Abstract
8-oxo-7,8-dihydroadenine (8-oxoAde) is a major product of adenine modification by reactive oxygen species. So far, only one mammalian DNA glycosylase, 8-oxoguanine-DNA-glycosylase 1 (OGG1), has been shown to excise 8-oxoAde, exclusively from pairs with Cyt. We have found that endonuclease VIII-like protein 1 (NEIL1), a mammalian homolog of bacterial endonuclease VIII, can efficiently remove 8-oxoAde from 8-oxoAde:Cyt pairs but not from other contexts. In an in vitro reconstituted system, reactions containing OGG1 produced a fully repaired product, whereas NEIL1 caused an abortive initiation of repair, stopping after 8-oxoAde removal and DNA strand cleavage. This block was partially relieved by polynucleotide kinase/3′-phosphatase. Thus, two alternative routes of 8-oxoAde repair may exist in mammals.
Keywords: DNA repair, DNA glycosylase, Endonuclease VIII homolog, 8-Oxoadenine
1. Introduction
Reactive oxygen species present a constant threat to the genome integrity [1]. All nucleobases are prone to oxidation, generating DNA lesions that may cause mutations, block DNA polymerases, or interfere with binding of regulatory proteins. 8-Oxo-7,8-dihydroadenine (8-oxoAde) is formed when DNA is attacked by hydroxyl radicals produced by ionizing radiation, Fenton chemistry, or other exogenous and endogenous factors [1]. 8-OxoAde is formed in the DNA of living mammals after γ-irradiation, and increased levels of 8-oxoAde have been detected in human cancers [2].
When present in a DNA template, 8-oxoAde weakly to moderately blocks DNA polymerases, mainly directing incorporation of non-mutagenic TMP. However, a limited amount of dGMP and dAMP incorporation have been reported [3,4]. In vivo, 8-oxoAde in DNA is weakly mutagenic in mammalian cells (~1%), preferentially causing A → C and A → G substitutions, which indicates transient formation of 8-oxoAde:Gua and 8-oxoAde:Cyt mispairs, respectively [4,5]. In addition, Ade in various monomers is oxidized easier than in DNA, most likely due to shielding of the base in DNA, raising a possibility of 8-oxoAde appearance through oxidation of the dATP pool [6]. The incorporation of 8-oxoAde into DNA from the oxidized dATP pool has not been studied in detail; only very inefficient incorporation of 8-oxodAMP opposite Thy by the Klenow fragment has been reported [7]. The detrimental effects of 8-oxoAde are not limited to its mutagenic action: 8-oxoAde inhibits eukaryotic RNA polymerase II [8] and the exonuclease activity of WRN helicase [9].
The mechanisms of 8-oxoAde repair remain poorly studied. So far, only one eukaryotic enzyme, 8-oxoguanine-DNA glycosylase (OGG1), has been reported to excise 8-oxoAde [10-12]. Interestingly, this excision is efficient only when 8-oxoAde is paired with Cyt [10-12]. This repair reaction could only be sensible if it removes 8-oxoAde incorporated from the oxidized dATP pool opposite Cyt by erroneous action of DNA polymerases. Bacterial formamidopyrimidine-DNA glycosylase (Fpg), which is not homologous to OGG1, removes 8-oxoAde very poorly from any nucleo-base pair [10]. Additionally, an unidentified enzyme distinct from OGG1 has been reported to excise 8-oxoAde when paired with Gua in mammalian cells [12].
Here, we report that endonuclease VIII-like protein 1 (NEIL1) protein, a mammalian DNA glycosylase homologous to bacterial endonuclease VIII (Nei), is proficient in the removal of 8-oxoAde from pairs with Cyt but not from other pairs. The existence of two enzymes for repairing 8-oxoAde:Cyt pairs suggests that this mispair may be of biological importance.
2. Materials and methods
2.1. Oligonucleotides and enzymes
The modified oligodeoxyribonucleotide (ODN) 5′-d(CTCTCCCTTCXCTCCTTTCCTCT)-3′ (X = 8-oxoAde) and the complementary ODNs placing Ade, Cyt, Gua, or Thy opposite to X were synthesized from the precursors purchased from Glen Research. The modified strand was 32P-labeled using T4 polynucleotide kinase (New England Bio-labs) and annealed to the complementary strand in a 1:2 molar ratio. NEIL1, OGG1, AP endonuclease 1 (APEX1), polynucleotide kinase/3′-phosphatase (PNKP), and DNA ligase IIIα (LIG3α)were purified as described [13-15]. Concentrations of the active form of NEIL1 and OGG1 were determined by sodium borohydride crosslinking [16]. Wild-type DNA polymerase β (POLβ), mutant POLβ (mPOLβ)containing three mutations (K35A K68A K72A), and XRCC1 were a generous gift from Dr. Svetlana Khodyreva (ICBFM).
2.2. 8-OxoAde excision assay
The reaction mixtures (20 μl) contained 200 nM of the labeled substrate and 20 nM NEIL1 or OGG1 in a buffer containing 25 mM potassium phosphate (pH 7.4), 5 mM MgCl2, 1 mM dithiothreitol, 0.25 mg/ml bovine serum albumin, and 2.5% glycerol. In kinetic measurements, the substrate concentration was 20–400 nM and the enzyme concentrations were 10 nM (NEIL1) or 1 nM (OGG1). If necessary, the mixture also contained APEX1, POLβ (75 nM each), and dGTP (0.5 mM). The reactions were initiated by adding NEIL1 or OGG1 and were incubated at 25 °C for 15 min. Then 10 μl of a formamide dye was added and the mixtures were heated for 1 min at 95 °C. The products were separated by electro-phoresis in 20% polyacrylamide gel with 8 M urea and quantified by phosphorimaging (Molecular Imager FX, Bio-Rad).
2.3. DNA repair reconstitution assay
The reaction mixtures (20 μl) contained 20 nM of the labeled substrate in a buffer containing 50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 10 mM dithiothreitol, 0.5 mM dGTP, 1 mM ATP, and 0.025 mg/ml bovine serum albumin. Different combinations of NEIL1 (15 nM), OGG1 (12 nM), APEX1 (15 nM), POLβ (6 nM), PNKP (10–300 nM), and LIG3α (10 nM) were added as required. The mixtures were incubated at 25 °C for 20 min and analyzed as above.
3. Results and discussion
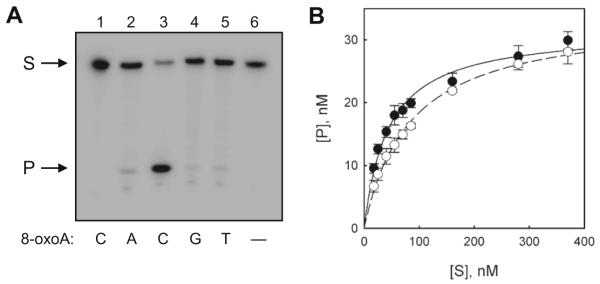
NEIL1 belongs to the Fpg/Nei family of DNA glycosylases [17]. The primary substrates for NEIL1 include oxidized pyrimidines, such as thymine glycols [18], dihydrouracil [19], and 5-hydroxycytosine [20], as well as formamidopyrimidine derivatives of purines[13,18-20]. In order to analyze whether NEIL1 can cleave DNA containing 8-oxoAde, we incubated the enzyme with an 8-oxoAde-containing ODN or with ODN duplexes bearing an 8-oxoAde residue with Ade, Cyt, Gua, or Thy placed opposite the lesion. Whilst we observed low levels of substrate cleavage of 8-oxoAde:Ade, 8-oxoAde:Gua and 8-oxoAde:Thy pairs, and no cleavage at all for 8-oxoAde in single-stranded DNA, NEIL1 very efficiently cleaved 8-oxoAde:Cyt (Fig. 1A). In this respect, NEIL1 was similar to OGG1, which also cleaves ODNs containing 8-oxoAde only when the lesion is opposite Cyt [10,11]. Therefore, we compared the catalytic efficiency of these two enzymes in the cleavage of the 8-oxoAde:Cyt substrate (Fig. 1B). When these data were fitted to the Michaelis–Menten model, the KM value for NEIL1 was ~2-fold lower than for OGG1, although the kcat value for OGG1 was ~10-fold higher than for NEIL1 (Table 1).
Fig. 1.
(A) Excision of 8-oxoAde from different base pairs by NEIL1. The reactions contained the following substrates: 1 and 3, 8-oxoAde:Cyt; 2, 8-oxoAde:Gua; 4, 8-oxoAde: Thy; 5, 8-oxoAde:Ade; 6, 8-oxoAde in single-stranded DNA. S, oligonucleotide substrate, P, cleavage product. (B) Kinetics of 8-oxoAde:Cyt cleavage by NEIL1 (●) and OGG1 (○). Mean and S.E.M. are shown.
Table 1.
Kinetic parameters of 8-oxoAde:Cyt cleavage by NEIL1 and OGG1
| KM (nM) | kcat(min−1) | kcat/kM × 103 (nM−1 × min−1) | |
|---|---|---|---|
| NEIL1 | 44 ± 5 | 0.21 ± 0.01 | 4.8 ± 0.6 |
| OGG1 | 84 ± 8 | 2.3 ± 0.1 | 27 ± 3 |
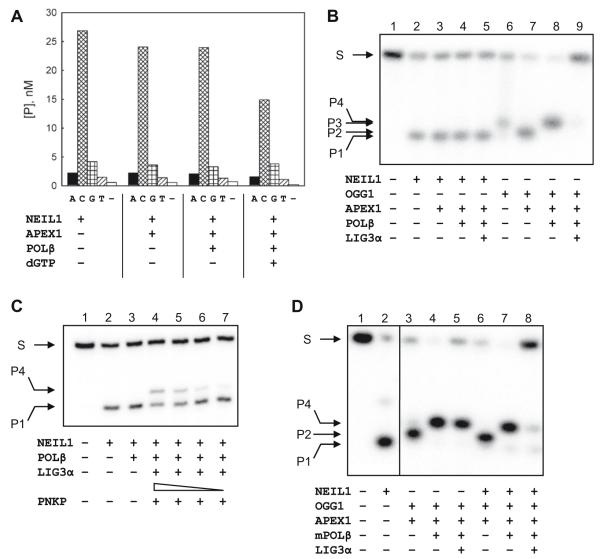
Other base excision repair (BER) components may markedly affect the activity of DNA glycosylases. For instance, OGG1 is stimulated by AP endonuclease APEX1, which dislodges OGG1 from the abasic site product; this process is as efficient on 8-oxoAde:Cyt as on 8-oxoGua:Cyt pairs, the primary OGG1 substrate [14]. In contrast to OGG1, the cleavage of 8-oxoAde:Cyt by NEIL1 was not stimulated by APEX1 (Fig. 2A), nor did APEX1 stimulate the cleavage of dihydrouracil-containing DNA by NEIL1 (data not shown). The addition of POLβ, with or without dGTP, also did not improve the excision efficiency (Fig. 2A). Therefore, it is unlikely that these BER proteins are required for the maximal activity of NEIL1 on the 8-oxoAde:Cyt substrate.
Fig. 2.
(A) NEIL1 excision of 8-oxoAde in the presence of other BER proteins. The components present in different reaction mixtures and the base opposite 8-oxoAde are indicated. (B) Reconstitution of the BER cycle for 8-oxoAde:Cyt. 1, substrate only; 2–5, NEIL1-initiated repair; 6–9, OGG1-initiated repair. S, ODN substrate or religated product (i and ix in Fig. 3); P1, β,δ-elimination product (iii); P2, β-elimination product (ii); P3, product of base excision, with or without β-elimination, followed by nicking by APEX1 (iv and v); P4, product of dGMP insertion (vi, vii, and viii). (C) Generation of POLβ-extendable 3′-ends by PNKP after NEIL1 excision of 8-oxoAde. The concentrations of PNKP in lanes 4–7 were 300, 100, 30, and 10 nM, respectively. (D) Reconstitution of the BER cycle for 8-oxoAde:Cyt using both OGG1 and NEIL1. Arrows indicating the mobilities of different species in (C) and (D) are in the same as in (B).
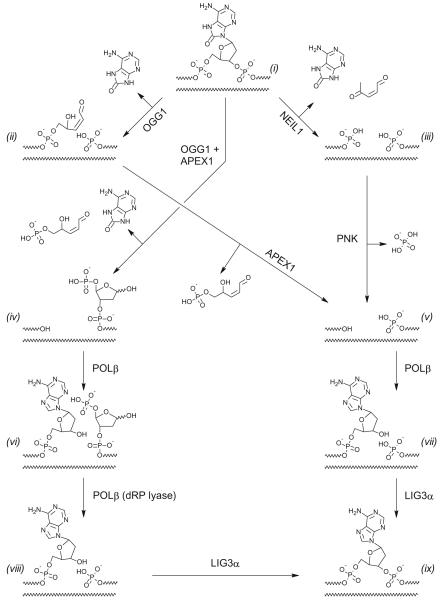
Unlike OGG1, which catalyzes elimination of the 3′-phosphate (β-elimination) after base removal, NEIL1 catalyzes elimination of both the 3′- and 5′-phosphates (β,δ-elimination), leaving a single-nucleotide gap flanked by two phosphates [18-20] (Fig. 3). The 3′-end at this gap requires processing to generate a 3′-hydroxyl that could be used by DNA polymerases in further BER steps; such processing has previously been shown to involve PNKP [21] (Fig. 3). On the other hand, the joint action of OGG1 and APEX1 directly generates a free 3′-hydroxyl. We therefore analyzed the repair of 8-oxoAde:Cyt in a reconstituted minimal BER system consisting of a DNA glycosylase (OGG1 or NEIL1), APEX1, POLβ, and a DNA ligase (LIG3α; Fig. 2B). The repair initiated by OGG1 fully restored the intact DNA (Fig. 2B, lanes 6–9) and generated the products of the expected lengths at each intermediate stage (P1–P3 in Fig. 2B). However, when the repair was initiated by NEIL1, the reaction proceeded to β,δ-elimination (P4 in Fig. 2B) but the 3′-terminus was not processed further in the minimal BER system. The addition of increasing amounts of PNKP led to the appearance of the products of dGMP insertion by POLβ (Fig. 2C) but DNA ligation was not restored under the conditions used, possibly due to the competition between LIG3α and other proteins for DNA binding. The introduction of XRCC1, a scaffold protein that normally couples the action of APEX1, POLβ, and LIG3α, did not affect the efficiency of dGMP incorporation or DNA ligation (not shown). The requirement of other BER accessory factors for the completion of NEIL1-initiated repair of 8-oxoAde:Cyt remains possible. For example, Cockayne syndrome complementation group B protein (CSB) is known to increase the efficiency of 8-oxoAde repair in vivo [22] and to stimulate the activity of NEIL1 [23].
Fig. 3.
Two branches of 8-oxoAde repair in eukaryotes involving OGG1 (i→ii→v→vii→ix or i→iv→vi→viii→ix) and NEIL1 (i→iii→v→vii→ix).
Finally, we inquired what pathway of 8-oxoAde:Cyt repair will dominate when NEIL1 and OGG1 compete for this substrate. When both glycosylases were present together with APEX1, POLβ, and LIG3α, the repair pathway was almost exclusively initiated by OGG1 with little β,δ-elimination product observed (Fig. 2D). Here, we used POLβ bearing a triple substitution, K35A K68A K72A, which is totally deficient in the processing of the 2′-deoxyribo-5′-phosphate (step (vi) → (viii) in Fig. 3). Yet NEIL1, in agreement with earlier observations for other substrates [16], was able to supply this activity, indicating that it has access to the post-insertion BER intermediate. We conclude that the repair of 8-oxoAde:Cyt by NEIL1 is likely a back-up for the repair by OGG1 and presumably follows the pathway independent of APEX1 and involving PNKP (Fig. 3), which has been described for a more typical NEIL1 substrate, 5-hydroxyuracil [21].
The ability of OGG1 to process 8-oxoAde:Cyt pairs may be a fortuitous consequence of the structure of the OGG1 active site, which forms the same set of bonds with both 8-oxoGua and 8-oxoAde and is highly specific for the opposite Cyt [11]. Yet NEIL1 does not show strong opposite base-specificity for other substrates [13,18-20], making 8-oxoAde:Cyt the first example of the opposite base-specific substrate for this enzyme. The repair of 8-oxoAde by any other member of the Fpg/Nei family has never been reported. Under the conditions of efficient 8-oxoAde:Cyt cleavage by NEIL1 and OGG1, we observed that Fpg displayed very low excision of 8-oxoAde from pairs with Cyt and no excision of 8-oxoAde from other pairs (data not shown), in agreement with published data [10]. NEIL2, another eukaryotic Fpg/Nei homolog [17], also did not remove 8-oxoAde under any conditions (data not shown). In mouse liver nuclear extracts, 8-oxoAde is removed from pairs with Cyt mostly through the action of OGG1, since this activity is greatly reduced in ogg1−/− mice [12]. However, it cannot be excluded that NEIL1 may be required for 8-oxoAde repair in some specific circumstances, e.g. in certain tissues or developmental stages.
Neither OGG1 nor NEIL1 remove 8-oxoAde paired with Thy. The existence of two enzymes that can process 8-oxoAde:Cyt but not 8-oxoAde:Thy points to a possible biological importance of the former mispair. 8-OxoAde arising by DNA oxidation in the 8-oxoAde:Thy context is read by DNA polymerases mostly error-free [3-5]. Even if dCMP is incorporated opposite 8-oxoAde, as was observed in mammalian cells [4], removal of 8-oxoAde from this mispair would generate an A → G transition and thus be meaningless for preserving genetic information. However, 8-oxoAde:Cyt could also appear in DNA by misincorporation of 8-oxodAMP opposite a template Cyt from the oxidized dATP pool. Although very limited information is available for the use of 8-oxodATP by DNA polymerases [7], the ability of human pyrophosphatase MTH1 to efficiently hydrolyze 8-oxodATP [24] provides an indirect evidence for the pro-mutagenic potential of this oxidized precursor. Structurally, if not restrained by Watson–Crick bonding, 8-oxo-7,8-dihydro-2′-deoxyadenosine can assume a syn conformation, in which it can form hydrogen bonds to Cyt or Gua [25]. Clearly, a kinetic assessment of the ability of DNA polymerases to incorporate 8-oxodATP is required to evaluate the importance of various 8-oxoAde-containing mispairs and the mutagenic consequences of 8-oxoAde.
Acknowledgments
This research was supported by the RAS Presidium (22.14), RFBR (08-04-00596), and Russian Ministry of Education and Science (02.740.11.0079/NSh-652.2008.4). G.L.D. is supported by the Medical Research Council and Cancer Research UK.
Abbreviations
- BER
base excision repair
- LIG3α
DNA ligase IIIα
- NEIL1
endonuclease VIII-like protein 1
- OGG1
8-oxoguanine-DNA-glycosylase 1
- PNKP
polynucleotide kinase/3′-phosphatase
- POLβ
DNA polymerase β
References
- [1].Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. Oxford University Press; Oxford: 2007. [Google Scholar]
- [2].Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: induction, repair and significance. Mutat. Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- [3].Shibutani S, Bodepudi V, Johnson F, Grollman AP. Translational synthesis on DNA templates containing 8-oxo-7,8-dihydrodeoxyadenosine. Biochemistry. 1993;32:4615–4621. doi: 10.1021/bi00068a019. [DOI] [PubMed] [Google Scholar]
- [4].Kamiya H, et al. 8-Hydroxyadenine (7, 8-dihydro-8-oxoadenine) induces misincorporation in in vitro DNA synthesis and mutations in NIH 3T3 cells. Nucleic Acids Res. 1995;23:2893–2899. doi: 10.1093/nar/23.15.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kalam MA, Haraguchi K, Chandani S, Loechler EL, Moriya M, Greenberg MM, Basu AK. Genetic effects of oxidative DNA damages: comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res. 2006;34:2305–2315. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].von Sonntag C. Free-Radical-Induced DNA Damage and Its Repair: A Chemical Perspective. Springer; Berlin, Heidelberg: 2006. [Google Scholar]
- [7].Purmal AA, Kow YW, Wallace SS. 5-Hydroxypyrimidine deoxynucleoside triphosphates are more efficiently incorporated into DNA by exonuclease-free Klenow fragment than 8-oxopurine deoxynucleoside triphosphates. Nucleic Acids Res. 1994;22:3930–3935. doi: 10.1093/nar/22.19.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kuraoka I, et al. RNA polymerase II bypasses 8-oxoguanine in the presence of transcription elongation factor TFIIS. DNA Repair. 2007;6:841–851. doi: 10.1016/j.dnarep.2007.01.014. [DOI] [PubMed] [Google Scholar]
- [9].Machwe A, Ganunis R, Bohr VA, Orren DK. Selective blockage of the 3′→5′ exonuclease activity of WRN protein by certain oxidative modifications and bulky lesions in DNA. Nucleic Acids Res. 2000;28:2762–2770. doi: 10.1093/nar/28.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Girard PM, D’Ham C, Cadet J, Boiteux S. Opposite base-dependent excision of 7,8-dihydro-8-oxoadenine by the Ogg1 protein of Saccharomyces cerevisiae. Carcinogenesis. 1998;19:1299–1305. doi: 10.1093/carcin/19.7.1299. [DOI] [PubMed] [Google Scholar]
- [11].Zharkov DO, Rosenquist TA, Gerchman SE, Grollman AP. Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J. Biol. Chem. 2000;275:28607–28617. doi: 10.1074/jbc.M002441200. [DOI] [PubMed] [Google Scholar]
- [12].Jensen A, Calvayrac G, Karahalil B, Bohr VA, Stevnsner T. Mammalian 8-oxoguanine DNA glycosylase 1 incises 8-oxoadenine opposite cytosine in nuclei and mitochondria, while a different glycosylase incises 8-oxoadenine opposite guanine in nuclei. J. Biol. Chem. 2003;278:19541–19548. doi: 10.1074/jbc.M301504200. [DOI] [PubMed] [Google Scholar]
- [13].Jaruga P, Birincioglu M, Rosenquist TA, Dizdaroglu M. Mouse NEIL1 protein is specific for excision of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine from oxidatively damaged DNA. Biochemistry. 2004;43:15909–15914. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- [14].Sidorenko VS, Nevinsky GA, Zharkov DO. Specificity of stimulation of human 8-oxoguanine-DNA glycosylase by AP endonuclease. Biochem. Biophys. Res. Commun. 2008;368:175–179. doi: 10.1016/j.bbrc.2008.01.076. [DOI] [PubMed] [Google Scholar]
- [15].Parsons JL, Dianova II, Allinson SL, Dianov GL. DNA polymerase β promotes recruitment of DNA ligase IIIα-XRCC1 to sites of base excision repair. Biochemistry. 2005;44:10613–10619. doi: 10.1021/bi050085m. [DOI] [PubMed] [Google Scholar]
- [16].Grin IR, Khodyreva SN, Nevinsky GA, Zharkov DO. Deoxyribophosphate lyase activity of mammalian endonuclease VIII-like proteins. FEBS Lett. 2006;580:4916–4922. doi: 10.1016/j.febslet.2006.08.011. [DOI] [PubMed] [Google Scholar]
- [17].Zharkov DO, Shoham G, Grollman AP. Structural characterization of the Fpg family of DNA glycosylases. DNA Repair. 2003;2:839–862. doi: 10.1016/s1568-7864(03)00084-3. [DOI] [PubMed] [Google Scholar]
- [18].Rosenquist TA, Zaika E, Fernandes AS, Zharkov DO, Miller H, Grollman AP. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair. 2003;2:581–591. doi: 10.1016/s1568-7864(03)00025-9. [DOI] [PubMed] [Google Scholar]
- [19].Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl Acad. Sci. USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Morland I, Rolseth V, Luna L, Rognes T, Bjørås M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wiederhold L, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- [22].Tuo J, Jaruga P, Rodriguez H, Dizdaroglu M, Bohr VA. The Cockayne syndrome group B gene product is involved in cellular repair of 8-hydroxyadenine in DNA. J. Biol. Chem. 2002;277:30832–30837. doi: 10.1074/jbc.M204814200. [DOI] [PubMed] [Google Scholar]
- [23].Muftuoglu M, et al. Cockayne syndrome group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J. Biol. Chem. 2009;284:9270–9279. doi: 10.1074/jbc.M807006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fujikawa K, Kamiya H, Yakushiji H, Fujii Y, Nakabeppu Y, Kasai H. The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein. J. Biol. Chem. 1999;274:18201–18205. doi: 10.1074/jbc.274.26.18201. [DOI] [PubMed] [Google Scholar]
- [25].Cho BP, Evans FE. Structure of oxidatively damaged nucleic acid adducts. 3 Tautomerism, ionization and protonation of 8-hydroxyadenosine studied by 15N NMR spectroscopy. Nucleic Acids Res. 1991;19:1041–1047. doi: 10.1093/nar/19.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]