is no more than one-sixth of the point-spread function as determined by the objective lens and wavelength of excitation. Both the scan rate and the pixel size are chosen specifically to increase the probability that molecules can be “tracked” in the direction of the scan before they leave the illumination volume (Fig. 2, RICS panel). However, these impose certain limitations on the possible “zoom” setting and scan rates at which the scanning microscope needs to be set and should not to be confused with the optical resolution, which is given by the point-spread function. Once the scan rate and pixel size have been established, the minimal number of frames required to provide statistical reliability of the data is determined. The autocorrelation is calculated frame by frame and averaged over many frames to increase the fluorescent signal-to-noise ratio (Brown et al., 2008). The amplitude of the correlation function obtained by RICS, as in the case of FCS, is inversely proportional to the average number of molecules present in the illumination volume. Thus, the technique benefits from a lower concentration of fluorescent molecules where the possibility of altering the overall cellular concentration of the molecule under study is also minimized.
A distinct advantage of RICS is the ability to detect whether the system under examination is exhibiting motion that would influence the determination of an accurate diffusion coefficient. This could be motion due to the movement of the cell, the plasma membrane, or other intracellular organelles. All such movement can potentially contaminate the primary data for the determination of translational diffusion of molecules. Both FRAP and FCS measurements typically ignore such potential artifacts unless they happen to produce recognizable signatures in the raw data. In RICS, these factors can be identified and at least partially accounted for in the processing of the data for final analysis. For example, one can use a moving average algorithm to “detrend” the data to minimize fluctuations due to the collective motion of the cell or motions associated with slowed diffusion values, such as vesicle or organelle motion. Photobleaching that occurs during data collection can also be assessed and compensations applied. With these factors properly addressed, the faster diffusion component or components can be quantified in the data by applying spatial correlation algorithms within a subregion of the scanned area [see Brown et al. (2008) for a more comprehensive explanation]. By systematically moving the subregion across the image, a coarse map of diffusion values can be obtained (Digman et al., 2008; Sanabria et al., 2008). This quantitative spatial map provides critical information about the heterogeneity of translational mobility of a given fluorescent molecule within the cell. Thus, RICS is well suited to assess whether the motion of molecules is affected by spatial compartmentalization. A weakness of RICS is the limited spatial and temporal resolution. The spatial resolution is restricted to the subregion in which the spatial correlation is calculated (∼0.8 μm × 0.8 μm) inside the overall image (Brown et al., 2008), and the temporal resolution is limited by the scan rate.
SPT
SPT has been used since the early 1980s (Barak and Webb, 1982) as a tool to quantify molecular mobility in living cells. In the field of neuroscience, SPT has been mainly applied to investigate the diffusion of channels and receptors as they move within the plane of the plasma membrane. A recent Toolbox article (Groc et al., 2007) and the references therein described in more detail the SPT approach in this context. Thus we will only introduce the technique, compare how ensemble techniques, such as FRAP, FCS, and RICS, differ from SPT, and briefly describe new extensions of SPT using two-photon and fluorescence photoactivated localization microscopy (FPALM/PALM).
The principle behind SPT is based on obtaining the spatial coordinates of single tagged molecules as a function of time. Given adequate tracking time a mean squared displacement (MSD) can be constructed that can be used to calculate the diffusion coefficient of single molecules and possibly reveal the underlying transport mechanism (Saxton and Jacobson, 1997). For example, a linear function of MSD versus time reveals normal diffusion, while deviations in the slope suggest active transport, hindered diffusion, or other physical/chemical properties of the system that alter diffusion. Importantly, SPT is a single-molecule technique, and therefore it has the ability to resolve differences in the diffusional behavior of molecules that are averaged together by the ensemble techniques of FRAP, FCS, or RICS.
The most common implementation of SPT is via the use of fast electron-multiplying cameras (EM-CCD) that can collect enough photons per molecule to localize single particles with an accuracy of 10–100 nm at millisecond frame rates (Kubitscheck et al., 2000). To gather traces in three dimensions, most image-based techniques acquire z-stacks as a function of time (Bornfleth et al., 1999; Thomann et al., 2002). Fast postprocessing algorithms are used to extract traces of each particle moving in the field of view, where a Gaussian intensity function is used to determine the main position of the signal that is followed over consecutive frames (Saxton, 2008). Particles can be either single dye-labeled molecules or single particles, such as quantum dots, attached to the molecule of interest (Pinaud et al., 2010). Like the other techniques described above, SPT is affected by photophysical properties of the dyes, such as bleaching and blinking, that may cause the molecule to disappear permanently, or more problematically, transiently and randomly over time. In addition to these problems, other pitfalls could be due to high particle density or particle merging or splitting (Jaqaman et al., 2008).
A recent alternative approach to image-based SPT is to scan a two-photon illuminated volume in orbital loops around single molecules (Levi et al., 2005). The particle is tracked in three dimensions by detecting changes in its intensity while making two orbital scans at two different focal planes. The position of the particle is determined by the phase and modulation of the signal after several orbits over and under the molecule. Then, the calculated position centers the next set of orbits and the process is repeated. As molecules diffuse away from the first location, the scanning orbital follows the position with suboptical resolution of ∼20 nm (Levi et al., 2005). Another promising new technique called sptPALM (Manley et al., 2008) combines the super-resolution technique of fluorescence photoactivated localization microscopy (FPALM/PALM) with SPT to resolve the dynamics of ensembles of many molecules simultaneously. The addition of super-resolution imaging allows for the localization and tracking of many previously overlapping trajectories, since the distance between the fluorescent molecules at any given time is greater than several times the width of their point-spread function.
Quantifying translational mobility in neurons
From all the optical methods described here, the motivation is to derive a quantitative value for the translational mobility of molecules within the intracellular milieu of the neuron. The diffusion coefficient describes the rate at which molecules move and, in ideal cases, obey the Stokes–Einstein relation, where diffusion is inversely proportional to the hydrodynamic radius of the probe and the viscosity of the intracellular milieu. However, the intracellular environment is not homogeneous, and specific and nonspecific interactions also affect molecular diffusion (Minton, 2001). A multiple diffusion model can be used to describe a situation where a molecule displays two or more diffusing species, each of which can be described using a single diffusion coefficient (e.g., a protein in the presence of a binding partner). When the diffusion coefficient is no longer a constant over time, then diffusion is said to be anomalous (Brown et al., 1999) and can either be characterized as subdiffusion or superdiffusion. Subdiffusion is the result of physical hindrances presented by other mobile or immobile obstacles or by complex physical/chemical interactions (Bouchaud and Georges, 1990). Superdiffusion appears when the molecule takes long “flights” and is most often associated with flow or directed motion as in the case of active transport (Solomon et al., 1993).
To distinguish which diffusion mode is present, SPT has an advantage over FRAP, FCS, or RICS because of the straight forward calculation of MSD versus time from single-particle traces. If these plots deviate from a straight line, then diffusion is anomalous (Fig. 2, lower right panel). FCS and FRAP are also suitable for distinguishing between free diffusion and anomalous diffusion through the use of complex fitting models (Magde et al., 1974; Brown et al., 1999; Wu and Berland, 2008); however, sometimes more than one model might appear to correctly describe the data (Goins et al., 2008). RICS is not necessarily suited for such tasks, but differences in the spatial heterogeneity of diffusion can be directly observed on 2D diffusion maps (Fig. 2, D panel). One caveat that needs to be considered in all the methodologies is the choice of an appropriate model for diffusion for fitting of the data. For example, whether 3D, 2D, or 1D diffusion, flow, or anomalous diffusion is expected should be determined and will significantly impact the choice of an appropriate model to fit the data. In neuroscience, this is of particular importance because neurons are highly structured, where each compartment might demand a distinct fitting model, and possess different kinds of mobility or transport, which are biologically significant.
Conclusions
In this article, we highlighted the strengths and weaknesses of four optical techniques to quantify translational mobility in living cells: FRAP, FCS, RICS, and SPT. Signaling at the cell surface, within the cytoplasm and nucleus or in dendrites and synapses in neurons are all mediated through the movement and interactions between ligands, proteins, lipids, and nucleic acids. Individually, or even better in combination, these techniques can provide unparalleled quantitative information needed to advance our understanding of cellular-level systems biology.
Footnotes
Editor's Note: Toolboxes are intended to describe and evaluate methods that are becoming widely relevant to the neuroscience community or to provide a critical analysis of established techniques. For more information, see http://www.jneurosci.org/misc/ifa_minireviews.dtl.
This work was supported by grants from the National Institutes of Health (NIH) [R01-NS26086 (M.N.W.), P41-RRO3155 and P50-GM076516 (E.G.), P41-RR04224 (W.R.Z.), and the Cell Migration Consortium U54-GM064346 (E.G. and M.A.D.)] and the Human Frontiers in Science Program (M.N.W. and P.S.). For portions of this work, S.A.K. was supported by a National Research Service Award from the National Institute of Mental Health/NIH and a Fulbright Commission Award, and H.S. was supported by the Keck Center Nanobiology Training Program of the Gulf Coast Consortia (R90-DK071054). Parts of the FRAP and FCS work were initiated in the laboratory of Prof. Watt W. Webb. We thank Mark Williams for helpful editorial comments on this manuscript.
References
- Akaaboune M, Grady RM, Turney S, Sanes JR, Lichtman JW. Neurotransmitter receptor dynamics studied in vivo by reversible photo-unbinding of fluorescent ligands. Neuron. 2002;34:865–876. doi: 10.1016/s0896-6273(02)00739-0. [DOI] [PubMed] [Google Scholar]
- Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J. 1976a;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D, Ravdin P, Koppel DE, Schlessinger J, Webb WW, Elson EL, Podleski TR. Lateral motion of fluorescently labeled acetylcholine receptors in membranes of developing muscle fibers. Proc Natl Acad Sci U S A. 1976b;73:4594–4598. doi: 10.1073/pnas.73.12.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak LS, Webb WW. Diffusion of low density lipoprotein-receptor complex on human fibroblasts. J Cell Biol. 1982;95:846–852. doi: 10.1083/jcb.95.3.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bats C, Groc L, Choquet D. The interaction between Stargazin and PSD-95 regulates AMPA receptor surface trafficking. Neuron. 2007;53:719–734. doi: 10.1016/j.neuron.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Berland KM, So PT, Gratton E. Two-photon fluorescence correlation spectroscopy: method and application to the intracellular environment. Biophys J. 1995;68:694–701. doi: 10.1016/S0006-3495(95)80230-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpied TA, Kerr JM, Ehlers MD. Structural plasticity with preserved topology in the postsynaptic protein network. Proc Natl Acad Sci U S A. 2008;105:12587–12592. doi: 10.1073/pnas.0711669105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blonk J, Don A, Van Aalst H, Birmingham J. Fluorescence photobleaching recovery in the confocal light scanning microscope. J Microsc. 1993;169:363–374. [Google Scholar]
- Bornfleth H, Edelmann P, Zink D, Cremer T, Cremer C. Quantitative motion analysis of subchromosomal foci in living cells using four-dimensional microscopy. Biophys J. 1999;77:2871–2886. doi: 10.1016/S0006-3495(99)77119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchaud JP, Georges A. Comment on “Stochastic pathway to anomalous diffusion”. Phys Rev A. 1990;41:1156–1157. doi: 10.1103/physreva.41.1156. [DOI] [PubMed] [Google Scholar]
- Brown EB, Wu ES, Zipfel W, Webb WW. Measurement of molecular diffusion in solution by multiphoton fluorescence photobleaching recovery. Biophys J. 1999;77:2837–2849. doi: 10.1016/S0006-3495(99)77115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CM, Dalal RB, Hebert B, Digman MA, Horwitz AR, Gratton E. Raster image correlation spectroscopy (RICS) for measuring fast protein dynamics and concentrations with a commercial laser scanning confocal microscope. J Microsc. 2008;229:78–91. doi: 10.1111/j.1365-2818.2007.01871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WC, Maxwell DJ, Gao X, Bailey RE, Han M, Nie S. Luminescent quantum dots for multiplexed biological detection and imaging. Curr Opin Biotechnol. 2002;13:40–46. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- Craig AM. Transfecting cultured neurons. In: Banker G, Goslin K, editors. Culturing nerve cells. Cambridge, MA: MIT Press; 1998. pp. 79–111. [Google Scholar]
- Digman MA, Brown CM, Horwitz AR, Mantulin WW, Gratton E. Paxillin dynamics measured during adhesion assembly and disassembly by correlation spectroscopy. Biophys J. 2008;94:2819–2831. doi: 10.1529/biophysj.107.104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Wiseman PW, Horwitz AR, Gratton E. Detecting protein complexes in living cells from laser scanning confocal image sequences by the cross correlation raster image spectroscopy method. Biophys J. 2009;96:707–716. doi: 10.1016/j.bpj.2008.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggeling C, Berger S, Brand L, Fries JR, Schaffer J, Volkmer A, Seidel CA. Data registration and selective single-molecule analysis using multi-parameter fluorescence detection. J Biotechnol. 2001;86:163–180. doi: 10.1016/s0168-1656(00)00412-0. [DOI] [PubMed] [Google Scholar]
- Eggeling C, Ringemann C, Medda R, Schwarzmann G, Sandhoff K, Polyakova S, Belov VN, Hein B, von Middendorff C, Schönle A, Hell SW. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- Ehrenberg M, Rigler R. Rotational Brownian motion and fluorescence intensity fluctuations. Chem Phys. 1974;4:390–401. [Google Scholar]
- Elson E, Magde D. Fluorescence correlation spectroscopy. I. Conceptual basis and theory. Biopolymers. 1974;13:1–27. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- Gennerich A, Schild D. Anisotropic diffusion in mitral cell dendrites revealed by fluorescence correlation spectroscopy. Biophys J. 2002;83:510–522. doi: 10.1016/S0006-3495(02)75187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen E, Smisdom N, vandeVen M, De Clercq B, Gratton E, Digman M, Rigo JM, Hofkens J, Engelborghs Y, Ameloot M. Measuring diffusion of lipid-like probes in artificial and natural membranes by raster image correlation spectroscopy (RICS): use of a commercial laser-scanning microscope with analog detection. Langmuir. 2009;25:5209–5218. doi: 10.1021/la8040538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goins AB, Sanabria H, Waxham MN. Macromolecular crowding and size effects on probe microviscosity. Biophys J. 2008;95:5362–5373. doi: 10.1529/biophysj.108.131250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groc L, Lafourcade M, Heine M, Renner M, Racine V, Sibarita JB, Lounis B, Choquet D, Cognet L. Surface trafficking of neurotransmitter receptor: comparison between single-molecule/quantum dot strategies. J Neurosci. 2007;27:12433–12437. doi: 10.1523/JNEUROSCI.3349-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinze KG, Costantino S, De Koninck P, Wiseman PW. Beyond photobleaching, laser illumination unbinds fluorescent proteins. J Phys Chem B. 2009;113:5225–5233. doi: 10.1021/jp8060152. [DOI] [PubMed] [Google Scholar]
- Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- Hess ST, Webb WW. Focal volume optics and experimental artifacts in confocal fluorescence correlation spectroscopy. Biophys J. 2002;83:2300–2317. doi: 10.1016/S0006-3495(02)73990-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N, George N, Johnsson K. Protein chemistry on the surface of living cells. Chembiochem. 2005;6:47–52. doi: 10.1002/cbic.200400290. [DOI] [PubMed] [Google Scholar]
- Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- Kim SA, Heinze KG, Schwille P. Fluorescence correlation spectroscopy in living cells. Nat Methods. 2007;4:963–973. doi: 10.1038/nmeth1104. [DOI] [PubMed] [Google Scholar]
- Kolin DL, Wiseman PW. Advances in image correlation spectroscopy: measuring number densities, aggregation states, and dynamics of fluorescently labeled macromolecules in cells. Cell Biochem Biophys. 2007;49:141–164. doi: 10.1007/s12013-007-9000-5. [DOI] [PubMed] [Google Scholar]
- Kubitscheck U, Kückmann O, Kues T, Peters R. Imaging and tracking of single GFP molecules in solution. Biophys J. 2000;78:2170–2179. doi: 10.1016/S0006-3495(00)76764-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi V, Ruan Q, Gratton E. 3-D particle tracking in a two-photon microscope: application to the study of molecular dynamics in cells. Biophys J. 2005;88:2919–2928. doi: 10.1529/biophysj.104.044230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Altan-Bonnet N, Patterson GH. Photobleaching and photoactivation: following protein dynamics in living cells. Nat Cell Biol. 2003;(Suppl):S7–S14. [PubMed] [Google Scholar]
- Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- Magde D, Elson E, Webb WW. Thermodynamic fluctuations in a reacting system—measurement by fluorescence correlation spectroscopy. Phys Rev Lett. 1972;29:705–708. [Google Scholar]
- Magde D, Elson EL, Webb WW. Fluorescence correlations spectroscopy. II. An experimental realization. Biopolymers. 1974;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- Majewska A, Tashiro A, Yuste R. Regulation of spine calcium dynamics by rapid spine motility. J Neurosci. 2000;20:8262–8268. doi: 10.1523/JNEUROSCI.20-22-08262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- Mavrakis M, Rikhy R, Lilly M, Lippincott-Schwartz J. Fluorescence imaging techniques for studying Drosophila embryo development. Curr Protoc Cell Biol. 2008;39:4.18.1–4.18.43. doi: 10.1002/0471143030.cb0418s39. [DOI] [PubMed] [Google Scholar]
- Meissner O, Häberlein H. Lateral mobility and specific binding to GABA(A) receptors on hippocampal neurons monitored by fluorescence correlation spectroscopy. Biochemistry. 2003;42:1667–1672. doi: 10.1021/bi0263356. [DOI] [PubMed] [Google Scholar]
- Meseth U, Wohland T, Rigler R, Vogel H. Resolution of fluorescence correlation measurements. Biophys J. 1999;76:1619–1631. doi: 10.1016/S0006-3495(99)77321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton AP. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J Biol Chem. 2001;276:10577–10580. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- Mitra K, Lippincott-Schwartz J. Analysis of mitochondrial dynamics and functions using imaging approaches. Curr Protoc Cell Biol. 2010;46:4.25.1–4.25.21. doi: 10.1002/0471143030.cb0425s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohe A, Petersen NO. Image correlation spectroscopy. Sci STKE. 2007;2007:pl7. doi: 10.1126/stke.4172007pl7. [DOI] [PubMed] [Google Scholar]
- Periasamy N, Bicknese S, Verkman AS. Reversible photobleaching of fluorescein conjugates in air-saturated viscous solutions: singlet and triplet state quenching by tryptophan. Photochem Photobiol. 1996;63:265–271. doi: 10.1111/j.1751-1097.1996.tb03023.x. [DOI] [PubMed] [Google Scholar]
- Pinaud F, Clarke S, Sittner A, Dahan M. Probing cellular events, one quantum dot at a time. Nat Methods. 2010;7:275–285. doi: 10.1038/nmeth.1444. [DOI] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Rittweger E, Han KY, Irvine SE, Eggeling C, Hell SW. STED microscopy reveals crystal colour centres with nanometric resolution. Nat Photonics. 2009;3:144–147. [Google Scholar]
- Sanabria H, Digman MA, Gratton E, Waxham MN. Spatial diffusivity and availability of intracellular calmodulin. Biophys J. 2008;95:6002–6015. doi: 10.1529/biophysj.108.138974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TR, Gray NW, Mainen ZF, Svoboda K. The functional microarchitecture of the mouse barrel cortex. PLoS Biol. 2007;5:e189. doi: 10.1371/journal.pbio.0050189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ. Single-particle tracking: connecting the dots. Nat Methods. 2008;5:671–672. doi: 10.1038/nmeth0808-671. [DOI] [PubMed] [Google Scholar]
- Saxton MJ, Jacobson K. Single-particle tracking: applications to membrane dynamics. Annu Rev Biophys Biomol Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- Sbalzarini IF, Mezzacasa A, Helenius A, Koumoutsakos P. Effects of organelle shape on fluorescence recovery after photobleaching. Biophys J. 2005;89:1482–1492. doi: 10.1529/biophysj.104.057885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwille P, Haupts U, Maiti S, Webb WW. Molecular dynamics in living cells observed by fluorescence correlation spectroscopy with one- and two-photon excitation. Biophys J. 1999;77:2251–2265. doi: 10.1016/S0006-3495(99)77065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seksek O, Biwersi J, Verkman AS. Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus. J Cell Biol. 1997;138:131–142. doi: 10.1083/jcb.138.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Solomon TH, Weeks ER, Swinney HL. Observation of anomalous diffusion and Lévy flights in a two-dimensional rotating flow. Phys Rev Lett. 1993;71:3975–3978. doi: 10.1103/PhysRevLett.71.3975. [DOI] [PubMed] [Google Scholar]
- Stagi M, Dittrich PS, Frank N, Iliev AI, Schwille P, Neumann H. Breakdown of axonal synaptic vesicle precursor transport by microglial nitric oxide. J Neurosci. 2005;25:352–362. doi: 10.1523/JNEUROSCI.3887-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Tank DW, Denk W. Direct measurement of coupling between dendritic spines and shafts. Science. 1996;272:716–719. doi: 10.1126/science.272.5262.716. [DOI] [PubMed] [Google Scholar]
- Tagawa A, Mezzacasa A, Hayer A, Longatti A, Pelkmans L, Helenius A. Assembly and trafficking of caveolar domains in the cell: caveolae as stable, cargo-triggered, vesicular transporters. J Cell Biol. 2005;170:769–779. doi: 10.1083/jcb.200506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada S, Kinjo M, Hirokawa N. Oligomeric tubulin in large transporting complex is transported via kinesin in squid giant axons. Cell. 2000;103:141–155. doi: 10.1016/s0092-8674(00)00094-5. [DOI] [PubMed] [Google Scholar]
- Thomann D, Rines DR, Sorger PK, Danuser G. Automatic fluorescent tag detection in 3D with super-resolution: application to the analysis of chromosome movement. J Microsc. 2002;208:49–64. doi: 10.1046/j.1365-2818.2002.01066.x. [DOI] [PubMed] [Google Scholar]
- Tsuriel S, Geva R, Zamorano P, Dresbach T, Boeckers T, Gundelfinger ED, Garner CC, Ziv NE. Local sharing as a predominant determinant of synaptic matrix molecular dynamics. PLoS Biol. 2006;4:e271. doi: 10.1371/journal.pbio.0040271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward TH, Polishchuk RS, Caplan S, Hirschberg K, Lippincott-Schwartz J. Maintenance of Golgi structure and function depends on the integrity of ER export. J Cell Biol. 2001;155:557–570. doi: 10.1083/jcb.200107045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Berland KM. Propagators and time-dependent diffusion coefficients for anomalous diffusion. Biophys J. 2008;95:2049–2052. doi: 10.1529/biophysj.107.121608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Lin AJ, Golan DE, Walsh CT. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat Protoc. 2006;1:280–285. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- Yoshii A, Constantine-Paton M. BDNF induces transport of PSD-95 to dendrites through PI3K-AKT signaling after NMDA receptor activation. Nat Neurosci. 2007;10:702–711. doi: 10.1038/nn1903. [DOI] [PubMed] [Google Scholar]
- Zipfel WR, Webb WW. In vivo diffusion measurements using multiphoton excitation fluorescence photobleaching recovery and fluorescence correlation spectroscopy. In: Periasamy A, editor. Methods in cellular imaging. New York: Oxford UP; 2001. pp. 216–235. [Google Scholar]