Abstract
For real-time 3D visualization of respiratory organ motion for MRI-guided therapy, a new adaptive 4D MR imaging method based on navigator echo and multiple gating windows was developed. This method was designed to acquire a time series of volumetric 3D images of a cyclically moving organ, enabling therapy to be guided by synchronizing the 4D image with the actual organ motion in real time. The proposed method was implemented in an open-configuration 0.5T clinical MR scanner. To evaluate the feasibility and determine optimal imaging conditions, studies were conducted with a phantom, volunteers, and a patient. In the phantom study the root mean square (RMS) position error in the 4D image of the cyclically moving phantom was 1.9 mm and the imaging time was ≈10 min when the 4D image had six frames. In the patient study, 4D images were successfully acquired under clinical conditions and a liver tumor was discriminated in the series of frames. The image quality was affected by the relations among the encoding direction, the slice orientation, and the direction of motion of the target organ. In conclusion, this study has shown that the proposed method is feasible and capable of providing a real-time dynamic 3D atlas for surgical navigation with sufficient accuracy and image quality.
Keywords: 4D MR imaging, MRI-guided therapy, respiratory motion
Intraoperative MRI has been recognized as a promising tool for surgical guidance over the past two decades. With the introduction of incision-free tumor heating by energy propagation, such as focused ultrasound and microwave, into MRI-guided therapy, the entire surgical process, including target localization, tumor treatment, and temperature monitoring, can be performed without damaging any surrounding normal tissues. Currently, MR-guided focused ultrasound surgery (MRgFUS) (1–8) is the clinically available option for such treatments by energy propagation from outside a patient’s body. Clinical cases for breast (4,6), uterine fibroid (5), brain (7), and bone (8) tumors have been reported in recent years. Another option for delivering energy to target tumor tissue is to use microwave radiation. Hyperthermia by microwave irradiation was proposed by Demura et al. (9) and they reported an animal experiment using an irradiation-type applicator with an experimental 7T MR scanner.
Integrating intraoperative MR imaging information and numerically controlled heat conduction devices improves the accuracy of treatment. MRI provides both a precise anatomical atlas and a temperature distribution. This information enables devices to target a heating area according to numerical position information and allows clinicians to monitor heat conduction in real time during therapy. Specifically, the lesion position and the treatment itself can be managed numerically throughout planning, treatment, and validation. For example, in clinical MRg-FUS systems the focal point of the ultrasound can be defined on an intraoperative MR image and controlled by computerized actuation of the applicator. The error between the planned heating area and the actual heated area has been reported to be within 1 mm for the liver tissue of a porcine model (10) and average 7.7 mm along the direction parallel to propagation of the ultrasound beam for the phantom (11).
Despite the capability of computerized targeting, however, its limited applicability to moving organs is an unresolved issue. Most organs in the abdominal cavity, such as the lungs and liver, move and deform during respiration, leading to a discrepancy between the target position on an intraoperative MR image and the actual target position in real space. Generally, the frame rate of intraoperative real-time MRI is less than 10 fps for 2D imaging, and at this frame rate these systems cannot capture motion of the target straying off the imaging slice. Integration of other imaging modalities, such as those using ultrasound (12,13) and x-ray radiation (14,15), with MRI has been discussed in the research community. These modalities have sufficiently high frame rates to track a moving target organ, and clinicians can choose a suitable imaging modality at any moment during therapy. From a technological point of view, integrating the coordinates used in each imaging modality is the key problem in this approach. There is also concern about finding space for additional imaging instruments in a narrow MRI gantry.
The problem of organ motion during therapy has also been raised in the field of radiotherapy planning and the use of 4D computed tomography (CT) has been proposed for lung radiotherapy (16–18). In those works the authors acquired a time series of 3D images throughout the period of a respiratory cycle in order to analyze the motion of a tumor. Another way to analyze the motion of a target is 4D MRI. Today’s fast imaging sequence and parallel imaging technologies allow the acquisition of 3D images within 1 sec and can provide a time series of images of a moving structure nearly in real time. Blackall et al. (19) proposed using dynamic 3D MRI to plan radiotherapy for lung cancer. They achieved an acquisition time of 330 ms/image by using fast field echo with echo planar imaging (FFE-EPI). Since the image quality of the dynamic 3D images was insufficient to show structures in detail, they employed image registration with a breath-holding 3D image. Instead of using fast 3D imaging, von Siebenthal et al. (20) proposed 4D imaging using internal respiratory gating. They used fast 2D imaging (180–190 ms/frame) both to detect the respiratory phase and to acquire multislice images and they reconstructed a 4D image by postprocessing.
Using 4D imaging would also be helpful in MRI-guided thermal therapy with energy propagation, and it might be even more suitable than with radiotherapy, since images could be acquired with the position in exactly the same posture as in the treatment. The clinical scenario for using 4D imaging in treatment is as follows: 1) A time series of 3D images of the target organ throughout the whole respiratory cycle is acquired just before treatment. 2) Real-time tracking of the target organ is performed during treatment to detect the patient’s respiratory phase. 3) A best-fit 3D image is selected at each moment during the operation according to the real-time respiratory phase parameter and a pseudo-real-time 3D image is provided to the clinicians.
In the research described here we developed and evaluated a 4D MR imaging method as a key technique for capturing organ motion and deformation in the clinical scenario described above. The method was designed to acquire multiple volumes during repetitive respiration by applying the navigator echo technique (21) with multiple gating windows. Unlike 4D imaging based on a fast imaging approach, the proposed method provides correspondence between the 3D frames and the respiratory phase and it allows us to synchronize the 4D image with the actual target motion in real time in order to show a best-fit 3D image during therapy. The idea of using multiple gating windows has been proposed for automatic selection of optimal gating windows (22,23), but only a few volumes were acquired at the peaks of the histogram of motion in the latter work (23). Furthermore, none of these previous works sought to capture and visualize organ motion specifically to guide therapy, or to evaluate the position reproducibility of the 4D image. We have thus developed an adaptive imaging method to acquire a series of 3D images covering the whole range of respiration by using the gating method. As described in this article, we evaluated the position accuracy, image quality, and imaging time of the 4D images obtained by the proposed method by conducting studies with a novel respiration-simulating phantom, volunteers, and a patient in a clinical setting.
MATERIALS AND METHODS
Adaptive 4D Imaging
The proposed adaptive 4D imaging technique is an extension of respiratory gating. In gating, the respiratory signal is monitored during image acquisition and MR imaging echoes are accepted only while the respiratory signal is within the range of a predefined gating window. In our proposed method, instead of using a single gating window the entire range of the respiratory signal is segmented into several partitions, called bins. Each bin is associated with a k-space for each frame in the final 4D image, and each k-space is filled with MR echo data acquired while the respiratory signal is in the associated bin. Therefore, the number of bins equals to the number of frames in the final 4D image, Nφ.
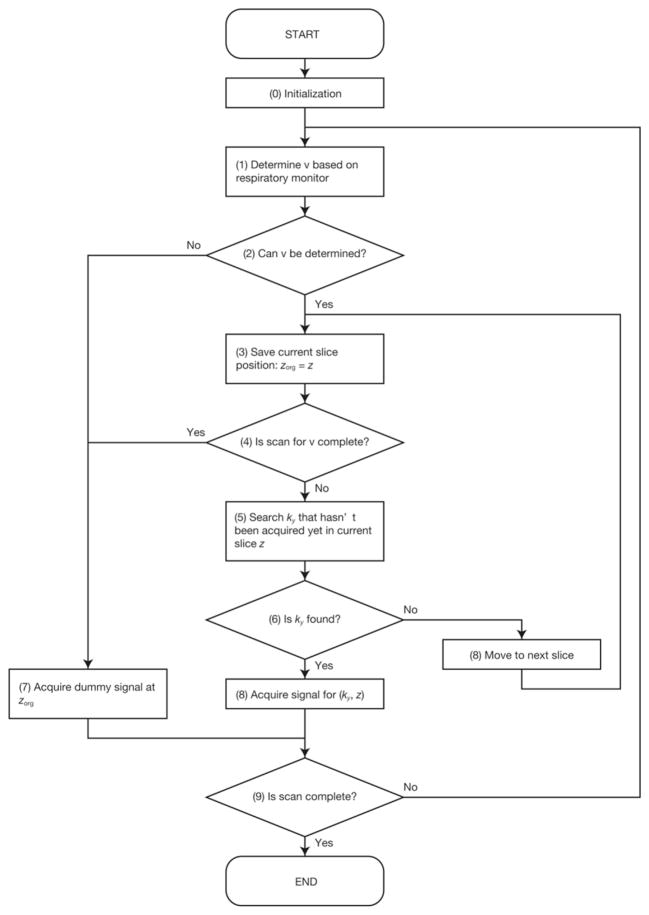
In conventional 2D multislice gradient echo or spin echo imaging, MR echoes are acquired by changing the phase encoding parameter ky and the slice offset z so as to scan a ky × z parameter space. Using multiple bins instead of a single gating window, however, makes it difficult to determine the trajectory to scan a ky × z parameter space before starting the scan, since the respiratory signal is unpredictable. Therefore, ky and z should be adaptively updated right before each MR echo acquisition according to the current respiratory signal. Figure 1 shows a flowchart illustrating the adaptive control of the scan parameters. Maintaining a steady state of magnetization during MR echo acquisition involves two important steps: 1) The slice position z is fixed as long as the scan of ky has not been completed. 2) If z has been updated from the previous slice excitation, several dummy MR echo signals are acquired. After all bins are filled with MR echoes, 2D image reconstruction is performed on the k-space for each frame and, finally, a 4D image is generated.
FIG. 1.
Procedure to determining the imaging parameters ky and z according to the current volume number ν.
Real-time Respiratory Monitoring
To monitor the respiratory signal in real time during a scan the shift of the liver and diaphragm of the subject was detected by the navigator echo technique (21). Navigator echoes were acquired from a sagittal slice, given reports that the liver moves mostly along the superior-to-inferior axis during respiration (24–26). A pulse sequence, in which navigator echoes were acquired every Nintv signal acquisitions, was developed. Nintv was configurable, and the sampling rate of respiratory monitoring was 1/(TR × Nintv) fps, where TR is the repetition time of the MR echo acquisitions. A lower Nintv is preferable in terms of the temporal resolution of respiratory signal monitoring, but on the other hand, frequent acquisition of navigator echoes prolongs the imaging time. In this study, Nintv = 5 was used. To minimize the respiratory signal error due to the temporal resolution of navigator echo acquisition and the duration of processing navigator echoes, the respiratory signal at each imaging echo acquisition was estimated by extrapolating the last three sampled signals. The bins were defined by partitioning the range of respiratory signal variation during preparatory monitoring, in which the respiratory signal was recorded during a few cycles of respiration. A template waveform was generated from the data recorded in this preparatory monitoring. As illustrated in Fig. 2, two partition methods to determine the widths of the bins were used: (A) Δsh = const., and (B) Th = const., for h = 1,2,…,Nφ, where Δsh is the width of each partition of respiratory signal s, and Th is the total duration for which s is in the hth bin. The advantage of using method (B) is that the echo signals are distributed to each partition evenly and, therefore, the imaging time can be reduced without compromising Nφ.
FIG. 2.
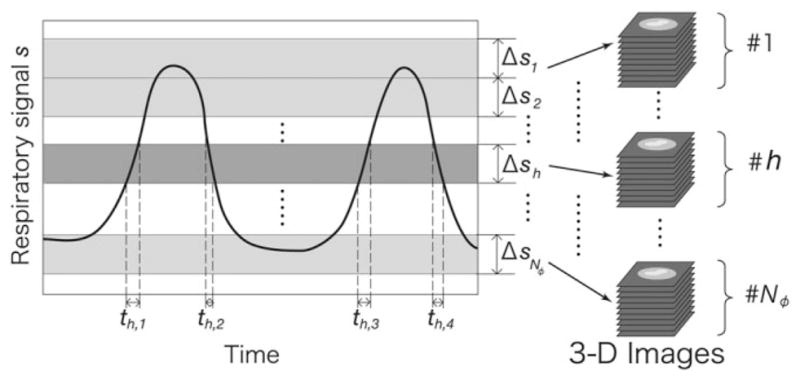
The range of respiratory phase variation is split into Nφ partitions. Two methods to determine the width of each partition were implemented: (A) Δsh = const., and (B) Th = const., for h = 1,2,…,Nφ, where Δsh is the width of each partition of the respiratory signal s, and Th is the total duration for which s is in the hth bin. Th is defined by Th = th,1 + th,2 + th,3 + … in the graph.
System Integration
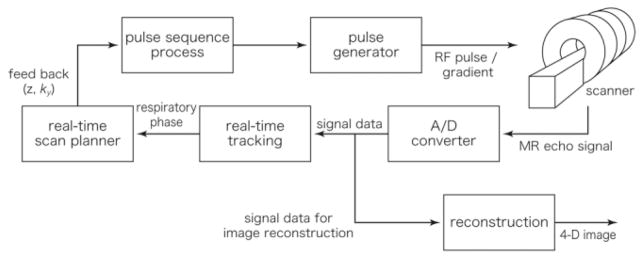
The proposed imaging method was implemented in an open-configuration, 0.5T clinical MR scanner system (GE Signa SP/i, GE Healthcare, Chalfont St. Giles, UK). Figure 3 shows a diagram of the system. A 2D pulse sequence based on fast gradient echo (FGRE) was developed for the adaptive 4D imaging by using the GE EPIC development environment. A data processing PC (OS: RedHat Linux 9.0; CPU: Intel Pentium 4, 2.8 GHz; Memory: 512 MB) was connected to a host workstation for the scanner system by 100 Base-T Ethernet. Both the imaging and navigator echoes were transferred from the scanner system to the data processing PC at every echo acquisition. The navigator echoes were processed on the fly to detect the subject’s respiratory signal by calculating the subject’s shift from the baseline position. This calculation included Fourier transformation of the echo data and matching of the profile data. Then the respiratory signal at the next signal acquisition was estimated from the last three sampled signals, stored in the data processing PC, and fed back to the scanner. With this system the respiratory signal data was obtained within 75 ms after each signal acquisition. The phase encoding parameter ky and the slice offset z were determined in a pulse sequence running on the scanner system, according to the estimated respiratory signal. The imaging echoes were stored in the data processing PC and the 4D image was reconstructed after all necessary echoes were acquired.
FIG. 3.
System diagram for real-time tracking and adaptive 4D MR imaging integrated with a clinical MR scanner. The z and ky parameters in the pulse sequence program are updated while the sequence is running, according to the feedback from the real-time tracking.
Phantom Study
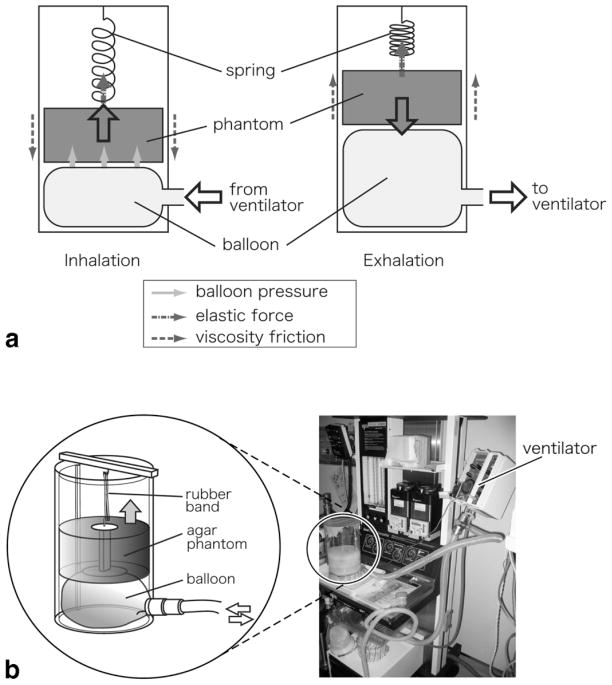
To determine optimal imaging parameters for MRI-guided therapy, a phantom study was performed to evaluate the quality, position accuracy, and acquisition time of the 4D images under several conditions. Since these factors are strongly affected by the motion profile of the subject we developed a moving phantom to simulate human liver motion due to respiration and to standardize a motion profile for comparison. To simulate the respiratory motion we roughly modeled the mechanism of human breathing according to the literature (27). In the human breathing process the liver is forced inferiorly as the diaphragm contracts during inhalation and it returns passively as a result of the elastic force from the abdomen during exhalation. At the same time, liver motion is also affected in a viscous manner by the inflow/outflow of air from the lungs. Therefore, the motion of the liver can be modeled as follows:
| [1] |
where x is the shift of the liver or diaphragm, Ml is the mass of the liver, Fd is the force from the diaphragm, k is the spring constant of the elastic force from the abdomen, and C represents the viscous effect from the lung. Figure 4 shows the phantom mechanism that we developed according to Eq. [1]. It consists of a solid agar phantom, an acrylic resin cylinder, a balloon, and a rubber band (acting as a spring). The phantom was inserted into the cylinder in the vertical direction and moved only along this direction. The phantom was driven by an MRI-compatible ventilator (ventiPAC 5, Pneupac, Luton, UK), which was connected to the balloon under the agar phantom, thus lifting the phantom while the balloon was inflating. This process corresponds to inhalation. For exhalation, the phantom dropped down while the balloon shrank. The speed of the phantom was restricted by the rubber band connecting the phantom and the top of the cylinder and by the friction between the phantom and the cylinder wall. The forces in the vertical direction can thus be described as:
| [2] |
where Fb is the lifting force from the balloon, M is the mass of the agar phantom, kr is the spring constant of the rubber band, and Cp is the “drag coefficient” for the friction between the phantom and the cylinder. x is the shift of the phantom, with the origin at the lowest position, and x0 is the point at which the rubber band is not stretched. Since Mg + krx0 = 0 in a steady state, Eq. [2] can be rewritten as:
| [3] |
FIG. 4.
Photograph and schematic illustrating the mechanism of the phantom simulating respiratory motion during the inhalation and exhalation phases. The agar phantom was inserted into an acrylic resin cylinder with an internal diameter of 140 mm and a height of ≈110 mm. A balloon was placed under the agar phantom and connected to a ventilator to lift up the phantom during inflation. In addition to the force from the balloon, the phantom hung from a spring (a rubber band) connected to the top of the cylinder in order to control the speed of the phantom while moving up and down.
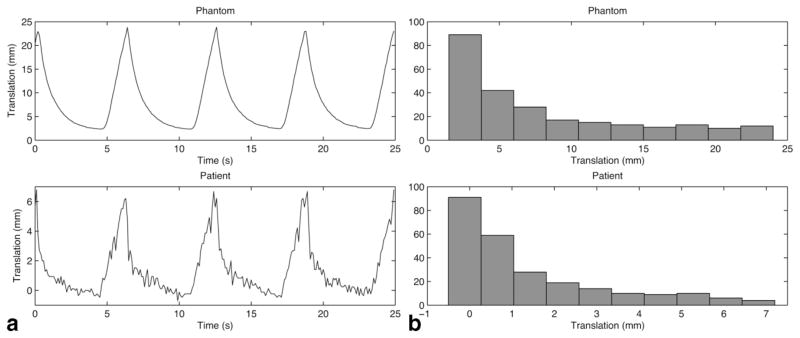
This is equivalent to the right side of Eq. [1]. Figure 5 shows the motion pattern and histogram of both the respiration-simulating phantom and the liver of a patient under general anesthesia. To obtain image contrast, four small vials filled with water were placed inside the solid agar phantom.
FIG. 5.
Respiratory patterns and histograms of the respiratory motion of the phantom (upper) and a patient under general anesthesia (lower). The translation of the liver in the superior–inferior direction, as measured by navigator echo, was plotted to observe the patient’s respiratory pattern. 250 points were sampled every 100 ms (corresponding to four cycles of respiration), and the translation distribution was calculated for each set of data.
The phantom was placed in the gantry as shown in Fig. 6 and it was driven during image acquisition with durations of 2.0 sec for inhalation and 4.0 sec for exhalation. The spring and the airflow of the ventilator were adjusted so that the stroke of the phantom motion was 25 mm. The imaging parameters were as follows: TR/TE/flip: 20 ms/9.3 ms/30°; bandwidth (BW): 12.5 kHz; field of view (FOV): 24 × 24 cm; matrix: 256 × 128. For each frame of a 4D image 25 slices were acquired, with a 5-mm slice thickness and 0-mm spacing. The navigator echoes to detect the 1D “simulated” respiratory motion of the phantom were acquired from the X-Y plane, so that the respiratory signal was equivalent to the translation of the phantom in this experiment. To evaluate the translation on each 2D slice image in each 4D image, the default imaging slice direction was in the X-Y plane. Three sets of 4D images were acquired: set 1 was acquired with varying encoding directions; set 2 was acquired with Nφ = 4, 6, 8, and 10 by using bins based on method (A); and set 3 was acquired using bins determined by both methods (A) and (B). The respiratory signals detected by the navigator echoes were recorded to the data processing PC to evaluate the error of respiratory signal extrapolation retrospectively. In addition to these image sets, a 3D image of the phantom at rest was acquired using a normal 2D multislice FGRE sequence with the same parameters given above in order to generate an “ideal” 4D image by shifting the 3D image according to the stroke of the phantom motion, the method of determining the bins, and Nφ. The position of the phantom in the hth frame of the “ideal” 4D image was defined by sh = (Sh + Sh+1)/2, where Sh is the border respiratory signal (equal to the translation) between the hth and (h+1)th bins. The generated 4D image was used as a “gold standard” for evaluating the image quality and image position error of each 4D image.
FIG. 6.
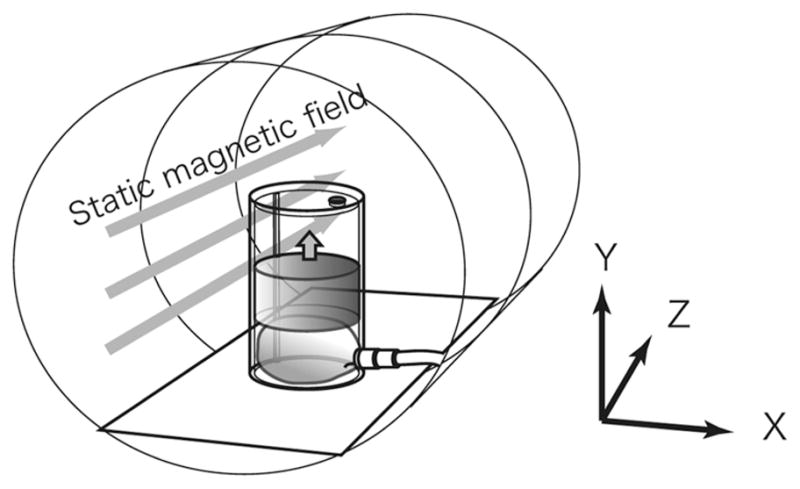
Coordinate system for the phantom study, which was based on the coordinate system for the MRI scanner, where the static magnetic field is in the Z direction. The phantom was placed at the center of the magnet bore, so that its motion was in the Y direction.
Volunteer Study
To evaluate the feasibility of the adaptive 4D imaging, 4D images of the livers of four healthy volunteers were acquired under near-clinical conditions while the volunteers were breathing freely. The imaging parameters were: TR/TE/flip: 20 ms/8.9 ms/30°; BW: 12.5 kHz; FOV: 30 × 30 cm; matrix: 256 × 128. The slice thickness and spacing were 7 mm and 0 mm, respectively, which were the same values as those usually used in clinical cases of liver imaging. The optimal number of frames Nφ and number of slices Nz were selected for each volunteer so that the 4D images covered enough volume to visualize the motion of the liver. The respiratory signals were assessed retrospectively to validate if extrapolation of the respiratory signal based on prior samples was acceptable for the volunteer’s respiration.
Clinical Study
To assess the feasibility of the proposed imaging method in a real clinical situation, a 4D image of a patient’s liver was acquired during a clinical case of MRI-guided microwave ablation therapy of liver tumors, which was approved by the Ethics Committee of Shiga University of Medical Science. Signed informed consent of this procedure including the use of images for research was obtained from the patient. The patient was under general anesthesia, with a ventilated duration of 2 sec for inhalation and 4 sec for exhalation. The imaging parameters were: TR/TE/flip: 20 ms/8.9 ms/30°; BW: 12.5 kHz; FOV: 30 × 30 cm; matrix: 256 × 128. Ten slices/frame were acquired, with a slice thickness of 7 mm and a spacing of 0 mm. The number of frames was Nφ = 4. For comparison, a single 3D image of the liver was also acquired using a 3D fast spoiled gradient echo (FSPGR) sequence, with an FOV of 30 × 30 × 18 cm, matrix of 256 × 128 × 36, and TR/TE/flip of 10 ms/2 ms/30°.
RESULTS
Phantom Study
The image quality of set 1 was evaluated by visual observation. Figure 7 shows that motion artifact appeared in the images when the phantom moved in the phase-encoded direction. The figure also indicates that the contrast between the agar area and the water in the vials was low when the imaging slice was perpendicular to the motion of the phantom. For sets 2 and 3, we calculated the position error of the imaged phantom in each slice between the acquired 4D images and the “ideal” 4D image, the correlation coefficient of the acquired and ideal 4D images, and the signal-to-noise ratio (SNR). Table 1 lists the imaging conditions and the results for the imaging time, image position error, and image quality of each 4D image. Throughout the phantom study the RMS error between the extrapolated respiratory signal and the true respiratory signal was 0.5 mm, where the true respiratory signal was obtained by linear interpolation of prior and posterior samples.
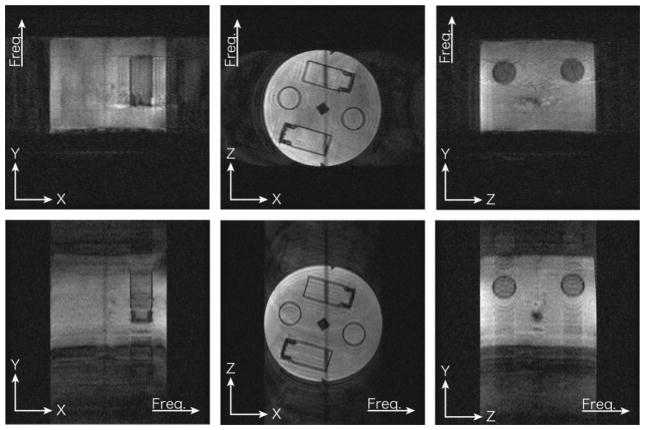
FIG. 7.
Parts of 4D images acquired from X-Y, X-Z, and Y-Z planes. For each plane orientation two datasets were acquired exchanging frequency and phase-encoded directions.
Table 1.
Imaging Time, Root Mean Square Error (RMSE) in Each 4D Image Slice, Signal-to-Noise Ratio (SNR), and Cross-Correlation (CC) with an “Ideal” 4D Image, Evaluated by Varying Nφ in the Partition Methods
| Nφ | Partition | Time | RMSE(mm) | SNR | CC |
|---|---|---|---|---|---|
| 4 | (A) | 12′ 01 | 2.2 | 80 | 0.952 |
| 6 | (A) | 21′ 57 | 1.9 | 79 | 0.953 |
| 8 | (A) | 29′ 15 | 1.8 | 73 | 0.949 |
| 10 | (A) | 38′ 79 | 1.9 | 71 | 0.949 |
| 6 | (B) | 10′ 12 | 1.7 | 76 | 0.942 |
Volunteer Study
Seven 4D datasets were successfully acquired for the four volunteers. Table 2 lists the imaging parameters (Nφ, Nz, and the slice and encoding directions), total acquisition time, and RMS error of the extrapolated respiratory signal for each 4D image. Figures 8 and 9 show parts of the 4D images of the livers in the sagittal and axial planes, respectively. The imaging time ranged from about 10–20 min, and the liver, blood vessels, and other structures in the abdominal area could be readily discriminated.
Table 2.
Imaging Conditions (Nφ, Nz, Slice Direction, and Frequency-Encoded Direction), Imaging Time, and RMSE Between Extrapolated Respiratory Signal (ERS) and True Respiratory Signal for Four Volunteers
| Volunteer no. | Nφ | Nz | Slice Dir. | Freq. Dir | Time | RMSE – ERS (mm) |
|---|---|---|---|---|---|---|
| 1 | 6 | 18 | SAG | S-I | 9′ 04 | 1.1 |
| 1 | 6 | 18 | SAG | A-P | 11′ 46 | 1.0 |
| 2 | 6 | 22 | SAG | A-P | 21′ 55 | 1.1 |
| 3 | 6 | 24 | AX | A-P | 15′ 01 | 1.2 |
| 3 | 6 | 24 | AX | R-L | 18′ 14 | 1.3 |
| 4 | 5 | 18 | SAG | S-I | 8′ 16 | 0.9 |
| 4 | 5 | 18 | SAG | S-I | 10′ 03 | 1.4 |
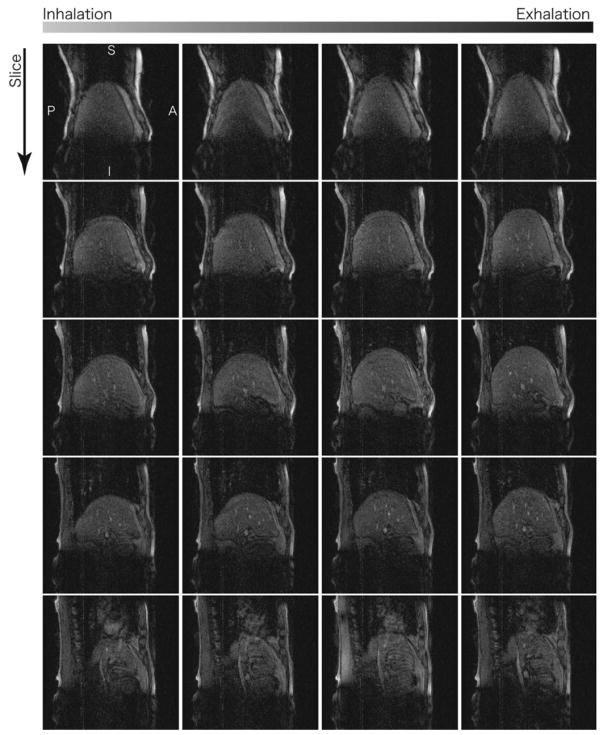
FIG. 8.
Subset of 4D images of a volunteer’s liver, acquired along sagittal planes by adaptive 4D imaging with Nφ = 6 and Nz = 18. The images in the bottom row include the area of the heart, and artifact due to the cardiac motion is observed.
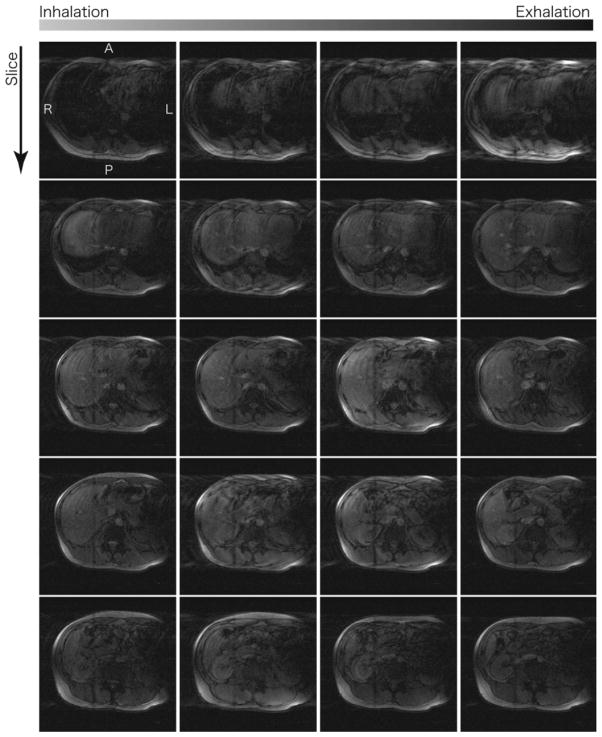
FIG. 9.
Subset of 4D images of the same volunteer’s liver, acquired along axial planes by adaptive 4D imaging with Nφ = 6 and Nz = 24. As observed in Fig. 8, the images in the top row shows strong artifact due to the cardiac motion.
Figure 10 illustrates the relationship between the encoding direction and the image quality. In the FOV there were two major moving parts, the liver and the chest/abdominal wall, which moved in different directions. These images show that more significant motion artifact appeared when the chest/abdominal wall moved along the phase-encoding direction than when the liver moved along the phase-encoding direction.
FIG. 10.
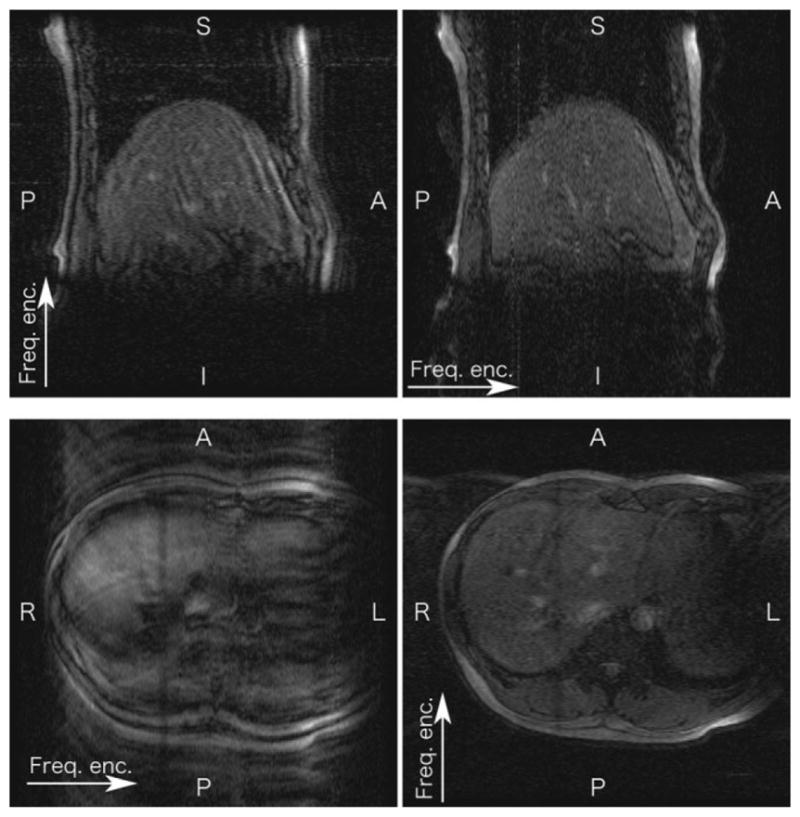
Images showing that the encoding direction affected the 4D image quality. Sagittal images were acquired by setting the frequency-encoding direction to S-I (upper left) and A-P (upper right). Axial images were acquired by setting the frequency-encoding direction to R-L (lower left) and A-P (lower right).
Clinical Study
A 4D image of the patient’s liver, including the area of the tumor, was successfully acquired in 10.5 min. Figure 11 shows this 4D image of the patient and the corresponding normal, breath-holding 3D image. The liver tumor was readily discriminated in each frame except for the second one.
FIG. 11.
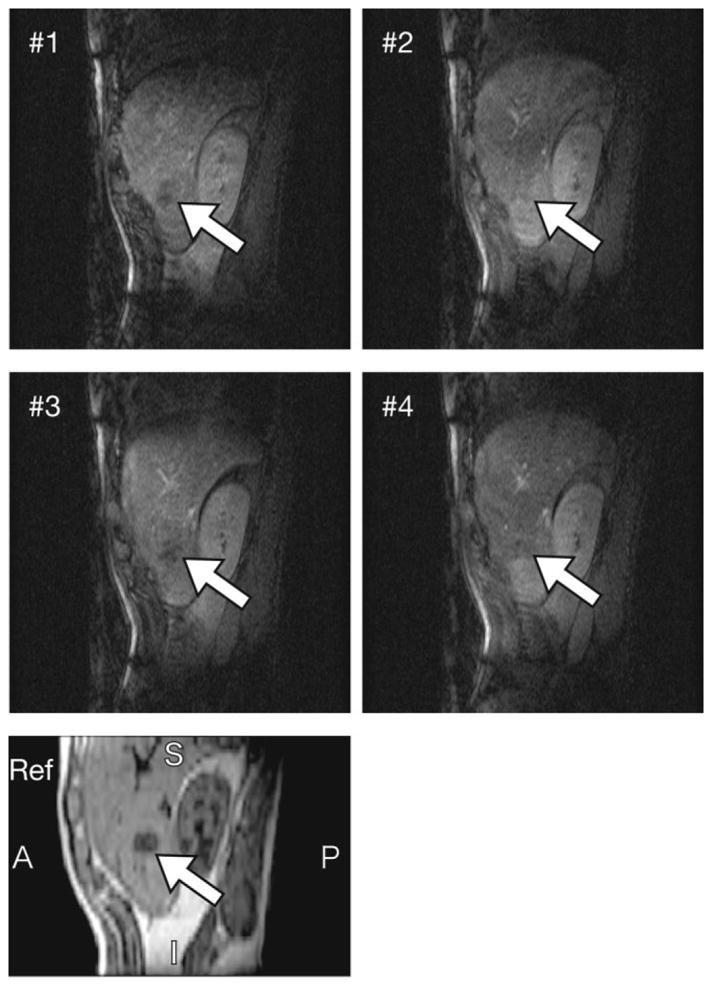
Subset of intraoperative 4D images of the patient, acquired during MRI-guided microwave coagulation therapy under general anesthesia. 2D images showing the liver tumor at four different respiratory phases are shown, with the tumor indicated in each image. In addition, part of a breath-holding 3D image is shown below as a reference.
DISCUSSION
We have demonstrated a new adaptive method for acquiring 4D images of a moving organ by using navigator echoes and multiple gating windows. This method has the scope to generate pseudo-real-time 3D images during therapy, especially tumor heating by energy propagation, by selecting the best-fit 3D image from the 4D image. A respiration-simulating phantom and the livers of four volunteers and one clinical patient were imaged by the proposed method. The image position accuracy, image quality, and imaging time were evaluated, with the focus on application to guiding therapy by obtaining a 4D image without requiring patients to hold their breath. The proposed method does not require any fast imaging technology (e.g., parallel imaging) and the 4D images were successfully acquired with an open-configuration 0.5T clinical MR scanner. This is an advantage over any previously proposed 4D MR imaging method and our method could expand the range of application of 4D imaging.
The RMS position error between the 4D images acquired with this method and an “ideal” 4D image was less than 1.9 mm when Nφ ≥6. This is thought to be a clinically acceptable error, because the typical size of tumors requiring treatment is on the order of tens of millimeters, the ablative focus area in MRgFUS is a jellybean-shaped region 5 mm in diameter and 25 mm in length (10), and the heated area in hyperthermia by microwave irradiation is 10–20 mm in diameter (9). To improve the accuracy of 4D imaging, a larger Nφ is theoretically preferable, since the position error of a 4D image is affected by the width of the bins and the stroke of motion. In the proposed method, each frame in a 4D image is reconstructed from MR echo signals acquired while the subject was in the range of the corresponding bin. Assuming a uniform distribution of positions at which MR echoes are acquired, the RMS error can be estimated as , where Δs is the bin width. When the respiratory signal is equivalent to the translation of the subject, and bin width is determined by method (A), as described above, Δs is calculated as L/Nφ, where L is the stroke of the subject’s motion. In contrast to this estimation, the position accuracy cannot be improved by increasing Nφ if it is greater than a certain value (6 in this study). The results indicate that there were other sources of error besides the bin width. Other possible sources of error would be spatial and temporal resolution of navigator-echo-based tracking. Spatial resolution of navigator echo is inherently limited by pixel size, which was 0.94 mm/pixel for the phantom study. Insufficient temporal resolution prolongs the time gap between the respiratory signal sampling and imaging signal acquisition and leads to misselection of bin. The time resolution was compensated by simple linear extrapolation of the navigator position based on three prior samples, but still the RMS error between extrapolated respiratory signal (signal for scanner control) and interpolated signal (actual respiratory signal) was 0.5 mm in the phantom study.
Irregularity of respiratory motion could cause several problems. Irregularity of the respiratory cycle affects the accuracy of extrapolated respiratory signal, and it should be validated when the method is applied to free-breathing subjects. For the volunteer study, retrospective assessment of the recorded respiratory signal showed that the RMS errors of the extrapolation ranged from 0.9 to 1.4 mm. These errors were much less than the size of the bins, where the stroke of liver motion is 10–20 mm and Nφ is 6, and therefore it is safe to say that the respiratory signal error due to the extrapolation was within the acceptable level for this study. Another irregularity issue is irregular orientation of motion. Since a plane to acquire navigator echoes is optimized before imaging to obtain reasonable respiratory data, unpredictable motion causes a change in waveform of navigator echoes, and results in a large error of respiratory signal. In most cases irregular signal could be excluded by out-of-range check, but further assessment is needed to know if the method is applicable for individual cases.
The imaging time was almost proportional to Nφ and could be reduced nearly 40% by changing the bin partitioning to method (B), as seen from Table 1. In the case of human liver imaging under free breathing, the histograms of motion were quite different from subject to subject, and we obtained no strong relations among Nφ, Nz, and the imaging time. The volunteer and clinical studies showed, however, that 4D images could be acquired in practical time, and images with a matrix of 256 × 128 × 24 × 5 were acquired in 15–18 min. Compared to past related work on 4D imaging based on fast 2D imaging (180–190 ms/image for 2D) with retrospective sorting (20), in which obtaining a 4D image with a matrix of 192 × 192 × 30 and temporal resolution of 2.8 Hz took 15 min to 1 hr, our proposed adaptive 4D imaging method achieved much more effective signal collection, despite using conventional slow imaging (2560 ms/image for 2D).
The phantom and volunteer studies showed that the slice and phase-encoding directions affected the image quality. For better image contrast, the slice direction should be the same as the target organ’s direction of motion. This is because different slices are selected for each excitation, and since the imaging slice in this case would be perpendicular to the motion, the magnetization would be almost recovered by the next time the same area is excited. When the slice orientation is parallel to the motion, the TR for each slice in the target organ can be kept constant. The phase-encoding direction affected the SNR of the image, with increasing motion artifact when the subject was moving along this direction. This can be explained by the inconsistency of the signal phase along the phase-encoding direction because of the bin width. In human imaging there are generally several objects moving in different directions, and Fig. 10 shows that the motion artifacts in the 4D image were mainly caused by the motion of the chest/abdominal wall, which is the highest intensity area in the FOV under these imaging conditions. Consequently, a suitable condition for the liver is imaging along the sagittal plane with phase-encoding in the superior–inferior direction.
In the clinical study the liver tumor was successfully discriminated, except in one frame. The problem we experienced in this study, however, was that we did not have enough time to find the optimal slice for navigator echo and the template waveform for the respiratory signal in the real clinical situation. This is the reason why Nφ was set at 4 to reduce the imaging time, since an inaccurate template waveform prolongs the imaging time. Therefore, methods for finding an optimal navigator echo setting and generating a template waveform are important issues for clinical application of the proposed 4D imaging approach.
Currently, Nφ and the slice thickness are the two major factors limiting the accuracy of MRI-guided therapy. Assuming that the range of a target’s motion is 20 mm, and that the target moves with constant speed, the error between the actual target and the image of the target, resulting from the discreteness of the frames, reaches a maximum of more than 3 mm when Nφ is 6. Therefore, the 3D frame interpolation technique (28) is expected to be introduced to reduce this kind of error. As for the slice thickness, it must be more than 7 mm for the MR scanner used in this study to obtain a sufficiently high SNR to observe anatomical structure, and the thickness limits the resolution of detecting motion in the direction perpendicular to the imaging slice. This problem could be solved by developing an adaptive 4D imaging method based on 3D imaging. In contrast to 2D multislice imaging, which was used in this study, 3D imaging introduces slice encoding to decompose signals in the slice direction, and a thinner slice thickness can be used without compromising the SNR.
CONCLUSION
We have proposed an adaptive 4D MR imaging technique for intraoperative MRI. The image position accuracy, image quality, and imaging time were assessed by applying this technique with a respiration-simulating phantom. Volunteer and clinical studies showed that the proposed method is feasible for clinical use and capable of providing a real-time, dynamic 3D atlas for surgical navigation.
Acknowledgments
We thank Drs. Yoshimasa Kurumi and Shigeyuki Naka for supporting the intraoperative imaging in the clinical study. We also thank Dr. Nobuhiko Hata for initiating this joint project.
Grant sponsor: Research Fellowships of the Japan Society for the Promotion of Science (JSPS) for Young Scientists.
References
- 1.Hynynen K, Darkazanli A, Unger E, Schenck JF. MRI-guided noninvasive ultrasound surgery. Med Phys. 1993;20:107–115. doi: 10.1118/1.597093. [DOI] [PubMed] [Google Scholar]
- 2.Cline HE, Hynynen K, Watkins RD, Adams WJ, Schenck JF, Ettinger RH, Freund WR, Vetro JP, Jolesz FA. Focused US system for MR imaging-guided tumor ablation. Radiology. 1995;194:731–737. doi: 10.1148/radiology.194.3.7862971. [DOI] [PubMed] [Google Scholar]
- 3.Graham SJ, Chen L, Leitch M, Peters RD, Bronskill MJ, Foster FS, Henkelman RM, Plewes DB. Quantifying tissue damage due to focused ultrasound heating observed by MRI. Magn Reson Med. 1999;41:321–328. doi: 10.1002/(sici)1522-2594(199902)41:2<321::aid-mrm16>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 4.Huber PE, Jenne JW, Rastert R, Simiantonakis I, Sinn HP, Strittmatter HJ, von Fournier D, Wannenmacher MF, Debus J. A new noninvasive approach in breast cancer therapy using magnetic resonance imaging-guided focused ultrasound surgery. Cancer Res. 2001;61:8441–8447. [PubMed] [Google Scholar]
- 5.Hindley J, Gedroyc WM, Regan L, Stewart E, Tempany C, Hynyen K, McDannold N, Inbar Y, Itzchak Y, Rabinovici J, Kim HS, Geschwind JF, Hesley G, Gostout B, Ehrenstein T, Hengst S, Sklair-Levy M, Shushan A, Jolesz F. MRI guidance of focused ultrasound therapy of uterine fibroids: early results. AJR Am J Roentgenol. 2004;183:1713–1719. doi: 10.2214/ajr.183.6.01831713. [DOI] [PubMed] [Google Scholar]
- 6.Hynynen K, Pomeroy O, Smith DN, Huber PE, McDannold NJ, Kettenbach J, Baum J, Singer S, Jolesz FA. MR imaging-guided focused ultrasound surgery of fibroadenomas in the breast: a feasibility study. Radiology. 2001;219:176–185. doi: 10.1148/radiology.219.1.r01ap02176. [DOI] [PubMed] [Google Scholar]
- 7.Hynynen K, McDannold N, Clement G, Jolesz FA, Zadicario E, Killiany R, Moore T, Rosen D. Pre-clinical testing of a phased array ultrasound system for MRI-guided noninvasive surgery of the brain—a primate study. Eur J Radiol. 2006;59:149–156. doi: 10.1016/j.ejrad.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Catane R, Beck A, Inbar Y, Rabin T, Shabshin N, Hengst S, Pfeffer RM, Hanannel A, Dogadkin O, Liberman B, Kopelman D. MR-guided focused ultrasound surgery (MRgFUS) for the palliation of pain in patients with bone metastases—preliminary clinical experience. Ann Oncol. 2007;18:163–167. doi: 10.1093/annonc/mdl335. [DOI] [PubMed] [Google Scholar]
- 9.Demura K, Morikawa S, Murakami K, Sato K, Shiomi H, Naka S, Kurumi Y, Inubushi T, Tani T. An easy-to-use microwave hyperthermia system combined with spatially resolved MR temperature maps: phantom and animal studies. J Surg Res. 2006;135:179–186. doi: 10.1016/j.jss.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 10.Kopelman D, Inbar Y, Hanannel A, Freundlich D, Castel D, Perel A, Greenfeld A, Salamon T, Sareli M, Valeanu A, Papa M. Magnetic resonance-guided focused ultrasound surgery (MRgFUS): ablation of liver tissue in a porcine model. Eur J Radiol. 2006;59:157–162. doi: 10.1016/j.ejrad.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Gorny KR, Hangiandreou NJ, Hesley GK, Gostout BS, McGee KP, Felmlee JP. MR guided focused ultrasound: technical acceptance measures for a clinical system. Phys Med Biol. 2006;51:3155–3173. doi: 10.1088/0031-9155/51/12/011. [DOI] [PubMed] [Google Scholar]
- 12.Piron CA, Causer P, Jong R, Shumak R, Plewes DB. A hybrid breast biopsy system combining ultrasound and MRI. IEEE Trans Med Imaging. 2003;22:1100–1110. doi: 10.1109/TMI.2003.816951. [DOI] [PubMed] [Google Scholar]
- 13.Gunther M, Feinberg DA. Ultrasound-guided MRI: preliminary results using a motion phantom. Magn Reson Med. 2004;52:27–32. doi: 10.1002/mrm.20140. [DOI] [PubMed] [Google Scholar]
- 14.Fahrig R, Butts K, Rowlands JA, Saunders R, Stanton J, Stevens GM, Daniel BL, Wen Z, Ergun DL, Pelc NJ. A truly hybrid interventional MR/X-ray system: feasibility demonstration. J Magn Reson Imaging. 2001;13:294–300. doi: 10.1002/1522-2586(200102)13:2<294::aid-jmri1042>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 15.Fahrig R, Butts K, Wen Z, Saunders R, Kee ST, Sze DY, Daniel BL, Laerum F, Pelc NJ. Truly hybrid interventional MR/X-ray system: investigation of in vivo applications. Acad Radiol. 2001;8:1200–1207. doi: 10.1016/S1076-6332(03)80702-X. [DOI] [PubMed] [Google Scholar]
- 16.Low DA, Nystrom M, Kalinin E, Parikh P, Dempsey JF, Bradley JD, Mutic S, Wahab SH, Islam T, Christensen G, Politte DG, Whiting BR. A method for the reconstruction of four-dimensional synchronized CT scans acquired during free breathing. Med Phys. 2003;30:1254–1263. doi: 10.1118/1.1576230. [DOI] [PubMed] [Google Scholar]
- 17.Vedam SS, Keall PJ, Kini VR, Mostafavi H, Shukla HP, Mohan R. Acquiring a four-dimensional computed tomography dataset using an external respiratory signal. Phys Med Biol. 2003;48:45–62. doi: 10.1088/0031-9155/48/1/304. [DOI] [PubMed] [Google Scholar]
- 18.Pan T, Lee TY, Rietzel E, Chen GT. 4D-CT imaging of a volume influenced by respiratory motion on multi-slice CT. Med Phys. 2004;31:333–340. doi: 10.1118/1.1639993. [DOI] [PubMed] [Google Scholar]
- 19.Blackall JM, Ahmad S, Miquel ME, McClelland JR, Landau DB, Hawkes DJ. MRI-based measurements of respiratory motion variability and assessment of imaging strategies for radiotherapy planning. Phys Med Biol. 2006;51:4147–4169. doi: 10.1088/0031-9155/51/17/003. [DOI] [PubMed] [Google Scholar]
- 20.von Siebenthal M, Szekely G, Gamper U, Boesiger P, Lomax A, Cattin P. 4D MR imaging of respiratory organ motion and its variability. Phys Med Biol. 2007;52:1547–1564. doi: 10.1088/0031-9155/52/6/001. [DOI] [PubMed] [Google Scholar]
- 21.Ehman RL, Felmlee JP. Adaptive technique for high-definition MR imaging of moving structures. Radiology. 1989;173:255–263. doi: 10.1148/radiology.173.1.2781017. [DOI] [PubMed] [Google Scholar]
- 22.Jhooti P, Gatehouse PD, Keegan J, Bunce NH, Taylor AM, Firmin DN. Phase ordering with automatic window selection (PAWS): a novel motion-resistant technique for 3D coronary imaging. Magn Reson Med. 2000;43:470–480. doi: 10.1002/(sici)1522-2594(200003)43:3<470::aid-mrm20>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 23.Kolmogorov VN, Watts R, Prince MR, Zabih R, Wang Y. Simultaneous multiple volume (SMV) acquisition algorithm for real-time navigator gating. Magn Reson Imaging. 2003;21:969–975. doi: 10.1016/s0730-725x(03)00203-0. [DOI] [PubMed] [Google Scholar]
- 24.Harauz G, Bronskill MJ. Comparison of the liver’s respiratory motion in the supine and upright positions: concise communication. J Nucl Med. 1979;20:733–735. [PubMed] [Google Scholar]
- 25.Weiss PH, Baker JM, Potchen EJ. Assessment of hepatic respiratory excursion. J Nucl Med. 1972;13:758–759. [PubMed] [Google Scholar]
- 26.Shimizu S, Shirato H, Xo B, Kagei K, Nishioka T, Hashimoto S, Tsuchiya K, Aoyama H, Miyasaka K. Three-dimensional movement of a liver tumor detected by high-speed magnetic resonance imaging. Radiother Oncol. 1999;50:367–370. doi: 10.1016/s0167-8140(98)00140-6. [DOI] [PubMed] [Google Scholar]
- 27.Tomlinson SP, Tilley DG, Burrows CR. Computer simulation of the human breathing process. IEEE Eng Med Biol. 1994;13:115–124. [Google Scholar]
- 28.Schreibmann E, Chen GT, Xing L. Image interpolation in 4D CT using a BSpline deformable registration model. Int J Radiat Oncol Biol Phys. 2006;64:1537–1550. doi: 10.1016/j.ijrobp.2005.11.018. [DOI] [PubMed] [Google Scholar]