Abstract
We previously proposed that endogenous siRNAs may regulate synaptic plasticity and long-term gene expression in the mammalian brain. Here, a hippocampal-dependent task was employed in which adult mice were trained to execute a nose-poke in a port containing one of two simultaneously present odors in order to obtain a reward. Mice demonstrating olfactory discrimination training were compared to pseudo-training and nose-poke control groups; size-selected hippocampal RNA was subjected to Illumina deep sequencing. Sequences that aligned uniquely and exactly to the genome without uncertain nucleotide assignments, within exons or introns of MGI annotated genes, were examined further. The data confirm that small RNAs having features of endogenous siRNAs are expressed in brain; that many of them derive from genes that regulate synaptic plasticity (and have been implicated in neuropsychiatric diseases); and that hairpin-derived endo-siRNAs and the 20- to 23-nt size class of small RNAs show a significant increase during an early stage of training. The most abundant putative siRNAs arose from an intronic inverted repeat within the SynGAP1 locus; this inverted repeat was a substrate for dicer in vitro, and SynGAP1 siRNA was specifically associated with Argonaute proteins in vivo. Unexpectedly, a dramatic increase with training (more than 100-fold) was observed for a class of 25- to 30-nt small RNAs derived from specific sites within snoRNAs and abundant noncoding RNAs (Y1 RNA, RNA component of mitochondrial RNAse P, 28S rRNA, and 18S rRNA). Further studies are warranted to characterize the role(s) played by endogenous siRNAs and noncoding RNA-derived small RNAs in learning and memory.
Keywords: RNA interference, synaptic plasticity, learning, Lrrtm1, SynGAP1, Dgcr8, dicer, snoRNAs
INTRODUCTION
RNA interference (RNAi) pathways are ubiquitous throughout the animal (and plant) kingdoms and comprise two main pathways—one that generates siRNAs from sense-antisense hybrid transcripts or other types of double-stranded RNA (dsRNA) structures (including inverted repeats or “hairpins”), and one that generates microRNAs from small stem–loop pre-miRNA precursors (for review, see Carthew and Sontheimer 2009). The siRNA pathway generates ∼22-nt dsRNAs, of which one strand is incorporated into the so-called RISC protein complex that binds to complementary mRNAs; if the siRNA is a perfect match to its target, the target mRNA is cleaved and thus rapidly destroyed.
We have proposed that RNA interference has features that theoretically make it an attractive mechanism for regulating synaptic plasticity and other long-term changes in gene expression in the mammalian brain, and predicted that endogenous siRNAs (endo-siRNAs) should be formed within neurons at the onset of learning (Smalheiser et al. 2001). However, early cloning studies in numerous species failed to detect endo-siRNAs against mRNA target sequences. With the advent of deep sequencing technologies, it is now appreciated that endo-siRNAs complementary to mRNAs are expressed in Caenorhabditis elegans and Drosophila as well as mouse oocytes (fore reviews, see Okamura and Lai 2008; Ghildiyal and Zamore 2009). Endo-siRNAs against mRNA sequences are expressed in ES cells (Babiarz et al. 2008), in HepG2 liver carcinoma cells (Kawaji et al. 2008), and in developing skin as well (Yi et al. 2009). However, apart from two reports regarding the vertebrate slc34a gene (Carlile et al. 2008, 2009), there is still virtually no evidence that endo-siRNAs play dynamic roles in any physiological process.
In order to test whether small RNAs having the features of endo-siRNAs are expressed in the brain and whether they are modulated during the learning process, we employed a hippocampal-dependent task in which adult mice were trained to execute a nose-poke in a port containing one of two simultaneously-present odors in order to obtain a reward (Larson and Sieprawska 2002). Mice demonstrating discrimination learning were compared to two yoked control groups: (1) mice that were exposed to the same two odors but reward was not contingent upon discriminative responding (pseudo-training), and (2) mice that were not exposed to odor pairs at all (nose-poke). In all groups, mice performed the same number of trials, with the same motor responses (nose-pokes). Any changes observed in the training vs. pseudo-training comparison are likely to reflect the process of learning to associate a specific odor with reward. We chose 70% responses correct as a criterion for learning because that is the earliest point at which mice behave significantly different from chance at P = 0.05. The training group consisted of seven mice that reached criterion after three sessions of 20 trials (requiring ∼40 min of training); each trained mouse was yoked to a pseudo-trained mouse and a mouse simply performing nose-pokes. Thus, the experimental design examines changes that occur near the onset of the learning process.
We have previously characterized miRNA expression in these mice and found that training up-regulated miRNA levels and reorganized miRNA co-expression modules (Smalheiser et al. 2010). In the present study, total RNA was pooled (three or four mice per pool, giving two pooled samples per treatment group), and Illumina deep sequencing of size-selected small RNAs was carried out. We report that endo-siRNAs and other ncRNA-derived RNAs are robustly expressed in the adult mouse hippocampus, some of which showed surprisingly large changes that were specific to the training group.
RESULTS
Using the Illumina system, each pooled sample provided 12–13 million raw sequence reads or “counts” (Supplemental Table S1). Here, we consider the data set of small RNAs that aligned (“mapped”) exactly and uniquely to the reference mouse genome within exons and introns of annotated MGI (Mouse Genome Informatics) gene entries. Excluded were sequences that contained uncertain nucleotide assignments or that mapped to more than one locus, as well as those that mapped to annotated miRNA pre-miR sequences found in miRBase. The number of “raw” sequence counts for each unique sequence (filtered and cleansed but non-normalized) was tabulated for each sample. Across the filtered data set under consideration here, a total of 65,516 unique RNA sequences mapped uniquely and exactly to 14,583 known genes (Supplemental File 1).
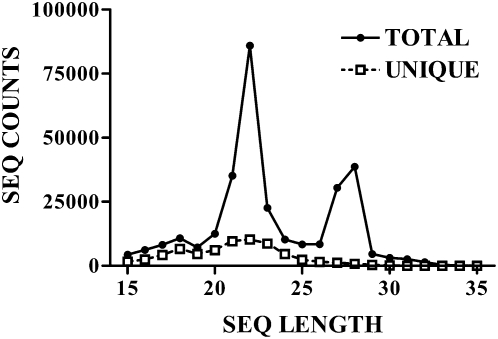
The resulting small RNA sequences exhibited a sharp peak in abundance at the 21- to 22-nt size class (Fig. 1). The vast majority of these can be considered candidates to represent endo-siRNAs, given that (1) the deep sequencing method was designed to amplify RNAse III cleavage products selectively (since adaptors were added selectively to RNAs that have a free –OH group on the 3′-end and a monophosphate on the 5′-end), (2) we excluded sequences that map to known miRNA genes, and (3) the sequences all mapped uniquely to exons or introns of MGI genes, most of which encode protein-coding mRNAs. Indeed, as shown below, several of the putative endo-siRNAs exhibited characteristic features strongly suggesting that they arise from dicer processing of RNA hairpin-like inverted repeats or sense-antisense transcript hybrids.
FIGURE 1.
Length distribution of small RNAs in the data set. Shown are the total number of sequence reads, and the total number of unique sequences, observed in the entire data set (across all samples and treatment groups). A more detailed breakdown of the data set is presented in Table 1.
In contrast, the data set appears to be depleted of other known small RNAs such as random mRNA degradation products (these would not be expected to produce a tight peak at 21-22 nt or to be acceptors for adaptors during deep sequencing), TSSa-RNAs (these should comprise overlapping sense and antisense positions near the transcriptional start site, generally upstream of the mRNA transcript) (Seila et al. 2008), piRNAs (these do not appear to be expressed in brain, are not RNAse III products, are >23 nt, often map to genomic repeats, and sometimes form overlapping pairs with a 10-nt offset), or tRNA-derived cleavage products.
Putative endo-siRNAs that map to predicted hairpin RNA inverted repeats
To begin examining the data set in detail, we looked for sequences whose features suggested that they derived from hairpin inverted repeats. Each genomic locus in the data set was examined to identify cases in which multiple small RNAs mapped to closely adjacent sites in sense orientation, to regions characteristic of RNA hairpins or linear double-stranded stems, which were not annotated as being contained within ncRNAs or genomic repeats. A total of eight gene loci satisfied these criteria (Abca2, Arhgef17, Camk2a, Gap43, Rab40b, Slc17a7, Syn1, and SynGAP1). Almost all (>99%) of the sequences mapping to these loci were 20–23 nt in length. All of these mapped within introns, but they do not appear to represent intronic miRNAs, since the small RNAs did not map to ESTs or annotated transcripts of the size of pre-miRs, did not map to genomic regions that show high cross-species conservation between mouse and man, and did not map near any known miRNAs. Interestingly, half of the hairpin endo-siRNAs are major synaptic components and/or regulators of synaptic plasticity, including SynGAP1, GAP43, CAMK2a, and synapsin I, and several are regulators of signaling (Arhgef17, Rab40b, Slc17a7).
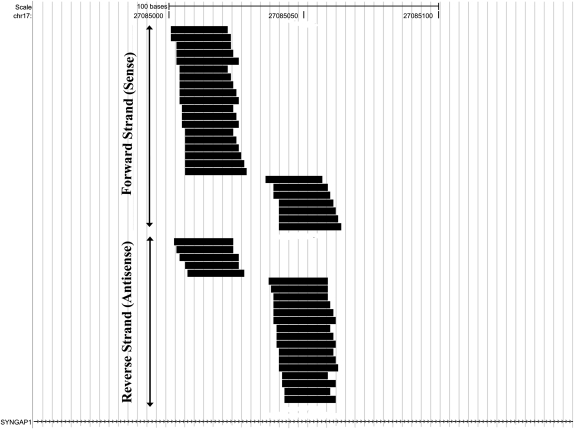
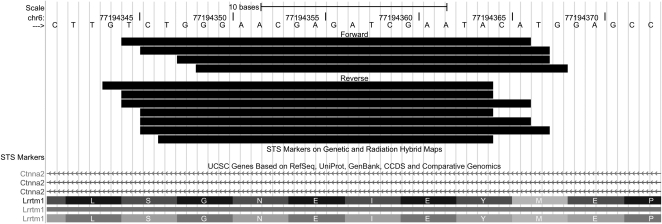
Of these hairpin-derived gene loci, the SynGAP1 locus (17qA3.3; MGI:3039785) gave rise to the most abundant set of small RNAs: about 50 unique sequences comprising several hundred counts in each sample (Supplemental Table S2), all of which mapped to two closely spaced sites within a single intron of the gene (Fig. 2). All sequences mapping to SynGAP1 were 19–24 nt in length, and >98% were in the 20- to 23-nt range (Supplemental Table S2); the average length was 21.67 nt, as expected for endo-siRNAs. The small RNAs mapped to two closely adjacent sites on both forward (sense) and reverse (antisense) strands in this region (Fig. 2).
FIGURE 2.
Small RNAs aligned to the SynGAP1 locus. Shown are all unique sequences that mapped to SynGAP1, including those that aligned to the forward or plus strand (placed on top) and to the reverse or minus strand (placed below the forward sequences).
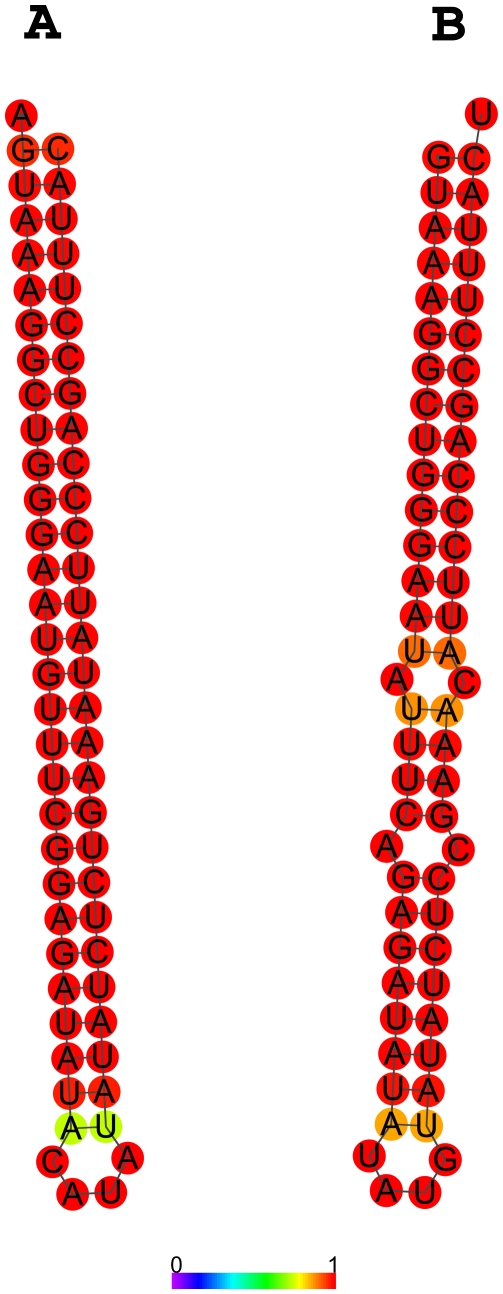
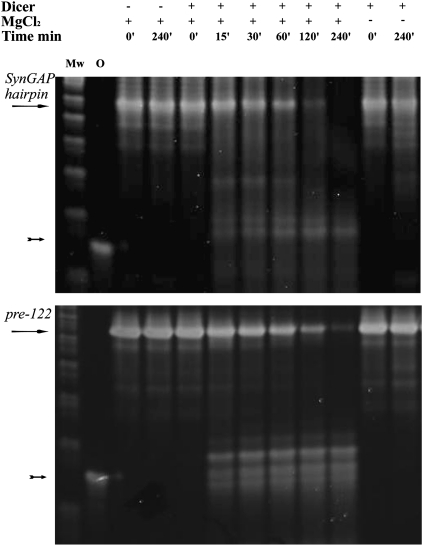
The RNAfold Webserver (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) predicted that the RNA transcribed on the sense strand forms a perfect hairpin (Fig. 3A); the sequence is predicted to form a near-perfect hairpin in the antisense direction as well (Fig. 3B). Such inverted repeats are thought to represent excellent dicer substrates but do not resemble typical pre-miR stem–loop precursor hairpins. We confirmed that a synthetic RNA transcript containing the inverted repeat sequence is a substrate for recombinant dicer in vitro and gives rise to ∼22-nt small RNAs in a Mg-dependent manner (Fig. 4).
FIGURE 3.
Predicted secondary structure of RNA corresponding to the region within the SynGAP1 locus that aligns with small RNAs. (A) The RNA encoded on the forward strand in the region covered by small RNAs (see Fig. 2) is predicted to form a perfect hairpin inverted repeat. (B) RNA encoded on the reverse strand forms an almost-perfect hairpin as well. Colors indicate the probability of base-pairing at each particular residue.
FIGURE 4.
Processing of synthetic SynGAP1 intronic hairpin RNA by recombinant dicer. SynGAP1 intronic hairpin RNA (arrow in top panel) or hsa-miR-122 small hairpin precursor RNA (arrow in bottom panel) was incubated with recombinant dicer for up to 4 h. As negative controls, some sampled lacked dicer or lacked MgCl2. A series of small RNAs were formed in a time- and Mg-dependent manner (arrows). Although the DNA ladder (Mw) does not allow a direct calibration of RNA sizes, these processed small RNAs were similar in size in both the SynGAP1 and miRNA precursor experiments (as well as the small RNAs formed by dicer cleavage of 700-bp-long dsRNA; data not shown), and they ran just slightly higher than a synthetic 21-nt siRNA used as a size standard (lane O).
The SynGAP1 locus is noteworthy because small RNAs mapped abundantly to both sense and antisense strands at the same location. Because the SynGAP1 locus was not previously known to have an antisense transcript, it is important to be sure that the antisense small RNAs are not simply small RNAs that really map to the sense strand but contain sequence errors or RNA editing changes. This is very unlikely, both because the antisense small RNAs are abundant and because there are multiple sequence mismatches between the sense and antisense small RNAs. Nevertheless, we confirmed that both sense and antisense RNA transcripts are expressed in mouse hippocampus, covering the regions both upstream of and downstream from the intronic hairpin inverted repeat (Supplemental Table S3).
Conceivably the sense and antisense transcripts could interact with each other to form a double-stranded hybrid that could be processed by dicer. However, because none of the small RNA sequences aligned to the loop regions of the predicted hairpins, it is more likely that the sense and antisense RNA hairpins are each separately processed to form small RNAs. In either case, the SynGAP1 small RNAs are excellent candidates to be endo-siRNAs. SynGAP1 protein is a major post-synaptic component that binds directly to NMDA receptors and PSD-95 protein, modulates synaptic signaling, and plays a critical role in learning (Komiyama et al. 2002). SynGAP1 mutant mice show behavioral abnormalities that model the symptoms of schizophrenia (Guo et al. 2009), and in humans, de novo truncating mutations have been found in nonsyndromal intellectual disability (mental retardation) (Hamdan et al. 2009).
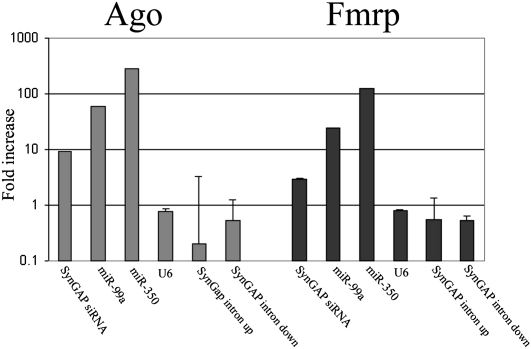
To test whether the SynGAP1 siRNA is associated with Argonaute homolog proteins, the mouse forebrain homogenates were immunoprecipitated using 4F9 monoclonal antibody (which recognizes all eIF2c isoforms) (Ikeda et al. 2006) and compared to an irrelevant antibody (synapsin I) used as a negative control. As shown in Figure 5, SynGAP1 siRNA was specifically brought down by 4F9 antibody. As a positive control, several selected miRNAs (mir-99a and mir-350) were also specifically brought down, and as a negative control, U6 RNA was not (Fig. 5). SynGAP1 siRNA and the selected miRNAs were also specifically brought down by an antibody against FMRP, which is a RISC-associated protein in brain (Fig. 5; e.g., Lugli et al. 2005). We did not detect specific association of SynGAP1 intronic sequences (on the sense strand, just upstream of and downstream from the intronic hairpin) with either the 4F9 or Fmrp antibodies (Fig. 5).
FIGURE 5.
Coimmunoprecipitation of SynGAP1 siRNA with Argonaute homolog proteins. The adult mouse forebrain S1 supernatants were immunoprecipitated using equal amounts of 4F9 anti-Ago antibody, anti-Fmrp antibody, or an irrelevant antibody against synapsin I. Equal fractions of the immunoprecipitates were measured for RNA content by qPCR as described in Materials and Methods. Each experiment was carried out on duplicate samples (error bars, SDs across duplicate samples), and each sample was assayed in duplicate. For each RNA, we plotted the ratio of the RNA abundance detected in the 4F9 or Fmrp immunoprecipitates relative to the synapsin I immunoprecipitates. SynGAP1 siRNA binding to 4F9 was impressively almost 10 times above baseline, although not quite as enriched as several miRNAs (mir-99a and mir-350). In contrast, U6 RNA did not show any specific binding nor did SynGAP1 intronic sequences derived from the sense strand just upstream or downstream from the intronic hairpin that gives rise to the siRNA. SynGAP1 and miRNAs also showed detectable binding to Fmrp, which in brain is a RISC-associated protein (e.g., Lugli et al. 2005; data not shown).
Small RNAs that map to loci co-expressing sense and antisense transcripts
The data set was then systematically examined to identify gene loci in which small RNAs mapped to overlapping regions on both sense and antisense strands. One prominent example was found: Fifty-one unique sequences mapped to Ctnna2 (catenin [cadherin associated protein], α2) in both sense and antisense orientations (eight to 13 sequences were detected in each sample). The majority of these mapped to the Lrrtm1 gene, which is encoded on the plus strand and lies entirely within an intron of Ctnna2 encoded on the minus strand. Most importantly, 11 distinct sense and antisense sequences overlapped each other extensively, exhibited overhangs of 0–3 nt, and were co-expressed in three of the samples (Fig. 6). The LRRTM gene family (Laurén et al. 2003) has been identified as having roles as synaptic organizers (Linhoff et al. 2009), and Lrrtm1 is a candidate gene for schizophrenia (Francks et al. 2007). Similarly, Ctnna2 is a candidate gene for schizophrenia (Mexal et al. 2008) and α-N-catenin protein has been described as participating in the stabilization of dendritic spines in rodent hippocampal neurons (Abe et al. 2004). In a second example, small RNAs mapped to opposite strands in an overlapping fashion within an intron of Rab40b, such that two 22-nt sequences exhibited a 1-nt overhang (Supplementary File 1).
FIGURE 6.
Small RNAs aligned to the Ctnna2 locus that putatively arise from processing of sense-antisense RNA hybrids. Multiple sequences align to a region of the Ctnna2 gene that also encodes the Lrrtm1 locus on the opposite strand. The small RNAs shown here align to both forward and reverse strands and exhibit a high degree of overlap.
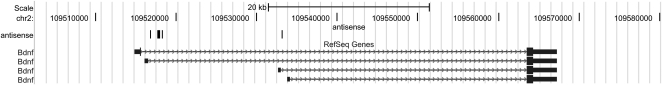
Whereas directly overlapping sense and antisense small RNA sequences were observed in only a few cases, it was quite common to observe small RNAs that map to both sense and antisense strands (at non-overlapping sites) of loci that are known to co-express both sense and antisense transcripts. For example, the human Bdnf locus has been shown to express a natural antisense transcript that forms an endogenous sense-antisense transcript hybrid within the brain (Liu et al. 2006; Pruunsild et al. 2007). Although a previous report failed to detect antisense transcripts in rodents (Liu et al. 2006), small RNA sequences mapped to Bdnf in both sense (five counts comprising four unique sequences) and antisense orientations (eight counts comprising eight unique sequences, seven of which mapped to a very small subregion of the Bdnf locus) (Fig. 7). Moreover, about 100 genes had previously been identified (in a noncomprehensive survey) as co-expressing sense and antisense transcripts within synaptic fractions of adult mouse hippocampus (Smalheiser et al. 2008). Small RNAs aligned to both sense and antisense strands of many of these genes, including the beta-site APP cleaving enzyme (Bace1), perlecan, Cdk4, sirtuin 3, nucleoporin 62, Rab6, ribosomal protein L31, FK506 binding protein 3, SNAP25, integrin-linked kinase, and activating transcription factor 5 (Supplementary File 1). Several of these genes are involved in synaptic signaling and/or in Alzheimer disease pathways.
FIGURE 7.
Small RNAs that aligned to the Bdnf locus in antisense orientation.
Learning-induced changes in expression of the 20- to 23-nt size class of small RNAs
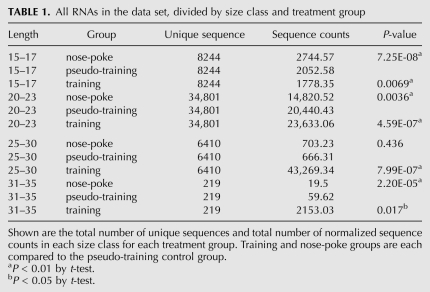
To examine whether the general class of siRNA-like small RNAs was modulated in the mouse learning paradigm, all RNAs detected in the data set were sorted by size and analyzed for global trends in expression across treatment groups. Values were normalized using mir-139-5p, which did not differ significantly across samples (see Materials and Methods). Among the 20- to 23-nt RNAs, a 38% increase in counts was observed in the pseudo-training vs. nose-poke comparison (P = 0.0036), and a further 16% increase occurred in the training vs. pseudo-training comparison (P = 4.6 × 10−7) (Table 1). When the set of hairpin endo-siRNAs (Abca2, Arhgef17, Camk2a, Gap43, Rab40b, Slc17a7, Syn1, and SynGAP1) was pooled and tested as a group, a 22% increase was observed in the training vs. pseudo-training comparison that was highly significant (P = 0.00012); the pseudo-training and nose-poke control groups were not significantly different from each other.
TABLE 1.
All RNAs in the data set, divided by size class and treatment group
Recently, Li et al. (2009) have characterized ultrashort RNAs (usRNAs) that are 15–17 nt in length, some of which appear to be short but potentially functional derivatives of miRNAs. Although miRNA-derived usRNAs were removed from our data set, other usRNAs were readily detectable and were about one-fifth as abundant as the 20- to 23-nt class in the nose-poke control group (Table 1). The usRNAs showed a 25% down-regulation in the pseudo-training group relative to the nose-poke controls (P = 7.25 × 10−8) and a further 16% decrease in the training group relative to the pseudo-training group (P = 0.007) (Table 1). Thus, the 15- to 17-nt usRNAs and the 20- to 23-nt siRNA-like RNAs responded in opposite directions in the mouse learning paradigm. Some of the usRNAs mapped to introns in the antisense orientation and/or mapped to regions containing SINEs or other genomic repeats (as defined by Repeatmasker). Although their biogenesis and heterogeneity remain uncertain, this suggests that they are not merely shorter versions of the 20- to 23-nt siRNA-like population.
Small RNAs with mixed miRNA/endo-siRNA features
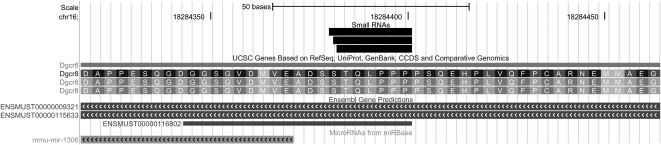
In the course of examining the data set, two additional classes of small RNAs were observed that exhibited atypical features of both miRNAs and endo-siRNAs: First, small RNAs derived from Dgcr8 (DiGeorge syndrome critical region gene 8) were well-expressed (30–50 counts per sample) and, intriguingly, mapped within the protein-coding portion of the gene in sense orientation. Of the 209 RNAs that mapped to this gene, all started at exactly the same genomic position and were 20–22 nt in length (Fig. 8). These mapped to one arm of a predicted RNA hairpin, and Ensembl annotates this region as containing a transcript (ENSMUST00000116802) that overlaps the pre-miR encoding mmu-mir-1306. Arguably, it is appropriate to regard this as an unusual novel miRNA sequence (possibly an offset miRNA) (Langenberger et al. 2009) rather than a hairpin endo-siRNA. Nevertheless, Dgcr8-derived small RNAs should bind to the Dgcr8 mRNA transcript (i.e., at the other arm of the RNA hairpin) and potentially may regulate its translation or stability. Furthermore, it is worth noting that their entire 5′-seed region is exactly complementary to mir-1224 in the mouse and rat (positions 1–10) and human (positions 1–9), as well as the human mir-30b* (positions 1–9). Thus, potentially the Dgcr8-derived small RNAs might bind to and regulate other miRNAs (Lai et al. 2004). Dgcr8 is a key co-factor of drosha that processes pri-miRs in the miRNA biogenesis pathway. An elevation in Dgcr8 mRNA levels (and a global increase in miRNA expression) has been observed in human post-mortem prefrontal cortex in schizophrenia (Beveridge et al. 2009). Dgcr8 haplo-insufficiency also contributes to phenotypic deficits in mice modeling the 22q11.2 microdeletion that confers high risk for schizophrenia (Stark et al. 2008).
FIGURE 8.
Small RNAs that aligned to the Dgcr8 locus. These lie within the protein-coding region of Dgcr8 and map within a short ENSEMBL transcript (ENSMUST00000116802) that overlaps with a nearby miRNA (mmu-mir-1306).
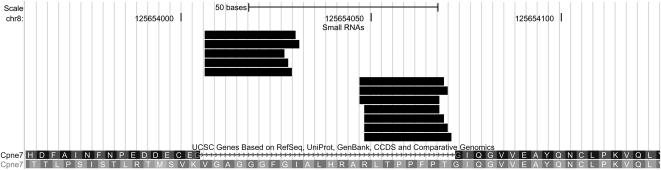
Second, in about a dozen cases, 20- to 23-nt intronic sequences were observed that started or terminated exactly at an exon/intron border (none continued past an exon/intron border). In three of these genes (Arhgef17, Acadvl, and Cpne7), sequences mapped to 5′- and 3′-ends of the same intron, which was predicted to fold into a RNA hairpin structure (e.g., Cpne7) (Fig. 9). Though these small RNAs do not correspond to known miRNAs, their localization is distinctive and corresponds to the situation described for miRNAs that are processed from mirtrons (Berezikov et al. 2007; Babiarz et al. 2008). Since mirtron processing is independent of drosha, which acts upon typical miRNA precursors (Berezikov et al. 2007), it may be appropriate to regard these as part of a spectrum of hairpin endo-siRNAs (Babiarz et al. 2008). This is further suggested insofar as a range of mapping patterns were observed: Sometimes small RNAs arose from a predicted RNA hairpin that encompassed an entire intron; sometimes the hairpin was entirely internal to the intron; and sometimes the hairpin terminated at an exon-intron border on one side but was internal on the other side (data not shown). Several Cpne7-derived RNAs (GTAGGAGCTGGAGGTGGGTTTG and longer related sequences) showed a 18-nt stretch of perfect complementarity (18/18; E = 0.02; includes the 5′-end of the small RNAs) with a site in the coding region of Begain (brain-enriched guanylate kinase-associated; NM_001163175). This represents a candidate trans target mRNA.
FIGURE 9.
Small RNAs aligned to the Cpne7 locus. These map to the 5′- and 3′-ends of a short intron.
Small RNAs derived from snoRNAs
In the data set, most of the small RNAs >25 nt in length were derived from snoRNAs (small nucleolar RNAs) or other abundant noncoding RNAs. Small RNAs mapped to about 100 different snoRNAs and scaRNAs (small Cajal body-specific RNAs) that are encoded within the introns of other (generally protein-coding) genes. Several recent publications have characterized this class of small RNA and shown that some H/ACA box snoRNAs exhibit typical pre-miRNA stem–loop secondary structures (Scott et al. 2009). Formation of the snoRNA-derived small RNAs is dependent upon dicer, even when the snoRNAs do not resemble typical pre-miRNAs and even when the small RNAs are shorter or longer than typical miRNAs (Ender et al. 2008; Taft et al. 2009). These small RNAs are associated with Argonaute homologs, and at least some snoRNA-derived RNAs can have miRNA-like functions to regulate other mRNAs (Ender et al. 2008). Small RNAs derived from C/D box snoRNAs (but not H/ACA box snoRNAs) are generally enriched in the nucleus (Taft et al. 2010).
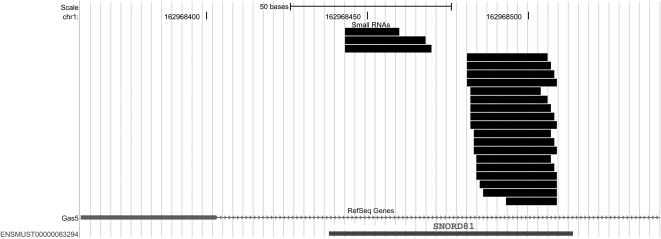
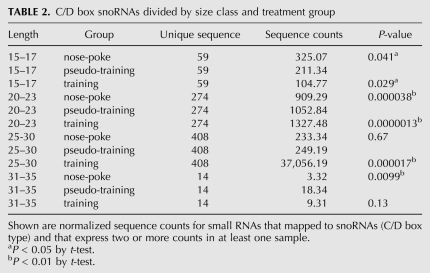
C/D box snoRNA-derived sequences tended to map to two or three discrete sites within the snoRNA, often near the two ends (e.g., Fig. 10). A variety of lengths ranging from 15–34 nt could map to a single site—and at a given site, sequences varied in their start or end positions by 1–4 nt. Thus, the localization of the sequences was well-defined, but their exact lengths and precise positioning varied (Fig. 10). As shown in Table 2, the different size classes of C/D box snoRNA sequences were differentially altered in the mouse learning paradigm, in a manner similar to that observed in the data set as a whole. Short sequences (15–17 nt) were down-regulated in the pseudo-training group by 41% (relative to the nose-poke control group, P = 0.041) and further down-regulated in the training group by 45% (relative to the pseudo-training group, P = 0.029). In contrast, the 20- to 23-nt sequences showed a 16% increase in pseudo-training (relative to nose-poke, P = 3.8 × 10−5) and a further 26% increase in the training vs. pseudo-training comparison (P = 1.3 × 10−6) (Table 2).
FIGURE 10.
Small RNAs aligned to SNORD81 (within the Gas5 locus). All sequences in the data set mapping to this locus aligned in sense orientation.
TABLE 2.
C/D box snoRNAs divided by size class and treatment group
Unexpectedly, the 25- to 30-nt sequences exhibited an extremely low level of expression in both the nose-poke and pseudo-training control groups (averaging less than one raw sequence read per distinct sequence per sample) and increased about 149-fold in the training group (P = 1.7 × 10−5) (Table 2). The two control groups did not differ significantly from each other (P = 0.67).
Since each snoRNA locus gives rise to numerous distinct small RNA sequences, the response to training is especially impressive when viewed on a per-snoRNA basis. For example, SNORD81 expressed 16 distinct small RNAs in the 25- to 30-nt size range that expressed an average of 21.5 raw sequence counts per sample (nose-poke group) vs. 32.5 counts per sample (pseudo-training group) vs. 7575 counts per sample (training group). It should also be noted that different sites within a given snoRNA could show quite different degrees of up-regulation. For example, the up-regulation of SNORD81-derived small RNAs occurred primarily at the 3′ site, while that of SNORD20 was observed primarily at the 5′ site (Fig. 10; data not shown).
The magnitude of the changes in C/D box snoRNA-derived small RNA expression greatly outweighed changes in host gene or snoRNA expression that have been reported in mouse plasticity paradigms (Rogelj et al. 2003; Meier et al. 2009). For example, the host Gas5 gene encodes 10 snoRNAs, and its mRNA expression has shown to be up-regulated by about 1.5- to twofold in various plasticity paradigms in mouse (Mei et al. 2005; Meier et al. 2009). Gas5, like all known host genes for snoRNAs, is a member of the 5′-TOP family that is involved in regulating protein synthesis (Smith and Steitz 1998). At the same time, synthesis of 5′-TOP proteins is stimulated in association with prolonged long-term synaptic potentiation (Gobert et al. 2008). Thus, it is not surprising that snoRNA host gene transcription should be stimulated in the olfactory discrimination training paradigm. Yet whereas small RNAs derived from SNORD68 and SNORD100 exhibited many-fold changes in expression, the host SNORD65, SNORD68, and SNORD100 genes showed only 2%–9% elevations in abundance in the training vs. pseudo-training comparison (Supplemental Table S4).
Ten H/ACA box snoRNAs were also included in the data set; in contrast to the C/D box snoRNAs, H/ACA-derived 25- to 30-nt sequences showed only about 10-fold up-regulation in the training vs. pseudo-training comparison (Supplemental File 1).
Small RNAs derived from other abundant noncoding RNAs
Very recently, it has become apparent that abundant cellular noncoding RNAs can give rise to small RNAs (e.g., Kawaji et al. 2008), not simply through random degradation but via a specific dicer-mediated process (e.g., Cole et al. 2009) or other specific enzymes (Fu et al. 2009; Lee et al. 2009; Yamasaki et al. 2009). Most of these RNAs (e.g., tRNAs and snRNAs) as well as newly described RNAs (e.g., piRNAs, centromeric RNAs, and TSSa-RNAs) were excluded from our data set, because they are encoded by multiple genomic loci, are not contained within MGI-annotated mRNAs, are not in the included size range, or are not expressed in brain. However, several cellular noncoding RNAs did give rise to small RNAs within our data set. These showed evidence of specific processing, and most importantly, the 25- to 30-nt small RNAs derived from these loci showed large, specific up-regulation with training.
Numerous small RNAs (300–600 counts per sample) mapped to the canonical Y1 RNA locus on chromosome 6. The small RNAs mapped to three discrete sites near the 5′-end, middle, and 3′-end of the Y1 RNA transcript (Supplemental Fig. S1). Those mapping to the 5′- and 3′-ends were most abundant, which is consistent with the fact that Y RNAs fold into a hairpin-loop structure in which the 5′- and 3′-ends form a single double-stranded stem (Perreault et al. 2007). The 25- to 30-nt RNA sequences derived from 5′- and 3′-ends showed very low expression in the control groups and an approximately 70-fold increase in expression that was specific to the training group (Supplemental Table S5). In contrast, 20- to 23-nt sequences did not show significant changes across treatment groups, even those that arose from the same site as the 25- to 30-nt up-regulated RNAs (Supplemental Table S5).
Small RNAs (most were >24 nt in length) mapped to two discrete sites at the extreme 5′- and 3′-ends of Rmrp, RNA component of mitochondrial RNAse P (Supplemental Fig. S2). Overall, an approximately 150-fold increase in expression was observed with training; RNAs derived from the 3′-end showed only an able fivefold elevation, whereas RNAs that derived from the 5′-end exhibited an approximately 250-fold increase.
Small RNAs mapped to two specific sites near the 5′- and 3′-ends within a locus encoding 28S rRNA within an intron of the Cdk8 host gene (Supplemental fig. S3). The 5′ site showed about 450-fold up-regulation of the 25- to 30-nt sequences with training; in contrast, the response was modest at the 3′ site (Supplemental Table S6). Finally, small RNAs mapped to a locus encoding 18S rRNA residing within a pseudo-gene. The 18S rRNA-derived small RNAs showed a broad size distribution and localization, suggesting that many of them may have arisen from random degradation (Supplemental Fig. S4). Still, the 25- to 30-nt small RNAs mapping to this locus increased by approximately fourfold in the training vs. pseudo-training comparison, which was statistically significant (P = 0.002) and specific to this size class (i.e., the 15- to 17-nt, 20- to 23-nt, and 31- to 35-nt species did not exhibit up-regulation).
To check whether miRNA loci also generated 25- to 30-nt species with training, we examined the set of sequences mapping uniquely and exactly to miRNA loci within known genes (Supplemental File 2). Overall, only ∼5% of the unique sequences were 25–30 nt in length (and only one sequence read in one sample was >29 nt long). Most miRNAs exhibited no 25- to 30-nt sequences in any treatment group, and in most cases, sequences in the 25- to 30-nt size class failed to show the kind of striking up-regulation with training that was observed for snoRNAs and other ncRNAs. A few exceptions were noted, however: mir-330, mir-342, mir-434, mir-541, mir-877, mir-1224, and mir-1944. Of these, the pre-miR for mir-342 lies entirely within a tRNA repeat; mir-877 and mir-1224 are mirtrons; and mir-1944 is encoded within SNORD43. Thus, the striking up-regulation of the 25- to 30-nt species with training did not affect typical miRNA loci (nor endo-siRNA generating loci). Rather, this phenomenon was characteristic of host noncoding RNAs and mirtrons.
DISCUSSION
In search of endo-siRNAs
We previously proposed that endo-siRNAs may serve as a physiological mechanism for regulating synaptic plasticity and other long-term changes in gene expression in the mammalian brain (Smalheiser et al. 2001). This hypothesis languished for several years because earlier cloning strategies failed to detect small RNAs that resemble endo-siRNAs in adult mammalian tissues. However, the present study has employed Illumina deep sequencing to demonstrate that adult mouse hippocampus expresses a diverse set of non-miRNA small RNAs, including a large population that has the characteristic size and features of endo-siRNAs.
One class of 20- to 23-nt small RNAs has particularly good candidate endo-siRNAs. These mapped robustly and discretely to two closely adjacent sites within introns, corresponding to the two arms of a predicted RNA hairpin inverted repeat. Interestingly, about half of the identified hairpin-derived siRNAs are encoded within major synaptic components and/or regulators of synaptic plasticity, such as SynGAP1, Camk2a (calcium/calmodulin-dependent protein kinase II α), GAP43, and synapsin I. Genomic analyses by Wang and Leung (2009) have reported that intronic hairpin-like long inverted repeats are significantly over-represented in genes indexed by the Gene Ontology term synapse (in man) and nervous system development (in both the mouse and man). The present study shows experimentally that these hairpin-like regions are processed in vivo to form small RNAs that are expressed in the brain. Moreover, the hairpin-derived endo-siRNAs considered as a group showed a specific increase in abundance with olfactory discrimination training relative to the pseudo-training group that was carefully matched for task novelty, odor exposure, and motor behavior.
Hairpin-derived endo-siRNAs have unique and intriguing features as potential regulatory RNAs, insofar as they would be expected to bind both the sense mRNA from which they derive, as well as any antisense transcript that may be expressed on the opposite strand. Thus, the magnitude (and direction) of their actions at a given moment may depend on the relative expression levels of sense and antisense transcripts. As a further complication, antisense transcripts may either reduce or enhance expression of their sense counterparts (Faghihi and Wahlestedt 2009; Werner et al. 2009). Therefore, one should not assume that the net effects of endo-siRNAs, particularly hairpin-derived siRNAs, are necessarily to silence host genes. Furthermore, hairpin endo-siRNAs may possibly have additional actions by interacting with other RNA targets in trans.
Another class of 20- to 23-nt RNAs observed in our data set mapped to both sense and antisense strands of plasticity-related genes that are known to co-express sense and antisense RNA transcripts, where they mapped predominantly to discrete hot-spots. These are candidates to arise from dicer processing of sense-antisense transcript hybrids. The case of Lrrtm1/Ctnna2 was particularly striking because small RNA sense and antisense sequences were highly overlapping. Studies of bona fide siRNA pairs, known to arise from sense-antisense transcript hybrids, have shown that only one member may be detectable within cells (Carlile et al. 2008, 2009; Tam et al. 2008; Watanabe et al. 2008), and the relative abundance of sense vs. antisense siRNAs can vary as a function of developmental stage or tissue type (Carlile et al. 2008, 2009). Thus, even gene loci that gave rise to small RNAs detected from only one strand may still be a source of siRNAs, especially if they derive from a region that is known to express both sense and antisense mRNAs.
The overall size class of 20- to 23-nt RNAs in our filtered data set mapped to hundreds of different gene loci, but as a class, they were modulated in the mouse learning paradigm in a characteristic manner: increased in the pseudo-training group (hence sensitive to task novelty, odor exposure and/or motor behavior) and further increased with training (hence sensitive to learning the specific association between correct response and reward). In contrast, the 15- to 17-nt usRNAs were decreased with pseudo-training and further decreased with training, and the 25- to 30-nt class showed a distinctive response as well (see below). That the 20- to 23-nt RNAs are up-regulated with training is congruent with the observations that dicer RNAse III activity is activated in a calpain-dependent manner within NMDA-stimulated hippocampal slices (Lugli et al. 2005, 2008) and that miRNA expression is increased globally with training in the same mice examined here (Smalheiser et al. 2010). This further supports the notion that the 20- to 23-nt small RNAs in our data set are siRNA-like and may be processed by dicer. Small RNAs (siRNAs and/or miRNAs) probably regulate learning in an active RISC-dependent manner since knocking-down Argonaute 2 expression in mouse hippocampus interferes with a contextual memory task (Batassa et al. 2010).
Endo-siRNAs and other ncRNA-derived RNAs were expressed at levels that fall within the same range as other miRNAs that were robustly expressed in the same samples (see Materials and Methods). Bearing in mind that miRNAs are far less potent in their target interactions than are siRNAs that bind perfectly to their targets, we believe that the small RNAs studied here may be regarded as potentially functional—and possibly even as influential as the miRNA system. However, many questions remain unanswered in this first report and warrant further investigation: (1) Although SynGAP1 siRNAs appear to be processed by dicer and to be associated with Argonaute proteins, it remains to be shown that the overall population of putative endo-siRNAs is indeed processed by dicer and associated with functional RISC. (2) Where in the cell are they formed? Most of the siRNA-like small RNAs mapped to intronic locations and might be expected to arise from processing within the nucleus. Yet a dendritic location for processing should also be considered since mRNAs with retained introns are transported to, and spliced within, dendrites (Glanzer et al. 2005); as well, some sense and antisense mRNA transcripts are co-expressed within synaptic fractions (Smalheiser et al. 2008). (3) Do they actively regulate long-term gene expression of their host genes and possibly other in trans target mRNAs (Smalheiser et al. 2001)? For example, will modulating SynGAP1 or other endo-siRNAs in neural systems alter learning or memory in vivo?
Because dsRNAs can elicit an interferon response that causes a nonspecific shut-down of protein synthesis rather than specific gene silencing, it may appear surprising or paradoxical to observe expression of endo-siRNAs in brain or other healthy differentiated mammalian tissues. However, this objection would not apply to the endo-siRNAs described in the present report that are derived from hairpin-like inverted repeats. More importantly, whether a long dsRNA will trigger nonspecific inhibition of protein synthesis may depend not only on its length but also on its concentration, its subcellular localization, and its end modifications. Many published reports have documented that exogenous long dsRNA can, indeed, elicit specific gene silencing via RNA interference without having global effects on irrelevant targets. These reports include mammalian systems: hypothalamic neurons transfected with dsRNA in vivo (Bhargava et al. 2004), neuroblastoma cells transfected with dsRNA in vitro (Gan et al. 2002), myotubes transfected with plasmids that express a 1-kb dsRNA in vitro (Yi et al. 2003), and adult muscle fibers injected with dsRNA in vivo (Kong et al. 2004). Similar results have also been reported for dsRNA introduced into neurons in invertebrates (Dzitoyeva et al. 2001, 2003; Korneev et al. 2002).
Small RNAs derived from snoRNAs and other abundant noncoding RNAs
Recently, snoRNA-derived small RNAs have been shown experimentally to be dicer-dependent and associated with Argonaute proteins/RISC, and at least one appears to have miRNA-like functions in regulating target mRNAs (Ender et al. 2008; Taft et al. 2009). In our data set, small RNAs mapped to about 100 different C/D box and H/ACA box snoRNAs and scaRNAs (i.e., snoRNA-like RNAs that localize to Cajal bodies). In C/D box snoRNAs, four different size classes of small RNAs mapped in a highly overlapping fashion to the same sites, yet each showed a different pattern of modulation in the mouse learning paradigm. Strikingly, the 25- to 30-nt sequences exhibited almost no expression in the control groups but rose by about 100-fold or more in the training group. The magnitude of up-regulation varied among different sites within the same snoRNA. These changes were not linked to changes in expression of the host genes encoding the snoRNAs, nor of the snoRNAs themselves.
Small RNAs of the 25- to 30-nt size class also mapped to discrete sites within several abundant cellular noncoding RNAs, including Y1 RNA, the RNA component of mitochondrial RNAse P, and 28S rRNA—and these too showed nearly no expression in the control groups but exhibited a striking and selective up-regulation with training, the magnitude of which varied from site to site within the same ncRNA. Why should a very similar type of up-regulation observed in snoRNAs also occur across such a variety of noncoding RNAs? It is plausible that Y1 RNA should be cleaved by dicer, given its stem–loop structure; and at least one type of abundant ncRNA, tRNA(Gln), is cleaved by dicer in vitro and in vivo and can associate detectably with RISC proteins (Cole et al. 2009). Why are the 25- to 30-nt species formed only with training? The activation of dicer RNAse III activity that accompanies synaptic stimulation and calcium entry is accompanied by the formation of discrete enzymatically active dicer fragments (Lugli et al. 2005). Dicer fragments might conceivably associate differentially with co-factors (e.g., TRBP and PACT) and/or process ncRNAs to form small RNAs of different characteristic lengths than full-length dicer protein.
Do all processing events occur in the same cellular compartment(s)? Full-length dicer protein contains several cryptic nuclear localization signals (Nicholson and Nicholson 2002), including a bipartite signal that is retained within calpain-processed dicer fragments (Lugli et al. 2005). Conceivably, certain dicer fragments may be formed and transported into the nucleus selectively in the training group, where they could process nuclear ncRNAs to form 25- to 30-nt species, whereas full-length dicer protein forms 20- to 23-nt products. Importin α, the protein that transports bipartite signal-bearing proteins into the nucleus, is known to be anchored at the post-synaptic face of excitatory synapses and to be released upon neuronal stimulation (Jeffrey et al. 2009). Importin α provides a potential mechanism by which dicer fragments, formed locally near synapses with training, could be transported back to the nucleus. Though this idea is speculative, it is testable and provides a roadmap for further experimentation. Moreover, it is congruent with current models of synaptic plasticity, in which a variety of transcription factors and other proteins are anchored near synapses and become transported to the nucleus upon neuronal stimulation, where they regulate long-term gene expression (e.g., Cohen and Greenberg 2008).
The physiological role(s) of ncRNA-derived small RNAs, if any, are not clear at present. They may arise as a byproduct of ncRNA metabolism that may have no functions at all or may only regulate the ncRNA host genes from which they arise. It is also conceivable that they lack specific target mRNAs yet bind to RISC and compete with miRNAs or other small RNAs, thus regulating mRNAs indirectly. Long-term potentiation is associated with increased synthesis of proteins having 5′-TOP motifs (including snoRNA host genes), which in turn up-regulate protein synthesis within neurons (Gobert et al. 2008). It will be interesting to learn whether the 25- to 30-nt small RNAs from snoRNAs and other noncoding RNAs are induced whenever 5′-TOP protein synthesis is stimulated. If so, ncRNA-derived small RNAs might be viewed as part of a system that supports protein synthesis and long-term cellular growth (in the case of neurons, long-term memory involves dendritic growth, spine stabilization, and new synapse formation). Alternatively, it is worth exploring whether at least some noncoding RNA-derived small RNAs may have specific mRNA targets (or may regulate other small RNAs) and possibly play more specific roles in regulating synaptic plasticity and learning.
In this study, we have restricted analysis of the data set only to those small RNAs that map exactly and uniquely to the reference mouse genome, within known MGI annotated genes (these are predominantly protein-coding genes). Future analyses should consider small RNAs that map to more than one locus and to other genomic regions, as well as sequences that undergo RNA editing and other modifications. Because several of the endo-siRNAs map to genes that regulate synaptic plasticity and that have been implicated in schizophrenia, autism, mental retardation, and other human neuropsychiatric diseases, it will be very important to characterize the expression and function of these small RNAs in human brain as well.
MATERIALS AND METHODS
Subjects
Subjects were male C57Bl/6J mice (Jackson Laboratory), at 2 mo of age. They were housed in groups of four in plastic cages in a climate-controlled animal colony on a normal 14:10 light:dark cycle. The mice were maintained on a water-deprivation schedule with access to 1.0–2.0 mL water once per day for at least 5 d prior to and throughout training. This schedule reduced body weight by ∼20% in the first few days but maintained the mice at a stable weight throughout the study. All testing was done during the light phase. The care and use of animals in this research followed protocols approved by the Institutional Animal Care and Use Committee of the University of Illinois at Chicago and were in accordance with the guidelines of the National Institutes of Health.
Olfactory discrimination training
This has been described in detail (Smalheiser et al. 2010). Briefly, all procedures were fully automated and controlled by computer within each training session. Mice were first trained to execute nose-poke responses for water reinforcement in two 20-trial sessions per day. Each trial was signaled by the extinction of a lamp at the West end of the test box; a nose-poke in either West sniff port within 60 sec terminated the trial and triggered the delivery of 12.5 μL water to the water cup. The intertrial interval was 10 sec. Mice were run in two cohorts; nose-poke training continued until all mice in the same cohort reached a criterion of nose-pokes on at least 80% of trials in both sessions on at least three days. This required 7 d of training for cohort 1 and 5 d for cohort 2. Mice were given a 5-d rest period after the last nose-poke training session before olfactory discrimination training.
Mice were then randomly assigned to three experimental groups: The first group received olfactory discrimination training. Mice were trained in a series of 20-trial sessions in which each trial began with the simultaneous presentation of two discriminative odors (S+ and S−) to the West sniff ports. The spatial position of the two odors on any given trial was randomly determined except that no more than three identical trials could occur in succession. A nose-poke response at the port carrying the S+ odor (L-carvone) terminated the trial, was scored as correct, and was rewarded with a drop of water; a response at the port carrying the S− odor (α-phellandrene) terminated the trial, was scored as incorrect, and was not rewarded. Each trial had a maximum duration of 60 sec and was followed by a 10-sec intertrial interval. The learning criterion was 14 or more correct trials in a 20-trial session.
The second group received pseudo-training with the same two odors and trial events as in the training group, except that rewards were not contingent upon responding at the correct odor port. A nose-poke response to either odor terminated the trial and was rewarded with a drop of water. Each mouse in the pseudo-training group was yoked to a mouse in the training group in terms of number of training sessions.
The third group simply continued nose-poke training and had no odors presented. Each mouse in this group was yoked to a mouse in the training group in terms of number of training sessions.
A total of nine yoked experiments were carried out. We analyzed data from seven experiments in which mice achieved criterion at the end of the third session (taking ∼40 min), since they exhibited lower variability in miRNA expression than in the two cases in which the mice achieved criterion within a single session.
RNA isolation
Mice were sacrificed immediately after their last training session. Mice were anesthetized with halothane and decapitated. The brains were rapidly removed and rinsed in PBS. Both hippocampi (including dorsal and ventral regions) were dissected free from each brain, pooled, and homogenized in Trizol buffer. RNA isolation was carried out using methods optimized for recovery of tiny RNAs as described (Lugli et al. 2008). In the present study, each treatment group was pooled into samples A and B. That is, the nose-poke A, pseudo-training A, and training A samples contained 5 μg total RNA from each of three mice (the first three yoked experiments) so that they could be directly compared to each other using paired statistics; and the three B samples were pooled from the remaining four experiments.
Library construction
All subsequent steps were performed at the W.M. Keck Center for Comparative and Functional Genomics, University of Illinois at Urbana-Champaign, under the supervision of Dr. Alvaro G. Hernandez. Small RNA libraries were prepared with the “Small RNA Alternative v1.5 Sample Prep Kit” following the manufacturer's instructions (Illumina). Briefly, the 18- to 30-bp RNA fragments were purified from total RNA in each sample on a 15% Novex TBE-UREA PAGE (Invitrogen). The RNA was ligated with 3′ RNA adaptor (5′-/5rApp/ATCTCGTATGCCGTCTTCTGCTTG/3ddC/) which is specifically modified to target miRNAs and other small RNAs that have a 3′ hydroxyl group resulting from cleavage by Dicer and other RNA processing enzymes, and then with 5′ RNA adaptor (5′-GUUCAGAGUUCUACAGUCCGACGAUC) at the 5′ end of RNA with phosphate group. The 5′ adaptor also includes the sequencing primer. RT-PCR amplification is then done using these adaptors as primers, and this selectively enriches the fragments that have adaptors on both ends. The resulting double-stranded DNA libraries were PAGE purified (6% Novex TBE PAGE, Invitrogen) to eliminate dimerized adaptors.
Sequencing
The libraries were loaded on individual lanes of a flow-cell at a concentration of 5 pM and were sequenced on a Genome Analyzer II for 37 cycles following the manufacturer's protocols (Illumina).
Analysis pipeline
Genome Analyzer Pipeline software is a customizable analysis engine that takes the raw image data generated by the Genome Analyzer and produces intensity scores, base calls, quality metrics, and quality-scored alignments. The pipeline software maximizes the number of clusters used to generate sequence data with automated image calibration, filters for high-quality reads using accurate cluster intensity scoring algorithms, and minimizes the propagation of downstream sequencing errors with quality-calibrated base calls.
Data acquisition and cleansing of raw sequence reads
The image analysis and base calling were done using Illumina's GA Pipeline and sequence files were generated for each sample. Six samples were sequenced on six different lanes of a flow-cell along with PhiX174 as control on a different lane. Based on the alignment of reads to the genome from control lane, the error rate is ∼0.21%. Yield from each of the sample lanes is about 12–13 million reads, resulting in 257K–350K unique sequences after adaptor trimming. Further analysis was done using Flicker 2.7, which is an add-on software to the GA Pipeline from Illumina. The 3′ adaptor sequence was trimmed from small RNA reads, and the trimmed reads are aligned to the mouse genome build 37 from NCBI using iterated ELAND short read aligner, allowing up to three mismatches. The reads <15 nt in length after adaptor trimming were not used for alignment. The results from the alignments were loaded into an Oracle database, and SQL/PLSQL queries were used to generate reports of the results. About 175K–210K unique sequences aligned to the genome. After removing reads with Ns, reports were generated for reads aligning uniquely without any mismatches to known genes and miRNAs using their chromosome coordinates from NCBI and miRBase respectively. The sequences aligned to 14,583 known genes and 384 miRNAs.
Data analysis and statistics
To compare abundance across treatment groups, raw small RNA sequence counts in each sample were normalized by endogenous miRNAs. Mir-139-5p was chosen as the normalizer because it was robustly expressed and showed no difference in expression between training and pseudo-training groups, as measured in the same mice using RT-PCR high-throughput plates (Smalheiser et al. 2010) and separately confirmed by carrying out manual RT-PCR on all samples. (A small difference in expression was observed relative to the nose-poke control group, which was taken into account when normalizing values in that group.) The normalization values were similar whether using only the canonical miRBase sequence for mir-139-5p or including all mir-139-5p related sequences 19–24 nt in length and were similar when sequences were pooled with two other miRNAs examined as well (mir-323-3p and mir-543-3p) that showed no change between training and pseudo-training groups. The three endogenous miRNAs (mir-139-5p, 323-3p, and 543-3p) were all robustly expressed as measured using high-throughput RT-PCR plates (Smalheiser et al. 2010), exhibiting mean Ct values of about 21, 25, and 28, respectively, and were detected by deep sequencing at about 2500, 250, and 25 counts per sample, respectively when filtered as for the other small RNAs in the data set. Raw sequence counts of mir-139-5p varied from 1994–3583 across the different groups; thus, the normalized counts in each group were obtained by dividing raw counts by a factor of 1.994–3.583. Differences in abundance (of a pooled set of sequences across treatment groups) were calculated using t-test, two-tailed, paired. In selected cases, t-test results were compared with nonparametric Wilcoxon sign-rank test, and similar results were obtained.
Processing of synthetic SynGAP1 intronic hairpin RNA by recombinant dicer
RNA substrates
To make synthetic SynGAP1 intronic hairpin RNA, synthetic oligos (forward and reverse) were custom made (IDT DNA) and annealed in 20 mM Tris-HCl and 1 mM EDTA to form double-stranded DNA. This corresponds to the sequence of the intronic hairpin (Fig. 3a) plus 22 bases of upstream flanking genomic sequence, 22 bases of downstream flanking genomic sequence, and a T7 promoter sequence. The forward sequence is 5′-CAGAGATGCATAATACGACTCACTATAGGGTATATATATATATATATATAAAGTAAAGGCTGGGAATATTTCAGAGATATATATGTATATCTCCGAAACATTCCCAGCCTTTACTAAGCCCCAATACAGTCAGTAA-3′. The annealed oligo was agarose gel purified with QIAEX II Gel extraction Kit (Qiagen). The DNA was transcribed into RNA using the MEGAshortscript kit (Ambion) following the manufacturer's instructions and DNAse treated using TURBO DNA-free DNase (Ambion) for 30 min. The RNA was gel purified in a 15% TBE polyacrylamide gel and eluted overnight in elution buffer as described in the kit. After Trizol purification of the precipitated RNA, OD was measured and purification confirmed by running an aliquot on a 15%TBE polyacrylamide gel (Bio-Rad) and on a 15% TBE-UREA gel (Bio-Rad). As positive controls, synthetic human pre-miR-122 RNA and 700 bp dsRNA were prepared as described by Lugli et al. (2005, 2008).
RNAse III assay
RNase III assays were performed at 37°C using the purified RNA sequence in 50 mM NaCl, 30 mM tris-HCl (pH 7.0), 0.5 mM EDTA, 1 mM ATP, 0.1% Triton X-100, 10% Glycerol, 1 mM DTT, 7.5 mM MgCl2. Recombinant human dicer (BLOCK-IT Dicer RNAi kit, Invitrogen, 1 U/5 μL reaction) was used as recommended by the manufacturer's protocol. As negative controls, some samples were run omitting dicer or omitting MgCl2 from the reaction. Aliquots were taken at the indicated times and the reaction stopped with stop reaction buffer. An aliquot from each time point and 10-bp DNA ladder (Invitrogen) was mixed in Gel Loading buffer II (Ambion), heated for 5 min at 99°C, and loaded in a Criterion polyacrylamide 15% TBE-Urea gel and run at 25 mV. Gels were stained with Syber Gold (Molecular Probes-Invitrogen; 3 μL/10 mL TBE) for 20 min and rinsed in TBE for 1 h. Pictures were taken with the EC3 Imaging system (UVP).
Coimmunoprecipitation of SynGAP1 siRNA with Argonaute homolog proteins
Homogenization
Two-month-old male C57Bl/6 mice forebrain (including cortex and hippocampus) was rapidly dissected, placed in RNAlater (Ambion), and immediately homogenized using a Dounce pestle in ice-cold homogenization buffer (HB) containing a cocktail of protease and Rnase inhibitors (20 nM Tris-Hcl at pH.7.5, 150 mM NaCl, 5 mM EDTA 2 mM phenylmethylsulfonyl fluoride, 10 mM N-ethylmaleimide, 10 μg/mL leupeptin, 1 μg/mL pepstatin A, 2 μg/mL aprotinin, 160 U/mL Superase-In [Ambion], 160 U/mL Rnase-OUT [Invitrogen Life Technologies]). Samples were spun down twice at 1500g for 10 min at 4°C. NP-40 was added to the supernatant to a final concentration of 0.5% and spun down again; the final supernatant (S1) was used for immunoprecipitation studies.
Immunoprecipitation
Mouse monoclonal anti-eIF2c2 (4F9) (Santa Cruz Biotechnology), which cross-reacts with all Ago isoforms (Ikeda et al. 2006); mouse monoclonal anti-FMRP (7G1-1; UC Davis); and, as a negative control, rabbit polyclonal anti-Synapsin 1 (Sigma) were used. Antibodies were concentrated with the Amicon Ultra 10K centrifugal filters, buffer exchanged in PBS, and coupled to the magnetic Dynabeads utilizing the Dynabeads Antibody coupling kit (Invitrogen) following the manufacturer's instructions. Antibody-coupled magnetic beads (1 mg) were coated with bovine serum albumin in HB with NP-40 (0.5%) for 1 h and incubated with S1 supernatant (600 μL at 5.8 μg/μL) overnight at 4°C on a rotor. Each sample was run in duplicate tubes. Beads were then washed twice with HB plus NP-40, once with HB, and once with 50 mM Tris-HCl (pH 7.0). RNA was extracted with Trizol as previously described (Lugli et al. 2008), and purified RNA was treated with RNAsecure (Ambion) and with DNAse (TURBO DNA-free DNase, Ambion). RT-qPCR was performed using equal fractions of the immunoprecipitated RNA as described below. (Using separate beads, we also isolated bound proteins and carried out Western blotting to confirm that antibody 4F9 brought down Argonaute homolog proteins and that Fmrp antibody brought down both Fmrp protein as well as Argonaute homolog proteins, as expected (e.g., Lugli et al. 2005; data not shown).
Detection of bound RNAs
To detect bound U6 RNA and SynGAP1 intronic RNA (lying on the sense strand, just upstream and just downstream of the intronic hairpin that gives rise to the siRNA), S1 supernatant total RNA (1.0 μg) or equal fractions of RNA purified from immunoprecipitated beads were reversed transcribed with short gene-specific primers with TM around 38°C–42°C with Superscript III (Invitrogen) as per the manufacturer's protocol with a few modifications. Denaturation was performed for 5 min at 95°C, and then samples were quickly placed on ice. Reverse transcription was performed for five min at 38°C, for 30 min at 42°C, and for the last 30 min at 55°C. Real-time quantitative PCR was performed on a Stratagene MX 3005P Real-Time PCR Instrument (Stratagene). Each sample was run in duplicate. All products were confirmed and distinguished from primer dimers by examining melting curves and by running the PCR product on 3.5% Agarose gels. As negative controls, parallel samples lacked reverse transcriptase or lacked added RNA sample. The PCR mix, 20 μL, contained 10 μL of SYBER GREEN PCR master mix (Applied Biosystems, Foster City), 0.6 μM of each pair of primers, and 5 μL of dilute cDNA (1:10). PCR was performed for 20 sec at 95°C, 25 sec at 56°C, and 25 sec at 72°C for 50 cycles followed by the thermal denaturation protocol. U6 primers are described by Lugli et al. (2008).
Primers
Short specific primers are as follows:
SynGAP1 mRNA Re-short: TCGTGAGGTCTG (TM = 4.09)
Intron Sense-Upstream Re-short: GGTTAGCCTCAG (TM = 38.6)
Intron sense-Downstream Re-short: AACCAACTAGGC (TM = 37.7)
PCR primers are as follows:
SynGAP1 mRNA Fo: CAAGGTGGTCAACTCCCATT (TM = 55.06)
SynGAP1 mRNA Re: TCGTGAGGTCTGCTCATCTG (TM = 56.42)
Intron Sense-Upstream Fo: GACCCCTTTATCCCACCATT (TM = 54.46)
Intron Sense-Upstream Re: GGTTAGCCTCAGCGTGCTAC (TM = 58.05)
Intron Sense-Downstream Fo: GCAGGGGACATAAAGGGTTT (TM = 55.18)
Intron Sense-Downstream Re: AACCAACTAGGCCTCCCATC (TM = 56.76)
To detect SynGAP1 siRNA, we chose the most abundant siRNA sequence arising on the sense strand (AAGGCTGGGAATGTTTCGGAGA) and made a specific TaqMan MicroRNA Reverse Transcription and Assay kit (Applied Biosystems) based on this sequence. Reverse transcription was performed in total RNA (S1 supernatant) using 100 ng of RNA or equal fractions of immunoprecipitated samples. A calibration curve was performed using S1 supernatant RNA (10, 100, and 1,000 ng) to ensure linearity of the assay. To validate that the system recognized the siRNA selectively and not intronic sequences present within SynGAP1 mRNA, total RNA was isolated from mouse N2A neuroblastoma cells and fractionated twice with the mirVANA miRNA isolation kit (Ambion) according to the manufacturer's directions, giving a nominal cut-off of 200 nt. Equal fractions of eluted RNA were loaded in a 1% Agarose gel and stained with ethidium bromide to confirm that the size separation was adequate. Over 90% of the siRNA was detected in the small RNA fraction, whereas the intronic RNA sequences were only detected in the large RNA fraction (data not shown). mmu-mir-99a and mmu-mir-350 were measured using TaqMan miRNA specific assay kits (Applied Biosystems).
For each RNA measured, we compared the Ct values of the samples immunoprecipitated by 4F9 or Fmrp antibodies to the Ct values immunoprecipitated by an irrelevant antibody, synapsin I. As a positive control, we verified that two selected miRNAs were specifically brought down, and as a negative control, we verified that U6 RNA was NOT specifically brought down (see Results).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Angela L. Lenon for assistance in training mice. Work was supported by the Jean Young and Walden W. Shaw Foundation, NIH (Autism Center of Excellence P50 HD055751), and the Stanley Medical Research Institute.
Footnotes
Abbreviations: siRNA, small inhibitory RNA; miRNA, microRNA; endo-siRNA, endogenous siRNA; RISC, RNA induced silencing complex.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2123811.
REFERENCES
- Abe K, Chisaka O, Van Roy F, Takeichi M 2004. Stability of dendritic spines and synaptic contacts is controlled by αN-catenin. Nat Neurosci 7: 357–363 [DOI] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batassa EM, Costanzi M, Saraulli D, Scardigli R, Barbato C, Cogoni C, Cestari V 2010. RISC activity in hippocampus is essential for contextual memory. Neurosci Lett 471: 185–188 [DOI] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC 2007. Mammalian mirtron genes. Mol Cell 28: 328–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ 2009. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry (in press) doi: 10.1038/mp.2009.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava A, Dallman MF, Pearce D, Choi S 2004. Long double-stranded RNA-mediated RNA interference as a tool to achieve site-specific silencing of hypothalamic neuropeptides. Brain Res Brain Res Protoc 13: 115–125 [DOI] [PubMed] [Google Scholar]
- Carlile M, Nalbant P, Preston-Fayers K, McHaffie GS, Werner A 2008. Processing of naturally occurring sense/antisense transcripts of the vertebrate Slc34a gene into short RNAs. Physiol Genomics 34: 95–100 [DOI] [PubMed] [Google Scholar]
- Carlile M, Swan D, Jackson K, Preston-Fayers K, Ballester B, Flicek P, Werner A 2009. Strand selective generation of endo-siRNAs from the Na/phosphate transporter gene Slc34a1 in murine tissues. Nucleic Acids Res 37: 2274–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Greenberg ME 2008. Communication between the synapse and the nucleus in neuronal development, plasticity, and disease. Annu Rev Cell Dev Biol 24: 183–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G 2009. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzitoyeva S, Dimitrijevic N, Manev H 2001. Intra-abdominal injection of double-stranded RNA into anesthetized adult Drosophila triggers RNA interference in the central nervous system. Mol Psychiatry 6: 665–670 [DOI] [PubMed] [Google Scholar]
- Dzitoyeva S, Dimitrijevic N, Manev H 2003. γ-Aminobutyric acid B receptor 1 mediates behavior-impairing actions of alcohol in Drosophila: Adult RNA interference and pharmacological evidence. Proc Natl Acad Sci 100: 5485–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G 2008. A human snoRNA with microRNA-like functions. Mol Cell 32: 519–528 [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C 2009. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francks C, Maegawa S, Laurén J, Abrahams BS, Velayos-Baeza A, Medland SE, Colella S, Groszer M, McAuley EZ, Caffrey TM, et al. 2007. LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia. Mol Psychiatry 12: 1129–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X 2009. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 583: 437–442 [DOI] [PubMed] [Google Scholar]
- Gan L, Anton KE, Masterson BA, Vincent VA, Ye S, Gonzalez-Zulueta M 2002. Specific interference with gene expression and gene function mediated by long dsRNA in neural cells. J Neurosci Methods 121: 151–157 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanzer J, Miyashiro KY, Sul JY, Barrett L, Belt B, Haydon P, Eberwine J 2005. RNA splicing capability of live neuronal dendrites. Proc Natl Acad Sci 102: 16859–16864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert D, Topolnik L, Azzi M, Huang L, Badeaux F, Desgroseillers L, Sossin WS, Lacaille JC 2008. Forskolin induction of late-LTP and up-regulation of 5′ TOP mRNAs translation via mTOR, ERK, and PI3K in hippocampal pyramidal cells. J Neurochem 106: 1160–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Hamilton PJ, Reish NJ, Sweatt JD, Miller CA, Rumbaugh G 2009. Reduced expression of the NMDA receptor-interacting protein SynGAP causes behavioral abnormalities that model symptoms of schizophrenia. Neuropsychopharmacology 34: 1659–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Gauthier J, Spiegelman D, Noreau A, Yang Y, Pellerin S, Dobrzeniecka S, Côté M, Perreau-Linck E, Carmant L, et al. 2009. Mutations in SYNGAP1 in autosomal nonsyndromic mental retardation. N Engl J Med 360: 599–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Satoh M, Pauley KM, Fritzler MJ, Reeves WH, Chan EK 2006. Detection of the argonaute protein Ago2 and microRNAs in the RNA induced silencing complex (RISC) using a monoclonal antibody. J Immunol Methods 317: 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey RA, Ch'ng TH, O'Dell TJ, Martin KC 2009. Activity-dependent anchoring of importin α at the synapse involves regulated binding to the cytoplasmic tail of the NR1-1a subunit of the NMDA receptor. J Neurosci 29: 15613–15620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. 2008. Hidden layers of human small RNAs. BMC Genomics 9: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama NH, Watabe AM, Carlisle HJ, Porter K, Charlesworth P, Monti J, Strathdee DJ, O'Carroll CM, Martin SJ, Morris RG, et al. 2002. SynGAP regulates ERK/MAPK signaling, synaptic plasticity, and learning in the complex with postsynaptic density 95 and NMDA receptor. J Neurosci 22: 9721–9732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong XC, Barzaghi P, Ruegg MA 2004. Inhibition of synapse assembly in mammalian muscle in vivo by RNA interference. EMBO Rep 5: 183–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneev SA, Kemenes I, Straub V, Staras K, Korneeva EI, Kemenes G, Benjamin PR, O'Shea M 2002. Suppression of nitric oxide (NO)-dependent behavior by double-stranded RNA-mediated silencing of a neuronal NO synthase gene. J Neurosci 22: RC227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Wiel C, Rubin GM 2004. Complementary miRNA pairs suggest a regulatory role for miRNA:miRNA duplexes. RNA 10: 171–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenberger D, Bermudez-Santana C, Hertel J, Hoffmann S, Khaitovich P, Stadler PF 2009. Evidence for human microRNA-offset RNAs in small RNA sequencing data. Bioinformatics 25: 2298–2301 [DOI] [PubMed] [Google Scholar]
- Larson J, Sieprawska D 2002. Automated study of simultaneous-cue olfactory discrimination learning in adult mice. Behav Neurosci 116: 588–599 [PubMed] [Google Scholar]
- Laurén J, Airaksinen MS, Saarma M, Timmusk T 2003. A novel gene family encoding leucine-rich repeat transmembrane proteins differentially expressed in the nervous system. Genomics 81: 411–421 [DOI] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23: 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhoff MW, Laurén J, Cassidy RM, Dobie FA, Takahashi H, Nygaard HB, Airaksinen MS, Strittmatter SM, Craig AM 2009. An unbiased expression screen for synaptogenic proteins identifies the LRRTM protein family as synaptic organizers. Neuron 61: 734–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Kim SW, Lin Y, Moore PS, Chang Y, John B 2009. Characterization of viral and human RNAs smaller than canonical MicroRNAs. J Virol 83: 12751–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QR, Lu L, Zhu XG, Gong JP, Shaham Y, Uhl GR 2006. Rodent BDNF genes, novel promoters, novel splice variants, and regulation by cocaine. Brain Res 1067: 1–12 [DOI] [PubMed] [Google Scholar]
- Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR 2005. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem 94: 896–905 [DOI] [PubMed] [Google Scholar]
- Lugli G, Torvik VI, Larson J, Smalheiser NR 2008. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem 106: 650–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei B, Li C, Dong S, Jiang CH, Wang H, Hu Y 2005. Distinct gene expression profiles in hippocampus and amygdala after fear conditioning. Brain Res Bull 67: 1–12 [DOI] [PubMed] [Google Scholar]
- Meier I, Fellini L, Jakovcevski M, Schachner M, Morellini F 2009. Expression of the snoRNA host gene gas5 in the hippocampus is upregulated by age and psychogenic stress and correlates with reduced novelty-induced behavior in C57BL/6 mice. Hippocampus 20: 1027–1036 [DOI] [PubMed] [Google Scholar]
- Mexal S, Berger R, Pearce L, Barton A, Logel J, Adams CE, Ross RG, Freedman R, Leonard S 2008. Regulation of a novel αN-catenin splice variant in schizophrenic smokers. Am J Med Genet B Neuropsychiatr Genet 147B: 759–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson RH, Nicholson AW 2002. Molecular characterization of a mouse cDNA encoding Dicer, a ribonuclease III ortholog involved in RNA interference. Mamm Genome 13: 67–73 [DOI] [PubMed] [Google Scholar]
- Okamura K, Lai EC 2008. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol 9: 673–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault J, Perreault JP, Boire G 2007. Ro-associated Y RNAs in metazoans: Evolution and diversification. Mol Biol Evol 24: 1678–1689 [DOI] [PubMed] [Google Scholar]
- Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T 2007. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics 90: 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogelj B, Hartmann CE, Yeo CH, Hunt SP, Giese KP 2003. Contextual fear conditioning regulates the expression of brain-specific small nucleolar RNAs in hippocampus. Eur J Neurosci 18: 3089–3096 [DOI] [PubMed] [Google Scholar]
- Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ 2009. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput Biol 5: e1000507 doi: 10.1371/journal.pcbi.1000507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA 2008. Divergent transcription from active promoters. Science 322: 1849–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Manev H, Costa E 2001. RNAi and memory: Was McConnell on the right track? Trends Neurosci 24: 216–218 [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Torvik VI, Mise N, Ikeda R, Abe K 2008. Natural antisense transcripts are co-expressed with sense mRNAs in synaptoneurosomes of adult mouse forebrain. Neurosci Res 62: 236–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Lenon AL, Davis JM, Torvik VI, Larson JR 2010. Olfactory discrimination training up-regulates and reorganizes expression of microRNAs in adult mouse hippocampus. ASN Neuro 2: art e00028 doi: 10.1042/AN20090055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Steitz JA 1998. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol 18: 6897–6909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills AA, Karayiorgou M, et al. 2008. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet 40: 751–760 [DOI] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS 2009. Small RNAs derived from snoRNAs. RNA 15: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Simons C, Nahkuri S, Oey H, Korbie DJ, Mercer TR, Holst J, Ritchie W, Wong JJ, Rasko JE, et al. 2010. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat Struct Mol Biol. 17: 1030–1034 [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Leung FC 2009. A study on genomic distribution and sequence features of human long inverted repeats reveals species-specific intronic inverted repeats. FEBS J 276: 1986–1998 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543 [DOI] [PubMed] [Google Scholar]
- Werner A, Carlile M, Swan D 2009. What do natural antisense transcripts regulate? RNA Biol 6: 43–48 [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi CE, Bekker JM, Miller G, Hill KL, Crosbie RH 2003. Specific and potent RNA interference in terminally differentiated myotubes. J Biol Chem 278: 934–939 [DOI] [PubMed] [Google Scholar]
- Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O'Carroll D, Stoffel M, Tuschl T, et al. 2009. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci 106: 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]