Abstract
The selenoprotein S (SELS) is a putative receptor for serum amyloid A, and recent studies have suggested that SELS may be a link between type 2 diabetes mellitus and inflammation. Genetic studies of SELS polymorphisms have revealed associations with circulating levels of inflammatory markers and hard end points of cardiovascular disease. In this study, we analyzed SELS expression in subcutaneous adipose tissue and SELS genotype in relation to metabolic risk factors. DNA microarray expression analysis was used to study the expression of SELS in lean and obese siblings from the Swedish Obese Subjects Sib Pair Study. TaqMan genotyping was used to analyze 3 polymorphisms, previously found to be associated with circulating levels of inflammatory markers, in the INTERGENE case-control study of myocardial infarction and unstable angina pectoris. Possible associations between SELS genotype and/or expression with anthropometry and measures of metabolic status were investigated. Real-time polymerase chain reaction was used to analyze the SELS expression in isolated human adipocytes incubated with insulin. In lean subjects, we found correlations between SELS gene expression in subcutaneous adipose tissue and measures of obesity (waist, P = .045; sagittal diameter, P = .031) and blood pressure (diastolic, P = .016; systolic P = .015); and in obese subjects, we found correlations with measures of obesity (body mass index, P = .03; sagittal diameter, P = .008) and glycemic control (homeostasis model assessment of insulin resistance, P = .011; insulin, P = .009) after adjusting for age and sex. The 5227GG genotype was associated with serum levels of insulin (P = .006) and homeostasis model assessment of insulin resistance (P = .007). The expression of SELS increased after insulin stimulation in isolated human adipocytes (P = .008). In this study, we found an association between both SELS gene expression in adipose tissue and SELS genotype with measures of glycemic control. In vitro studies demonstrated that the SELS gene is regulated by insulin in human subcutaneous adipocytes. This study further supports a role for SELS in the development of metabolic disease, especially in the context of insulin resistance.
1. Introduction
Selenoprotein S (SELS; also known as SEPS1, AD-015, or SELENOS) was recently described as an endoplasmic reticulum (ER) and plasma membrane-located selenoprotein involved in the physiologic adaptation to ER stress [1-4]. The SELS gene is known to be expressed in a wide variety of tissues and cell types, including tissues important for glycemic control such as adipose tissue, muscle, and liver [1,4-6]. In omental adipose tissue from diabetic subjects, gene expression of SELS is increased compared with that in nondiabetic controls; and in both groups, SELS expression correlates with homeostasis model assessment of insulin resistance (HOMA-IR) [7]. In HepG2 cells, SELS expression is inhibited by glucose; and SELS has been suggested to be involved in glucose homeostasis in an animal model of type 2 diabetes mellitus [6] and in human diabetic subjects [5]. Furthermore, in HepG2 and intestinal epithelial cells, proinflammatory cytokines activate the transcription of SELS [8,9]. The activation of SELS expression is influenced by a promoter polymorphism (rs28665122); and human studies have shown that SELS genotype is associated with circulating levels of proinflammatory cytokines, such as tumor necrosis factor–α (TNF-α) and interleukin-1β (IL-1β), suggesting that SELS plays a role in inflammation [10]. SELS is within a region of chromosome 15q26.3 that exhibits copy number variation in approximately 5% of individuals of European descent [11], but the frequency of this variation in the Swedish population is not known. It has also been shown that SELS is a putative receptor for the acute phase protein serum amyloid A (SAA) [6]. Consequently, it has been speculated that SELS may be a link between type 2 diabetes mellitus and inflammation. Recently, it was shown that SELS is secreted from hepatoma cells and that it associates with low-density lipoprotein (LDL) particles [12], suggesting a role also in lipid metabolism. Genetic studies of SELS polymorphisms in relation to cardiovascular disease have revealed that variation in the SELS locus is associated with coronary heart disease (CHD) and ischemic stroke in women [13].
Previously, genetic analyses of SELS in different cohorts have been performed [9,10,13,14]; and associations between SELS polymorphisms and inflammation [10] and hard end points in cardiovascular disease [13] have been found. In this study, we wanted to also include measures of glycemic control, lipid metabolism, and other risk factors as well as investigate possible associations with adipose tissue expression of SELS to further explore the role of SELS and the association with metabolic disease.
2. Materials and methods
2.1. Subjects and samples
2.1.1. Ethics statement
Regional Ethical Review Boards approved the studies, and all participants gave written informed consent.
2.1.2. The INTERGENE study
INTERGENE is a population-based research program assessing the INTERplay between GENEtic susceptibility and environmental factors for the risk of chronic diseases in western Sweden. The study population consists of randomly selected women and men, aged 25 to 74 years (at time of sampling), living in the Västra Götaland Region. The subjects included in this study are part of the INTERGENE case-control study, comprising 1236 (906 men and 330 women) well-characterized patients, of which 618 have suffered acute coronary symptoms (myocardial infarction or unstable angina pectoris) and 618 are matched healthy controls. The study is described in detail elsewhere [15] and at http://www.sahlgrenska.gu.se/intergene/. Body weight and height were measured to the nearest 0.1 kg and 1 cm, respectively. Waist circumference was measured at a level midway between the lower rib margin and iliac crest, and hip circumference was measured as the maximum perimeter over the buttocks. Blood pressure (BP) was measured twice in each person, after a 10-minute rest, with a validated (Gohara) automatic device (Omron 711 Automatic IS; Omron Healthcare, Vernon Hills, IL) while the subjects were in a supine position. Blood samples were collected after 4-hour fasting for immediate serum lipid (total cholesterol, high-density lipoprotein [HDL] cholesterol, LDL cholesterol, and triglycerides) and glucose analysis. Serum total cholesterol and triglyceride concentrations were determined with enzymatic assays. Serum HDL cholesterol concentrations were measured after dextran sulfate-magnesium precipitation of apolipoprotein B–containing lipoproteins. Serum glucose was analyzed with a hexokinase method (Roche Hitachi 917 and Roche ModularP; Roche, Basel, Switzerland). Measurements of serum insulin and high-sensitivity C-reactive protein (hs-CRP) were performed at the Sahlgrenska University Hospital.
2.1.3. The Swedish Obese Subjects Sib Pair study
The Swedish Obese Subjects (SOS) Sib Pair study includes 154 nuclear families with body mass index (BMI)–discordant sibling pairs (BMI difference ≥10 kg/m2), resulting in a study population of 732 subjects. In this study, the most extreme siblings according to BMI were chosen in each family [16]. Sex discordant sib pairs were excluded, resulting in 78 pairs of sisters and 12 pairs of brothers. Measurements of anthropometry, fat mass (FM), fat-free mass (FFM), BP, fasting glucose, total cholesterol, triglycerides, HDL cholesterol, LDL cholesterol, serum insulin, serum C-peptide, and hs-CRP were performed at the Sahlgrenska University Hospital. Dual-energy x-ray absorptiometry was performed with LUNAR DPX-L (Scanexport Medical, Helsingborg, Sweden). The dual-energy x-ray absorptiometry generates a 3-compartment model consisting of FM, lean tissue mass, and bone mineral content. The FFM was calculated as lean tissue mass + bone mineral content.
2.2. Isolation and incubation of adipocytes
For isolation of adipocytes, subcutaneous adipose tissue was obtained from 5 women (BMI, 38 ± 17 kg/m2) and 4 men (BMI, 43 ± 10 kg/m2) [17]. One man and one woman received drug treatment for type 2 diabetes mellitus. Adipocytes were isolated from adipose tissue as previously described [18]. Subcutaneous adipocytes were diluted 10 times in modified Eagle medium and incubated with or without insulin (Sigma-Aldrich, St Louis, MO; 1 mU/mL, corresponding to 6 nmol/L) for 6 hours at 37°C in a gently shaking water bath. Total RNA was isolated from the adipocytes using the Lipid Tissue RNeasy Kit (Qiagen, Hilden, Germany).
2.3. Real-time polymerase chain reaction analysis
Reagents for real-time polymerase chain reaction (PCR) analysis of SELS and low-density lipoprotein receptor-related protein 10 (LRP10) (Assays-on-Demand, TaqMan Reverse Transcriptase reagents, and TaqMan GeneExpression Master mix) were purchased from Applied Biosystems (Foster City, CA), and conditions according to the manufacturer's protocol were used. Complementary DNA (cDNA) synthesis was performed in a total reaction volume of 20 μL using 200 ng total RNA. cDNA corresponding to 10 ng RNA per reaction was used for real-time PCR amplification. Amplification and detection of specific products were performed using the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems) using default cycle parameters. Working standards were prepared from a large pool of adipocyte RNA, and standard cDNA was synthesized in parallel with the sample cDNAs. A standard curve was plotted for each primer probe set with a serial dilution of the cDNA from the pooled adipose tissue RNA. Human LRP10 was used as reference to normalize the expression levels between samples [19].
2.4. Microarray analysis in the SOS Sib Pair study
Adipose tissue was obtained by needle aspirations in the paraumbilical area. Total RNA, cDNA, and hybridization (Human Genome U133 plus 2.0, Affymetrix, Santa Clara, CA) was performed as previously described [20-22]. Data were analyzed using robust multiarray average. SELS expression was analyzed using probe set 223209_s_at, and SAA expression was analyzed using probe set 214456_x_at.
2.5. Genotype analysis
Three SELS polymorphisms (Table 1), previously reported to be associated with serum levels of IL-1β, IL-6, and/or TNF-α [10], were selected for analysis. The C-105T polymorphism (rs28665122) is located within the SELS promoter, the C3705T polymorphism (rs4965814) is located in intron 5, and the A5227G polymorphism (rs4965373) is located within the 3′UTR of exon 6 [10]. Validated genotyping assays for the C3705T and the A5227G polymorphisms, a custom assay for the C-105T polymorphism (primer and probe sequences are available upon request), and TaqMan Universal PCR Master mix were purchased from Applied Biosystems and used according to the manufacturer's protocol. The ABI PRISM 7900HT Sequence Detection System (Applied Biosystems) was used for amplification and detection. To ensure genotyping quality control, negative controls were included in all genotyping plates. The DNA samples were transferred from 96-well stock plates to 384-well working plates using a Beckman Biomek FX (Beckman Coulter, CA) robotic system to minimize pipetting errors and to help eliminate sample plating errors. The genotyping success rate was 93%, which resulted in a total of 1150 subjects included in the full analysis and 535 matched case-control pairs available for paired analysis.
Table 1.
Information on the studied SELS polymorphisms
| SNP IDa | Location | dbSNP ID | SNP sequence [minor/major allele] | Minor allele frequency (%) |
|---|---|---|---|---|
| C-105T | 5′UTR | rs28665122 | GTCGTGGTCC[T/C]GGCCAATCGC | 14 |
| C3705T | Intron 5 | rs4965814 | TACAGCTCAG[C/T]GTTTAAGGTC | 18 |
| A5227G | 3′UTR | rs4965373 | AGTAATAGTT[A/G]GAGGTTGTAA | 33 |
National Center for Biotechnology Information SNP data used as reference.
2.6. Statistics
Statistical analyses were performed using SPSS (version 16.0; SPSS, Chicago, IL). Values are given as mean ± SD unless stated otherwise. In the INTERGENE study, parameter difference for recessive and dominant models was assessed using independent samples t test. Multiple linear regression adjusted for diabetes status (subjects treated with insulin and/or other diabetic drug treatment at inclusion, with self-reported diabetes in questionnaire, or with serum glucose greater than 6.7 mmol/L), sex, and age were used to investigate possible associations between genotype and measured parameters. Only control subjects without lipid-lowering medication (eg, statin treatment) were included in the regression analysis of genotype effect on serum lipid levels. The genotype was coded to fit an additive model in the regression analyses. The Mantel-Haenszel test was used to test for association between genotype and risk for acute coronary symptoms. Significant deviation from the Hardy-Weinberg equilibrium was tested for each single nucleotide polymorphism (SNP) using a χ2 test. Linkage disequilibrium and haplotype frequencies were estimated using Haploview software version 4.1 [23]. PHASE 2.1 [24] was used to estimate the most likely haplotypes for each subject and to test if haplotype frequencies differed between cases and controls.
In the SOS Sib Pair study, correlation between SELS expression, SAA expression, and anthropometric and biochemical markers was performed using the Spearman rank correlation test. Partial correlation was used to control for sex, age, and BMI when appropriate. To obtain approximate normal distributions of expression data, microarray signals in the whole SOS Sib Pair study offspring cohort (n = 359) were transformed using Box-Cox power transformations. Subsequently, expression data were standardized to mean = 0 and variance = 1. Differences in gene expression between lean and obese siblings as well as SELS gene expression in isolated adipocytes incubated with or without insulin were assessed using a paired t test. A P value < .05 (2-sided) was considered statistically significant. This study is explorative in nature. Hence, we did not adjust for multiple testing.
3. Results
3.1. Association between SELS expression and metabolic risk factors in the SOS Sib Pair study
To further investigate SELS in relation with metabolic risk factors, we used the SOS Sib Pair study [16] to study SELS expression in relation to measures of glycemic control, blood lipids, anthropometry, and body composition. In all subjects, we found significant correlation only with serum levels of HDL cholesterol (r = −0.21, P = .005) when adjusting for sex, age, and BMI. In the lean subjects, we found significant correlations between SELS gene expression in subcutaneous adipose tissue and waist circumference, sagittal diameter, FFM, as well as systolic and diastolic BP after adjusting for age and sex (Table 2). In the obese subjects, we found significant correlations with BMI, sagittal diameter, serum levels of HDL cholesterol, triglycerides, insulin, and HOMA-IR after adjusting for age and sex (Table 2). No association was found between SELS and the expression of its putative ligand SAA after adjusting for age and sex in either group. Obese subjects had slightly but significantly higher SELS expression levels than lean subjects (P = .047; microarray signal, 45.1 ± 6.9 and 42.9 ± 7.3, respectively).
Table 2.
Spearman correlation coefficients between SELS mRNA expression in adipose tissue and clinical parameters in the SOS Sib Pair studya
| Lean (n=90) |
Obese (n=90) |
|||||
|---|---|---|---|---|---|---|
| Mean ± SD | r | r adj | Mean ± SD | r | r adj | |
| BMI (kg/m2) | 22.0 ± 1.7 | 0.19 | 0.15 | 37.7 ± 5.3 | 0.17 | 0.25⁎ |
| Waist (cm) | 77.0 ± 6.1 | 0.24⁎ | 0.23⁎ | 113.8 ± 12.4 | 0.04 | 0.17 |
| WHR | 0.80 ± 0.06 | 0.15 | 0.10 | 0.94 ± 0.07 | −0.07 | 0.13 |
| Sagittal diameter (cm) | 18 ± 18 | 0.26⁎ | 0.24⁎ | 27 ± 37 | 0.15 | 0.31† |
| FM (kg) | 17.6 ± 4.0 | 0.16 | 0.16 | 46.2 ± 9.5 | 0.30† | 0.21 |
| FFM (kg) | 47.2 ± 7.5 | 0.32† | 0.36‡ | 57.8 ± 8.2 | −0.01 | 0.16 |
| Systolic BP (mm Hg) | 106.9 ± 9.5 | 0.27⁎ | 0.27⁎ | 121.3 ± 17.3 | −0.04 | −0.11 |
| Diastolic BP (mm Hg) | 65.3 ± 8.9 | 0.26⁎ | 0.27⁎ | 73.9 ± 11.4 | 0.11 | 0.19 |
| Total cholesterol (mmol/L) | 4.2 ± 0.9 | 0.04 | 0.02 | 4.6 ± 0.8 | −0.16 | −0.08 |
| Triglyceride (mmol/L) | 0.7 ± 0.4 | 0.15 | 0.12 | 1.3 ± 0.9 | 0.182 | 0.27⁎ |
| HDL cholesterol (mmol/L) | 1.4 ± 0.4 | −0.20 | −0.15 | 1.2 ± 0.3 | −0.30† | −0.35† |
| LDL cholesterol (mmol/L) | 2.5 ± 0.6 | 0.11 | 0.08 | 2.8 ± 0.6 | −0.13 | −0.07 |
| Glucose (mmol/L) | 4.7 ± 1.1 | 0.01 | 0.05 | 5.2 ± 1.0 | −0.08 | 0.02 |
| Insulin (mU/L) | 5.6 ± 2.8 | −0.08 | −0.05 | 13.7 ± 9.7 | 0.20 | 0.30† |
| HOMA-IR | 1.2 ± 0.7 | −0.07 | −0.04 | 3.3 ± 3.0 | 0.18 | 0.30⁎ |
| C-peptide (mmol/L) | 0.5 ± 0.2 | 0.21 | 0.21 | 0.9 ± 0.5 | 0.15 | 0.21 |
| Hs-CRP (mg/L) | 2.1 ± 5.5 | −0.01 | 0.01 | 6.8 ± 7.4 | 0.06 | 0.05 |
| SAA (microarray signal) | 367 ± 130 | 0.01 | 0.13 | 448 ± 163 | 0.21⁎ | 0.10 |
Spearman correlation (r) or partial correlation adjusted for sex and age (r adj) between SELS gene expression and clinical parameters was performed. Note that r adj was calculated for n = 79 and n = 73 in the lean and obese group, respectively.
P ≤ .05.
P ≤ .01.
P < .001.
3.2. SELS expression in insulin-stimulated adipocytes
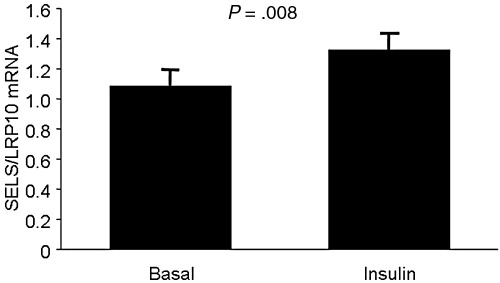
To further investigate the link between glycemic control and SELS gene expression, we analyzed the effect of insulin stimulation on isolated subcutaneous adipocytes and found that the expression of SELS increased after insulin stimulation (Fig. 1, P = .008). Note that all subjects displayed increased expression after insulin exposure, except for one of the diabetic subjects in which the expression was unchanged. The increase was still significant after both type 2 diabetes mellitus subjects were excluded (P = .01).
Fig. 1.
SELS expression in isolated human adipocytes stimulated with insulin in vitro (n = 9). LRP10 was used as a reference to normalize the expression levels between the samples. Values are presented as mean ± SEM.
3.3. Association between SELS genotype and cardiovascular and metabolic risk factors in the INTERGENE study
Using independent t tests, we found associations between the A5227G polymorphism and diastolic BP (P = .049), serum levels of HDL cholesterol (P = .049) and insulin (P = .006), and HOMA-IR (P = .007) for the dominant model for minor allele (AA + GA vs GG). For the recessive model for minor allele (AA vs AG + GG), an association was found between the A5227G polymorphism and diastolic BP (P = .019). For the C3705T and C-105T polymorphisms, no associations were found.
When the polymorphisms were included in a multiple linear regression model adjusting for diabetes status, sex, and age, the A5227G polymorphism was associated with diastolic BP (P = .008) and serum levels of insulin (P = .039) in all subjects. No association was found between the C-105T polymorphism and measured parameters, whereas the C3705T polymorphism was associated with serum levels of glucose (P = .043) and HOMA-IR (P = .034).
When analyzing the data using multiple linear regression in groups of cases and controls, we found associations only in the cases between the C-105T polymorphism with serum levels of insulin (P = .026) and HOMA-IR (P = .014), between the C3705T polymorphism with serum levels of insulin (P = .032) and HOMA-IR (P = .015), and between the A5227G polymorphism and serum levels of insulin (P = .038). In cases, we also found associations with diastolic BP (P = .048), BMI (P = .046), and waist-to-hip ratio (WHR) (P = .038) for the A5227G polymorphism, and an association with WHR (P = .036) for the C3705T polymorphism.
The associations between SELS genotype and BP in the whole cohort remained significant also when omitting subjects on antihypertensive treatment.
Using the Mantel-Haenszel test, we found that the 5227GG genotype was overrepresented among cases (odds ratio = 1.28; 95% confidence interval, 1.008 1.636; P = .043). No overrepresentation was found among cases for the other 2 polymorphisms.
The analyzed polymorphisms, A5227G, C3705T, and C-105T, were all in Hardy-Weinberg equilibrium and had minor allele frequencies comparable to those reported by Curran et al [10] (Table 1). Patient characteristics of the matched case-control pairs are shown in Table 3. Haplotype estimation identified 3 haplotypes with frequencies greater than 5% (CTG, 49%; CTA, 32%; and TCG, 13% in the order C-105T, C3705T, and A5227G); and together, these represented greater than 93% of haplotype diversity. No single haplotype was overrepresented among cases. See supplement for details on cohort and subject characteristics divided by genotype.
Table 3.
Characteristics of the subjects included in the paired SELS polymorphism analysis
| Cases (n=535) | Controls (n=535) | |
|---|---|---|
| Diabetic (n) | 125 | 51 |
| Male/female (n) | 383/152 | 383/152 |
| BMI (kg/m2) | 28.0 ± 4.3 | 26.8 ± 3.6 |
| WHR | 0.95 ± 0.08 | 0.91 ± 0.08 |
| Systolic BP (mm Hg) | 133.8 ± 21.1 | 142.6 ± 21.3 |
| Diastolic BP (mm Hg) | 81.8 ± 11.1 | 85.4 ± 10.5 |
| Triglyceride (mmol/L) | 1.7 ± 1.3 | 1.5 ± 0.8 |
| Total cholesterol (mmol/L) | 4.6 ± 1.1 | 5.8 ± 1.0 |
| HDL cholesterol (mmol/L) | 1.4 ± 0.4 | 1.6 ± 0.4 |
| LDL cholesterol (mmol/L) | 2.5 ± 0.9 | 3.5 ± 0.9 |
| Glucose (mmol/L) | 6.1 ± 2.3 | 5.5 ± 1.3 |
| Insulin (mU/L) | 14.1 ± 15.6 | 8.5 ± 6.7 |
| Hs-CRP (mg/L) | 4.7 ± 9.7 | 2.4 ± 4.0 |
4. Discussion
In this study, we analyzed SELS expression in subcutaneous adipose tissue and SELS genotype in relation to metabolic risk factors. SELS genotype was associated with measures of glycemic control and diastolic BP. Furthermore, SELS gene expression was associated with measures of glycemic control and serum levels of HDL cholesterol in obese subjects, whereas SELS gene expression and BP were associated in lean subjects. SELS gene expression was also correlated with measures of obesity in both lean and obese subjects. We have also shown that SELS gene expression is stimulated by insulin in isolated human adipocytes.
SELS is expressed in 3 primary insulin target tissues: liver, muscle, and adipose tissue [6]. In an animal model of type 2 diabetes mellitus, the sand rat Psammomys obesus, it has been shown that SELS (tanis) is expressed in the liver in inverse proportion to circulating glucose and insulin levels [6]. It has also been shown that SELS expression is inhibited by glucose in cultured hepatocytes and that overexpression of SELS causes a reduction in glucose uptake in cultured hepatoma cells [1,6]. In humans, it has been shown that, in type 2 diabetes mellitus patients, SELS messenger RNA (mRNA) levels in subcutaneous adipose tissue tended to increase after insulin infusion, whereas no effect was seen in nondiabetic subjects [5]. In contrast, we found that SELS mRNA was up-regulated after insulin stimulation of isolated human subcutaneous adipocytes in nondiabetic subjects. Furthermore, we found that SELS mRNA was correlated with levels of insulin in obese subjects.
No correlation was seen between SELS mRNA levels in adipose tissue and glucose, insulin, or blood lipid levels in the study by Karlsson et al [5]. However, that study only included 10 type 2 diabetes mellitus subjects and 11 controls. In this study, we found positive correlations between SELS mRNA levels in subcutaneous adipose tissue and sagittal diameter in both lean (n = 90) and obese (n = 90) subjects after adjustment for sex and age. Furthermore, we found positive correlations between SELS mRNA levels in subcutaneous adipose tissue and waist circumference, FFM, and systolic and diastolic BP in the lean siblings and positive correlations with BMI, FM, serum levels of triglycerides and HDL cholesterol, insulin, and HOMA-IR in the obese siblings. Previous studies suggest a difference in SELS gene regulation depending on type 2 diabetes mellitus status in both subcutaneous [5] and omental adipose tissue [7]. Unfortunately, the cohort used for expression analysis in this study consists of relatively young subjects; and very few have been diagnosed with type 2 diabetes mellitus. Furthermore, no omental adipose tissue biopsies were obtained in this study.
It has been speculated that the interaction between the SAA1 protein and the putative receptor SELS [6] could be involved in the development of insulin resistance, and a recent study in rats showed that the expression level of SELS in liver was higher in diabetic rats compared with both healthy rats and diabetic rats treated with the insulin sensitizer rosiglitazone [25]. We show in this study that SELS expression is correlated with both insulin and HOMA-IR in the obese siblings, but no correlation was found in the lean group. In obese subjects, the levels of SAA in adipose tissue are higher than those in lean subjects, a fact that could possibly have effects on the ligand-receptor signaling even without large effects on SELS expression. However, further experiments using in vitro or in vivo models are needed to investigate this hypothesis.
SELS, and its interaction with SAA, has been proposed to be a mechanistic link between type 2 diabetes mellitus, inflammation, and cardiovascular disease [6]. Previous studies have shown that, in humans, both SAA and SELS adipose tissue expression are correlated with serum levels of SAA [5,26]. However, the relation between SELS and SAA expression in adipose tissue has previously not been studied. Although there was a weak correlation in the obese sibling group, the correlation was lost after adjusting for sex and age and was not present in the lean group, suggesting that SELS and SAA gene expression levels are not closely related in subcutaneous adipose tissue in humans.
Obesity and metabolic disease including atherosclerosis and insulin resistance are associated with modest but chronically elevated levels of circulating inflammatory markers [27,28]. It has previously been reported that 3 different SELS polymorphisms are associated with increased levels of circulating proinflammatory cytokines [10]. In this study, we analyzed these 3 polymorphisms in the context of the metabolic syndrome including risk factors such as obesity, measures of glycemic control, lipid metabolism, and BP. Using multiple regression analysis, we found associations between the C3705T polymorphism and measures of glycemic control (HOMA-IR and serum levels of glucose) and between the A5227G polymorphism and serum levels of insulin as well as diastolic BP. Using independent t tests, we found that the 5227GG genotype was associated with serum levels of insulin and HOMA-IR. Previous studies have suggested that the SELS gene is dysregulated in diabetes, and the data presented here indicate that the C3705T and/or the A5227G polymorphisms may play a role in this context. However, the C-105T polymorphism, which showed the strongest association with circulating proinflammatory cytokines [10] but no direct association with inflammatory bowel disease [9] or rheumatoid arthritis [14], was not associated with metabolic risk factors analyzed in this study.
In the study by Alanne et al [13], 2 polymorphisms, rs8025174 and rs7178239 (not analyzed in the INTERGENE study), showed significant associations with CHD and stroke, respectively. These associations were most pronounced in women. In this study, we found a weak association between risk of CHD and the A5227G polymorphism. No association with CHD was found for this polymorphism in the study by Alanne et al; but it was associated with serum levels of TNF-α, IL-1β, and IL-6 in the study by Curran et al [10]. It has to be considered that the number of subjects included in this study is about half of the number of subjects included in the study by Alanne et al [13]; and unfortunately, there were too few women in the cohort to perform the analysis split by sex. This may explain why the results for association with cardiovascular disease are different in the INTERGENE cohort.
In conclusion, we found an association between SELS gene expression and SELS genotype with measures of glycemic control. In vitro studies showed that the SELS gene is regulated by insulin in subcutaneous adipocytes. This study further supports a role for SELS in the development of metabolic disease, especially in the context of glucose homeostasis.
Acknowledgment
We thank Margareta Jernås and Johanna Andersson for running the microarrays, Jenny Palming for help with adipocyte isolation and incubation, Elisabeth Strandhagen for providing INTERGENE samples and data, and the Genomics Core Facility platform at the Sahlgrenska Academy, University of Gothenburg, which was funded by a grant from the Knut and Alice Wallenberg Foundation.
This work was supported by grants from the Swedish Research Council (K2009-65X-15424-05, K2010-55X-11285-13), the Swedish foundation for Strategic Research to Sahlgrenska Center for Cardiovascular and Metabolic Research, the Swedish Diabetes foundation, the Åke Wiberg foundation, the foundations of the National Board of Health and Welfare, the Jeansson foundations, the Magnus Bergvall foundation, the Tore Nilson foundation, the Royal Physiographic Society (Nilsson-Ehle foundation), the VINNOVA-VINNMER program, the Swedish federal government under the LUA/ALF agreement, the Swedish Knowledge Foundation through the Industrial PhD program in Medical Bioinformatics at Corporate Alliances, Karolinska Institutet, and the Wellcome Trust (GR079534).
Footnotes
The authors have no competing interests to disclose. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Regional Ethical Review Boards approved the studies, and all participants gave written informed consent.
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.metabol.2010.05.011.
Appendix A. Supplementary data
Supplementary material
References
- 1.Gao Y., Walder K., Sunderland T. Elevation in Tanis expression alters glucose metabolism and insulin sensitivity in H4IIE cells. Diabetes. 2003;52:929–934. doi: 10.2337/diabetes.52.4.929. [DOI] [PubMed] [Google Scholar]
- 2.Kryukov G.V., Castellano S., Novoselov S.V. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 3.Ye Y., Shibata Y., Yun C., Ron D., Rapoport T.A. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 4.Kim K.H., Gao Y., Walder K., Collier G.R., Skelton J., Kissebah A.H. SEPS1 protects RAW264.7 cells from pharmacological ER stress agent-induced apoptosis. Biochem Biophys Res Commun. 2007;354:127–132. doi: 10.1016/j.bbrc.2006.12.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karlsson H.K., Tsuchida H., Lake S., Koistinen H.A., Krook A. Relationship between serum amyloid A level and Tanis/SelS mRNA expression in skeletal muscle and adipose tissue from healthy and type 2 diabetic subjects. Diabetes. 2004;53:1424–1428. doi: 10.2337/diabetes.53.6.1424. [DOI] [PubMed] [Google Scholar]
- 6.Walder K., Kantham L., McMillan J.S. Tanis: a link between type 2 diabetes and inflammation? Diabetes. 2002;51:1859–1866. doi: 10.2337/diabetes.51.6.1859. [DOI] [PubMed] [Google Scholar]
- 7.Du J.L., Sun C.K., Lu B. Association of SelS mRNA expression in omental adipose tissue with Homa-IR and serum amyloid A in patients with type 2 diabetes mellitus. Chin Med J (Engl) 2008;121:1165–1168. [PubMed] [Google Scholar]
- 8.Gao Y., Hannan N.R., Wanyonyi S. Activation of the selenoprotein SEPS1 gene expression by pro-inflammatory cytokines in HepG2 cells. Cytokine. 2006;33:246–251. doi: 10.1016/j.cyto.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Seiderer J., Dambacher J., Kuhnlein B. The role of the selenoprotein S (SELS) gene -105G>A promoter polymorphism in inflammatory bowel disease and regulation of SELS gene expression in intestinal inflammation. Tissue Antigens. 2007;70:238–246. doi: 10.1111/j.1399-0039.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- 10.Curran J.E., Jowett J.B., Elliott K.S. Genetic variation in selenoprotein S influences inflammatory response. Nat Genet. 2005;37:1234–1241. doi: 10.1038/ng1655. [DOI] [PubMed] [Google Scholar]
- 11.Redon R., Ishikawa S., Fitch K.R. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Y., Pagnon J., Feng H.C. Secretion of the glucose-regulated selenoprotein SEPS1 from hepatoma cells. Biochem Biophys Res Commun. 2007;356:636–641. doi: 10.1016/j.bbrc.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Alanne M., Kristiansson K., Auro K. Variation in the selenoprotein S gene locus is associated with coronary heart disease and ischemic stroke in two independent Finnish cohorts. Hum Genet. 2007;122:355–365. doi: 10.1007/s00439-007-0402-7. [DOI] [PubMed] [Google Scholar]
- 14.Marinou I., Walters K., Dickson M.C., Binks M.H., Bax D.E., Wilson A.G. Evidence of epistasis between interleukin-1 and selenoprotein-S with susceptibility to RA. Ann Rheum Dis. 2008 doi: 10.1136/ard.2008.090001. [DOI] [PubMed] [Google Scholar]
- 15.Strandhagen E., Berg C., Lissner L. Selection bias in a population survey with registry linkage: potential effect on socioeconomic gradient in cardiovascular risk. Eur J Epidemiol. 2010;25:163–172. doi: 10.1007/s10654-010-9427-7. [DOI] [PubMed] [Google Scholar]
- 16.Carlsson L.M., Jacobson P., Walley A. ALK7 expression is specific for adipose tissue, reduced in obesity and correlates to factors implicated in metabolic disease. Biochem Biophys Res Commun. 2009;382:309–314. doi: 10.1016/j.bbrc.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palming J., Gabrielsson B.G., Jennische E. Plasma cells and Fc receptors in human adipose tissue–lipogenic and anti-inflammatory effects of immunoglobulins on adipocytes. Biochem Biophys Res Commun. 2006;343:43–48. doi: 10.1016/j.bbrc.2006.02.114. [DOI] [PubMed] [Google Scholar]
- 18.Smith U., Sjöstrom L., Björntorp P. Comparison of two methods for determining human adipose cell size. J Lipid Res. 1972;13:822–824. [PubMed] [Google Scholar]
- 19.Gabrielsson B.G., Olofsson L.E., Sjögren A. Evaluation of reference genes for studies of gene expression in human adipose tissue. Obes Res. 2005;13:649–652. doi: 10.1038/oby.2005.72. [DOI] [PubMed] [Google Scholar]
- 20.Brazma A., Hingamp P., Quackenbush J. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 21.Gabrielsson B.G., Johansson J.M., Jennische E. Depot-specific expression of fibroblast growth factors in human adipose tissue. Obes Res. 2002;10:608–616. doi: 10.1038/oby.2002.83. [DOI] [PubMed] [Google Scholar]
- 22.Jernås M., Palming J., Sjöholm K. Separation of human adipocytes by size: hypertrophic fat cells display distinct gene expression. FASEB J. 2006;20:1540–1542. doi: 10.1096/fj.05-5678fje. [DOI] [PubMed] [Google Scholar]
- 23.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Stephens M., Smith N.J., Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J., Tang H., Niu L., Xu Y. Upregulation of Tanis mRNA expression in the liver is associated with insulin resistance in rats. Tohoku J Exp Med. 2009;219:307–310. doi: 10.1620/tjem.219.307. [DOI] [PubMed] [Google Scholar]
- 26.Sjöholm K., Palming J., Olofsson L.E. A microarray search for genes predominantly expressed in human omental adipocytes: adipose tissue as a major production site of serum amyloid A. J Clin Endocrinol Metab. 2005;90:2233–2239. doi: 10.1210/jc.2004-1830. [DOI] [PubMed] [Google Scholar]
- 27.Ridker P.M., Hennekens C.H., Buring J.E., Rifai N. C-reactive protein and other markers of inflammation in the prediction of cardiovascular disease in women. N Engl J Med. 2000;342:836–843. doi: 10.1056/NEJM200003233421202. [DOI] [PubMed] [Google Scholar]
- 28.Yudkin J.S., Stehouwer C.D., Emeis J.J., Coppack S.W. C-reactive protein in healthy subjects: associations with obesity, insulin resistance, and endothelial dysfunction: a potential role for cytokines originating from adipose tissue? Arterioscler Thromb Vasc Biol. 1999;19:972–978. doi: 10.1161/01.atv.19.4.972. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material