Abstract
The cardiorenal syndrome refers to the coexistence of kidney and cardiovascular disease, where cardiovascular events are the most common cause of death in patients with chronic kidney disease. Both, cardiovascular as well as kidney diseases have been extensively analyzed on a molecular level, resulting in molecular features and associated processes indicating a cross-talk of the two disease etiologies on a pathophysiological level. In order to gain a comprehensive picture of molecular factors contributing to the bidirectional interplay between kidney and cardiovascular system, we mined the scientific literature for molecular features reported as associated with the cardiorenal syndrome, resulting in 280 unique genes/proteins. These features were then analyzed on the level of molecular processes and pathways utilizing various types of protein interaction networks. Next to well established molecular features associated with the renin-angiotensin system numerous proteins involved in signal transduction and cell communication were found, involving specific molecular functions covering receptor binding with natriuretic peptide receptor and ligands as well known example. An integrated analysis of identified features pinpointed a protein interaction network involving mediators of hemodynamic change and an accumulation of features associated with the endothelin and VEGF signaling pathway. Some of these features may function as novel therapeutic targets.
1. Introduction
The risk of developing cardiovascular disease (CVD) is dramatically increased in patients with chronic kidney diseases (CKDs). Mortality as a consequence of cardiovascular events is 10 to 30 times higher in patients on dialysis treatment than in the general population [1]. Due to this recognition of CVD as the leading cause of morbidity and mortality in patients with reduced kidney function, a growing body of literature has become available regarding this link of CKD and CVD, termed as cardiorenal syndrome (CRS).
CRS can be classified into five subtypes depending on the origin of damage (either the cardiovascular system or the kidney) and the course of disease (either acute or chronic) [2, 3]. Major mechanisms leading to CRS1 and CRS2 (acute and chronic cardiorenal syndrome) include hemodynamically mediated damage, hormonal factors, immune-mediated damage, low cardiac output, endothelial dysfunction, and chronic hypoperfusion. Hallmarks of kidney dysfunction leading to CRS3 and CRS4 (acute and chronic renocardiac syndrome) on the other hand are volume expansion, drop of the glomerular filtration rate, humoral signaling, anemia, uremic toxins, and inflammation. The fifth subtype of the cardiorenal syndrome (CRS5) describes the secondary cardiorenal syndrome which refers to systemic diseases such as diabetes that ultimately lead to simultaneous cardiovascular and kidney dysfunction.
The multitude of cardiac risk factors in patients with chronic kidney disease is complex and increases with age, the stage of kidney disease, and the level of proteinuria. Another powerful risk factor is hypertension which goes along with sodium retention, and activation of the renin-angiotensin system. Atherosclerosis results from an impairment of endothelial function which, in turn, is associated with albuminuria. Changes in blood-lipid composition and oxidative stress as a consequence of inflammation due to renal dysfunction also contribute to endothelial dysfunction and subsequent CVD [4].
Management and therapy of the CRS is challenging since drugs in use for the treatment of cardiovascular diseases may go along with impairment of kidney function and vice versa. Examples include diuretics, ionotropes, angiotensin-converting enzyme inhibitors, angiotensin receptor blockers, or natriuretic peptides but treatment decision must be based on a combination of individual patient information and understanding of individual treatment options [5].
Biomarkers of relevance in the context of the CRS mainly hold proteins known either in the field of nephrology or cardiology, for the latter including, for example, the family of natriuretic peptides and troponins, whereas frequently reported renal-specific markers include neutrophil gelatinase-associated lipocalin (NGAL), kidney injury molecule 1 (KIM1), Cystatin C, interleukin 18 (IL18), and N-acetyl-β-D-glucosaminidase [6]. Levels of circulating fibroblast growth factor 23 (FGF-23) for example have been shown to be independently associated with left ventricular mass index and left ventricular hypertrophy in patients with CKD [7]. Chung and colleagues described the relationship between activation of matrix metalloproteinase 2 (MMP2) and elastic fiber degeneration, stiffening, medial calcification, and vasomotor dysfunction in macroarterial vasculature of dialyzed CKD patients [8]. Next to these proteins, a multitude of other molecular features is mentioned in the literature in the context of the cardiorenal syndrome. Perco et al. reported a list of 31 CVD biomarkers that were extracted from the literature and characterized with respect to biological function, gene expression in CKD, and known protein-protein interactions [9].
Literature mining approaches have the potential to reveal such biomarkers, thus providing a more global picture on genes, proteins, and metabolites associated with a specific disease. The biomedical literature can be seen as the condensed result of the combined effort of the scientific community, and as such represents the primary resource upon which further investigations may be based on. As such, it represents the primary resource upon which further investigations may be based on. PubMed, for instance, presently holds close to 20 million abstracts. Thus, computational literature mining tools assisting researchers in keeping pace with this ever-growing amount of fast changing information became indispensable [10, 11].
In the context of drug discovery, the most prevailing approach is based on concept cooccurrence [12, 13]. Here, a disease profile consisting of the concepts (e.g., drugs, genes, etc.) which are frequently mentioned together with the disease under analysis can be derived via text mining. Likewise, literature-based profiles for drugs or genes can be generated. Next to conveniently reaching an overview on biomarkers this information base may additionally be used to gain hints about yet undiscovered dependencies between diseases, drugs, and potential drug targets.
To further enhance text mining efforts, several “controlled vocabularies” (“ontologies”) have been developed to allow a precise definition of the employed concepts [14]. The most popular ones are maintained by the U.S. Library of Medicine, namely, the Unified Medical Language System (UMLS) and the Medical Subject Headings (MeSH). Given that the majority of PubMed articles are indexed with MeSH, a fast and accurate extraction of biomedical concepts has become feasible [13, 15]. With the advent of literature mining approaches also in combination with high-throughput Omics experiments, a number of bioinformatics tools and ontologies have been developed for the analysis of resulting large sets of genes or proteins. Analyzing extended sets of biomarker candidates on the level of molecular pathways and processes, represented as protein interaction networks, adds another layer of information for the interpretation of molecular feature (biomarker) sets.
A recent review by Lusis and colleagues summarized studies dealing with network analyses in cardiovascular disease [16]. Networks based on prior knowledge, such as existing pathway sources, literature cocitations, or other correlation measures as coexpression and sequence similarity were outlined by Ashley et al. [17], who mapped genes being differentially regulated between patients suffering from de-novo atherosclerosis and in-stent restenosis on a cocitation network obtained by literature mining of Medline abstracts. Similar concepts can be followed by utilizing networks derived from physical protein interactions, or networks generated from measuring the response to experimental perturbations. Further approaches include system genetics and detailed analyses at the level of dynamic systems such as flux balance analyses which are often used to characterize enzymatic reactions in dynamic models of metabolism. Some of these approaches, especially highly abstracted network models on the level of phenotypes, managed to predict comorbidity patterns for myocardial infarction using a “human disease network” thus closing the gap to clinical applications [18].
Diez et al. presented another application of the network paradigm to reveal the mechanisms of cardiovascular disease, identifying a set of differentially expressed genes separating asymptomatic from symptomatic carotid stenosis patients [19]. Based on these transcriptomics data, a correlation network was generated. Furthermore, an association network of the differentially regulated genes was derived by mining the literature for gene associations thus resulting in an interaction network combining Omics data and associated features extracted from the literature. Subnetworks were identified, characterized by enriched lipid-, immune-, and atherogenesis-related pathways and gene ontology terms. On this level of representation, the interplay of APOC1 (a gene that is linked to coronary heart disease) became evident. Weiss et al. investigated networks on cardiovascular metabolism pointing out aspects of network structure, namely, differences between designed networks in engineering and networks having undergone an evolutionary process [20]. Based on the level of abstraction, three types of network on cardiovascular metabolism were proposed: first, on the very abstract level of nodes and edges, metabolite networks described by using topological characteristics [21, 22], second physical, spatially compartmentalized networks including the description of energy fluxes in the network [23, 24], and on a third level dynamic networks [25–27].
The present knowledge regarding mechanisms leading to the formation of the CRS suggests a critical role for hemodynamic changes, originating either from the kidney or the cardiovascular system. In the following analysis, we used a literature mining approach to extract genes and proteins reported in the context of the cardiorenal syndrome, and analyzed these features on the level of protein interaction networks. Specific focus was laid on secreted proteins being specifically expressed in either renal or vascular tissue with the aim to identify molecular mediators potentially contributing to the cross-talk between the kidney and the cardiovascular system for allowing identification of novel therapeutic targets addressing both systems.
2. Materials and Methods
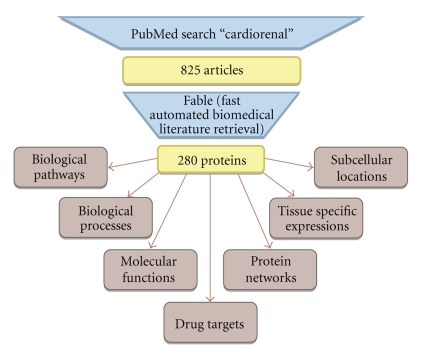
The general analysis strategy applied in this work is outlined in Figure 1. Major components include feature extraction via literature mining, followed by a range of bioinformatics analysis procedures for deciphering characteristics of individual features as well as joint interpretation on the level of protein interaction networks.
Figure 1.
Overview scheme on the analysis workflow: Literature mining was applied for identifying unique proteins associated with CRS. Bioinformatics included feature characterization as well as network analysis.
2.1. Literature Mining
The strength but also the challenge of biomedical text mining relies on the fact that the scientific literature embraces a variety of concepts (genes, drugs, diseases, etc.) which in turn are interrelated in a variety of ways. Thus, carefully designed text mining methods are needed to extract “meaningful” information and reduce the amount of noise present in the final results.
In general, text mining consists of two steps: Information Retrieval (IR) and Information Extraction (IE) [10]. The first consists in identifying documents which are of relevance for a certain research objective (e.g., a PubMed query for “cardiorenal”), whereas the later is used to extract facts from these documents. Named Entity Recognition (NER) can be seen as the most prevalent type of IE used in real world applications, aiming at the identification of biological entities like genes, cell types, or drugs.
Even though the concept of NER might appear almost trivial at a first glance, it actually represents a challenging computational problem as the existence of over fifty available tools demonstrates [28]. The key obstacle that needs to be addressed when extracting genes or proteins from free text relies in the term ambiguity present at multiple levels. Some genes are spelled like normal English words (e.g., “WAS” with the NCBI GeneID: 7454) and even a gene with the official Gene Symbol “T” exists (NCBI GeneID: 6862). The same gene may additionally be referred to in various ways due to different naming conventions.
Ultimately, this ambiguities lead to two different types of errors which all methods are confronted with: erratically assuming that a certain gene was mentioned in a paper (false positive) or erratically assuming that it was not mentioned, even though it actually was given (false negative) [29]. Based on the trade-off between these two types of errors, the precision of a method (i.e., how much of the predicted genes were actually mentioned in the document) and its recall (i.e., how much of all actually mentioned genes were also identified as such) are determined.
We chose a method favoring precision over recall for mining genes/proteins in Medline/PubMed abstracts. The Fast Automated Biomedical Literature Extraction (FABLE) tool available at http://fable.chop.edu/ was used in order to fulfill this task. The algorithm basically consists of two steps: first, a statistical classifier was used to train a probabilistic model, which served as basis for gene tagging, that is, to identify possible occurrences of a gene, taking the textual context into account. Given that such an occurrence exhibits a sufficient likelihood of actually representing a gene, this occurrence was normalized in a second step to the official Gene Symbol. This normalization step was based on gene synonym lists, which were compared to the predicted occurrence using both exact and relaxed pattern matching procedures. It has been shown that this approach is competitive to alternative methods such as standard information extraction techniques and direct pattern matching both in terms of precision and recall [30, 31]. We applied this procedures for all papers retrieved from PubMed associated with “cardiorenal” (PubMed status as of March 2010).
2.2. Functional Annotation of Identified Genes/Proteins
The list of genes and proteins identified on the basis of the literature mining approach was in a first step annotated using the Stanford Source tool [32]. The set of genes was assigned to biological processes, pathways, and molecular functions using the PANTHER (Protein Analysis through Evolutionary Relationships) Classification System [33, 34]. Significantly enriched categories were identified using the whole human genome as a reference dataset. Biological processes, pathways, and molecular functions showing P-values below .0001 were considered as statistically significant in terms of feature enrichment.
The subcellular location of proteins was determined using experimental data provided by SwissProt [35]. For proteins not covered in SwissProt, in-silico predictions using WoLF PSORT were done [36]. WoLF PSORT computes probabilities based on the protein sequence of a given protein for ten subcellular locations. Subcellular location tags from SwissProt were mapped to the ten locations defined by WoLF PSORT. Only assignments that were either reported in SwissProt or showed a probability value of 1 according to WoLF PSORT were considered for subcellular location enrichment analysis. Based on a reference dataset of 45,008 proteins assigned to one of the WoLF PSORT categories, the significance of enrichment was calculated using the Fisher's exact test. P-values below .01 were considered as statistically significant.
Information on tissue-specific expression patterns was extracted from NCBI UniGene EST profiles. EST counts of in total 45 tissues were extracted for each gene. Tissue-specific expression patterns for each single tissue for each single gene were calculated based on the normalized transcripts per million counts as provided by UniGene [37].
2.3. Network Analysis Framework
For network analysis, we used an extended version of the protein dependency network “omicsNET” as described in Bernthaler et al. [38]. The network is comprised of information from protein-protein interactions, tissue-specific reference coexpression, shared pathway information, gene ontology distance, and subcellular colocalization, and was extended by networks generated from shared transcription factor binding sites and shared miRNA target sites. In omicsNET, these sources were consolidated into a single human protein reference interaction network, where edges represent pairwise dependencies between proteins.
Protein-protein dependencies were calculated between proteins in the list resulting from the literature mining approach. Furthermore, highly connected subgraphs were identified and functionally annotated. We only considered dependencies with high confidence in the network construction process and focused on genes reported at least twice in the scientific literature in the context of the cardiorenal syndrome in order to reduce the number of false positive assignments.
2.4. Identification of Drug Targets
Drug targets were identified in our set of 280 literature-derived proteins using information from DrugBank [39, 40]. DrugBank combines information on drugs and their molecular targets and currently contains around 4800 drug entities with more than 1350 FDA-approved small molecule drugs and more than 2500 protein drug targets.
3. Results and Discussion
3.1. Literature Mining
825 papers associated with the term “cardiorenal” were identified in PubMed. In this set of 825 papers, 280 genes could be extracted utilizing FABLE, with 132 genes being reported at least twice. The top ranked gene, mentioned in 156 articles, was the aspartyl protease renin (REN), followed by the natriuretic peptide precurser A (NPPA), and angiotensinogen (AGT), with 122 and 64 reports, respectively.
The list of 54 genes mentioned in at least 5 articles along with the term cardiorenal is provided in Table 1 (see supplementary Table 1 for the total list of 280 genes in Supplementary Material available online at doi:10.4061/2011/809378). Next to the number of articles, the relative expression levels in the four tissues blood, heart, vascular, and kidney are provided based on data from the UniGene expressed sequence tag counts.
Table 1.
List of identified genes/proteins, number of articles identified for cardiorenal, and relative expression levels based on UniGene EST counts for blood, heart, vascular, and kidney, and tissue showing maximum expression of a specific feature.
| Symbol | Articles | Expression in blood (%) | Expression in heart (%) | Expression in vascular (%) | Expression in kidney (%) | Max. expression (%) | |
|---|---|---|---|---|---|---|---|
| REN | 156 | 0 | 0 | 0 | 19,27 | 39,58 | intestine |
| NPPA | 122 | 88,04 | 0 | 0 | 0 | 88,04 | heart |
| AGT | 64 | 1,79 | 18,54 | 0 | 5,71 | 29,74 | liver |
| ADM | 55 | 0,95 | 1,38 | 1,09 | 3,11 | 15,3 | adipose tissue |
| ACE | 39 | 0,86 | 2,37 | 4,09 | 4,53 | 15,63 | parathyroid |
| EDN1 | 39 | 0 | 4,12 | 15,82 | 2,77 | 32,68 | umbilical cord |
| NPPB | 31 | 85,93 | 0 | 0 | 1,2 | 85,93 | heart |
| RAPGEF5 | 28 | 0 | 0 | 0 | 0,76 | 76,62 | parathyroid |
| NOS3 | 27 | 3,92 | 2,69 | 2,33 | 2,2 | 20,32 | spleen |
| EPO | 22 | 0 | 0 | 0 | 0 | 58,82 | prostate |
| CNP | 21 | 0,85 | 1,74 | 3,58 | 5,4 | 18,03 | brain |
| TGFB1 | 20 | 8,67 | 0,99 | 0 | 1,79 | 17,67 | salivary gland |
| MME | 19 | 0,26 | 3,59 | 0 | 11,63 | 12,06 | lymph node |
| PTGS2 | 19 | 16,39 | 0 | 29,1 | 0,59 | 29,1 | vascular |
| INS | 18 | 0 | 0 | 0 | 0 | 100 | pancreas |
| NPR1 | 17 | 0 | 1,32 | 2,29 | 2,83 | 23,69 | mammary gland |
| NOS2 | 13 | 4,23 | 0 | 0 | 0 | 25,4 | pharynx |
| DDR1 | 13 | 0 | 0,94 | 0 | 0,46 | 20,12 | trachea |
| KNG1 | 10 | 0 | 0 | 0 | 33,18 | 57,18 | liver |
| PLEK | 10 | 11,02 | 0,34 | 1,77 | 0,87 | 16,81 | lymph |
| NCF1 | 10 | 10,88 | 0 | 0 | 0,76 | 32,38 | lymph node |
| HESX1 | 10 | 0 | 0 | 0 | 0 | 43,18 | ovary |
| FOS | 9 | 19,04 | 2,09 | 4,31 | 0,77 | 19,04 | blood |
| CALCA | 9 | 0 | 0 | 0 | 0 | 100 | prostate |
| S100A6 | 9 | 1,2 | 0,87 | 5,16 | 1,18 | 20,08 | umbilical cord |
| NOS1 | 8 | 0 | 0 | 0 | 1,68 | 65,97 | muscle |
| AVP | 8 | 0 | 0 | 0 | 80 | 80 | kidney |
| RHOA | 7 | 2,5 | 1,57 | 2,02 | 1,72 | 5,28 | cervix |
| CYBB | 7 | 19,44 | 0 | 2,55 | 3,15 | 27,68 | lymph node |
| MAPK1 | 7 | 1,84 | 1,35 | 2,36 | 1,44 | 10,94 | mouth |
| AKT1 | 7 | 1,14 | 1,57 | 0,45 | 1,51 | 13,52 | salivary gland |
| ICAM1 | 7 | 3,19 | 0,55 | 2,39 | 1,62 | 15,19 | spleen |
| CALCRL | 7 | 0 | 2,55 | 14,85 | 1,39 | 25,06 | trachea |
| SERPINE1 | 7 | 0,17 | 0,12 | 14,5 | 0,69 | 27,77 | umbilical cord |
| EDNRA | 7 | 0 | 6,4 | 2,21 | 1,63 | 10,94 | uterus |
| SHBG | 7 | 0 | 0 | 0 | 0 | 36,84 | eye |
| RAMP2 | 7 | 5,09 | 0 | 0 | 1,85 | 28,7 | thyroid |
| UTS2 | 7 | 0 | 0 | 0 | 3,88 | 35,92 | spleen |
| OLR1 | 6 | 1,23 | 0 | 0 | 2,15 | 81,05 | esophagus |
| AGTR1 | 6 | 0 | 5,19 | 0 | 3,3 | 19,1 | larynx |
| NFKB1 | 6 | 4,69 | 0,76 | 0,66 | 1,62 | 8,69 | nerve |
| UTS2R | 6 | 0 | 0 | 0 | 0 | 100 | ovary |
| NR3C2 | 6 | 0 | 0 | 6,41 | 7,08 | 20,74 | stomach |
| EPHB2 | 6 | 6,73 | 0 | 0 | 2,85 | 14,78 | umbilical cord |
| ISYNA1 | 6 | 1,49 | 0,43 | 0,52 | 3,31 | 17,72 | umbilical cord |
| GPR182 | 5 | 0 | 0 | 0 | 0 | 38,67 | adrenal gland |
| COX8A | 5 | 0,77 | 11,02 | 1,48 | 0,98 | 11,02 | heart |
| CPOX | 5 | 9,24 | 3,63 | 0 | 5,28 | 11,06 | liver |
| EGFR | 5 | 0 | 2,2 | 1,69 | 2,49 | 14,89 | mouth |
| COX5A | 5 | 0 | 0 | 0 | 0 | 100 | muscle |
| CCL2 | 5 | 0 | 0 | 0 | 0 | 100 | placenta |
| PPARG | 5 | 0 | 1,46 | 2,52 | 3,72 | 12,08 | placenta |
| CYBA | 5 | 2,25 | 6,82 | 1,67 | 3,43 | 15,46 | tonsil |
| RAMP3 | 5 | 7,76 | 0 | 0 | 2,54 | 21,44 | adipose tissue |
The top ranked feature in the list of 280 literature derived genes is renin (REN) which is secreted by cells of the juxtaglomerular apparatus of the kidney and plays a key role in the blood pressure and water balance-regulating renin-angiotensin system (RAS). The connection between CRS and an increased activity of this hormone system was first reported in 1971 [41] and its consequences like renal hypoxia, vasoconstriction, intraglomerular hypertension, glomerulosclerosis, tubulointerstitial fibrosis, and proteinuria continue to be demonstrated in clinical practice. Conservative therapy for blocking the RAS activity is the administration of angiotensin-converting enzyme inhibitors and angiotensin receptor blockers, but recent studies demonstrate the benefit of a combination with direct renin inhibitors [42].
Further genes frequently reported in association with CRS are the components of the natriuretic peptide system (NPS) NPPA and NPPB, as well as their receptors NPR1, NPR2, and NPR3. Functions of the NPS include the counter-regulation of RAS, and it is suggested that its activation provides organ protection in cardiorenal disease, especially in diabetic patients [43].
3.2. Functional Annotation
According to the PANTHER Classification System, the biological processes of “signal transduction” and “cell communication” were identified as most significantly enriched, with 135 and 136 genes assigned to these categories, respectively. In total, 28 processes showed a P-value > .0001 in terms of enrichment, including “blood circulation”, “regulation of vasoconstriction”, and “angiogenesis”. The most significantly enriched molecular functions are “receptor binding” and “protein binding” (Table 2).
Table 2.
List of enriched biological processes and molecular functions. Given are the total number of genes assigned to a process/function, the number of genes assigned as derived from literature mining, the number of genes expected from a statistical perspective, and the significance level of enrichment.
| Biological process | No. genes total | No. genes CRS | No. genes CRS expected | P-value |
|---|---|---|---|---|
| Signal transduction | 4191 | 135 | 57,67 | 4.55E-25 |
| Cell communication | 4365 | 136 | 60,07 | 6.84E-24 |
| Cell surface receptor linked signal transduction | 2235 | 91 | 30,76 | 3.80E-22 |
| Immune system process | 2628 | 97 | 36,16 | 9.70E-21 |
| Blood circulation | 210 | 28 | 2,89 | 5.11E-19 |
| Regulation of biological process | 59 | 18 | 0,81 | 1.01E-18 |
| Regulation of vasoconstriction | 59 | 18 | 0,81 | 1.01E-18 |
| Molecular function | No. genes total | No. genes CRS | No. genes CRS expected | P-value |
| Receptor binding | 1233 | 64 | 16,97 | 2.46E-20 |
| Protein binding | 3157 | 103 | 43,44 | 2.71E-18 |
| Catalytic activity | 5336 | 128 | 73,43 | 1.44E-12 |
| Oxidoreductase activity | 703 | 33 | 9,67 | 1.21E-09 |
| Binding | 6751 | 140 | 92,9 | 3.65E-09 |
| Kinase activity | 695 | 28 | 9,56 | 5.18E-07 |
The two enriched categories “receptor binding” and “receptor activity” indicate that numerous receptors and ligands are involved in the cardiorenal syndrome. These receptors form the first line of molecules in a number of signaling cascades, which as such is another category enriched in genes associated with the cardiorenal syndrome. We therefore took a closer look at receptor-ligand interactions. We searched for receptors mainly expressed in the cardiovascular system having ligands predominantly secreted by the renal tissue, and vice versa.
The natriuretic peptide receptor NPR3 showed high expression in kidney tissue, whereas the ligands NPPA and NPPB were found to be almost exclusively expressed in the heart. Thus, a deregulation of blood pressure maintenance and extracellular fluid volume by heart-derived ligands of the natriuretic peptide system directly affect the kidney and may contribute to the formation of CRS.
Enrichment of the process “regulation of vasoconstriction” reflects the consequences of impaired heart function including a decreased cardiac output, and thus the hypoperfusion of organs. Since glomerular filtration is controlled by blood pressure, hypoperfusion of the kidney leads to the activation of the RAS and subsequent vasoconstriction, which, in turn, causes systemic hypertension and an increased heart preload [2].
22 PANTHER pathways could be identified as significantly enriched in the list of 280 literature-derived genes. 28 genes could be assigned to “angiogenesis”, 21 genes to “endothelin mediated signaling”, and 15 genes to the “VEGF signaling pathway” (Table 3).
Table 3.
List of enriched biological pathways. Given are the total number of genes assigned to a process/function, the number of genes assigned as derived from literature mining for CRS, the number of genes expected from a statistical perspective, and the significance level of enrichment.
| Pathway | No. genes total | No. genes CRS | No. genes CRS expected | P-value |
|---|---|---|---|---|
| Angiogenesis | 191 | 28 | 2,63 | 4.51E-20 |
| Endothelin signaling pathway | 91 | 21 | 1,25 | 3.33E-19 |
| VEGF signaling pathway | 75 | 15 | 1,03 | 3.33E-13 |
| Inflammation mediated by chemokine and cytokine signaling pathway | 283 | 24 | 3,89 | 2.76E-12 |
| PDGF signaling pathway | 159 | 18 | 2,19 | 1.68E-11 |
| T cell activation | 102 | 14 | 1,4 | 2.72E-10 |
| Apoptosis signaling pathway | 123 | 15 | 1,69 | 3.10E-10 |
The connection between angiogenic processes and cardiovascular disorders is well understood, since decreased cardiac output goes along with decreased organ perfusion, and vascularization is the natural response to diminution of blood supply. Apart from negative effects on organ function due to hypoperfusion, microvascularization is extensively performed at sites of inflammation which explains the role of angiogenesis in diseased kidney tissue. On the other hand, decreased vascularization and loss of capillaries lead to kidney fibrosis. However, deregulation of angiogenesis seems to be crucial for kidney function and a key regulatory mechanism of angiogenic processes is the VEGF signaling pathway [44–46]. A third enriched pathway is the “endothelin signaling pathway” which is known to regulate the renin-angiotensin system thus being a further player in the hemodynamic cross-talk between the kidney and the cardiovascular system.
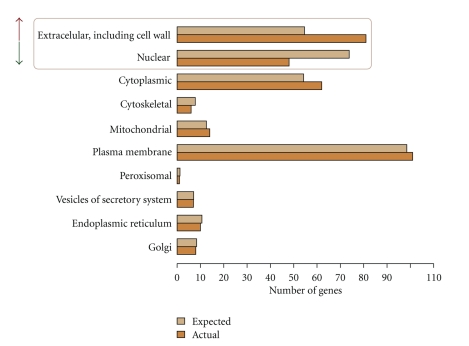
Following the rationale that features secreted from kidney cells may lead to damage in vessels and vice versa, literature-derived proteins were classified in terms of subcellular location. The most significantly enriched compartment was “extracellular, including cell wall” with 81 genes being assigned to this category, whereas “nuclear” was significantly depleted with 48 genes as indicated in Figure 2.
Figure 2.
Subcellular location of literature-derived proteins. Presented are categories of subcellular location, the expected number of proteins in a particular category using the total set of human proteins, and the actual number of proteins found as being associated with CRS.
The list of 81 secreted genes included components of the renin-angiotensin system (REN, AGT, ACE) and the natriuretic peptide system (NPPA, NPPB), as well as some other regulators of vasoconstriction. Kininogen 1 (KNG1) for example is essential for the assembly of the blood pressure regulating kallikrein-kinin system. Another molecule serving as a vasodilator is the peptide hormone calcitonin-related polypeptide alpha (CALCA).
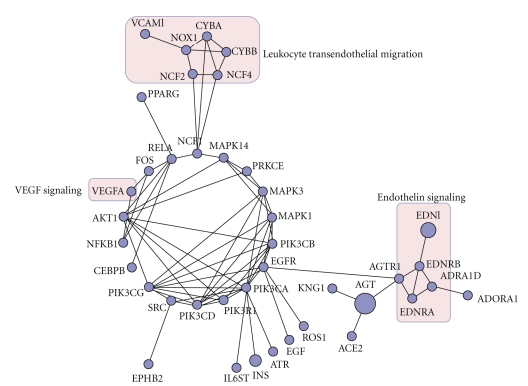
3.3. Network Analysis
A subset of 40 proteins out of the list of 132 proteins mentioned in at least two publications in the context of the cardiorenal syndrome formed a highly connected protein interaction network as given in Figure 3. The main components of this protein network are mediators of hemodynamic change. An accumulation of features involved in previously described signaling pathways like the endothelin signaling pathway or the VEGF signaling pathway is evident. Next to these two pathways, a number of members of the blood pressure regulating kallikrein-kinin system and the renin-angiotensin system are part of this network.
Figure 3.
Protein interaction network of highly connected proteins associated with the cardiorenal syndrome. Nodes represent genes (gene symbols); edges indicate functional associations. Highlighted nodes represent proteins that are specific for either the VEGF signaling, the leukocyte transendothelial migration, or the endothelin signaling pathway.
Another highly connected cluster holds genes associated with leukocyte transendothelial migration. The process of leukocyte migration from blood into tissues is vital for inflammation, and it is known that inflammation is an important cardiorenal connector and a hallmark of kidney and heart diseases [5].
3.4. Identification of Drug Targets
116 out of the 280 proteins associated with the CRS were listed as drug target for at least one drug in DrugBank (see supplementary Table 1). The proteins with the most number of drugs were PTGS1, PTGS2, and NOS3 with 49, 43, and 41 drugs associated. The drug with the most drug targets in our list of 280 proteins was NADH.
Standard therapeutic regimes in the context of cardiovascular and kidney disease included aliskiren, irbesartan, or ramipril. Another drug candidate is nesiritide, a recombinant B-type natriuretic peptide that counter-regulates the RAS, as used in the treatment of acute decompensated heart failure (ADHF). However, on the basis of a prospective, randomized, double-blinded, placebo-controlled clinical trial, Witteles et al. concluded that nesiritide therapy does not impact renal function in patients with ADHF and pre-existing renal dysfunction [47].
It is known that reducing blood pressure has beneficial effects on renal function and there is a multitude of antihypertensive agents acting on the RAS. Administration of angiotensin receptor antagonists in combination with angiotensin-converting enzyme inhibitors showed a significant reduction of urine albumin creatinine ratio in patients with hypertension and microalbuminuria and thus, a reduction of the risk for myocardial infarction [48].
Further potential targets for regulation of hemodynamics are members of the endothelin signaling pathway. Endothelin receptor antagonists are used in the treatment of a variety of cardiovascular conditions but less is known about the effects on combined kidney dysfunction. Ding et al. showed in animal models that chronic endothelin receptor blockade with endothelin receptor antagonists is beneficial in the treatment of progressive renal dysfunction and sodium retention associated with chronic heart failure [49]. Studies in humans are required to fully elucidate the effects and risks of endothelin receptor antagonist treatment in patients with CRS.
4. Conclusions
In this work, we provide a comprehensive list of genes/proteins associated with the cardiorenal syndrome identified on the basis of a literature mining approach. On the basis of 825 articles identified in the context of CRS, 280 unique genes could be identified and were further characterized with respect to molecular function, biological processes, cellular pathways, subcellular location, tissue-specific expression, as well as on the level of protein interaction networks.
The most frequently reported genes are involved in blood pressure regulating systems, particularly in the renin-angiotensin system (REN, AGT, ACE), as well as in the antagonistic natriuretic peptide system (NPPA, NPPB). Enriched molecular functions include “receptor binding” and “receptor activity”. Of special note in this context are again players of the natriuretic peptide system, namely, the two ligands NPPA and NPPB and its receptor NPR3. Tissue-specific expression patterns of these molecules showed that NPPA and NPPB are mainly expressed in the heart, whereas their receptor NPR3 is highly expressed in kidney tissue, suggesting that this regulatory system is part of the cross-talk between the kidney and the cardiovascular system.
Therapy of the CRS is largely focused on natriuretic peptides or the renin-angiotensin system with a number of other molecular targets like the endothelin signaling pathway holding promise for future therapeutic strategies.
Altogether, the results of the present study strongly indicate the critical role of hemodynamic changes, blood pressure regulating hormone systems, and inflammatory processes in the formation of the CRS. Our analyses led to a comprehensive picture of molecular features involved in the functional interplay between the kidney and the cardiovascular system. One limitation of this automated literature mining approach is that we do not have experimental data on the expression levels of the reported molecules in the process of disease development. An obvious next step would therefore be to integrate the findings of this work with Omics datasets on kidney disease as well as vascular diseases. Such a combined approach has the potential to identify deregulated features for potentially identifying novel players for diagnostic or therapeutic approaches in the field of kidney and cardiovascular diseases.
Supplementary Material
List of 280 identified genes/proteins, number of articles identified for cardiorenal, and relative expression levels based on UniGene EST counts for blood, heart, vascular, and kidney, and tissue showing maximum expression of a specific feature.
Acknowledgment
This research was supported by Fresenius Medical Care Austria GmbH.
References
- 1.Sarnak MJ, Levey AS, Schoolwerth AC, et al. Kidney disease as a risk factor for development of cardiovascular disease: a statement from the American Heart Association Councils on Kidney in Cardiovascular Disease, High Blood Pressure Research, Clinical Cardiology, and Epidemiology and Prevention. Hypertension. 2003;42(5):1050–1065. doi: 10.1161/01.HYP.0000102971.85504.7c. [DOI] [PubMed] [Google Scholar]
- 2.Ronco C, Haapio M, House AA, Anavekar N, Bellomo R. Cardiorenal Syndrome. Journal of the American College of Cardiology. 2008;52(19):1527–1539. doi: 10.1016/j.jacc.2008.07.051. [DOI] [PubMed] [Google Scholar]
- 3.Ronco C, McCullough PA, Anker SD, et al. Cardiorenal syndromes: an executive summary from the consensus conference of the acute dialysis quality initiative (ADQI) Contributions to Nephrology. 2010;165:54–67. doi: 10.1159/000313745. [DOI] [PubMed] [Google Scholar]
- 4.Schiffrin EL, Lipman ML, Mann JFE. Chronic kidney disease: effects on the cardiovascular system. Circulation. 2007;116(1):85–97. doi: 10.1161/CIRCULATIONAHA.106.678342. [DOI] [PubMed] [Google Scholar]
- 5.Mahapatra HS, Lalmalsawma R, Singh NP, Kumar M, Tiwari SC. Cardiorenal syndrome. Iranian Journal of Kidney Diseases. 2009;3(2):61–70. [PubMed] [Google Scholar]
- 6.Soni SS, Fahuan Y, Ronco C, Cruz DN. Cardiorenal syndrome: biomarkers linking kidney damage with heart failure. Biomarkers in Medicine. 2009;3(5):549–560. doi: 10.2217/bmm.09.59. [DOI] [PubMed] [Google Scholar]
- 7.Gutiérrez OM, Januzzi JL, Isakova T, et al. Fibroblast growth factor 23 and left ventricular hypertrophy in chronic kidney disease. Circulation. 2009;119(19):2545–2552. doi: 10.1161/CIRCULATIONAHA.108.844506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung AWY, Clarice Yang HH, Kim JM, et al. Upregulation of matrix metalloproteinase-2 in the arterial vasculature contributes to stiffening and vasomotor dysfunction in patients with chronic kidney disease. Circulation. 2009;120(9):792–801. doi: 10.1161/CIRCULATIONAHA.109.862565. [DOI] [PubMed] [Google Scholar]
- 9.Perco P, Wilflingseder J, Bernthaler A, et al. Biomarker candidates for cardiovascular disease and bone metabolism disorders in chronic kidney disease: a systems biology perspective. Journal of Cellular and Molecular Medicine. 2008;12(4):1177–1187. doi: 10.1111/j.1582-4934.2008.00280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen LJ, Saric J, Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nature Reviews Genetics. 2006;7(2):119–129. doi: 10.1038/nrg1768. [DOI] [PubMed] [Google Scholar]
- 11.Krallinger M, Valencia A, Hirschman L. Linking genes to literature: text mining, information extraction, and retrieval applications for biology. Genome Biology. 2008;9(2, article S8) doi: 10.1186/gb-2008-9-s2-s8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y, Adelstein SJ, Kassis AI. Target discovery from data mining approaches. Drug Discovery Today. 2009;14(3-4):147–154. doi: 10.1016/j.drudis.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Agarwal P, Searls DB. Literature mining in support of drug discovery. Briefings in Bioinformatics. 2008;9(6):479–492. doi: 10.1093/bib/bbn035. [DOI] [PubMed] [Google Scholar]
- 14.Altman RB, Bergman CM, Blake J, et al. Text mining for biology—the way forward: opinions from leading scientists. Genome Biology. 2008;9(2, article S7) doi: 10.1186/gb-2008-9-s2-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jani SD, Argraves GL, Barth JL, Argraves WS. GeneMesh: a web-based microarray analysis tool for relating differentially expressed genes to MeSH terms. BMC Bioinformatics. 2010;11, article 166 doi: 10.1186/1471-2105-11-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lusis AJ, Weiss JN. Cardiovascular networks: systems-based approaches to cardiovascular disease. Circulation. 2010;121(1):157–170. doi: 10.1161/CIRCULATIONAHA.108.847699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashley EA, Ferrara R, King JY, et al. Network analysis of human in-stent restenosis. Circulation. 2006;114(24):2644–2654. doi: 10.1161/CIRCULATIONAHA.106.637025. [DOI] [PubMed] [Google Scholar]
- 18.Goh K-I, Cusick ME, Valle D, Childs B, Vidal M, Barabási A-L. The human disease network. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(21):8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diez D, Wheelock AM, Goto S, et al. The use of network analyses for elucidating mechanisms in cardiovascular disease. Molecular BioSystems. 2010;6(2):289–304. doi: 10.1039/b912078e. [DOI] [PubMed] [Google Scholar]
- 20.Weiss JN, Yang L, Qu Z. Thematic review series: systems biology approaches to metabolic and cardiovascular disorders: network perspectives of cardiovascular metabolism. Journal of Lipid Research. 2006;47(11):2355–2366. doi: 10.1194/jlr.R600023-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Barabási A-L, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nature Reviews Genetics. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 22.Jeong H, Tombor B, Albert R, Oltval ZN, Barabásl A-L. The large-scale organization of metabolic networks. Nature. 2000;407(6804):651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 23.Rizzuto R, Pinton P, Carrington W, et al. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280(5370):1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 24.Vendelin M, Béraud N, Guerrero K, et al. Mitochondrial regular arrangement in muscle cells: a “crystal-like” pattern. American Journal of Physiology. 2005;288(3):C757–C767. doi: 10.1152/ajpcell.00281.2004. [DOI] [PubMed] [Google Scholar]
- 25.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402(6761):C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 26.O’Rourke B, Ramza BM, Marban E. Oscillations of membrane current and excitability driven by metabolic oscillations in heart cells. Science. 1994;265(5174):962–966. doi: 10.1126/science.8052856. [DOI] [PubMed] [Google Scholar]
- 27.Aon MA, Cortassa S, O’Rourke B. Percolation and criticality in a mitochondrial network. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(13):4447–4452. doi: 10.1073/pnas.0307156101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krallinger M, Valencia A. Text-mining and information-retrieval services for molecular biology. Genome Biology. 2005;6(7, article 224) doi: 10.1186/gb-2005-6-7-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Y, Hines LM, Weng H, et al. Analysis of genomic and proteomic data using advanced literature mining. Journal of Proteome Research. 2003;2(4):405–412. doi: 10.1021/pr0340227. [DOI] [PubMed] [Google Scholar]
- 30.Crim J, McDonald R, Pereira F. Automatically annotating documents with normalized gene lists. BMC Bioinformatics. 2005;6(1, article S13) doi: 10.1186/1471-2105-6-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonald R, Pereira F. Identifying gene and protein mentions in text using conditional random fields. BMC Bioinformatics. 2005;6(1, article S6) doi: 10.1186/1471-2105-6-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diehn M, Sherlock G, Binkley G, et al. SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Research. 2003;31(1):219–223. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas PD, Campbell MJ, Kejariwal A, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Research. 2003;13(9):2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas PD, Kejariwal A, Guo N, et al. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Research. 2006;34(supplement 2):W645–W650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Apweiler R, Bairoch A, Wu CH, et al. UniProt: the universal protein knowledgebase. Nucleic Acids Research. 2004;32(supplement 1):D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horton P, Park KJ, Obayashi T, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Research. 2007;35(supplement 2):W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sehuler GD. Pieces of use puzzle: expressed sequence tags and the catalog of human genes. Journal of Molecular Medicine. 1997;75(10):694–698. doi: 10.1007/s001090050155. [DOI] [PubMed] [Google Scholar]
- 38.Bernthaler A, Mühlberger I, Fechete R, Perco P, Lukas A, Mayer B. A dependency graph approach for the analysis of differential gene expression profiles. Molecular BioSystems. 2009;5(12):1720–1731. doi: 10.1039/b903109j. [DOI] [PubMed] [Google Scholar]
- 39.Wishart DS, Knox C, Guo AC, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Research. 2006;34(supplement 1):D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wishart DS, Knox C, Guo AC, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Research. 2008;36(supplement 1):D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Judson WE, Helmer OM. Relationship of cardiorenal function to renin-aldosterone system in patients with valvular heart disease. Circulation. 1971;44(2):245–253. doi: 10.1161/01.cir.44.2.245. [DOI] [PubMed] [Google Scholar]
- 42.Ferrario CM. Addressing the theoretical and clinical advantages of combination therapy with inhibitors of the renin-angiotensin-aldosterone system: antihypertensive effects and benefits beyond BP control. Life Sciences. 2010;86(9-10):289–299. doi: 10.1016/j.lfs.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McFarlane SI, Winer N, Sowers JR. Role of the natriuretic peptide system in cardiorenal protection. Archives of Internal Medicine. 2003;163(22):2696–2704. doi: 10.1001/archinte.163.22.2696. [DOI] [PubMed] [Google Scholar]
- 44.Kang D-H, Hughes J, Mazzali M, Schreiner GF, Johnson RJ. Impaired angiogenesis in the remnant kidney model: II. Vascular endothelial growth factor administration reduces renal fibrosis and stabilizes renal function. Journal of the American Society of Nephrology. 2001;12(7):1448–1457. doi: 10.1681/ASN.V1271448. [DOI] [PubMed] [Google Scholar]
- 45.Risau W. Mechanisms of angiogenesis. Nature. 1997;386(6626):671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- 46.Rudnicki M, Perco P, Enrich J, et al. Hypoxia response and VEGF-A expression in human proximal tubular epithelial cells in stable and progressive renal disease. Laboratory Investigation. 2009;89(3):337–346. doi: 10.1038/labinvest.2008.158. [DOI] [PubMed] [Google Scholar]
- 47.Witteles RM, Kao D, Christopherson D, et al. Impact of nesiritide on renal function in patients with acute decompensated heart failure and pre-existing renal dysfunction. A randomized, double-blind, placebo-controlled clinical trial. Journal of the American College of Cardiology. 2007;50(19):1835–1840. doi: 10.1016/j.jacc.2007.03.071. [DOI] [PubMed] [Google Scholar]
- 48.Menne J, Farsang C, Deák L, et al. Valsartan in combination with lisinopril versus the respective high dose monotherapies in hypertensive patients with microalbuminuria: the VALERIA trial. Journal of Hypertension. 2008;26(9):1860–1867. doi: 10.1097/HJH.0b013e32830508aa. [DOI] [PubMed] [Google Scholar]
- 49.Ding S-S, Qiu C, Hess P, Xi J-F, Clozel J-P, Clozel M. Chronic endothelin receptor blockade prevents renal vasoconstriction and sodium retention in rats with chronic heart failure. Cardiovascular Research. 2002;53(4):963–970. doi: 10.1016/s0008-6363(01)00558-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of 280 identified genes/proteins, number of articles identified for cardiorenal, and relative expression levels based on UniGene EST counts for blood, heart, vascular, and kidney, and tissue showing maximum expression of a specific feature.