Abstract
Neurons in the peripheral nervous system (PNS) display a higher capacity to regenerate after injury than those in the central nervous system, suggesting cell specific transcriptional modules underlying axon growth and inhibition. We report a systems biology based search for PNS specific transcription factors (TFs). Messenger RNAs enriched in dorsal root ganglion (DRG) neurons compared to cerebellar granule neurons (CGNs) were identified using subtractive hybridization and DNA microarray approaches. Network and transcription factor binding site enrichment analyses were used to further identify TFs that may be differentially active. Combining these techniques, we identified 32 TFs likely to be enriched and/or active in the PNS. Twenty-five of these TFs were then tested for an ability to promote CNS neurite outgrowth in an overexpression screen. Real-time PCR and immunohistochemical studies confirmed that one representative TF, STAT3, is intrinsic to PNS neurons, and that constitutively active STAT3 is sufficient to promote CGN neurite outgrowth.
Keywords: Dorsal Root Ganglion, Transcription Factor, High Content Analysis, Screen, Systems Biology, Cerebellar Granule Neuron, STAT3
Introduction
Adult central nervous system (CNS) axons fail to regenerate after injury due to inhibition by myelin and glial scar associated molecules (Silver and Miller, 2004; Yiu and He, 2006), as well as a loss of developmentally regulated intrinsic growth ability (Cohen et al., 1986; Goldberg et al., 2002). In contrast, neurons in the peripheral nervous system (PNS) regenerate under a variety of experimental paradigms, including stimulation with cAMP (Qiu et al., 2002), treatment with a small molecule inhibitor of protein kinase C (Sivasankaran et al., 2004), and after priming the peripheral nerve with a conditioning lesion (Richardson and Issa, 1984). Furthermore, DRG neurons have the ability to regenerate long distances through intact and degenerating CNS white matter (Davies et al., 1997; Davies et al., 1999), whereas CNS neurons transplanted under similar conditions display relatively modest outgrowth (Tom et al., 2004).
Pharmacological blockade of RNA synthesis (Smith and Skene, 1997), as well as genetic ablation of specific factors such as Jun (Raivich et al., 2004), strongly suggest that the intrinsic growth ability of PNS neurons is dependent upon transcription. Comparisons of gene expression changes after a peripheral nerve lesion have led to the identification of several regeneration associated transcription factors (TFs) including AKRD1 (Stam et al., 2007), SMAD1 (Zou et al., 2009), and NFIL3 (MacGillavry et al., 2009). Much less clear, however, are the identities of intrinsic, constitutively expressed TFs that are upstream of injury mechanisms and allow PNS neurons to respond to injury in situations where CNS neurons cannot.
Altering intrinsic gene expression of adult CNS neurons has led to improvements of axonal outgrowth after injury. Deletion of genes in postnatal CNS neurons such as Pten (Park et al., 2008), Klf4 (Moore et al., 2009) or Socs3 (Smith et al., 2009) results in significant regeneration of optic nerve fibers. Likewise, overexpression of developmentally downregulated genes such as Klf6 and Klf7 can boost the growth of cortical spinal motor neurons in vitro by 50% (Blackmore et al.; Moore et al., 2009). As the identities and roles of these proteins in growth regulatory pathways become better appreciated, so too should our ability to target therapies to improve functional outcomes.
The accelerating pace of biomedical annotation has lead to several novel techniques for probing the biological functions of transcription factors. Among these are the ability to query the promoters of an expression set to identify overrepresented transcription factor bindings sites (TFBSs) compared to a control expression set (Li et al., 2008). The pathway analysis tool Metacore (http://www.genego.com/metacore.php), which is able to draw upon an extensive set of curated protein-protein and protein-DNA interactions, has also proven useful in identifying differences in transcription networks between expression sets using closest-paths algorithms (Birmachu et al., 2007). These techniques are essential because the enrichment of a TF in a particular cell type is not necessarily a predictor of function. The NF-κB transcriptional complex provides an excellent example for this principle in that it can either enhance or diminish sensory axon growth based on the phosphorylation state of RelA/p65 (Gutierrez et al., 2008).
In this report we outline a combinatorial strategy in which we utilize these new methods of biomedical annotation to identify 32 TFs that exhibit intrinsic differences between postnatal mouse PNS and CNS neurons, hypothesizing that some might be associated with the intrinsic ability of PNS neurons to regenerate. We then performed a high content analysis screen to identify potential roles of those factors in regulating neurite outgrowth of primary CNS neurons. Further, we demonstrate that the TF signal transducer and activator of transcription 3 (STAT3) is constitutively enriched and active in the PNS and can enhance the outgrowth of cerebellar granule neurons.
Results
Identification of PNS specific genes
To identify TFs preferentially expressed in the PNS, we generated a cDNA library of mRNAs enriched in DRG neurons using subtractive hybridization (Bonaldo et al., 1996; Diatchenko et al., 1996). This was accomplished by generating two separate cDNA libraries: the first from cultured postnatal DRG neurons, the second from autologous cerebellar tissue, followed by a hybridization step to remove cerebellar (driver) sequences that were also present in the DRG mRNA pool (Figure 1A). Subtractive hybridization is advantageous as it allows detection of rare and novel mRNA populations and has greater sensitivity than microarrays (Cao et al., 2004).
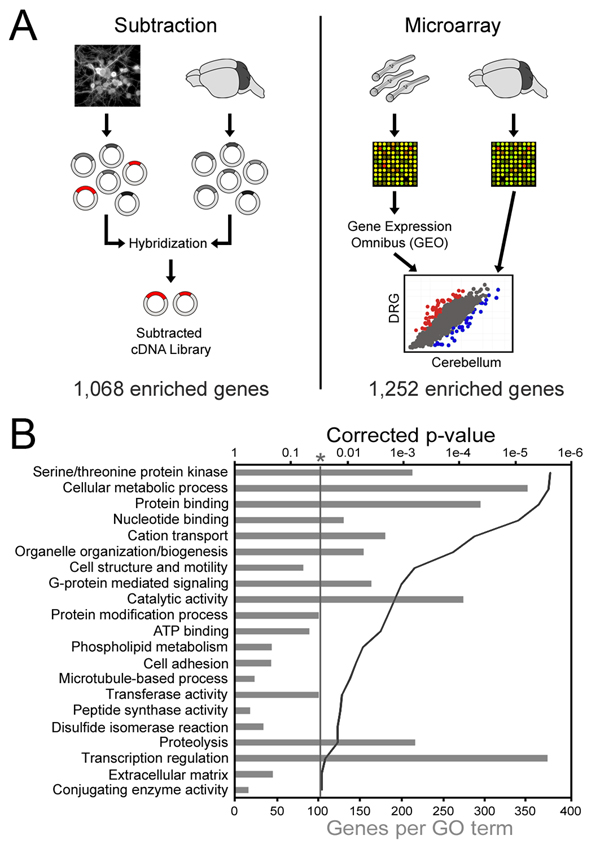
Figure 1. Identification of DRG enriched genes.
(a) Cultures of postnatal DRG neurons and whole cerebellum were used to generate cDNA libraries representing the respective mRNA populations. The cerebellum library driver was then hybridized to the DRG library tester in great excess, followed by recovery of non-bound DRG cDNAs by HAP chromatography. The resulting “subtracted” library was sequenced and BLASTed against the UniGene, whereupon 1,068 unique genes were identified. For the microarray analysis, a data set consisting of 5 replicates of LCM DRG neurons from adult mice was obtained from GEO and compared to our own set of 3 replicates of P11 cerebellum that was collected on the same microarray platform (MG-U74Av2). (b) DAVID was used to identify gene ontology terms describing subtraction genes. Grey bars represent the number of subtraction genes with the labeled annotation, and the solid line denotes the Benjamini-Hochberg corrected EASE p-value. Only terms with a corrected p-value less than 0.05 were considered.
Over 2000 clones from the DRG enriched cDNA library were sequenced and BLASTed against the Mus musculus UniGene database using the software package EST Express (Smith et al., 2008) to produce a list of 1,068 distinct genes (Table S1). These sequences were deposited in GenBank and are publicly available (http://www.ncbi.nlm.nih.gov/UniGene/library.cgi?LID=24175). We used DAVID (Dennis et al., 2003) to identify overrepresented Gene Ontology (GO) (Ashburner et al., 2000) and Panther (Thomas et al., 2003) terms to describe the DRG enriched gene list (Table S1). Using this approach we found many genes associated with cell motility (BP00285), adhesion (BP00124) and microtubule dynamics (GO:0007017). Interestingly, the DRG enriched gene list also included 378 genes (45.7% of annotated genes in the library) involved in the transcriptional regulation of mRNA synthesis (BP00044; Figure 1B).
To complement the list of DRG enriched genes from the subtraction, we made use of publicly available microarray data from the Gene Expression Omnibus (GEO) (Edgar et al., 2002). Data series GSE2917 includes 5 replicates of laser capture microdissected (LCM) adult mouse DRG neurons obtained after intraperitoneal injection of the retrograde tracer cholera toxin B (Peeters et al., 2006). We combined this data with our own set of 3 replicate chips measuring P11 cerebellum mRNA levels using the same microarray platform. Because the microarray data sets were obtained in separate locations, we processed the DRG and cerebellum CEL files together using the Robust Multi-array Average (RMA) technique (Irizarry et al., 2003) and performed hierarchical clustering to verify replicate consistency (Figure S1). The RMA technique performs background correction and quantile normalization to equally distribute probe intensities across arrays (Bolstad et al., 2003).
Normalized probe intensities representing 12,488 genes were used to determine a relative DRG/cerebellum expression measure, and a two-tailed Student’s t-test was used to determine the significance. To correct for multiple comparisons, we made use of the Benjamini-Hochberg false discovery rate (FDR) technique (Hochberg and Benjamini, 1990). By selecting genes that were 2-fold enriched with corrected p-values of less than 0.01, we identified 1,252 and 892 genes that were significantly enriched in DRG neurons and cerebellum respectively (Table S2).
The subtractive hybridization and microarray approaches produced complementary data sets representing mRNA from two populations of sensory neurons separated by age (P7-10 vs. adult) and by experimental conditions (cultured vs. LCM). In addition, only 485 genes identified by subtractive hybridization were represented on the U74AV2 microarray platform (45%), creating the potential for only moderate overlap between the two data sets. 108 genes identified by subtractive hybridization were also found to be enriched in LCM DRG neurons, in contrast to 51 subtraction genes that were enriched in the cerebellum. Several factors may have contributed to the latter discrepancy, including the inherent differences between approaches mentioned above, misaligned ESTs and/or the presence of alternative transcripts not accounted for in the microarray probe set. Taking these limitations into consideration, we decided to take advantage of the availability of these two distinct experimental data sets to identify common intrinsic transcriptional regulators in the PNS.
PNS enriched transcription factor networks
We employed the curation-based pathway analysis tool Metacore (GeneGo Inc.) to identify networks of known interacting proteins and genes within the subtractive hybridization and microarray data sets (Figure 2A). We hypothesized that any TF preferentially expressed and/or active in DRG neurons would be co-expressed with gene products that are enriched within those neurons. Metacore's "Transcriptional Regulation" tool linked DRG enriched genes with 1,773 known transcriptional regulators. The resulting networks were then scored based on permutations of all possible network configurations, generating a p-value that was further corrected for multiple comparisons. We performed this analysis for three lists of genes: the subtractive hybridization list as well as the list of DRG enriched genes and the list of cerebellum enriched genes from the microarray data set. Any transcriptional regulator that was significantly linked to the cerebellum-enriched list (e.g. p53, CREB1) was then filtered from the other two lists.
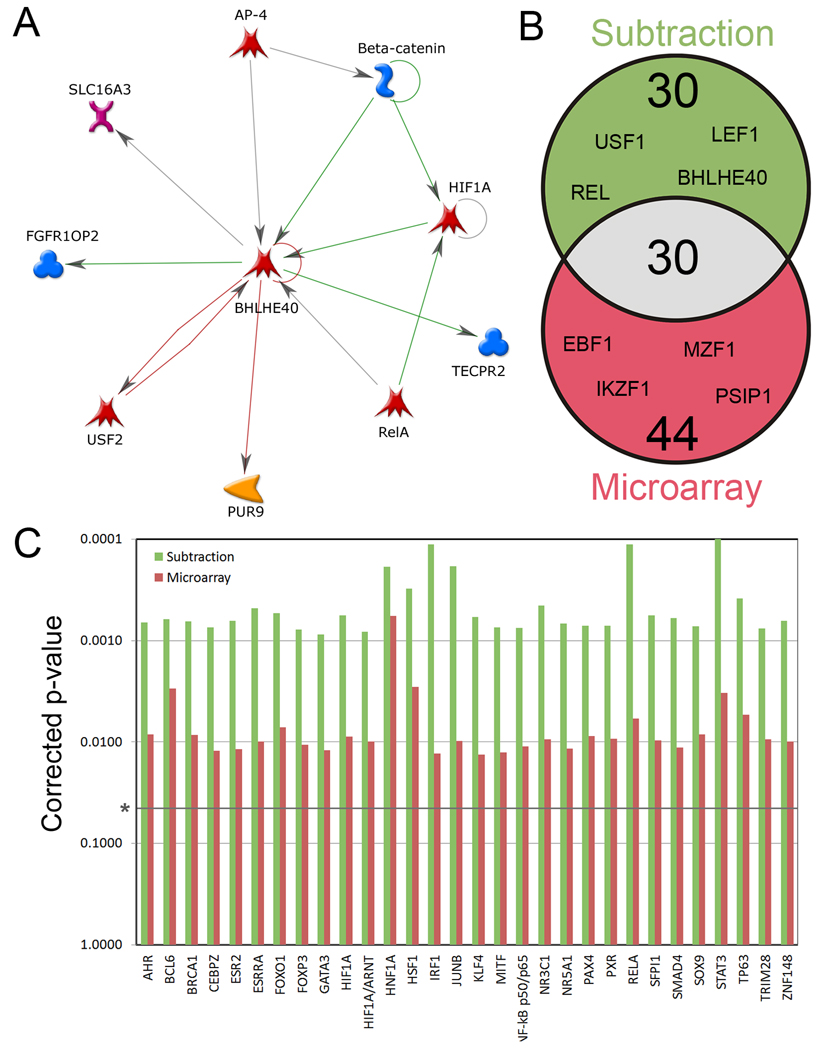
Figure 2. Transcription factor (TF) networks enriched in DRG neurons.
(a) Metacore generated screenshot of nearest neighbors network for transcription factor BHLHE40. Each node represents a protein or gene, and connecting arrows represent a curated interaction between the two nodes. Outward arrows denote regulation by BHLHE40, whereas inward arrows denote regulation of the factor. (b) Venn Diagram of transcription factor networks found to be significantly overrepresented in DRG neurons by Metacore. This analysis considered all known TFs within Metacore and was not biased towards the expression level of the TFs. Using this approach, we expect to identify differentially active TFs in addition to factors that may have been missed by the Subtraction or Microarray. (c) 30 TF networks were found to be significantly overrepresented in both the Subtraction and the Microarray data sets. P-values were determined by a method developed by Metacore (discussed in the methods section), followed by a Benjamini-Hochberg correction for multiple comparisons.
Using this approach we identified 104 TFs that interacted significantly with our DRG enriched data sets (Table S3), including 31 factors that interacted with only the subtraction list, 44 factors that interacted only with the microarray list, and a further 30 factors that interacted with both lists (Figure 2B, C). By comparison, the overlap between the 61 subtraction linked TFs and those linked to cerebellum-enriched genes was 5.
PNS enriched transcription factor binding sites
Next we sought to determine whether our DRG enriched data sets exhibited signatures of regulation by specific factors. We hypothesized that if a TF was preferentially active in DRG neurons, its binding sites would be overrepresented in the promoters of DRG enriched genes (Figure 3A). Consensus binding sites from TRANSFAC 10.2 were combined with known conserved 5’ sequence motifs (Xie et al., 2005) to create a list of 1,042 transcription factor binding sites (TFBSs). Custom software was then used to match each TFBS against the promoter regions of all DRG enriched genes with genomic annotations (847 from subtractive hybridization and 1,075 from the microarray). A promoter region was defined by the sequence of DNA 1,000bp upstream and 200bp downstream of the transcription start site (TSS). A background list of 41,116 Entrez Gene IDs with TSS annotations was used to determine whether representation of binding sites was non-random. Significance was determined by a Fisher Exact Test followed by a correction for multiple comparisons. As in the case of the network analysis, a third promoter scan was also carried out on the list of cerebellar enriched genes from the microarray, and any common matches were filtered from those matching the DRG enriched data sets.
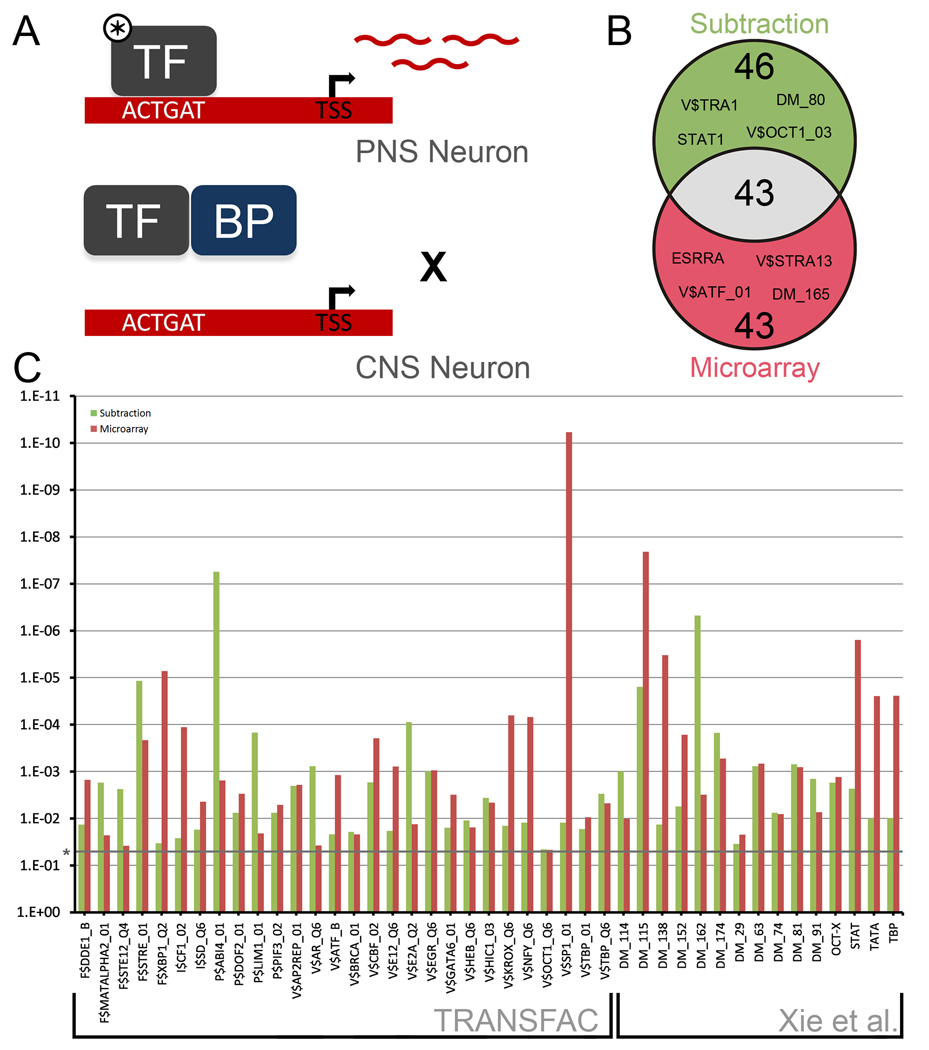
Figure 3. Overrepresented mammalian transcription factor binding sites in the promoters of DRG enriched genes.
(a) Model demonstrating how differentially active TFs may be detected by the analysis of DRG enriched gene promoters. In PNS neurons, an active TF (marked by *) is able to bind target sequences, inducing the transcription of mRNAs that can be detected by the Subtraction and Microarray. In CNS neurons, a TF may be unable to drive expression of target sequences due to a lack of upstream signals or co-activators, the presence of binding proteins dampening activity (BP), or an altered genomic landscape. (b) Venn Diagram of 132 transcription factor binding consensuses found to be significantly overrepresented in the promoter regions of DRG enriched genes. 1200 bp promoter regions (1000bp upstream and 200 bp downstream of TSS) were scanned for consensus binding sites from TRANSFAC 10.2 and from a paper by Xie et al. (2005). (c) 28 TF binding consensuses from TRANSFAC and 15 from Xie et al. (2005) were found to be significantly overrepresented in both the Subtraction and the Microarray data sets. Significance was determined by a two-tailed Fisher’s Exact Test, followed by a Benjamini-Hochberg correction for multiple comparisons.
The promoter analysis yielded 132 TFBSs that were overrepresented in the DRG enriched data sets (Table S4), including 43 TFBSs that matched both sets (Figure 3B, 3C). This high degree of overlap between the two datasets suggests that there may in fact be constitutive DRG transcriptional regulators that do not change postnatally and specify cell function independently of downstream injury mechanisms.
High-content analysis of TF overexpression on neurite outgrowth
By combining differentially expressed TFs from the subtractive hybridization and microarray data sets with the network and TFBS enrichment analyses, we identified 32 transcription factors that were identified by at least two techniques (Table 1). Given the many differences in function between DRG and cerebellar granule neurons, it is highly unlikely that a large fraction of these 32 factors would play a role in neurite outgrowth. It was our belief, however, that this set constituted a good starting point for an overexpression screen.
Table 1. Candidate DRG enriched transcription factors.
Listing of transcription factors identified by at least two different techniques. “SUB” = found in the subtraction. “MA” = found in the microarray. “PS/PM” = found in the promoter analysis of the subtraction (S) or microarray (M) genes. “NS/NM” = found in the interaction network analysis of the subtraction (S) or microarray (M) genes. “RT” = found to be significantly upregulated by real-time PCR in dorsal root ganglia (DRG) or cerebellum (CGN). "SCR" = whether or not the transcription factor was screened for the ability to promote outgrowth in cerebellar granule neurons. KLF4 and KLF7 are included as controls (CNTRL).
| Transcription Factor | Symbol | SUB | MA | NS | NM | PS | PM | RT | SCR |
|---|---|---|---|---|---|---|---|---|---|
| Kruppel-like factor 7 | Klf7 | X | CNTRL | ||||||
| Signal transducer and activator of transcription 3 | Stat3 | X | X | X | X | X | X | DRG | Yes |
| Interferon regulator factor 1 | Irf1 | X | X | X | X | DRG | Yes | ||
| Breast cancer 1, early onset | Brca1 | X | X | X | X | Yes | |||
| CCAAT/enhancer binding protein zeta | Cebpz | X | X | X | X | Yes | |||
| GATA binding protein 3 | Gata3 | X | X | X | CGN | Yes | |||
| Jun oncogene | Jun | X | X | X | DRG | Yes | |||
| Myelocytomatosis oncogene | Myc | X | X | X | DRG | Yes | |||
| Signal transducer and activator of transcription 6 | Stat6 | X | X | X | DRG | Yes | |||
| Upstream transcription factor 1 | Usf1 | X | X | X | CGN | Yes | |||
| SMAD family member 4 | Smad4 | X | X | CGN | No | ||||
| Signal transducer and activator of transcription 1 | Stat1 | X | X | DRG | Yes | ||||
| Activating transcription factor 3 | Atf3 | X | X | X | Yes | ||||
| Basic helix-loop-helix family, member e40 | Bhlhe40 | X | X | X | Yes | ||||
| Hypoxia inducible factor 1, alpha subunit | Hif1a | X | X | X | Yes | ||||
| Glucocorticoid receptor | Nr3c1 | X | X | X | Yes | ||||
| Steroidogenic factor 1 | Nr5a1 | X | X | X | Yes | ||||
| V-rel reticuloendotheliosis viral oncogene homolog A (avian) | Rela | X | X | X | Yes | ||||
| SFFV proviral integration 1 (PU.1) | Sfpi1 | X | X | CGN | Yes | ||||
| upstream transcription factor 2 | Usf2 | X | X | X | Yes | ||||
| Chromodomain helicase DNA binding protein 4 | Chd4 | X | X | No | |||||
| Early B-cell factor 1 | Ebf1 | X | X | No | |||||
| FtsJ methyltransferase domain containing 2 | Ftsjd2 | X | X | No | |||||
| IKAROS family zinc finger 1 | Ikzf1 | X | X | No | |||||
| Myeloid zinc finger 1 | Mzf1 | X | X | No | |||||
| RD RNA-binding protein | Rdbp | X | X | No | |||||
| Ring finger protein 38 | Rnf38 | X | X | Yes | |||||
| hepatoma-derived growth factor, related protein 2 | Hdgfrp2 | X | X | Yes | |||||
| Kruppel-like factor 6 | Klf6 | X | X | Yes | |||||
| PC4 and SFRS1 interacting protein 1 | Psip1 | X | X | Yes | |||||
| replication factor C (activator 1) 1 | Rfc1 | X | X | Yes | |||||
| Transcription factor AP4 | Tcfap4 | X | X | Yes | |||||
| YY1 transcription factor | Yy1 | X | X | Yes |
To provide support for the idea that CGNs are a reasonable surrogate for other CNS neurons important in regeneration we examined the expression levels of the factors in Table 1 using publicly available GNF Mouse GeneAtlas V3 microarray data (Lattin et al., 2008). This analysis revealed that of the 32 TFs, 10 were enriched more than two fold in adult mouse DRG compared to cerebral cortex, and 11 were enriched more than two fold compared to hippocampus (Table S5). By contrast, only 2 TFs were enriched in cortex and 1 in the hippocampus compared to DRG. Considering that our approach sought to identify factors that differed in their activity in addition to expression, these data are consistent with the assertion that our method selects for PNS vs CNS enrichment.
We transfected P7-10 mouse cerebellar granule neurons (CGNs) with full length TF cDNAs with the goal of identifying factors that had an effect on neurite outgrowth. A majority of the TFs were available from our laboratory’s mammalian expression open reading frame collection from Open Biosystems. TFs that were not available within our lab were obtained from the plasmid-sharing repository, Addgene (http://www.addgene.com). Using the Amaxa Nucleofector Shuttle system we analyzed 25 of the 32 TFs identified in Table 1. Successfully transfected neurons were identified by simultaneous transfection of a reporter gene (pMAX-eGFP, Amaxa). The amount of TF to reporter gene ratio was optimized such that co-transfection rates typically yielded >70%, similar to other larger high content analysis screens conducted by our laboratory (Blackmore, et al., 2010). CGNs were allowed to grow for 48 hours after which they were fixed and immunostained for neuronal specific BIII-tubulin (Figure S4A). Image acquisition and neurite length data was automatically measured using the Cellomics ArrayScanVTI Neuronal Profiling Algorithm, allowing unbiased quantification analysis of neuronal image data (Figure S4A–C). In each experiment, neurite length for at least 100 eGFP positive neurons was quantified across 6 replicate wells. We included KLF7, a known promoter of neurite outgrowth (Moore et al., 2009) as a positive control, and 3 negative “control” genes (OXR, Oxr1 and Sparcl1) that have consistently shown in our hands not to effect neurite outgrowth. To facilitate comparisons across multiple and repeated experiments, we used mean total neurite length relative to the average of the three control genes for each TF (Figure S4D). Although several TFs produced consistent trends in this screen, no single factor was found to significantly affect CGN neurite outgrowth after one-way ANOVA post-hoc tests. While this could suggest that DRG intrinsic TFs simply have no bearing on CNS neurite outgrowth, another interpretation of this data is that CNS neurons lack additional factors that are necessary for these TFs to successfully drive transcription of key growth promoting genes.
STAT3 is constitutively enriched and active in DRG neurons
Signal transducer and activator of transcription 3 (STAT3), found to be enriched in DRG neurons by subtractive hybridization, microarray (4.2 fold) as well as by the network and TFBS enrichment analyses, produced the largest increase in neurite outgrowth in the overexpression screen and was thus identified as a top candidate for further study. To probe STAT3 expression and activity in vivo, untreated autologous DRG, cerebellum and cerebral cortex lysates were probed for total STAT3 by Western blotting using neuronal specific βIII-tubulin as a loading control. The alpha (89kDa) and beta (80kDa) isoforms of STAT3 were detected at higher levels in DRG compared to the two CNS tissues (Figure 4A).
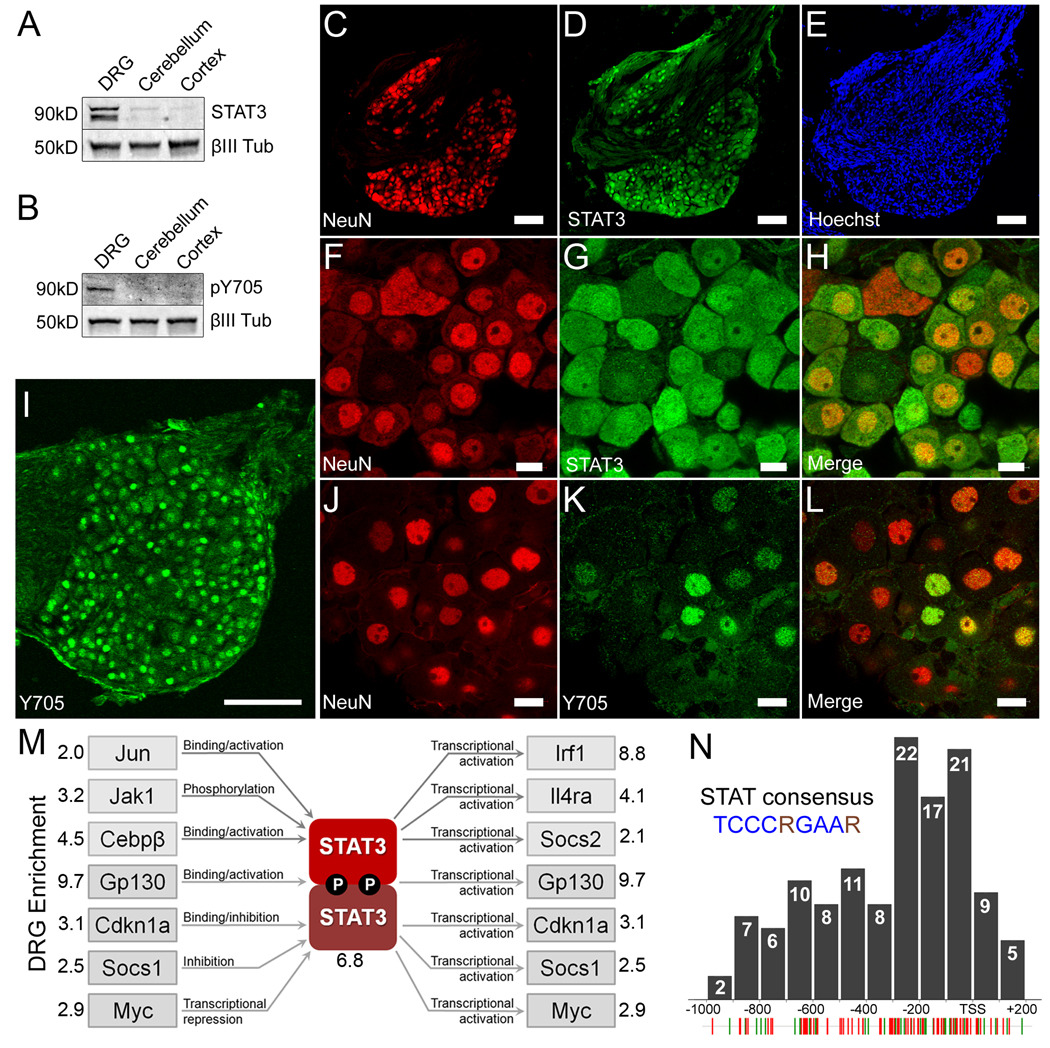
Figure 4. STAT3 is constitutively enriched and active in PNS neurons but not CNS neurons.
(a–b) Representative Western blot of lysates from P7-10 mouse dorsal root ganglia, cerebellum, and cerebral cortex probed for (a) total STAT3 and (b) Y705 phosphorylated STAT3. Neuronal specific βIII-tubulin was used as a loading control. (c–e) STAT3 co-localizes with NeuN but not Hoechst positive Schwann cells in the nerve root. (c) NeuN, (d) total STAT3, (e) the nuclear dye Hoechst. (f–h) High magnification confocal micrograph of (f) NeuN, (g) total STAT3, and the (h) merged image showing co-localization. (i) DRG neurons express nuclear Y705 phosphorylated STAT3 constitutively. (j–l) High resolution confocal micrographs showing (j) Islet 1 staining neuronal nuclei, (k) Y705 STAT3 and (l) the merged image. Scale bars: (c,d,e,i), 100µm; (f,g,h,j,k,l), 10µm. (m) The relative expression of 84 JAK-STAT pathway members in P7-10 dorsal root ganglia and cerebellum was probed using real-time PCR. Multiple factors that regulate and are regulated by STAT3 activity were found to be enriched in DRGs, suggesting the presence of at least one PNS specific STAT3 signaling pathway. (n) Distribution of the STAT consensus binding motif (TCCCRGAAR) within the promoter regions of 126 DRG enriched genes. Individual STAT consensus matches for genes identified by the subtraction (green hatches) and microarray (red hatches) are shown below. Many sites cluster within 200bp upstream of the transcription start site (TSS).
Immunohistochemical analysis revealed strong, specific staining of STAT3 in DRG sections and virtually no staining in autologous cerebellum sections (Figure S2A–B). STAT3 immunoreactivity was observed in cerebellar cultures, however, only in GFAP positive astrocytes (Figure S2E), consistent with previous reports (Xia et al., 2002). Within the DRG, STAT3 is selectively expressed in neurons, as evidenced by Hoechst staining of non-neuronal cells in the nerve rootlet (Figure 4C–E), as well as colabeling with NeuN (Figure 4F–H) and near identical co-expression with the TF ISL1 (Figure S2C). STAT3 expression in both the cytoplasm and in the nucleus of DRGs is consistent with previous observations in other cell types (Figure 4F–H and data not shown) (Liu et al., 2005).
To determine whether the STAT3 was constitutively active in DRGs, we made use of a phospho-specific antibody that recognizes tyrosine 705 (Y705). Phosphorylation of this residue is necessary for STAT3 dimerization and transcriptional activity, and is used extensively to assay STAT3 activity (Levy and Lee, 2002). Probing lysates of DRG, cerebellum and cortex using the Y705 antibody revealed STAT3 activation in DRG neurons, but not in cerebellum nor in cortex (Figure 4B). Interestingly, only the alpha isoform (89kDa) was found to be phosphorylated, suggesting differential roles for the alpha and beta isoforms in DRG neurons. Labeling of sections with the Y705 antibody revealed a neuronal specific signal in the DRG (Figure 4I–L) but not cerebellum (Figure S3). Unlike total STAT3 staining, however, Y705 staining was observed only in nuclei. Additionally, only 60% of ISL1 positive neurons exhibited detectable levels of Y705 phosphorylated STAT3, with a large range of intensities observed among cells.
To further characterize the activity of the JAK/STAT pathway in DRG neurons, we performed real-time PCR on 84 related genes using Percoll purified DRG neurons and cerebellar granule neurons (Table S6). Genes assayed include upstream activators of the STATs such as GP130 (also known as IL6ST), JAK1-3, STAT1-6, as well as downstream targets of the STATs such as IRF1. Four housekeeping genes (β-glucuronidase, HSP90, GAPDH and β-actin) were used to normalize threshold cycles. In total, 19 genes were found to be significantly enriched in DRG neurons (corrected p-value <0.05), including several genes regulated by and gene products that regulate STAT3 (Figure 4M). These results suggest the existence of constitutive STAT3 pathways that are enriched in DRG neurons.
To further probe the idea that STAT3 is a constitutive regulator of DRG cell function, we examined the distribution of 126 STAT TFBSs (TCCCRGAAR) in the promoters of the genes identified by subtractive hybridization and microarray. The majority of STAT regulatory elements (REs) were found to be within 400 hundred base pairs upstream of the TSS (Figure 4N), a phenomenon widely observed in promoter studies (Tharakaraman et al., 2008), which is also consistent with a role for STAT3 in DRG cellular function.
STAT3 activity enhances CNS neurite outgrowth
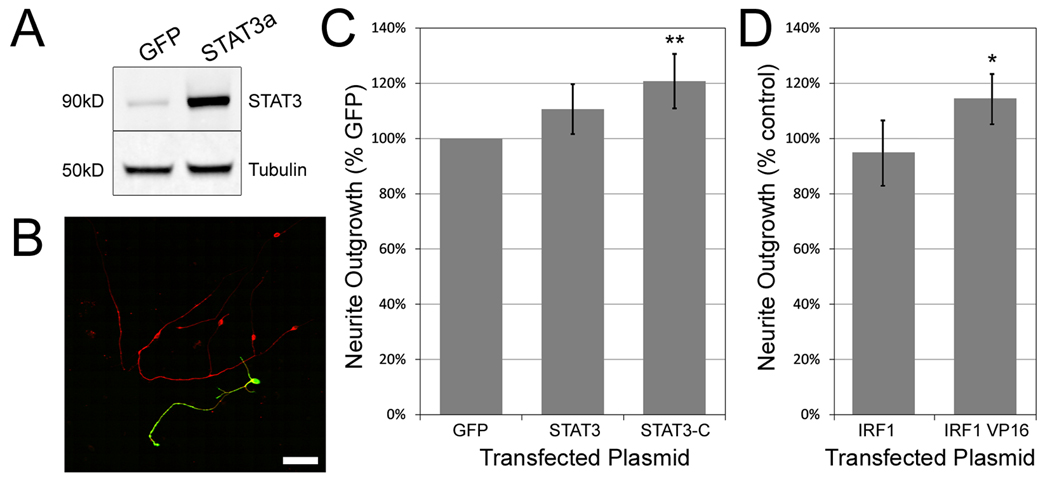
Since we had evidence that STAT3 promotes neurite outgrowth in the CNS (Figure S4D), and evidence that STAT3 may be a constitutive regulator of function in DRG neurons (Figure 4M) we conducted a second, targeted neurite outgrowth experiment to determine whether STAT3 activity would augment neurite outgrowth in cerebellar granule neurons. Postnatal day 7–10 CGNs were transfected with eGFP, wildtype full length STAT3 alpha (Figure 5A), or constitutively active A662C/N664C STAT3-C (Bromberg et al., 1999), and allowed to grow for 48 hours on poly-D-lysine or poly-D-lysine with laminin. Because STAT3 expression was virtually undetectable in non-transfected CGNs, we were able to use the pan-STAT3 antibody to identify transfected neurons (Figure 5B), using non-transfected wells to establish a threshold for background fluorescence.
Figure 5. Exogenous expression of constitutively active STAT3 increases outgrowth of CGNs.
(a) Western blot of COS7 cells transfected with GFP and full length STAT3 cDNA stained for total STAT3 and βIII-tubulin. (b) Confocal micrograph showing a single cerebellar granule neuron transfected with the STAT3 construct. After transfection, cultures were probed with antibodies for βIII-tubulin (red) and total STAT3 (green). Scale bar: 50µm. (c) Mean total neurite length of CGNs transfected with GFP, wild type STAT3 and constitutively active STAT3-C, and grown on poly-D-lysine for 48 hours. Only STAT3-C significantly improved outgrowth. At least 100 STAT3 positive neurons were quantified in each experiment, n=4. Error bars = 95% confidence intervals. Significance determined by one way ANOVA followed by Dunnett’s post-hoc test, ** = p<0.01. (d) Overexpression of a constitutively active form of IRF1, IRF1-VP16, increased mean total neurite outgrowth compared to the wildtype form of the gene. IRF1 is a major downstream target of STAT3. Significance was determined by an unpaired two-tailed Student’s t-test, * = p<0.05.
Transfection of STAT3-C, but not wildtype STAT3, resulted in a significant (20%, p<0.01) increase in total neurite outgrowth over GFP controls (Figure 5C). This effect was not mirrored by a significant increase in the fraction of neurons with neurites, suggesting that it reflects a change in outgrowth rather than survival (data not shown). These data suggest that STAT3 activity, presumably through the transcriptional regulation of specific gene targets, increases the rate of growth of postnatal CNS neurons.
One of these potential targets is interferon regulatory factor 1 (IRF1), a transcription factor induced by JAK-STAT signaling and specifically STAT3 (Harroch et al., 1994). We identified IRF1 as a candidate DRG intrinsic factor in Table 1, owing to its presence in the Subtraction as well as the network and TFBS analyses. Furthermore, our real-time PCR experiment revealed IRF1 mRNA to be nearly 9-fold enriched in DRG neurons. This result is supported by Allen Spinal Cord Atlas (http://mousespinal.brain-map.org) in-situ hybridization data from postnatal day 4 mice that clearly shows an enrichment of IRF1 mRNA in the DRG compared to neighboring spinal cord. To determine whether IRF1 activity affects neurite outgrowth, we overexpressed wild-type mouse IRF1 in addition to a constitutively active form of the factor fused to a VP16 activation domain. We observed that the constitutively active form significantly enhanced neurite outgrowth by 27% compared to the wild-type factor (p<0.05, Figure 5D). Together these findings support the idea that TF activity is key to process of axon regeneration and that overexpression screens may miss many important factors due to the absence of key signaling molecules and co-activators.
Discussion
PNS intrinsic transcription factors
Our systems-based discovery effort identified TFs predicted to be enriched and active in postnatal PNS neurons. Lists of DRG enriched genes relative to CGN were generated by two independent expression analyses were scanned for the overrepresentation of TF interaction networks and TFBSs. Combining these approaches, we identified 32 TFs, many of which were identified neither by subtraction nor by microarray methods, suggesting preferential activity in the PNS rather than enrichment of expression levels. Interestingly, analysis of microarray data on expression of these TFs in DRG compared to cortex and hippocampus showed a strong bias for higher expression in the DRG, arguing that our combined bioinformatic approach successfully identified TFs enriched in the PNS.
Our list of 32 candidate TFs contains several previously known regeneration associated gene (RAG) products, including STAT3, JUN, ATF3, and KLF6. STAT3 is phosphorylated in the axons and somas of DRG neurons after peripheral nerve injury (Sheu et al., 2000). JUN expression is elevated in PNS neurons after injury, regulates other regeneration associated genes, and acts to promote regeneration after injury (Herdegen et al., 1991; Pradervand et al., 2004; Raivich et al., 2004). ATF3 is induced after peripheral nerve injury (Tsujino et al., 2000) and is intrinsically linked to the growth rate of adult DRG neurons (Seijffers et al., 2007). KLF6 was recently identified as a promoter of mouse cortical neuron outgrowth from a screen conducted in our laboratory (Blackmore et al., 2010; Moore et al., 2009). The identification of these factors in part validates the bioinformatics approaches used to construct Table 1.
Seven transcriptional regulators stand out due to their identification in multiple approaches listed in Table 1. These include STAT3, JUN, CCAAT/enhancer binding protein zeta (C/EBPζ), breast cancer 1 (BRCA1), interferon regulator factor 1 (IRF1), the myelocytomatosis viral oncogene (MYC), and STAT6. Each of these factors has been shown to physically interact or transcriptionally regulate at least one of the other factors. IRF1 is a major downstream target of the STAT family of TFs and is regulated by STAT3 (Harroch et al., 1994). JUN is a binding partner of STAT3, and the two cooperate in the induction of several genes, including α2-macroglobulin (Ito et al., 1989) and vasoactive intestinal polypeptide (Lewis et al., 1994). MYC is a target of both IRF1 (Dror et al., 2007) and STAT3 (Kiuchi et al., 1999), and can cooperate with STAT3 in the transcription of CDC25A (Barre et al., 2005). MYC binds to the promoters of STAT3 (Zeller et al., 2006) and STAT6 (Fernandez et al., 2003). C/EBPζ, whose own transcriptional activity can be enhanced by BRCA1 (MacLachlan et al., 2000), can bind to JUN and enhance the activity of the AP-1 complex (Ubeda et al., 1999). The high degree of co-operation among these TFs, many which are regeneration associated, and our finding that many of them are enriched in DRGs (Figure 4M) suggests they may contribute to a larger transcriptional complex regulating intrinsic growth ability.
We performed a real-time PCR analysis of 84 genes related to the JAK-STAT pathway to shed light on possible PNS specific networks. Although our goal was to determine the possibility and extent of constitutive STAT signaling in the postnatal PNS, this group of genes contained 10 of 32 TFs from Table 1. The majority of these factors (6/10) were confirmed to be enriched in the PNS by real time PCR, whereas the remaining four factors were found to be enriched in CGNs (n=3, corrected p<0.05). These four factors were identified by the network and TFBS enrichment analyses only, suggesting the possibility of transcriptional repression, rather than activation, in DRG neurons.
High-Content Analysis of TF overexpression in CGNs
We performed an overexpression screen of 25 transcription factors from Table 1 in cerebellar granule neurons. Although we identified trends within this data set leading to further experiments characterizing STAT3, no single factor was found to significantly increase or decrease neurite outgrowth. Notably, we found that KLF7 had qualitatively similar effects on CGN neurite outgrowth as in cortical neurons (Moore et al., 2009), but its effects on neurite length in CGNs was not as robust as in cortical neurons (±15% in CGNs vs. ±50% in cortical neurons), possibly reflecting cell specific mechanisms for promoting/decreasing axon growth. Interestingly, KLF6 was not sufficient to promote CGN neurite outgrowth, suggesting distinct growth mechanisms in different populations of CNS neurons. ATF3, which has been shown to mediate intrinsic growth capacity in DRG neurons, produced a reduction in CGN neurite outgrowth, furthering the idea that PNS enriched regeneration associated factors do not necessarily recapitulate their effects in CNS neurons.
STAT3 as a constitutive regulator of DRG neuron function
Given that STAT3 was the single largest modulator of neurite outgrowth in the overexpression screen, we conducted a series of follow-up experiments to both characterize STAT3 expression and to determine the effect of its activity on neurite outgrowth. STAT3 is vital for the survival of DRG neurons during murine development (Alonzi et al., 2001), and may play a role in the induction of key genes after a conditioning lesion (Qiu et al., 2005), although this report uses an inhibitor of JAK2 (AG490) as a proxy for STAT3 loss of function. We report here that STAT3 is both enriched and preferentially active in DRG neurons compared to CNS neurons, and that STAT3 Y705 is phosphorylated constitutively, independent of a peripheral injury. Furthermore, overexpression of constitutively active STAT3 leads to a significant increase in CNS neurite outgrowth not attributable to an increase in survival.
Immunohistochemical and western blot analysis of postnatal DRG neurons consistently showed enrichment of STAT3 compared to cortex and cerebellum in vivo. The only time STAT3 expression was observed in postnatal CNS tissues was in GFAP positive cerebellar astrocytes in vitro (Figure S2E). This data is consistent with published reports showing induction of STAT3 in astrocytes after a lesion to the entorhinal cortex (Xia et al., 2002). Although adult cortical neurons appear not to express STAT3, even after an injury, several reports have shown expression and phosphorylation of STAT3 in the retina after an optic nerve crush or lens injury, and after treatment with ciliary neurotrophic factor (CNTF) (Leibinger et al., 2009; Muller et al., 2007; Park et al., 2004). Furthermore, these reports show both total and phospho-STAT3 in the ganglion cell layer (GCL) after injury, suggesting that retinal ganglion cells may have distinct molecular mechanism governing regeneration.
STAT3 activates more than 100 downstream targets, several of which play key roles in neuronal outgrowth. Small proline rich protein 1a (SPRR1A), which is activated in response to IL-6 and GP130 (Pradervand et al., 2004), is upregulated in the PNS after a conditioning lesion and is necessary for the outgrowth of DRG axons from explants in vitro (Bonilla et al., 2002). WNT5A, which has been shown to regulate axon specification through PKC (Zhang et al., 2007), and p21/Cip1/Waf1, which can increase neurite outgrowth threefold through the inhibition of Rho-kinase (Tanaka et al., 2002), are also a direct targets of STAT3 (Coqueret and Gascan, 2000; Fujio et al., 2004). Another downstream target of STAT3 is IRF1, which we showed independently to be enriched and potentially active in DRG neurons compared to CGNs. We further report here that the overexpression of IRF1 containing a VP16 activation domain significantly enhanced neurite outgrowth, suggesting a potential mechanism that is consistent with STAT3-C’s effects. This is to our knowledge the first report both of preferential IRF1 expression in the PNS and a role for the factor in CNS neurite outgrowth.
Although STAT3 is constitutively expressed and activated in DRG neurons, its mRNA is upregulated as much as 3 fold after injury (Schwaiger et al., 2000). Furthermore, STAT3 is phosphorylated on Y705 subsequent to injury, first in distal axons and then in the cell bodies (Lee et al., 2004). That STAT3 can upregulate GP130 (O'Brien and Manolagas, 1997) suggests the potential for a positive feedback loop that relies on basal levels of STAT3. Our findings are consistent with the idea that STAT3 may serve as an early response factor in the cascade of events leading to successful axon regeneration.
We have demonstrated that overexpression of an active form of STAT3 enhances CGN neurite outgrowth by 20%. While this effect is clearly significant, it may be possible to improve STAT3 mediated neurite outgrowth in future experiments. STAT3 is known to synergize with many of the factors in Table 1, suggesting that combinations of TFs could be overexpressed to boost CNS neurite outgrowth. Furthermore, exogenously expressed STAT3 staining in CGNs differs from endogenous DRG patterns, with notably less nuclear localization (data not shown). Assuming that STAT3 is acting in the nucleus to increase growth, it may be possible to alter nuclear import pathways to boost target gene expression. To enter the nucleus STAT3 must first bind to α3 (Liu et al., 2005), α5 or α7 importin (Ma and Cao, 2006). The STAT3/α-importin complex binds to β1-importin, which shuttles STAT3 into the nucleus. While we observed no differences in the level of α importin mRNA between DRG and CGN by microarray analysis, β1 importin mRNA is more than 3 fold upregulated in DRG neurons. This is consistent with a report showing that β1 importin mRNA is present at high levels in DRG axons and is locally translated after a peripheral injury (Hanz et al., 2003).
In summary, we took a systems approach to identify TFs preferentially enriched and active in PNS neurons, which have the capacity to extend axons through myelin rich environments and to regenerate after injury. Unexpectedly, high content analysis of TF overexpression in primary CNS neurons revealed that none of these factors could individually produce a significant effect on neurite outgrowth. However, constitutively active forms of STAT3 and its target, IRF1, are sufficient to promote neurite growth in the same model, confirming that transcription factor activity is a driving force of functional outcomes, at least in vitro. Furthermore, these results suggest that combinatorial TF transfection (of co-activators, for example) could be a valuable potential therapeutic approach for tackling CNS injuries.
Materials and Methods
Construction of Directionally Cloned cDNA Libraries
Approximately 1µg of P7-10 wild type C57BL/6 mouse cerebellum and DRG poly(A)+ purified mRNA was directionally cloned into the Stratagene Lambda Zap-CMV XR vector (Agilent Technologies) and packaged into lambda particles using Gigapack III Gold Packaging extracts (Stratagene). Mass library-wide excision of the phagemid resident in the lambda library was carried out using reagents and protocols provided by the Lambda Zap-CMV XR vendor (Stratagene), yielding a pCMV-Script EX mammalian-expression phagemid library.
DRG Library Subtraction
Excised mammalian expression libraries in pCMV-Script EX were used to carry out subtractions (DRG – cerebellum) essentially as previously described (Bonaldo et al., 1996). In brief, purified double-stranded plasmid from the DRG and cerebellum libraries were converted to single-stranded circles by the sequential enzymatic action of Gene II (Invitrogen) and Exo III. Subtractions are carried out to Cot = 50 by hybridization of 100ng of DRG single-stranded phagemid circles with 2.5µg of purified PCR cerebellum driver at 30°C for 88 hours. Sequences present in all members of the PCR driver population such as flanking MCS, flanking vector, etc. were prevented from hybridizing to the single-stranded DRG circles. Subtracted hybridizations were processed by Hydroxyapatite (HAP) chromatography to remove double-stranded cDNAs resulting from hybridization of DRG single-stranded circles to complementary cerebellum PCR products. Recovered single-stranded DNA was desalted, concentrated and used to generate partially double-stranded plasmids which were electroporated into DH10B bacterial cells to generate the subtracted library (Bonaldo et al., 1996).
Library sequencing
Diluted library glycerol stocks were inoculated in deep 96 well blocks (Qiagen’s HiSpeed Miniprep kit) filled with 1.4mL of 2xYT media + 50µg/ml kanamycin and shaken at 310RPM for 16 hours at 37°C. The Qiagen BioRobot 3000 was used purify and elute library sequences in 100µl of EB, after which the average concentration of three wells was determined (usually 300 ng/µl). Sequencing reactions were carried using the BigDye Terminator 3.1 kit (Applied Biosystems), then sealed and shipped to the W.M. Keck Center at the University of Illinois (http://www.biotech.uiuc.edu/centers/Keck) for sequencing.
Microarray analysis
CEL data from laser capture microdissected DRG neurons (Peeters et al., 2006) was generously provided to us by Pieter Peeters of Johnson & Johnson. Microarray data from P11 mouse cerebellum using the same platform (Affymetrix Murine Genome U74AV2) was obtained by our lab separately and has been described previously (Tapanes-Castillo et al., 2010).
All samples were normalized using the Robust Multi-Array average method (Irizarry et al., 2003) using the software package RMA Express (http://rmaexpress.bmbolstad.com). Normalized intensities were pooled in Spotfire DecisionSite (TIBCO Software Inc.) and mean DRG/Cerebellum ratios were calculated. Significance was determined by an unpaired two tailed Student’s T-test, followed by a correction of p-values for multiple comparisons using the Benjamini-Hochberg False Discover Rate (FDR) technique (Hochberg and Benjamini, 1990). Significantly upregulated genes (i.e. >2 fold differentially regulated, p<0.01) were included in further analyses.
For the analysis of GNF GeneAtlas V3 data, the data matrix file containing gcrma computed expression measures was downloaded from GEO (accession: GSE10246), whereupon replicate values were averaged. DRG/Cortex and DRG/Hippocampus ratios were calculated for each probe set, and that value was then averaged for each gene (if multiple probe sets were available) and log2 transformed.
Gene ontology clustering
1,086 UniGene IDs from the subtraction along with 1,252 and 892 Affymetrix probe IDs upregulated in DRGs and cerebellum respectively were imported into DAVID (Dennis et al., 2003) (http://david.abcc.ncifcrf.gov). Annotations overrepresented in the two DRG enriched gene lists were then determined using the EASE algorithm, which uses a modified Fisher’s Exact Test to determine significance. A list of all annotations and their associated p-values was determined for the cerebellar enriched microarray list for comparison. DAVID requires that a reference gene list be used to establish a background for annotations. For the subtraction, the default DAVID Mus Musculus list was used, whereas for the microarray a list of all genes represented on the U74Av2 gene chip was used.
Network analysis
Subtraction and microarray gene lists were imported into Metacore (GeneGo) and converted into Metacore network objects. List genes were then connected to each of 1,773 transcriptional regulators in the Metacore database using the “Transcriptional Regulation” tool. To determine the significance of connectivity between a gene list and a given transcription factor, Metacore considers the problem as a selection without replacement. In this test the intersection between a user’s list and a generic network is assumed to follow a hypergeometric distribution and the probability that a subset of a network of size n to include r genes from a user’s gene list is:
Where R is the total number of genes with the users list and N is the total number of genes in the database. From this equation, the mean and dispersion of the distribution can be calculated and used to generate a p-value, equivalent to the probability that the intersection of two randomly selected subsets of N would have a size n or larger. In the case of a transcription factor centered network, Metacore builds an arbitrary network of size n around the factor and then compares the network with the genes in R. Metacore p-values were corrected for multiple comparisons using the Benjamini-Hochberg technique.
Promoter analysis
The transcriptional start site (TSS) for each gene from the subtraction and microarray list was determined from Entrez Gene (http://www.ncbi.nlm.nih.gov/gene). Custom software was then used to extract sequence 1000bp upstream and 200bp downstream of the TSS for each gene using genomic contig files (Mus Musculus genome build 36.1). Promoters were then matched against two consensus lists: Transfac 10.2 (Biobase GmbH) and a list of conserved mammalian promoter elements reported previously(Xie et al., 2005). To establish a background match rate, a list of all mouse Entrez Gene genes containing a transcription start site annotation was used (ftp://ftp.ncbi.nih.gov/gene/DATA/). This background list was used for both the subtraction and microarray gene lists because it was assumed there was no bias towards transcription start site locations in the microarray gene sample. A two-tailed Fisher’s Exact Test was used to determine the significance of a consensus matching a gene list, followed by a correction for multiple comparisons as described above.
Antibodies
The following antibodies were used for experiments in this paper: Total and Y705 phosphorylated STAT3 antibodies (#9132, Cell Signaling Technology), 1:250 for immunohistochemistry and 1:500 for Western blotting; E7 monoclonal tubulin antibody (Developmental Studies Hybridoma Bank) was used at 1:1000 to stain neurites for automatic tracing; chick anti-BIII tubulin (PRB-435P/TuJ1, Covance Inc.) was used at 1:10,000 to determine neuronal loading by Western blot; monoclonal anti-GFAP (#G3893, Sigma-Aldrich-Aldrich) was used at 1:500 to identify astrocytes; NeuN antibody (MAB377, Chemicon) was used at 1:1000; neural crest marker anti-Islet1 (clone 39.4D5, DHSB) was used at 1:500.
Cell culture
P7-10 C57BL/6 mice were euthanized and their DRGs and cerebellum were removed. Neurons were dissociated and plated on 18mm glass coverslips (Carolina Biological Supply) or 96 well Packard plates (Perkin Elmer) coated with 100µg/ml poly-D-lysine (Sigma-Aldrich) alone or with 4µg/ml laminin (Sigma-Aldrich). Neurons were cultured in defined Neurobasal medium containing 100 U/ml penicillin and 100µg/ml streptomycin, 5µg/ml insulin , 100µM sodium pyruvate, 200µM L-glutamine , B27 supplement (Invitrogen); 4µg/ml triiodo-thyronine, 50µg/ml N-acetyl-L-cysteine, 100µg/ml transferrin, 100µg/ml bovine-serum albumen, 63ng/ml progesterone, 16µg/ml putrescine and 400ng/ml sodium selenite (Sigma-Aldrich). When making comparisons between DRGs and CGNs, 50ng/ml of NGF was added to each well (Sigma-Aldrich). To improve CGN transfection efficiency and cell survival after transfection, we added 10ng/ml forskolin to the plating media (Sigma-Aldrich). Transfected CGNs also received 25mM KCl (Sigma-Aldrich) to improve cell viability. Cells were grown at 37°C and 5% CO2, prior to fixation with 4% paraformaldehyde for 48 hours.
High Content Analysis of transcription factor overexpression
Cells were transfected using the Amaxa Rat Neuron 96-well Nucleofector® Kit according the manufacturers recommendations (Amaxa Biosystems). Briefly, 70,000 cells per condition were resuspended in 20µl of rat neuron Nucleofector solution and mixed with 0.2µg of reporter (pmaxEGFP) and 1.2µg of plasmid DNA (OpenBiosystems, Thermo Scientific) using the rat neuron high efficiency program within the 96-well Shuttle™ system. Approximately 4900 cells per well were plated in 96-well tissue culture dishes (Perkin-Elmer) coated with poly-d-lysine and laminin (Sigma-Aldrich). Cells were grown for 48 hours and then fixed for 30 minutes at room temperature with a 4% formaldehyde and 4% sucrose solution (Sigma-Aldrich), incubated in blocking/permeabilization buffer containing 20% normal goat serum (Invitrogen), 0.3% Triton X-100, 18mg/ml L-lysine and 10mg/ml BSA (Sigma-Aldrich). The Cellomics ArrayScanVTI performed automated image acquisition and neurite tracing (Neuronal Profiling BioApplication v3.5; Thermo Scientific). Tracing data was imported into Spotfire DecisionSite for further analysis (Tibco Inc.). The ratio of reporter gene to plasmid DNA (1:6) typically yielded co transfection rates of ≥70%.
Western blotting
Freshly dissected whole DRGs, cerebellum and cortex from P7-10 C57BL/6 mice were suspended in lysis buffer containing 1% NP-40, 200µM sodium vanadate and 50µM sodium fluoride, followed by sonication. Dilutions of each sample were run on an 8–15% gradient gel (Lonza Inc.) and probed for BIII tubulin to determine neuronal loading. Densitometric analysis was then used to adjust sample loading for a second gel. STAT3 and BIII tubulin signals were read simultaneously using the Odyssey infrared imager (LI-COR Inc.).
Immunohistochemistry
P7-10 C57BL/6 mice were perfused with saline followed by 4% formaldehyde solution, whereupon their DRG and cerebellum were removed. The tissue was then post-fixed overnight, followed by equilibration in 30% sucrose. The cerebellum and DRGs of the same mouse were mounted into the same OCT block and sectioned by cryostat at a thickness of 20µm and places on slides. Because formaldehyde masks the Y705 antigen, we used a simple antigen retrieval technique described previously (Jiao et al., 1999) , consisting of a 30 minute incubation in 25mM sodium citrate at 80°C. Sections were incubated for 2 hours in blocking/permeabilization buffer containing 20% normal goat serum (Invitrogen), 0.2% Triton X-100, 18mg/ml L-lysine and 10mg/ml BSA (Sigma-Aldrich). Slides were then incubated in primary antibody overnight and then secondary antibody for a second night, followed by mounting with ProLong Gold anti-fade reagent (Invitrogen). Sections were imaged by confocal microscopy.
Real-time PCR
Dorsal root ganglia and cerebellum from P7-10 wildtype C57BL/6 mice were removed using standard protocols. Dissociated DRG cells were purified by layering dissociated cells on top of a 28% isotonic Percoll solution, as described previously (Tucker et al., 2006), followed by centrifugation for 10 minutes at 400g and 4°C. To determine purity, neurons were cultured for 18 hours and then fixed and stained for anti neurofilament-medium (160 kDa), followed by quantification using the Cellomics Kineticscan plate reader. For real-time PCR, cell pellets were resuspended in Buffer RLT from the Qiagen RNAeasy micro kit. One microgram of total RNA was then reverse transcribed into first strand cDNA using the “ReactionReady” First Strand cDNA Synthesis Kit from Superarray (SA Biosciences), followed by incubation at 95°C to hydrolyze any remaining RNA. Diluted first strand product was then combined with SYBR-Green master mix from Superarray and aliquoted into Jak/Stat RT² Profiler plates, containing primers for 84 genes of interest and 5 housekeeping genes, as well as genomic contamination and quality controls. PCR reactions were run using an Eppendorf MasterCycler Realplex (Eppendorf AG), and threshold cycles were determined by the machine. After the reaction was complete, a melt curve was determined to confirm the presence of a single PCR product. The fold change for a gene of interest (GOI) using a housekeeping gene (HKG) for normalization was determined using the “ΔΔCt” method (Livak, 1997). Significance was determined using an unpaired two-tailed Student’s T-test on normalized ΔCt values, followed by Benjamini-Hochberg correction for multiple comparisons.
STAT3 outgrowth assay
1 million cells per condition were resuspended in 100µl mouse neuron Nucleofector solution (Amaxa GmbH) and 3µg of plasmid DNA was added. Transfected plasmids include: wildtype STAT3 alpha isoform (3593588, OpenBiosystems), A662C/N664C mutant “STAT3-C”, and pmaxGFP as a negative control. Cells were electroporated using program G-013 on the Amaxa Nucleofector device, after which 900µl of Neurobasal media with added KCL (described earlier) was added to each cuvette to dilute the Nucleofector solution and improve survival. Cells were then plated at a density of 20–30,000 cells per 96 well, and processed as described above; with the exception that anti-STAT3 was used to identify transfected cells.
IRF1 outgrowth assay
Wildtype Irf1 (3600525, OpenBiosystems) was compared to a mutant Irf1 construct containing an N-terminal VP16 activation domain fusion. Total neurite length was determined as described in the high content neurite outgrowth screen.
Supplementary Material
Acknowledgements
This work was supported by the Miami Project to Cure Paralysis, the Buoniconti Fund, DOD W81XWH-05-1-0061, grant no. 2396 from the Paralyzed Veterans of America Research Foundation and NIH HD057632 and NS059866. W. Buchser is a recipient of Lois Pope LIFE Scholar award. V. Lemmon holds the Walter G. Ross Distinguished Chair in Developmental Neuroscience at the University of Miami. The authors would like to thank Pieter J. Peeters for his generous gift of the raw LCM DRG microarray data files.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen Spinal Cord Atlas [Internet] Seattle (WA): ©2009. Available from: http://mousespinal.brain-map.org. [Google Scholar]
- Alonzi T, Middleton G, Wyatt S, Buchman V, Betz UA, Muller W, Musiani P, Poli V, Davies AM. Role of STAT3 and PI 3-kinase/Akt in mediating the survival actions of cytokines on sensory neurons. Molecular and cellular neurosciences. 2001;18:270–282. doi: 10.1006/mcne.2001.1018. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barre B, Vigneron A, Coqueret O. The STAT3 transcription factor is a target for the Myc and riboblastoma proteins on the Cdc25A promoter. The Journal of biological chemistry. 2005;280:15673–15681. doi: 10.1074/jbc.M413203200. [DOI] [PubMed] [Google Scholar]
- Birmachu W, Gleason RM, Bulbulian BJ, Riter CL, Vasilakos JP, Lipson KE, Nikolsky Y. Transcriptional networks in plasmacytoid dendritic cells stimulated with synthetic TLR 7 agonists. BMC Immunol. 2007;8:26. doi: 10.1186/1471-2172-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmore MG, Moore DL, Smith RP, Goldberg JL, Bixby JL, Lemmon VP. High content screening of cortical neurons identifies novel regulators of axon growth. Mol Cell Neurosci. 2010;44:43–54. doi: 10.1016/j.mcn.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. Bioinformatics. Vol. 19. Oxford, England: 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias; pp. 185–193. [DOI] [PubMed] [Google Scholar]
- Bonaldo MF, Lennon G, Soares MB. Normalization and subtraction: two approaches to facilitate gene discovery. Genome research. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- Bonilla IE, Tanabe K, Strittmatter SM. Small proline-rich repeat protein 1A is expressed by axotomized neurons and promotes axonal outgrowth. J Neurosci. 2002;22:1303–1315. doi: 10.1523/JNEUROSCI.22-04-01303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Cao W, Epstein C, Liu H, DeLoughery C, Ge N, Lin J, Diao R, Cao H, Long F, Zhang X, Chen Y, Wright PS, Busch S, Wenck M, Wong K, Saltzman AG, Tang Z, Liu L, Zilberstein A. Comparing gene discovery from Affymetrix GeneChip microarrays and Clontech PCR-select cDNA subtraction: a case study. BMC genomics. 2004;5:26. doi: 10.1186/1471-2164-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J, Burne JF, Winter J, Bartlett P. Retinal ganglion cells lose response to laminin with maturation. Nature. 1986;322:465–467. doi: 10.1038/322465a0. [DOI] [PubMed] [Google Scholar]
- Coqueret O, Gascan H. Functional interaction of STAT3 transcription factor with the cell cycle inhibitor p21WAF1/CIP1/SDI1. J Biol Chem. 2000;275:18794–18800. doi: 10.1074/jbc.M001601200. [DOI] [PubMed] [Google Scholar]
- Davies SJ, Fitch MT, Memberg SP, Hall AK, Raisman G, Silver J. Regeneration of adult axons in white matter tracts of the central nervous system. Nature. 1997;390:680–683. doi: 10.1038/37776. [DOI] [PubMed] [Google Scholar]
- Davies SJ, Goucher DR, Doller C, Silver J. Robust regeneration of adult sensory axons in degenerating white matter of the adult rat spinal cord. J Neurosci. 1999;19:5810–5822. doi: 10.1523/JNEUROSCI.19-14-05810.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:3. [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror N, Alter-Koltunoff M, Azriel A, Amariglio N, Jacob-Hirsch J, Zeligson S, Morgenstern A, Tamura T, Hauser H, Rechavi G, Ozato K, Levi BZ. Identification of IRF-8 and IRF-1 target genes in activated macrophages. Molecular immunology. 2007;44:338–346. doi: 10.1016/j.molimm.2006.02.026. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes & development. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujio Y, Matsuda T, Oshima Y, Maeda M, Mohri T, Ito T, Takatani T, Hirata M, Nakaoka Y, Kimura R, Kishimoto T, Azuma J. Signals through gp130 upregulate Wnt5a and contribute to cell adhesion in cardiac myocytes. FEBS Lett. 2004;573:202–206. doi: 10.1016/j.febslet.2004.07.082. [DOI] [PubMed] [Google Scholar]
- Goldberg JL, Klassen MP, Hua Y, Barres BA. Science. Vol. 296. New York, N.Y.: 2002. Amacrine-signaled loss of intrinsic axon growth ability by retinal ganglion cells; pp. 1860–1864. [DOI] [PubMed] [Google Scholar]
- Gutierrez H, O'Keeffe GW, Gavalda N, Gallagher D, Davies AM. Nuclear factor kappa B signaling either stimulates or inhibits neurite growth depending on the phosphorylation status of p65/RelA. J Neurosci. 2008;28:8246–8256. doi: 10.1523/JNEUROSCI.1941-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, Fainzilber M. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- Harroch S, Revel M, Chebath J. Induction by interleukin-6 of interferon regulatory factor 1 (IRF-1) gene expression through the palindromic interferon response element pIRE and cell type-dependent control of IRF-1 binding to DNA. The EMBO journal. 1994;13:1942–1949. doi: 10.1002/j.1460-2075.1994.tb06463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdegen T, Kummer W, Fiallos CE, Leah J, Bravo R. Expression of c-JUN, JUN B and JUN D proteins in rat nervous system following transection of vagus nerve and cervical sympathetic trunk. Neuroscience. 1991;45:413–422. doi: 10.1016/0306-4522(91)90237-i. [DOI] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Statistics in medicine. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Biostatistics. Vol. 4. Oxford, England: 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data; pp. 249–264. [DOI] [PubMed] [Google Scholar]
- Ito T, Tanahashi H, Misumi Y, Sakaki Y. Nuclear factors interacting with an interleukin-6 responsive element of rat alpha 2-macroglobulin gene. Nucleic acids research. 1989;17:9425–9435. doi: 10.1093/nar/17.22.9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Sun Z, Lee T, Fusco FR, Kimble TD, Meade CA, Cuthbertson S, Reiner A. A simple and sensitive antigen retrieval method for free-floating and slide-mounted tissue sections. Journal of neuroscience methods. 1999;93:149–162. doi: 10.1016/s0165-0270(99)00142-9. [DOI] [PubMed] [Google Scholar]
- Kiuchi N, Nakajima K, Ichiba M, Fukada T, Narimatsu M, Mizuno K, Hibi M, Hirano T. STAT3 is required for the gp130-mediated full activation of the c-myc gene. The Journal of experimental medicine. 1999;189:63–73. doi: 10.1084/jem.189.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, Saijo K, Glass CK, Hume DA, Kellie S, Sweet MJ. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Neitzel KL, Devlin BK, MacLennan AJ. STAT3 phosphorylation in injured axons before sensory and motor neuron nuclei: potential role for STAT3 as a retrograde signaling transcription factor. The Journal of comparative neurology. 2004;474:535–545. doi: 10.1002/cne.20140. [DOI] [PubMed] [Google Scholar]
- Leibinger M, Muller A, Andreadaki A, Hauk TG, Kirsch M, Fischer D. Neuroprotective and axon growth-promoting effects following inflammatory stimulation on mature retinal ganglion cells in mice depend on ciliary neurotrophic factor and leukemia inhibitory factor. J Neurosci. 2009;29:14334–14341. doi: 10.1523/JNEUROSCI.2770-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DE, Lee CK. What does Stat3 do? J Clin Invest. 2002;109:1143–1148. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SE, Rao MS, Symes AJ, Dauer WT, Fink JS, Landis SC, Hyman SE. Coordinate regulation of choline acetyltransferase, tyrosine hydroxylase, and neuropeptide mRNAs by ciliary neurotrophic factor and leukemia inhibitory factor in cultured sympathetic neurons. Journal of neurochemistry. 1994;63:429–438. doi: 10.1046/j.1471-4159.1994.63020429.x. [DOI] [PubMed] [Google Scholar]
- Li M, Paik HI, Balch C, Kim Y, Li L, Huang TH, Nephew KP, Kim S. Enriched transcription factor binding sites in hypermethylated gene promoters in drug resistant cancer cells. Bioinformatics. 2008;24:1745–1748. doi: 10.1093/bioinformatics/btn256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, McBride KM, Reich NC. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. Comparative Ct method, ABI Prism 7700 Sequence Detection System. PE Applied Biosystems. 1997 [Google Scholar]
- Ma J, Cao X. Regulation of Stat3 nuclear import by importin alpha5 and importin alpha7 via two different functional sequence elements. Cellular signalling. 2006;18:1117–1126. doi: 10.1016/j.cellsig.2005.06.016. [DOI] [PubMed] [Google Scholar]
- MacGillavry HD, Stam FJ, Sassen MM, Kegel L, Hendriks WT, Verhaagen J, Smit AB, van Kesteren RE. NFIL3 and cAMP response element-binding protein form a transcriptional feedforward loop that controls neuronal regeneration-associated gene expression. J Neurosci. 2009;29:15542–15550. doi: 10.1523/JNEUROSCI.3938-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLachlan TK, Somasundaram K, Sgagias M, Shifman Y, Muschel RJ, Cowan KH, El-Deiry WS. BRCA1 effects on the cell cycle and the DNA damage response are linked to altered gene expression. The Journal of biological chemistry. 2000;275:2777–2785. doi: 10.1074/jbc.275.4.2777. [DOI] [PubMed] [Google Scholar]
- Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, Goldberg JL. KLF family members regulate intrinsic axon regeneration ability. Science. 2009;326:298–301. doi: 10.1126/science.1175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Hauk TG, Fischer D. Astrocyte-derived CNTF switches mature RGCs to a regenerative state following inflammatory stimulation. Brain. 2007;130:3308–3320. doi: 10.1093/brain/awm257. [DOI] [PubMed] [Google Scholar]
- O'Brien CA, Manolagas SC. Isolation and characterization of the human gp130 promoter. Regulation by STATS. J Biol Chem. 1997;272:15003–15010. doi: 10.1074/jbc.272.23.15003. [DOI] [PubMed] [Google Scholar]
- Park K, Luo JM, Hisheh S, Harvey AR, Cui Q. Cellular mechanisms associated with spontaneous and ciliary neurotrophic factor-cAMP-induced survival and axonal regeneration of adult retinal ganglion cells. J Neurosci. 2004;24:10806–10815. doi: 10.1523/JNEUROSCI.3532-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, He Z. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters PJ, Aerssens J, de Hoogt R, Stanisz A, Gohlmann HW, Hillsley K, Meulemans A, Grundy D, Stead RH, Coulie B. Molecular profiling of murine sensory neurons in the nodose and dorsal root ganglia labeled from the peritoneal cavity. Physiological genomics. 2006;24:252–263. doi: 10.1152/physiolgenomics.00169.2005. [DOI] [PubMed] [Google Scholar]
- Pradervand S, Yasukawa H, Muller OG, Kjekshus H, Nakamura T, St Amand TR, Yajima T, Matsumura K, Duplain H, Iwatate M, Woodard S, Pedrazzini T, Ross J, Firsov D, Rossier BC, Hoshijima M, Chien KR. Small proline-rich protein 1A is a gp130 pathway- and stress-inducible cardioprotective protein. The EMBO journal. 2004;23:4517–4525. doi: 10.1038/sj.emboj.7600454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Cafferty WB, McMahon SB, Thompson SW. Conditioning injury-induced spinal axon regeneration requires signal transducer and activator of transcription 3 activation. J Neurosci. 2005;25:1645–1653. doi: 10.1523/JNEUROSCI.3269-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Cai D, Dai H, McAtee M, Hoffman PN, Bregman BS, Filbin MT. Spinal axon regeneration induced by elevation of cyclic AMP. Neuron. 2002;34:895–903. doi: 10.1016/s0896-6273(02)00730-4. [DOI] [PubMed] [Google Scholar]
- Raivich G, Bohatschek M, Da Costa C, Iwata O, Galiano M, Hristova M, Nateri AS, Makwana M, Riera-Sans L, Wolfer DP, Lipp HP, Aguzzi A, Wagner EF, Behrens A. The AP-1 transcription factor c-Jun is required for efficient axonal regeneration. Neuron. 2004;43:57–67. doi: 10.1016/j.neuron.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Richardson PM, Issa VM. Peripheral injury enhances central regeneration of primary sensory neurones. Nature. 1984;309:791–793. doi: 10.1038/309791a0. [DOI] [PubMed] [Google Scholar]
- Schwaiger FW, Hager G, Schmitt AB, Horvat A, Hager G, Streif R, Spitzer C, Gamal S, Breuer S, Brook GA, Nacimiento W, Kreutzberg GW. Peripheral but not central axotomy induces changes in Janus kinases (JAK) and signal transducers and activators of transcription (STAT) Eur J Neurosci. 2000;12:1165–1176. doi: 10.1046/j.1460-9568.2000.00005.x. [DOI] [PubMed] [Google Scholar]
- Seijffers R, Mills CD, Woolf CJ. ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration. J Neurosci. 2007;27:7911–7920. doi: 10.1523/JNEUROSCI.5313-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu JY, Kulhanek DJ, Eckenstein FP. Differential patterns of ERK and STAT3 phosphorylation after sciatic nerve transection in the rat. Experimental neurology. 2000;166:392–402. doi: 10.1006/exnr.2000.7508. [DOI] [PubMed] [Google Scholar]
- Silver J, Miller JH. Regeneration beyond the glial scar. Nat Rev Neurosci. 2004;5:146–156. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- Sivasankaran R, Pei J, Wang KC, Zhang YP, Shields CB, Xu XM, He Z. PKC mediates inhibitory effects of myelin and chondroitin sulfate proteoglycans on axonal regeneration. Nature neuroscience. 2004;7:261–268. doi: 10.1038/nn1193. [DOI] [PubMed] [Google Scholar]
- Smith DS, Skene JH. A transcription-dependent switch controls competence of adult neurons for distinct modes of axon growth. J Neurosci. 1997;17:646–658. doi: 10.1523/JNEUROSCI.17-02-00646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PD, Sun F, Park KK, Cai B, Wang C, Kuwako K, Martinez-Carrasco I, Connolly L, He Z. SOCS3 deletion promotes optic nerve regeneration in vivo. Neuron. 2009;64:617–623. doi: 10.1016/j.neuron.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RP, Buchser WJ, Lemmon MB, Pardinas JR, Bixby JL, Lemmon VP. EST Express: PHP/MySQL based automated annotation of ESTs from expression libraries. BMC bioinformatics. 2008;9:186. doi: 10.1186/1471-2105-9-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam FJ, MacGillavry HD, Armstrong NJ, de Gunst MC, Zhang Y, van Kesteren RE, Smit AB, Verhaagen J. Identification of candidate transcriptional modulators involved in successful regeneration after nerve injury. Eur J Neurosci. 2007;25:3629–3637. doi: 10.1111/j.1460-9568.2007.05597.x. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Yamashita T, Asada M, Mizutani S, Yoshikawa H, Tohyama M. Cytoplasmic p21(Cip1/WAF1) regulates neurite remodeling by inhibiting Rho-kinase activity. J Cell Biol. 2002;158:321–329. doi: 10.1083/jcb.200202071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapanes-Castillo A, Weaver EJ, Smith RP, Kamei Y, Caspary T, Hamilton-Nelson KL, Slifer SH, Martin ER, Bixby JL, Lemmon VP. A modifier locus on chromosome 5 contributes to L1 cell adhesion molecule X-linked hydrocephalus in mice. Neurogenetics. 2010;11:53–71. doi: 10.1007/s10048-009-0203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharakaraman K, Bodenreider O, Landsman D, Spouge JL, Marino-Ramirez L. The biological function of some human transcription factor binding motifs varies with position relative to the transcription start site. Nucleic acids research. 2008;36:2777–2786. doi: 10.1093/nar/gkn137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome research. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tom VJ, Doller CM, Malouf AT, Silver J. Astrocyte-associated fibronectin is critical for axonal regeneration in adult white matter. J Neurosci. 2004;24:9282–9290. doi: 10.1523/JNEUROSCI.2120-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujino H, Kondo E, Fukuoka T, Dai Y, Tokunaga A, Miki K, Yonenobu K, Ochi T, Noguchi K. Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: A novel neuronal marker of nerve injury. Mol Cell Neurosci. 2000;15:170–182. doi: 10.1006/mcne.1999.0814. [DOI] [PubMed] [Google Scholar]
- Tucker BA, Rahimtula M, Mearow KM. Laminin and growth factor receptor activation stimulates differential growth responses in subpopulations of adult DRG neurons. The European journal of neuroscience. 2006;24:676–690. doi: 10.1111/j.1460-9568.2006.04963.x. [DOI] [PubMed] [Google Scholar]
- Ubeda M, Vallejo M, Habener JF. CHOP enhancement of gene transcription by interactions with Jun/Fos AP-1 complex proteins. Molecular and cellular biology. 1999;19:7589–7599. doi: 10.1128/mcb.19.11.7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia XG, Hofmann HD, Deller T, Kirsch M. Induction of STAT3 signaling in activated astrocytes and sprouting septal neurons following entorhinal cortex lesion in adult rats. Mol Cell Neurosci. 2002;21:379–392. doi: 10.1006/mcne.2002.1180. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu G, He Z. Glial inhibition of CNS axon regeneration. Nat Rev Neurosci. 2006;7:617–627. doi: 10.1038/nrn1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, Fu Y, Weng Z, Kuznetsov VA, Sung WK, Ruan Y, Dang CV, Wei CL. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhu J, Yang GY, Wang QJ, Qian L, Chen YM, Chen F, Tao Y, Hu HS, Wang T, Luo ZG. Dishevelled promotes axon differentiation by regulating atypical protein kinase C. Nat Cell Biol. 2007;9:743–754. doi: 10.1038/ncb1603. [DOI] [PubMed] [Google Scholar]
- Zou H, Ho C, Wong K, Tessier-Lavigne M. Axotomy-induced Smad1 activation promotes axonal growth in adult sensory neurons. J Neurosci. 2009;29:7116–7123. doi: 10.1523/JNEUROSCI.5397-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.