Summary
Blastomycosis is a serious and potentially fatal infection by the thermally dimorphic fungus Blastomyces dermatitidis. PCR assays targeting the BAD-1 virulence gene promoter have been developed to aid in the detection of the pathogen in clinical and environmental samples. However, little is known regarding the genetic diversity of B. dermatitidis and how this might affect the performance characteristics of these assays. We explored the genetic relatedness of 106 clinical and environmental isolates of B. dermatitidis using a previously described rDNA PCR RFLP assay. In addition, we looked for polymorphisms in the promoter region upstream of BAD-1. RFLP analysis showed that all isolates fell into one of five genotypic groups, designated A through E. Genotypic groups A and B predominated, comprising 50/106 (47.2%) and 51/106 (48.1%) of isolates, respectively. Three of 106 (2.8%) isolates were genotype C. Genotypes D and E represented novel genotypes and were each associated with single clinical isolates. PCR of the BAD-1 promoter revealed significant size differences among amplification products. Fifty one of 106 isolates (50/50 RFLP genotypic group A and 1/51 genotypic group B) had amplicons of 663 bp – nearly twice the size of the expected product. Sequence analysis of amplification products from 17 representative isolates revealed four haplotypes and showed that the size disparity was due to two large insertions. Because these insertions were present in a high percentage of isolates, the utility of the PCR assays for diagnostic purposes could be affected. It is possible that the larger PCR product may have amplified less efficiently and that the larger amplicon could have been misinterpreted as a non-specific product. However, the novel RFLP genotypes and multiple BAD-1 haplotypes may prove useful as markers in population genetic studies.
Keywords: Blastomyces dermatitidis, PCR, RFLP, BAD-1, genetic diversity
Introduction
Blastomycosis is a serious and potentially fatal infection due to the thermally dimorphic fungus Blastomyces dermatitidis. The organism exists as a mold at ambient temperatures in the environment, but transforms into a pathogenic yeast form after infectious spores are inhaled by susceptible mammalian hosts. In the U.S., most cases occur in the Ohio and Mississippi river valleys, the southeastern states, and around the Great Lakes (1).
A variety of laboratory techniques have been used to aid in the clinical diagnosis of blastomycosis. These include microscopic detection of characteristic broad-based budding yeast forms in body fluids or tissue biopsies, isolation and identification of the organism in culture, detection of B. dermatitidis-specific antigens in urine, and detection of specific immunologic responses to infection (2,3). It has proven much more difficult to isolate B. dermatitidis from environmental sources. Isolation techniques have ranged from animal inoculations to in vitro cultivation with a variety of selective and non-selective artificial media (4,5,6). Despite intense efforts, less than two dozen environmental isolates have been reported in the medical literature.
Recently, two groups of investigators developed PCR assays for B. dermatitidis with the goal of improving the sensitivity and specificity of organism detection in paraffin-embedded tissues and from natural soil samples (7,8). Both assays used identical primer pair sequences targeting the putative promoter region of BAD-1 (referred to previously as WI-1), a gene which codes for an important adhesin molecule and virulence factor (9). Because little is known about the extent of genetic diversity that exists for this fungus, we evaluated a panel of clinical and environmental strains according to a previously published rDNA PCR restriction fragment length polymorphism (RFLP) assay and looked for polymorphisms in the promoter region of BAD-1. We observed two novel RFLP genotypic groups in addition to three genotypes described previously (10). Additionally, a high percentage of isolates had polymorphisms in the BAD-1 promoter when compared to an ATCC reference strain. These polymorphisms could undermine the utility of the PCR assays for detection of B. dermatitidis for diagnostic purposes, but may prove useful for population genetic studies.
Materials and Methods
Blastomyces dermatitidis Strains
One hundred and six isolates of B. dermatitidis were evaluated in this study. These included: clinical isolates from human (n=75), feline (n=3), canine (n=8), and equine (n=1) sources obtained between 2005–2008 from Marshfield Laboratories (n=87), clinical isolates from human (n=6) and canine (n=1, ATCC# MYA-2585) (11) sources obtained between 2007 and 2008 from Aurora Health Care, Inc. (n= 7); an in vitro environmental isolate from Wisconsin (ATCC# MYA-2586) (6) obtained in 1997 (n=1), human clinical (n=2) and environmental (n=3) isolates from the Eagle River (5) and Tomorrow River (12) Wisconsin outbreaks in 1984 and 1985, environmental isolates (isolation dates unknown) from Georgia (13) (n= 4), a human clinical isolate (isolation date unknown) from Africa (n=1), and the ATCC (American Type Culture Collection, Rockville, MD) strain #26199, deposited in 1970. All isolates were identified using standard methods which included conversion to the yeast form when incubated in Middlebrook 7H9 broth at 35° C (14).
DNA extraction
B. dermatitidis DNA was extracted using the QIAamp® DNA Mini Kit Tissue Protocol (QIAGEN, Valencia, CA), with the volumes of buffer ATL, proteinase K, buffer AL, and ethanol altered to 540 µl, 60 µl, 500 µl, and 400 µl respectively. Samples were incubated overnight at 56 °C and then split onto two spin columns and eluted with a total of 100 µl buffer AE.
rDNA PCR RFLP
Isolates were classified into genotypic groups by a PCR restriction fragment length polymorphism (RFLP) assay for the rDNA repeat region as described previously (10). Briefly, DNA was amplified with the Expand Long Template PCR System (Roche, Indianapolis, IN) with primers BD28S and NS2. The resulting ~5.5 kb product was purified with the QIAQuick® PCR Purification Kit (QIAGEN) and digested with Dde I. The fragments were separated through a 1.5% (wt/vol) SeaKem® LE agarose (Lonza, Rockland, ME) gel in TBE buffer for 6 hrs at 83 volts. Bands were visualized by ultraviolet transillumination after ethidium bromide staining using a Gel Doc 2000 (Bio-Rad Laboratories, Hercules, CA). DNA band patterns were analyzed with BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium) and matched for percentage similarities by the Dice method (1.5% interband tolerance). Dendrograms were generated using the unweighted pair-group method with arithmetic average (UPGMA) method (15).
PCR Amplification, Sequencing and analysis of the BAD-1 promoter
A 363 bp region of the BAD-1 promoter region was amplified by PCR using the Blasto I and Blasto II (Blasto I/II) primers as described previously (8). PCR products were analyzed by gel electrophoresis on a 1.5% NuSieve® 3:1 agarose gel (Lonza) with 0.5 mg/L ethidium bromide and visualized using a Gel Doc 2000 (Bio-Rad Laboratories). PCR products were purified using the QIAQuick® PCR Purification Kit (QIAGEN) with the elution volume altered to 10 µl. The purified PCR product was sequenced using the BigDye® Terminator v3.1 cycle Sequencing Kit (Applied Biosystems, Warrington, United Kingdom) according to the manufacturer’s recommendations. Dye terminators were removed from each reaction using the DyeEx 2.0 Spin Kit (QIAGEN). The reactions were dried at 70 °C for 30 min, resuspended in 10 µl Hi-Di formamide, and denatured at 95 °C for 2 min. Sequencing was performed on automated DNA sequencer model 3130xl (Applied Biosystems). DNA sequences were aligned using Lasergene99 software (DNASTAR Inc, Madison, WI) based upon 4-fold coverage of the amplified region. Confirmation of sequence identity was performed using the Basic Local Alignment Search Tool (BLAST) program for database searches at the National Center for Biotechnology Information (NCBI) website (16). DNA sequences were used to determine haplotypes for the sequenced region. Sequence data were imported into BioNumerics and the sequences aligned and analyzed using pairwise alignment and UPGMA analysis to produce dendrograms displaying sequence relatedness.
Results
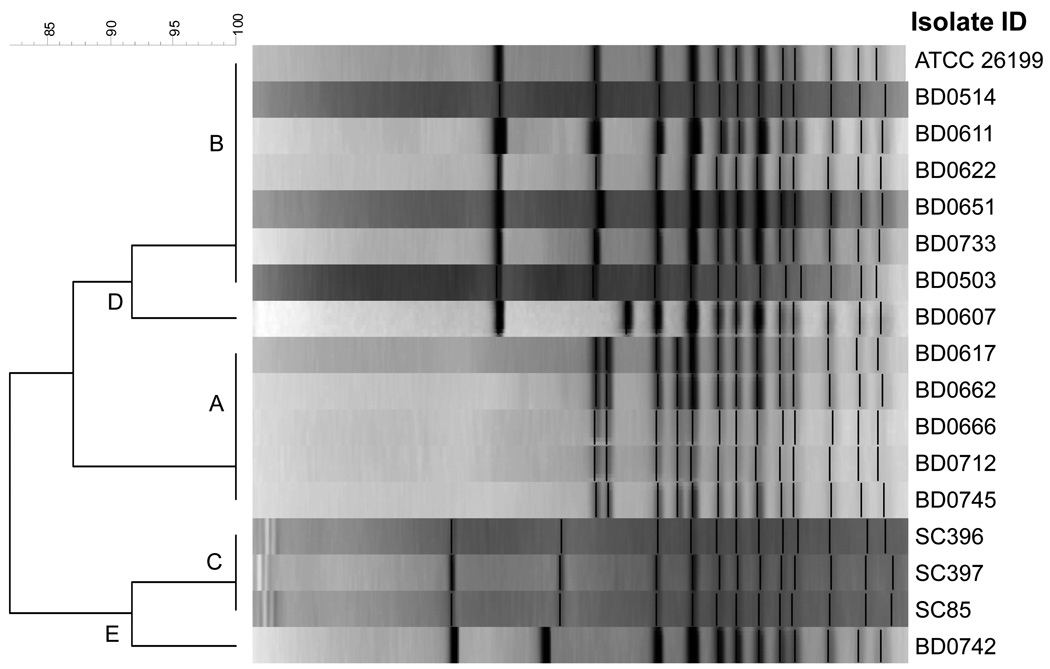
Based on rDNA PCR RFLP analysis, each of the 106 B. dermatitidis isolates fell into one of five genotypic groups designated A through E (Figure 1). Three of the genotypic groups (A, n=50; B, n=51; and C, n=3) correspond to those published previously by McCullough et al. (10). Additionally, two novel genotypes, D (n=1) and E (n=1) were observed. Forty-nine of the group A isolates were from Wisconsin and one from Africa. Fifty of the B isolates were from Wisconsin with the ATCC strain the other B type. All group C isolates were from Georgia. The group D isolate was from the upper peninsula of Michigan. The group E isolate was from Wisconsin. A dendrogram showing representative isolates from each genotypic group is shown in Figure 1.
Figure 1.
Dendrogram produced from the rDNA PCR RFLP band patterns of 17 isolates of Blastomyces dermatitidis. Representative isolates of each genotypic group are shown from 106 isolates typed using this schema.
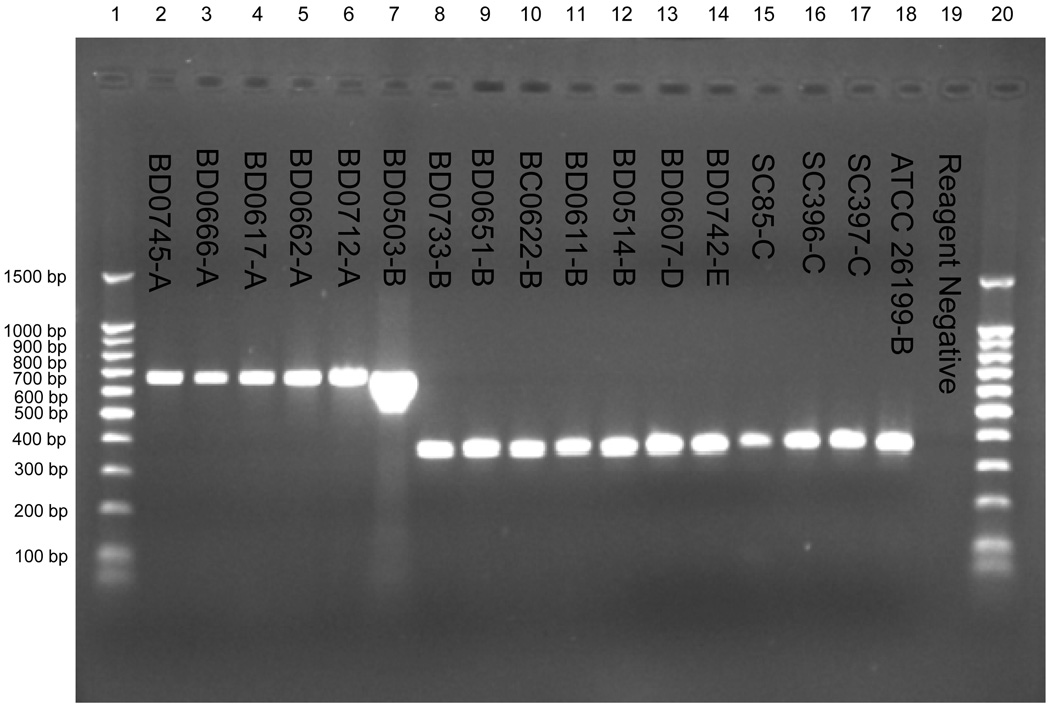
An unexpected finding was that the observed size of the Blasto I/II PCR products varied significantly between genotypic groups. All group C, D, and E isolates and 50 of 51 group B isolates had amplicons of the expected size of ~363 bp. However, 50 of 50 genotypic group A isolates and 1 of 51 genotypic group B isolates had amplicons of 663 bp (Figure 2). Sequencing of the amplicons from 17 representative isolates (5 group A, 7 group B, 3 group C, 1 group D, and 1 group E) confirmed the presence of the size polymorphism and revealed several additional smaller genetic changes among and within genotypic groups (Figure 3 Panel A). The larger amplicon size in the genotypic group A isolates and the single group B isolate was due to a 35 bp insertion after bp 1614 and a 251 bp insertion after base pair 1668 (numbering based on the published BAD-1 (WI-1) gene sequence, Genbank accession U37772).
Figure 2.
The amplification products from a subset of the Blastomyces dermatitidis isolates (with the rDNA PCR RFLP genotypic groups designated) using the BlastoI/II primers within BAD-1 (WI-1) putative promoter region (7). Lanes 1 and 20 contain 100 bp molecular weight ladder. Lanes 2 – 18 contain the amplification products from individual B. dermatitidis isolates. Lane 19 contains the PCR reagent negative control.
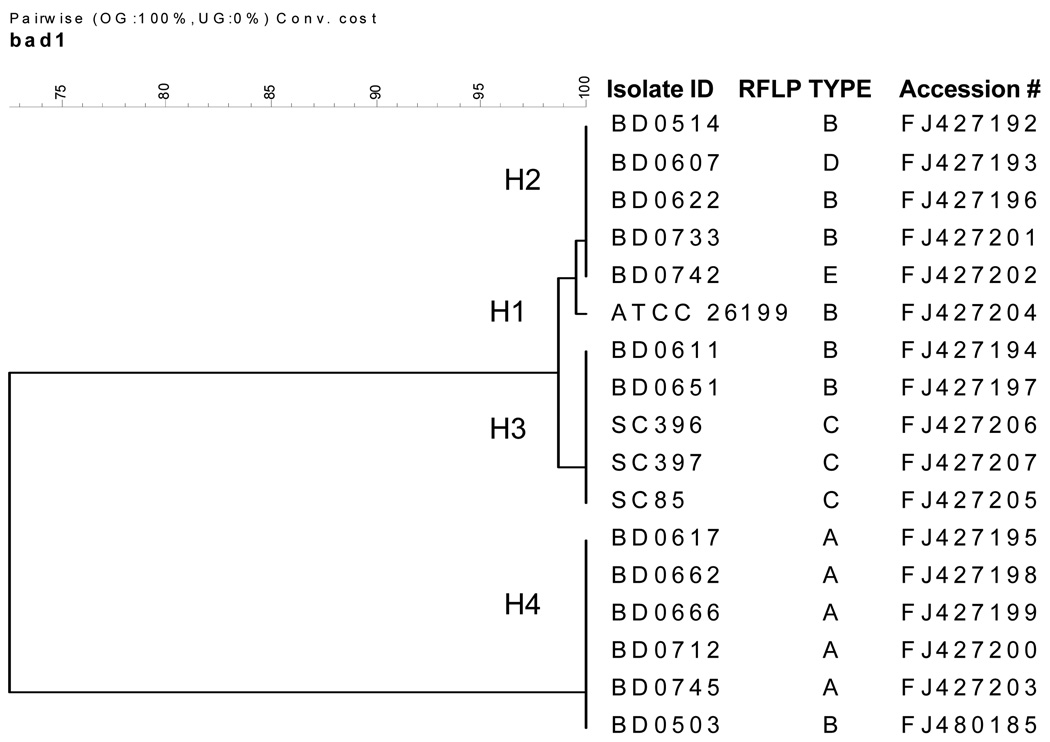
Figure 3.
(A) The DNA sequencing alignment of Blastomyces dermatitidis amplification product resulting from the Blasto I/II primers located within the putative promoter region of BAD-1. A representative isolate is shown for each of the 4 haplotypes (H1-H4) from the 17 sequenced isolates. H1 contains the ATCC 26199 isolate (RFLP genotypic group B); H2 contains isolates from the PCR RFLP genotypic groups B, D and E; H3 contains isolates from the rDNA PCR RFLP genotypic groups B and C; and H4 contains isolates from rDNA RFLP genotypic group A and a single genotypic group B isolate. Major insertions identified through sequencing are indicated by arrows (H4). (B) Phylogenetic relationship of 17 representative isolates of B. dermatitidis based upon 4-fold sequencing of the amplicon produced by the Blasto I/II primer set.
Based upon the sequence variation of the BAD-1 amplicon, four haplotypes were identified. The ATCC strain 26199, which is genotypic group B, was the only strain in haplotype 1 (H1). Haplotype 2 (H2) included isolates from PCR RFLP genotypic groups B, D and E. Haplotype 3 (H3) consisted of isolates from PCR RFLP genotypic groups B and C. Haplotype 4 (H4) was comprised of isolates from PCR RFLP genotypic group A and a single genotypic group B isolate. DNA sequences were submitted to Genbank and have been given the following accession numbers: FJ427192, FJ427193, FJ427194, FJ427195, FJ427196, FJ427197, FJ427198, FJ427199, FJ427200, FJ427201, FJ427202, FJ427203, FJ427204, FJ427205, FJ427206, FJ427207 and FJ480185. Phylogenetic relationships determined from sequence comparisons are shown in Figure 3 Panel B.
Discussion
Blastomycosis is an important mycosis of humans and other mammals because it can be associated with significant morbidity and mortality (1). Unfavorable outcomes are more likely when there is overwhelming infection or a delay in diagnosis and initiation of therapy. Fortunately, the laboratory diagnosis of blastomycosis is usually straightforward and can be accomplished by routine histology and conventional culture techniques (14). However, there are instances where histologic sections show scant and/or atypical fungal elements and there is no unfixed specimen available for culture confirmation. For these cases the availability of molecular diagnostic assays, such as PCR, can be a valuable diagnostic tool. In addition, molecular detection assays hold great promise in helping to elucidate the ecology of B. dermatitidis (7). To date, efforts to identify the ecologic niche by organism detection using in vivo and in vitro culture methods have been largely unsuccessful (17,18). Therefore, most of our knowledge of the ecologic characteristics and distribution of the fungus is based on reported disease occurrence. A sensitive and specific PCR-based assay for the detection of B. dermatitidis from environmental samples could greatly enhance our understanding of factors modulating disease transmission dynamics in natural settings.
Two groups of investigators have published PCR assays for B. dermatitidis that target a 363 bp fragment of DNA upstream from the BAD-1 virulence gene (6,7). Initially considered to be a promoter region, subsequent deletion experiments suggest that the targeted gene segment has little or no influence on BAD-1 expression. Nevertheless, the stated goals of these studies were to develop molecular diagnostic tools for detection of B. dermatitidis DNA in tissue from paraffin-embedded blocks (7) and from natural soil samples (8). Both studies showed promising results, but neither included an evaluation of how genetic diversity might affect the performance characteristics of their assays. We explored this possibility as part of our ongoing studies of the genetic diversity of B. dermatitidis in Wisconsin. Analysis of a large panel of isolates by PCR RFLP of the rDNA repeat region revealed that all isolates fell into one of five genotypic groups. Three genotypic groups (A,B, and C) predominated and accounted for over 98% of the isolates. Groups D and E were interesting because they appear to represent novel genotypes but were uncommon in our panel of isolates. Of note, nine of the 106 isolates in our study were also included in the previous study by McCullough et al. (10). We assigned five of the nine isolates to different genotypic groups than were reported previously. This disparity in assigning genotypes may be due to differences in the gel running conditions, type and concentration of agarose, band tolerance settings, and the normalization of the gels between runs.
A significant finding of our study was that different genotypic groups of B. dermatitidis are associated with markedly different PCR amplicon sizes when using the Blasto I/II primer pair. Group C, D, E, and all but one of the B group isolates had products of the expected size, ~363 bp. However, all group A isolates and a single group B isolate produced a 663 bp product, nearly twice the expected size. This size disparity was due to the presence of two large insertions. Therefore, awareness of this size polymorphism is important when using the Blasto I/II primers, especially when working in geographic areas like Wisconsin where group A strains comprise ~ 50% of isolates, or in regions where the genetic diversity of B. dermatitidis is simply unknown. Results of previous studies using this detection method may need to be reevaluated because positive samples producing faint bands at sizes other than 363 bp may have been disregarded as non-specific amplification products. Additionally, amplification of a larger product may not have occurred if the assay was not optimized for amplification of the larger product. This is of special concern for environmental samples because of the wide range of inhibitory factors inherent to soil samples that may complicate nucleic acid extraction and amplification (19).
In addition to the large insertions, several additional polymorphisms were observed upstream from BAD-1. Four haplotype groups were identified among the 17 strains selected for sequencing (H1-H4). These haplotypes do not strictly correspond to the rDNA PCR RFLP groups with the exception of RFLP group A, of which all 5 isolates sequenced were H4. The haplotypes identified through sequencing of this region may prove useful as genetic markers in studies aimed at understanding the population genetics of B. dermatitidis. The lack of variation within the genotypic group A (H4) isolates may indicate an apparent lack or reduction in the genetic recombination between genotypic group A with genotypic groups B, C, D and E. Genotypic group A isolates clustered together in both typing schema, whereas the B, C, D and E strains showed different relatedness to each other in the two genotyping approaches. Isolates from genotypic groups A, B, D and E were obtained primarily from Wisconsin from human and veterinary clinical specimens from overlapping geographic regions. While genotypic group A (H4) isolates appear to be genetically dissimilar to the other groups, this does not appear to be a function of temporal or spatial isolation. Additional research is needed to determine if the genetic isolation of genotypic group A holds true by examining additional regions of the fungal genome and isolates from diverse geographic areas. If so, it will be important to understand whether the lack of genetic recombination is the result of the reduced biological ability to recombine or rather a function of ecological niche partitioning in the environment.
Acknowledgements
The authors wish to acknowledge the financial support of the Marshfield Clinic Research Foundation and the National Institutes of Health. We also wish to thank Alice Stargardt for assistance in the preparation of this manuscript.
References
- 1.Bradsher RW, Chapman SW, Pappas PG. Blastomycosis. Infect Dis Clin North Am. 2003;17:21–40. doi: 10.1016/s0891-5520(02)00038-7. [DOI] [PubMed] [Google Scholar]
- 2.Rippon JW. Medical Mycology: The pathogenic fungi and the pathogenic actinomycetes. 3rd ed. Philadelphia, PA: W.B. Saunders; 1988. Blastomycosis; pp. 474–505. [Google Scholar]
- 3.Mongkolrattanothai K, Peev M, Wheat LJ, Marcinak J. Urine antigen detection of blastomycosis in pediatric patients. Pediatr Infect Dis J. 2006;25:1076–1078. doi: 10.1097/01.inf.0000241144.89426.2a. [DOI] [PubMed] [Google Scholar]
- 4.Denton JF, McDonough ES, Ajello L, Ausherman RJ. Isolation of Blastomyces dermatitidis from soil. Science. 1961;133:1126–1127. doi: 10.1126/science.133.3459.1126. [DOI] [PubMed] [Google Scholar]
- 5.Klein BS, Vergeront JM, Weeks RJ, Kumar UN, Mathai G, Varkey B, et al. Isolation of Blastomyces dermatitidis in soil associated with a large outbreak of blastomycosis in Wisconsin. N Engl J Med. 1986;314:529–534. doi: 10.1056/NEJM198602273140901. [DOI] [PubMed] [Google Scholar]
- 6.Baumgardner DJ, Paretsky DP. The in vitro isolation of Blastomyces dermatitidis from a woodpile in north central Wisconsin, USA. Med Mycol. 1999;37:163–168. [PubMed] [Google Scholar]
- 7.Bialek R, Cirera AC, Herrmann T, Aepinus C, Shearn-Bochsler VI, Legendre AM. Nested PCR assays for detection of Blastomyces dermatitidis DNA in paraffin-embedded canine tissue. J Clin Microbiol. 2003;41:205–208. doi: 10.1128/JCM.41.1.205-208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgess JW, Schwan WR, Volk TJ. PCR-based detection of DNA from the human pathogen Blastomyces dermatitidis from natural soil samples. Med Mycol. 2006;8:741–748. doi: 10.1080/13693780600954749. [DOI] [PubMed] [Google Scholar]
- 9.Hogan LH, Josvai S, Klein BS. Genomic cloning, characterization and functional analysis of the major surface adhesin WI-1 on Blastomyces dermatitidis yeasts. J Biol Chem. 1995;270:30725–30732. doi: 10.1074/jbc.270.51.30725. [DOI] [PubMed] [Google Scholar]
- 10.McCullough MJ, DiSalvo AF, Clemons KV, Park P, Stevens DA. Molecular Epidemiology of Blastomyces dermatitidis. Clin Infect Dis. 2000;30:328–335. doi: 10.1086/313649. [DOI] [PubMed] [Google Scholar]
- 11.Baumgardner DJ, Paretsky DP. Identification of B dermatitidis in the stool of a dog with acute pulmonary blastomycosis. J Med Vet Mycol. 1997;35:419–421. [PubMed] [Google Scholar]
- 12.Klein BS, Vergeront JM, DiSalvo AF, Kaufman L, Davis JP. Two outbreaks of blastomycosis along rivers in Wisconsin. Am. Rev. Respir. Dis. 1987;136:1333–1338. doi: 10.1164/ajrccm/136.6.1333. [DOI] [PubMed] [Google Scholar]
- 13.Denton JF, DiSalvo AF. Isolation of Blastomyces dermatitidis from natural sites at Augusta, Georgia. Am J Trop Med Hyg. 1964;13:716–722. doi: 10.4269/ajtmh.1964.13.716. [DOI] [PubMed] [Google Scholar]
- 14.Brandt ME, Warnock DW. Manual of Clinical Microbiology. 9th ed. Washington D.C.: ASM Press; 2007. Apr, Histoplasma, Blastomyces, Coccidioides, and other dimorphic fungi causing systemic mycoses. Chapter 123; pp. 1857–1873. [Google Scholar]
- 15.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 16.Altschul SF, Gish W, Miller EW, Myers K, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. Available from: http://blast.ncbi.nlm.nih.gov/Blast.cgi. [DOI] [PubMed]
- 17.DiSalvo AF. The ecology of B. dermatitidis. In: Al-Doory Y, DiSalvo AF, editors. Blastomycosis. New York: Plenum; 1992. pp. 43–73. [Google Scholar]
- 18.Blondin N, Baumgardner DJ, Moore GE, Glickman LT. Blastomycosis in indoor cats: suburban Chicago, Illinois, USA. Mycopathologia. 2007;163:59–66. doi: 10.1007/s11046-006-0090-1. [DOI] [PubMed] [Google Scholar]
- 19.Wilson IG. Inhibition and facilitation of nucleid acid amplification. Appl. Environ. Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]