Abstract
Ricin produced by the castor bean plant and Shiga toxins produced by pathogenic Escherichia coli (STEC) and Shigella dysenteriae are type II ribosome inactivating proteins (RIPs), containing an enzymatically active A subunit that inhibits protein synthesis by removing an adenine from the α-sarcin/ricin loop (SRL) of the 28S rRNA. There are currently no known antidotes to Shiga toxin or ricin, and the ability to screen large chemical libraries for inhibitors has been hindered by lack of quantitative assays for catalytic activity that can be adapted to a high throughput format. Here, we describe the development of a robust and quantitative reverse transcription polymerase chain reaction (qRT-PCR) assay that can directly measure the toxins' catalytic activity on ribosomes and can be used to examine the kinetics of depurination in vivo. The qRT-PCR assay exhibited a much wider dynamic range than the previously used primer extension assay (500-fold vs. 16-fold) and increased sensitivity (60 pM vs. 0.57 nM). Using this assay, a 400-fold increase in ribosome depurination was observed in yeast expressing ricin A chain (RTA) relative to uninduced cells. Pteroic acid, a known inhibitor of enzymatic activity, inhibited ribosome depurination by RTA and Shiga toxin 2 with an IC50 of ∼100 μM, while inhibitors of ricin transport failed to inhibit catalytic activity. These results demonstrate that the qRT-PCR assay would enable refined kinetic studies with RIPs and could be a powerful screening tool to identify inhibitors of catalytic activity.
Keywords: ribosome inactivating protein, qRT-PCR, rRNA depurination, α-sarcin/ricin loop, ricin, Shiga toxin
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) is an emerging bacterial pathogen responsible for food-borne outbreaks of disease around the world, resulting in significant morbidity and mortality, including hemorrhagic colitis (HC) and hemolytic uremic syndrome (HUS) (Pickering et al. 1994; Paton and Paton 1998). Shiga toxins and ricin, produced by the castor bean (Ricinus communis) are of major concern for public health and bioterrorism and have been classified as category B select agents. There are currently no known antidotes to Shiga toxin or ricin and no specific therapeutics that could prevent lethality from exposure. Shiga toxin and ricin are AB toxins whose enzymatically active A subunit is an N-glycosidase that removes an adenine from the α-sarcin/ricin loop (SRL) of the 28S rRNA, resulting in inhibition of protein synthesis in the elongation phase (Endo et al. 1988). Depurination at the highly conserved SRL is necessary, though in itself insufficient for the cellular toxicity of ribosome inactivating proteins (RIPs) (Hudak et al. 2004; Li et al. 2007). Therefore, enzymatic activity of RIPs represents a fundamental target for preventing RIP toxicity.
The B subunit of ricin is a galactose-specific lectin that facilitates uptake of the toxin by endocytosis (Spooner et al. 2006). Ricin is then trafficked via the retrograde pathway to the Golgi and the endoplasmic reticulum (ER) (Sandvig and van Deurs 2000, 2002, 2005). Because the B subunit is required for endocytosis and retrograde trafficking of the toxin to the ER in mammalian cells, it has not been possible to study enzymatic activity of the A subunits in the absence of the B subunits in vivo. Furthermore, due to lack of quantitative assays, it has not been possible to examine the kinetics of depurination and compare the relative activity of different RIPs in vivo. Traditionally the activity of RIPs has been examined using an in vitro translation system that measures the incorporation of radioactive amino acids into protein (Endo and Tsurugi 1987). This assay is labor intensive and suffers from high background noise and a low dynamic range. Nonradioactive luminescence based protein synthesis assays have also been developed using luciferase as a reporter (Saenz et al. 2007; Wahome et al. 2010). Because the luciferase-based assays do not measure the depurination activity directly, they are prone to false positives that interfere with the translation machinery or affect the turnover of the luciferase reporter (Saenz et al. 2007).
Ribosome depurination activity has been examined by RIP treatment of ribosomes followed by aniline treatment, which causes cleavage of the RNA at the abasic site (Stirpe et al. 1988; Taylor and Irvin 1990). This method is not quantitative and suffers from poor sensitivity. A more sensitive primer extension method has been described (Iordanov et al. 1997). To quantify depurination by primer extension, we used a dual-oligo primer extension assay with a primer that hybridized downstream from the depurination site to measure the extent of depurination and a second control primer, which hybridized near the 5′ end of the 25S rRNA to quantify the total amount of rRNA (Parikh et al. 2002). The ratio of the depurination fragment compared with the control fragment allowed for quantification of the extent of depurination (Parikh et al. 2002, 2005; Baykal and Tumer 2007; Li et al. 2007; Chiou et al. 2008). Other methods to measure depurination include quantification of the adenine released from RIP depurination by HPLC, a calorimetric assay (Brigotti et al. 1998; Heisler et al. 2002) or an enzymatically coupled assay to continuously measure the release of adenine by RIPs (Sturm and Schramm 2009). More recently, a quantitative RT-PCR (qRT-PCR) assay, which was designed to detect low levels of ricin in environmental samples and in food, was described (Melchior and Tolleson 2010). This assay relied on the inherent property of reverse transcriptase of incorporating a deoxyadenosine at the position opposite the abasic site (Takeshita et al. 1987). The resulting T to A transversion on the cDNA strand could be detected by sequence-specific primers. This assay used total RNA from mammalian cells as the substrate, which is ∼105-fold less sensitive to ricin than intact ribosomes (Endo and Tsurugi 1988). The temperature of assay was elevated to 55°C and the pH was lowered to pH 5.0 for detection of RIP activity (Melchior and Tolleson 2010) because RTA and type I and type II RIPs have a low catalytic pH optimum (pH 4.0) with nucleic acid substrates, such as stem–loop RNA, poly A RNA, and herring sperm DNA, while natural ribosomal substrate is depurinated at physiological pH (Barbieri et al. 1997, 2004).
We were interested in adapting this assay for detection of depurination under physiological conditions in vivo using yeast as a model system and in vitro for use in high throughput screens with small molecules that are either labile or insoluble at neutral pH. Here we describe the development of a qRT-PCR assay for detection of the toxins' catalytic activity in vivo and in vitro using ribosomes as the substrate. Yeast was used as a model for kinetic analysis of RIP activity because by expressing the A subunits de novo in yeast we can bypass the need for the B subunit for endocytosis and retrograde transport, and focus more specifically on the catalytic activity of the A subunits on ribosomes (Hur et al. 1995; Hudak et al. 2004; Li et al. 2007). Therefore, the qRT-PCR assay described here can distinguish between the inhibitors that target the depurination activity of RIPs and those that target other steps in cytotoxicity and could be used to identify the compounds that have a direct effect on the catalytic activity. The advantages of the qRT-PCR assay over the previously described dual primer extension assay (Iordanov et al. 1997; Parikh et al. 2002) include increased sensitivity, a wider dynamic range of detection enabling refined kinetic studies with different RIPs and inhibitors.
RESULTS
qRT-PCR assay design, verification, and validation
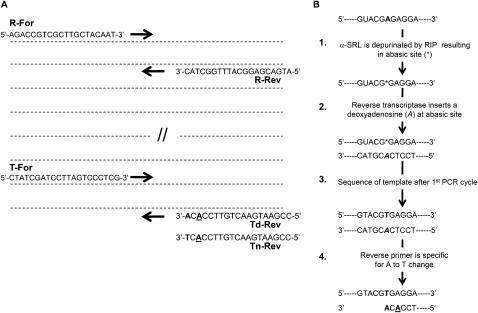
To quantify the extent of depurination by qRT-PCR, we used the comparative CT method (ΔΔCT) with two primer pairs, one amplifying the target of interest and the second amplifying a reference amplicon, corresponding to the total amount of input 25S rRNA. The target reactions were normalized to the reference amplicon to measure the amount of depurinated RNA relative to the total amount of RNA. Figure 1A shows a schematic diagram of the yeast 25S rRNA and the positions of the target and reference primers and amplicons. The forward target primer (T-For) anneals to nucleotides 2756 to 2777 of 25S rRNA sequence (271 nt 5′ to the depurination site). The reverse target primer (Td-Rev) anneals to nucleotides 3027 to 3045, with its 3′ terminus at the position of depurination (A3027). This primer pair will generate a 289-base pair (bp) amplicon. The forward reference primer (R-For) anneals to nucleotides 2071 to 2090 of 25S rRNA sequence and the reverse reference primer (R-Rev) anneals to nucleotides 2273 to 2292. This primer pair will yield a 221-bp amplicon. A general outline of the assay is depicted in Figure 1B. The SRL sequence is shown with the adenine that serves as a substrate for RIPs in capital and bold. Following treatment with the toxin (step 1) the adenine is depurinated leaving behind an abasic residue (*). Upon conversion, the reverse transcriptase preferentially inserts a dAMP opposite the abasic site (step 2), which following the first round of amplification in the qPCR step (step 3) will result in a complementary dTMP. This adenine to thymine change is detected using a reverse primer whose 3′ terminus contains an adenine (step 4). Specificity for detection of the A to T change is enhanced by the inclusion of a secondary mismatch two base pairs 5′ to the terminus (underlined A).
FIGURE 1.
Schematics of primer and assay design. (A) Schematic representation of 25S rRNA showing relative positioning of forward and reverse primers for target and reference amplicons used in qRT-PCR. Sequences shown above and below the line are forward and reverse primers, respectively. (B) Overview of the assay depicting the SRL sequence with the depurinated adenine shown in bold. Depurination by an RIP (step 1) leaves behind an abasic site (*). Reverse transcriptase incorporates an adenosine at the position opposite the abasic site (step 2), resulting in a T to A transversion in the cDNA, which is propagated during PCR (step 3). The base change at this position is detected using reverse primer with and adenine at the 3′ terminus (in bold) and specificity is enhanced by a secondary mismatch (underlined).
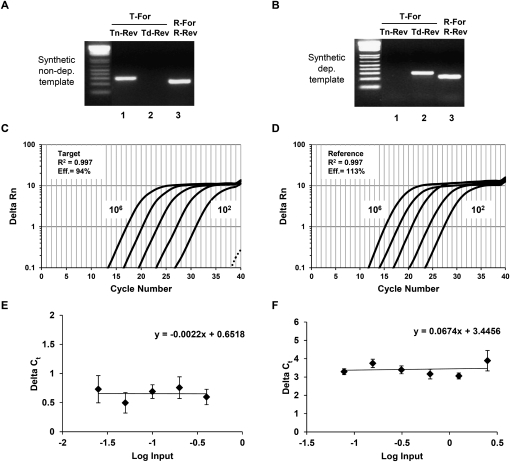
To determine if the target primer pairs were specific in their detection of depurinated rRNA, we constructed synthetic DNA targets that mimic the depurinated and nondepurinated rRNA templates. To construct a DNA template that mimics the depurinated rRNA, we used site-directed mutagenesis to change the target adenosine (A3027) to a thymidine. The nondepurinated DNA template contained a dAMP at the same position. Pairwise PCR reactions were set up using ∼7.25 × 108 copies of synthetic target mimicking nondepurinated and depurinated templates with target primers, T-For with Tn-Rev, which are specific for nondepurinated RNA; T-For and Td-Rev, which are specific for depurinated rRNA, and reference primers R-For and R-Rev (Fig. 1). End point reactions were resolved on a 1.5% agarose gel stained with ethidium bromide and are shown in Figure 2A,B. The reference primer pair amplified the correct size product from both synthetic targets (Fig. 2A,B, lane 3). The target primer pair, T-For and Tn-Rev, amplified a product only from the synthetic target corresponding to the nondepurinated rRNA (Fig. 2A, lane 1), but not the depurinated rRNA (Fig. 2B, lane 1). Likewise, T-For when used with Td-Rev amplified product only from the synthetic target that mimics the depurinated rRNA (Fig. 2B, lane 2), but not the nondepurinated rRNA (Fig. 2A, lane 2). The forward and reverse primers for target and reference amplicons were used in qPCR with 10-fold serially diluted synthetic target that corresponds to depurinated rRNA ranging from 106 copies to 102 copies per reaction. Figure 2C,D shows representative amplification profiles for the target and reference primers, respectively demonstrating robust amplification even at 100 copies of starting template. With the CT set at Delta Rn of 1 the R2 values for T-For/Td-Rev and R-For/R-Rev were both 0.997 and their calculated amplification efficiencies were 94% and 113%, respectively. To verify that template discrimination observed for primer pair T-For/Td-Rev in end point PCR is also observed in quantitative PCR, we used 105 copies of synthetic target corresponding to nondepurinated rRNA as the template. This reaction is represented as the dashed line in the lower right side of the amplification profile in Figure 2C and shows no detectable amplification above the CT set at Delta Rn of 1. These results, coupled with the end point PCR data, indicate that the designed target and reference primer pairs are suitable for real time PCR and that the target primers are highly specific for the template containing the base substitution that results from depurination.
FIGURE 2.
Verification and validation of forward and reverse target and reference primers used in qRT-PCR. Ethidium bromide stained 1.5% agarose gel showing end point PCR reactions using primers that amplify the target and the reference amplicons with ∼7.25 × 108 copies of synthetic template representing cDNA corresponding to nondepurinated rRNA (A) and depurinated rRNA (B). Representative amplification profiles from qPCR reactions using depurination specific target primers (C) or reference primers (D) with synthetic template mimicking 106 − 102 copies of depurinated rRNA. The amplification profile shown with a dashed line in C is a reaction using depurination-specific target primers with 105 copies of synthetic template mimicking nondepurinated rRNA to demonstrate primer specificity. (E) Plot of ΔCT between target and reference reactions versus log input amount of synthetic template corresponding to depurinated rRNA with target primers specific for depurinated rRNA. The regression line exhibits a slope with an absolute value <0.1. Data are the mean ±SD of triplicate reactions. (F) Plot of ΔCT between the target and reference reactions versus log input amount of template, which is cDNA corresponding to rRNA extracted from RTA treated ribosomes. The regression line exhibits a slope with an absolute value <0.1. Data are the mean ±SD of triplicate reactions.
For valid ΔΔCT quantitation it is crucial that target and reference PCR amplification efficiencies are approximately equal. To determine if the target and reference primer pairs have comparable PCR efficiencies, we used the validation criteria and assay outlined by Applied Biosystems (Applied Biosystems 2001). In this assay, the target and reference primers are used in triplicate qRT-PCR reactions with decreasing amounts of starting template. The CT values for the target and reference reactions are determined separately. The CT, threshold cycle, is the cycle number at which the fluorescence generated within a reaction crosses the detection threshold. The ΔCT, or difference in CT between the target and reference (CT target – CT reference), is then plotted versus the log input template amount to give a semi-log regression line. According to the validation criteria set forth by Applied Biosystems, if the absolute value of the slope of the semi-log regression line is <0.1, then the efficiencies of the two PCR reactions are approximately equal and therefore the assay is valid. The validation assay was carried out using the synthetic target that represents the depurinated rRNA and cDNA prepared from rRNA isolated from RTA-treated ribosomes. Figure 2E,F shows the graphs of ΔCT as a function of log input. The absolute values of each slope resulting from semi-log regression were determined to be 0.002 and 0.06 for the synthetic and cDNA targets, respectively, which are <0.1. Based on these results, we concluded that the primer design and assay conditions allowed quantitation of depurination by the ΔΔCT method.
Comparison of the qRT-PCR and the dual primer extension assay
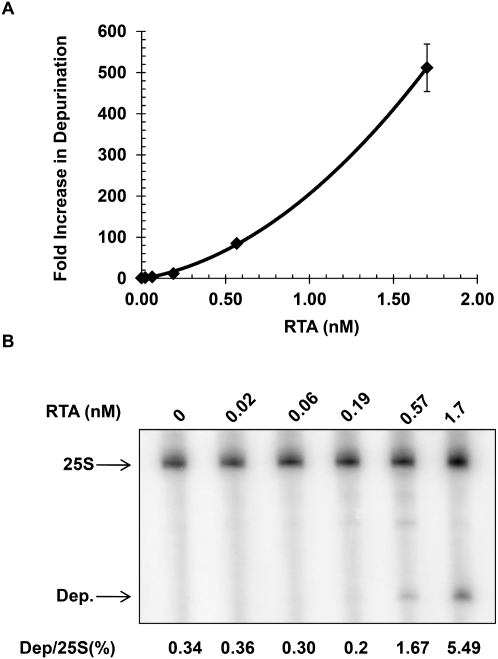
To compare the qRT-PCR method to the previously described dual primer extension assay (Iordanov et al. 1997; Parikh et al. 2002), we examined the dose dependence of depurination in the presence of increasing concentrations of RTA. Yeast ribosomes (10 pmol) were incubated for 10 min at 30°C with RTA at final concentrations of 0.02, 0.06, 0.19, 0.57, and 1.7 nM; rRNA was extracted and equal amounts were used in qRT-PCR. The fold increase in depurination relative to non-RTA treated ribosomes versus RTA concentration was plotted in Figure 3A. Very little depurination was observed in reactions containing RTA up to 0.06 nM at which point a 3.3-fold increase in depurination was detected relative to non-RTA treated ribosomes. When the RTA concentration was increased to 0.19 nM, an 11-fold increase in depurination was seen and this raised sharply with higher RTA concentrations, resulting in a >500-fold increase in depurination at 1.7 nM RTA. These results demonstrated a dosage dependent response in depurination with a minimum level of detection at 0.06 nM by qRT-PCR.
FIGURE 3.
Analysis of depurination as a function of RTA concentration in vitro. (A) Ten pmoles of yeast ribosomes were incubated for 10 min at 30°C with RTA at final concentrations of 0.02, 0.06, 0.19, 0.57, and 1.7 nM; rRNA was extracted and precipitated. Equal amounts of rRNA were used for qRT-PCR and the fold increase in depurination relative to nontreated ribosomes was determined by the comparative CT method. Data are the mean ±SD of triplicate reactions. (B) Dual primer extension analysis with the same rRNA used for the qRT-PCR assay. The percentage of depurination was calculated by establishing a ratio between the intensity of depurination product (Dep) to the intensity of the 25S rRNA product (25S) and is shown below each lane.
The same rRNA samples were used in the dual primer extension assay to compare the sensitivity of the two different assays. Figure 3B shows the results of the primer extension analysis with the 25S and depurination primer (Dep) products marked by the arrows. The percentage of depurinated rRNA relative to the 25S control at each RTA concentration was calculated and is shown underneath each lane in Figure 3B. Depurination was not detected in reactions containing RTA at 0.02 nM to 0.19 nM. At 0.57 nM RTA the primer extension assay detected 1.67% depurination, which is a fivefold increase compared to no RTA treatment. Finally, when RTA was at a concentration of 1.7 nM 5.49% depurination was detected, which was a 16-fold increase compared to no RTA treatment. We conclude that with the dual primer extension assay the minimum level of RTA detection was 0.57 nM and the dynamic range of detection over the RTA titration was 16-fold. These results represented a striking contrast compared to the results obtained using the qRT-PCR assay where the minimum level of RTA detection was almost 10-fold lower at 0.06 nM. In addition, the qRT-PCR assay exhibited a larger dynamic range of detection (3.3-fold at the low end to 500-fold at the high end) compared to the dual primer extension assay (fivefold at the low end and 16-fold at the high end). Therefore, qRT-PCR was capable of detecting depurination with greater sensitivity and over a broader range than the dual primer extension assay.
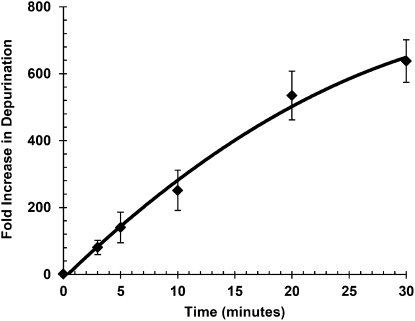
Analysis of the rate of ribosome depurination by RTA in vitro
To assess the ability of the qRT-PCR assay to detect depurination over time and to establish the conditions of the assay, we conducted a time course in which 7 pmol (70 nM final concentration) of yeast ribosomes were incubated with 0.25 nM RTA. RNA was extracted and converted to cDNA using equal amounts of rRNA at each time point. The resulting cDNA was used in real time PCR quantitation carried out by ΔΔCT. Figure 4 shows the fold increase in depurination relative to non-RTA treated ribosomes over time. The reaction was nearly linear up to 20 min at 30°C, while 0.5 and 1 nM RTA reached maximal activity at shorter times (data not shown). Since prolonging the linear phase is desirable for rate determinations and for inhibitor assays, 0.25 nM RTA was chosen as standard for these applications.
FIGURE 4.
Time course of depurination of ribosomes treated with RTA in vitro. Seven pmoles of yeast ribosomes were incubated at 30°C for 3, 5, 10, 20, and 30 min in the presence of RTA at a final concentration of 0.25 nM. The rRNA was extracted and equal amounts were used for qRT-PCR. The fold increase in depurination relative to nontreated ribosomes was determined by the comparative CT method. Data are the mean ±SD of triplicate reactions.
Analysis of the rate of ribosome depurination by RTA in vivo
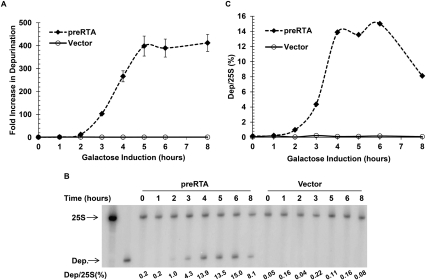
To examine depurination in vivo, we transformed wild-type yeast with plasmid NT1403, which contains coding sequence for the precursor form of RTA (pre-RTA), or with a vector control plasmid without pre-RTA (pRS416). Transformants were maintained in selective dextrose media, then shifted to selective galactose media. Figure 5A shows the results of this experiment in which the fold increase in depurination relative to the 0-h time point was plotted as a function of time in galactose for cells expressing pre-RTA (black diamonds; dashed line) or harboring the vector (open circles; solid line). Vector control samples showed no increase in depurination over the course of induction. Yeast cells expressing pre-RTA exhibited modest levels of depurination at 2 h post-induction (∼13-fold increase). However, by 3 h post-induction, we observed a >100-fold increase and by 5 h, depurination further increased to >400-fold and did not change through the 8-h time point. These results demonstrated that depurination levels could be monitored by qRT-PCR over time in cells expressing pre-RTA in vivo.
FIGURE 5.
Analysis of depurination in yeast expressing pre-RTA from the galactose inducible GAL1 promoter in vivo. (A) Yeast carrying pre-RTA under a galactose inducible promoter or a vector control plasmid was grown to mid-log phase in dextrose-containing media. The carbon source was then switched to galactose to induce expression of pre-RTA. Equal amounts of total RNA was used for qRT-PCR and the fold increase in depurination relative to the 0 h time point was determined by the comparative CT method. The fold increase in depurination for cells expressing pre-RTA (black diamond; dashed line) or vector control (open circle; solid line) were then plotted as a function of time. Data are the mean ±SD of triplicate reactions. (B) The dual primer extension assay using the same rRNA as used in the qRT-PCR assay. The percent depurination was calculated by establishing a ratio between the intensity of the depurinated band to the intensity of the 25S band and is shown under each lane and is plotted in C.
The rRNA samples analyzed by qRT-PCR were used in the dual primer extension assay to compare the relative sensitivity of in vivo depurination detection by the two different assays. Figure 5B shows the gel image of primer extended products for 25S and depurination primers marked by the arrows in samples expressing pre-RTA or harboring the vector. The percentage of depurinated rRNA relative to the 25S control product for each time point was calculated and is shown underneath each lane in Figure 5B and graphically in Figure 5C. Depurination detected by primer extension showed a similar profile as seen with the qRT-PCR method in which there was a steady increase in depurination with increasing time. However, we observed that after 4 h there was no further increase in depurination using the dual primer extension assay, while the maximum level of depurination was not seen until 5 h using the qRT-PCR assay. This suggested that the qRT-PCR assay was more sensitive to subtle changes in the depurination state of the ribosome. A second difference between the two assays was the drop in depurination detected by dual primer extension at the 8-h time point. A similar decrease was observed in yeast expressing pokeweed antiviral protein (PAP) using dual primer extension analysis (Parikh et al. 2002). This apparent decrease might be attributed to the cleavage of the rRNA containing an abasic site, which would be detected by primer extension, but not by qRT-PCR. These results demonstrated that the qRT-PCR provided a powerful tool for examining the kinetics of ribosome depurination in vivo.
Inhibition of depurination by small molecule inhibitors
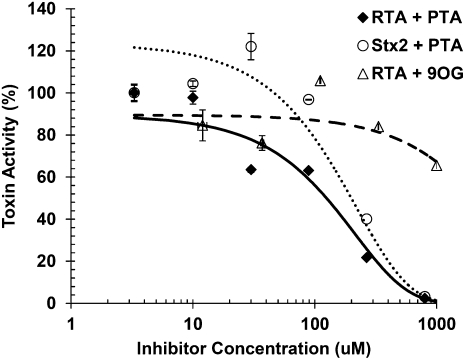
To determine if the qRT-PCR assay could be used to identify inhibitors of ribosome depurination by different RIPs we examined the effect of known inhibitors of the enzymatic activity of RTA. Pteroic acid (PTA) is an adenine analog which was shown by X-ray crystallography to bind to the active site of RTA and was kinetically shown to inhibit RTA with an IC50 of 600 μM when assayed by in vitro translation using Artemia salina ribosomes (Yan et al. 1997). 8-methyl-9-oxo-guanine (9OG) was previously reported as a more potent inhibitor than PTA in the Artemia translation system with an IC50 of 400 μM (Miller et al. 2002). We tested the ability of PTA and 9OG to inhibit the activity of RTA and Shiga toxin 2 (Stx2) using the qRT-PCR assay. Figure 6 shows the plot of RTA activity (solid diamond; solid line) and Stx2 activity (open circle; small dashed line) in the presence of increasing concentrations of PTA. A clear inhibition of activity for both RTA and Stx2 was observed with IC50 values of ∼105 μM and ∼110 μM, respectively. The qRT-PCR assay showed a ∼6-fold decrease, from 600 μM to 100 μM, in the measured IC50 for PTA compared to the IC50 measurements made using in vitro translation (Yan et al. 1997). We failed to see significant inhibition of RTA depurination by 9OG at any concentration <1 mM (open triangle; large dashed line), and observed only 16% inhibition of depurination at 333 μM and 35% inhibition at 1 mM. In addition, we tested golgicide A, and compounds 75 and 134, which have been shown to inhibit toxin transport (Saenz et al. 2007) and found essentially no effect in the qRT-PCR assay at levels up to 1 mM (data not shown). These results demonstrated the potential of the qRT-PCR assay in screening for small molecule inhibitors of catalytic activity of RIPs without interference by inhibitors that target other steps in ricin cytotoxicity.
FIGURE 6.
Inhibition of RTA and Stx2 by pteroic acid (PTA) and 8-methyl-9-oxoguanine (9OG). Yeast ribosomes (7 pmol) were incubated with RTA or Stx2 at 1 nM for 5 min at 30°C in the presence of PTA at final concentrations of 3.3, 10, 30, 89, 266, and 800 μM or with RTA at 1 nM in the presence of 9OG at 12, 37, 111, 333, and 1000 μM. rRNA was extracted, precipitated, and equal amounts were used in qRT-PCR. The fold increase in depurination relative to the no inhibitor reactions was determined by the comparative CT method and used to set the activity for each toxin to 100%. RTA and Stx2 inhibition by PTA is depicted with closed diamonds and open circles, respectively. RTA inhibition by 9OG is depicted with open triangles. Data are the mean ±SD of triplicate reactions.
DISCUSSION
Here, we describe a qRT-PCR assay that exhibited excellent dynamic range and sensitivity, demonstrating robustness for quantifying ribosome depurination activity of RIPs in vivo and in vitro. The optimal conditions of the qRT-PCR assay using isolated yeast ribosomes differed substantially from the qRT-PCR assay reported previously (Melchior and Tolleson 2010) and more closely resembled the physiological conditions. Similar conditions were used in the previously described primer extension assay (Iordanov et al. 1997; Parikh et al. 2002). The qRT-PCR assay showed no background when cells harboring the vector were used (Fig. 5). The background observed using a synthetic target corresponding to nondepurinated rRNA as the template showed no detectable amplification with the primer pair specific for the depurinated template (Fig. 2C), demonstrating the specificity of this assay for detecting a single nucleotide change. The qRT-PCR assay results were very consistent with the dual primer extension results, but with significantly increased sensitivity because of the PCR amplification and a wider dynamic range of detection. Both methods use reverse transcriptase, which either inserts a dAMP in the cDNA at the position opposite the abasic site (Takeshita et al. 1987) or stalls at the abasic site (Iordanov et al. 1997). The qRT-PCR assay detects only the rRNA that contains the abasic site. In contrast, the primer extension analysis would detect not only the rRNA containing the abasic site where the reverse transcriptase has stalled (Iordanov et al. 1997), but also the rRNA that may be cleaved at the abasic site, which would not be detected by the qRT-PCR assay described here. This could explain the difference observed in depurination after 6 h of RTA expression by qRT-PCR or primer extension using the same RNA (Fig. 5). The yeast model system coupled with the quantitative nature of qRT-PCR allowed for in vivo kinetic analyses of ribosome depurination with greater sensitivity than the primer extension analysis, indicating that both methods have certain advantages and disadvantages. The qRT-PCR assay will be a useful tool to compare the kinetics of ribosome depurination by different RIPs and for resolving kinetic activities of RTA mutants in vivo without having to purify each protein.
The qRT-PCR assay can be used not only for examining the kinetics of ribosome depurination by RIPs, but also as a screening tool to identify small molecule inhibitors of RIPs. Current screening methods for small molecule inhibitors of ricin and Shiga toxin include the use of high throughput cell-based assays using luciferase (Zhao and Haslam 2005; Saenz et al. 2007; Wahome et al. 2010) or radioactive translation assays (Stechmann et al. 2010). The majority of compounds identified from the cell-based assays interfered with intracellular trafficking of the toxins (Saenz et al. 2007; Stechmann et al. 2010). Structure-based screening has been employed where a chemical library is screened in silico against a resolved structure to identify small molecules that potentially provide a best “fit” into the active site and most likely disrupt enzymatic function (Yan et al. 1997; Bai et al. 2009, 2010). RTA is a very difficult virtual screening target because of its very large polar active site (Bai et al. 2010). Thus, to date small molecule RTA inhibitors identified have IC50 values mostly in the milimolar range. Many compounds, especially those modeled after adenine at the active site are significantly inhibitory by themselves in cell free translation systems presenting serious difficulties for measurement of their IC50 values. In addition, the fairly sharp pH optimum of translation around pH 7.4 also presents problems for molecules, such as PTA, which are not water soluble (Yan et al. 1997; Miller et al. 2002). Furthermore, cell free translation assays cannot distinguish between compounds that affect translation by inhibiting the catalytic activity of the toxins and those that interfere with translation by toxin-independent mechanisms.
There is clearly an urgent need for alternative assays that can be used to identify inhibitors of ricin and Shiga toxin that target catalytic activity. The assay reported here represents a platform upon which to build a screen for small molecule inhibitors of these toxins using Saccharomyces cerevisiae as a model system. To demonstrate its validity and sensitivity, we treated purified ribosomes with RTA and Stx2 in the presence and absence of the active site inhibitor, PTA (Yan et al. 1997) and showed that depurination by both toxins decreased in a dose-dependent manner. In contrast, 9OG, reported as more potent than PTA by in vitro translation (Miller et al. 2002), failed to cause a similar level of inhibition of depurination activity. The qRT-PCR assay can be adapted to a multiplex format to enable target and reference detection in a single reaction, allowing the analysis of 30 compounds per 96-well plate. Since the qRT-PCR assay measures toxin activity directly, it could also be used as a confirmatory assay to demonstrate that the candidate molecules identified in high throughput screens act directly on the catalytic activity. Moreover, the qRT-PCR assay can be used in refined kinetic studies to distinguish among inhibitors that affect substrate binding, catalytic activity, and protein stability.
Ricin or α-sarcin, which damage rRNA either by depurination or cleavage of the phosphodiester backbone within the α-SRL, activate the ribotoxic stress response, leading to transcription of genes that will either promote cell survival or cause apoptotic cell death (Iordanov et al. 1997). To determine how rRNA damage by α-sarcin correlates with ribotoxic stress we are adapting this method to measure rRNA cleavage at the α-SRL in real time using forward and reverse primers that enable quantitation of the loss of intact rRNA. In addition, we are working on extending the assay to mammalian ribosomes to monitor depurination of mammalian rRNA.
MATERIALS AND METHODS
Expression of RTA in yeast and RNA isolation
Wild-type S. cerevisiae strain W303 (MATa ade2-1 trp1-1 ura3-1 leu2-3, 112 his3-11, 15 can1-100) was transformed with plasmids NT1403 (GAL1-preRTA-V5-His6) or pRS416 (vector control) and selected for growth on synthetic dropout (SD) medium (2% dextrose, 0.67% Bacto-yeast nitrogen base) supplemented with amino acids but lacking uracil (SD-ura). Transformants were grown overnight at 30°C in liquid SD-Ura, then used to inoculate SD-Ura with 2% galactose at a concentration of 0.5 OD/mL. At 0, 1, 2, 3, 4, 5, 6, and 8 h post-induction, 1.5 mL of cells (∼2.25 × 107 cells) were removed and pelleted by centrifugation. Cells were resuspended in 600 μL RLT buffer (Qiagen RNeasy Mini Kit) and added to 600 μL 425–600 micron glass beads (Sigma). Cells were lysed using a Mini-Bead Beater for 2 min at 4°C. Total RNA was extracted using the RNeasy Mini Kit (Qiagen) according to the manufacturer's yeast protocol and eluted in 40 μL of RNase-free water. Purified RNA was quantitated using a NanoDrop 1000 prior to cDNA synthesis.
cDNA synthesis
Total RNA or rRNA was converted to cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The 20-μL reactions were set up according to the manufacturer's protocol using 375–500 ng of total RNA or rRNA and incubated for 10 min at 25°C, 2 h at 37°C, then 5 min at 85°C. Reactions were stored at −20°C.
Primer design for qRT-PCR
Forward and reverse primers (Sigma) were designed to amplify a reference amplicon and a target amplicon. The forward and reverse reference primers annealed to the yeast 25S rRNA sequence 5′ to the depurination site at position 2071 to 2090 and position 2273 to 2292, respectively, and yielded a 221-bp fragment (R-For: 5′-AGACCGTCGCTTGCTACAAT-3′, and R-Rev: 5′-ATGACGAGGCATTTGGCTAC-3′). The forward primer to detect the depurinated target annealed 271 nucleotides 5′ to the depurination site at position 2756 to 2777 (T-For: 5′-CTATCGATCCTTTAGTCCCTC-3′). The 3′ terminus of the reverse primer to detect the depurinated target annealed at the site of depurination at position 3027 (Td-Rev: 5′-CCGAATGAACTGTTCCACA-3′) and when used with the T-For primer produced a 289-bp amplicon. The terminal base of Td-Rev contained an adenine (in bold) to detect the A to T transversion in the cDNA corresponding to the depurinated rRNA. To increase specificity for amplification of only the depurinated target template a T two bases 5′ to the terminus (underlined) was changed to an A. A second reverse primer, Tn-Rev (5′-CCGAATGAACTGTTCCACT-3′), contained sequence at the 3′ end that was specific for the nondepurinated template. This primer was used to confirm specificity of the target primer in end point PCR for detecting the depurination event.
For the validation experiment using synthetic template, the template was diluted in a twofold series so that resulting input amounts ranged from 400 pg to 25 pg DNA. Triplicate qPCR reactions were set up to calculate the average CT value at each template amount for each primer pair, and the ΔCT (CT target – CT reference) was plotted versus the log input template amount. For the validation experiment using cDNA, we first prepared cDNA from rRNA extracted from RTA treated ribosomes. Yeast ribosomes (8 pmol) were treated with 3 ng RTA for 15 min to allow for depurination of the SRL. cDNA synthesized from the extracted rRNA (500 ng) was diluted 50-fold and further diluted in twofold series down to 1600-fold. Triplicate qRT-PCR reactions were set up where relative input amounts of rRNA ranged from 2.5 ng to 80 pg. The average CT value for each template amount was determined for each primer pair, and the ΔCT (CT target – CT reference) was plotted versus the log input amount.
qRT-PCR and data analysis
qRT-PCR was performed using an ABI Prism 7000 Sequence Detection System (Applied Biosystems) using JumpStart Taq Ready Mix (Sigma) with SYBR Green I dye (Invitrogen) and ROX reference Dye (Invitrogen). All reactions were done in triplicate in a total volume of 20 μL using 4 μL of cDNA diluted 100-fold from the RT reaction. Primers were at a final concentration of 0.2 μM. Thermal cycling conditions were 95°C for 2 min followed by 40 cycles of 95°C for 30 sec, 58°C for 30 sec, 72°C for 30 sec. Data were analyzed using ABI SDS software allowing the software to set the baseline while the cycle threshold (CT) was set manually making sure it was within the exponential phase of amplification. Amplification plots were examined and obvious outliers were removed from further data analysis. The comparative CT method (ΔΔCT) was used for quantitation. This method does not require the use of a standard curve and relies on an arithmetic formula to achieve a relative quantitation between the target and reference. First, the average CT value for replicate reactions was determined and standard deviations were calculated. Next, the ΔCT between target (depurinated rRNA) and endogenous reference (total rRNA) was determined for a given sample. The standard deviation of ΔCT was calculated using the standard deviations for the target and reference CT in the formula, S = (Starget2 + Sreference2)0.5. Next, the ΔΔCT was established in which the ΔCT for a calibrator sample was subtracted from the ΔCT of the test sample. The calibrator samples were non-RTA treated ribosomes for the RTA titration and time course, the 0 h time point for in vivo expression time course, and no inhibitor reactions for the inhibitor studies. The standard deviation for ΔΔCT was the same value as that of the ΔCT for the test sample and is used in the equation 2−(ΔΔCT±S) allowing for the fold differences to be expressed as a range. For the purpose of clear presentation in graphical form, we represented the fold differences as an average of the range along with the standard deviation for that average.
Dual primer extension assay
rRNA was isolated from RTA treated ribosomes as previously described (Parikh et al. 2002). Two micrograms of rRNA was incubated with 106 cpm of γ-32P-labeled primer (APEX2) to detect depurination and 2 × 105 cpm of γ-32P-labeled primer (25S) to detect the total rRNA. The APEX2 primer hybridized downstream from the depurination site and generated a 73-nt primer extension product. The 25S primer hybridized near the 5′ end of the 25S rRNA and generated a 100-nt primer extension product (Parikh et al. 2002). Superscript II reverse transcriptase (Invitrogen) was used in the primer extension reaction and the products were separated on the 6% polyacrylamide gel containing 7 M urea and quantified with the Phosphoimager (Molecular Dynamics).
Treatment of ribosomes with RTA and Shiga toxin 2
Monomeric ribosomes were isolated from yeast cells as previously described (Chiou et al. 2008). Ribosome depurination was performed as previously described (Chiou et al. 2008) using 10 mM Tris pH 7.4, 10 mM MgCl2, 60 mM KCl, 2 mM DTT, ricin A chain (Vector Laboratories), and 10 pmol purified yeast ribosomes at 30°C in 100 μL. The reaction was stopped by the addition of 100 μL 2× extraction buffer (240 mM NaCl, 50 mM Tris-HCl pH 8.8, 20 mM EDTA, 2% SDS) and RNA was isolated by phenol extraction as described for the primer extension assay (Parikh et al. 2002). The final assay conditions after optimization were 10 mM MOPS pH 7.4, 10 mM Mg(OAC)2, 60 mM KCl, 2 mM DTT, 7 pmol ribosomes, 0.75 to 3.0 ng (0.25 to 1nM) RTA or 10 ng (1 nM) Stx2, which was preincubated with 20 mM DTT for 40 min at 30°C. PTA was obtained from Sigma and was dissolved in 0.5N NaOH (Miller et al. 2002). Dissolving PTA in NaOH prior to dilution for the qRT-PCR analysis had no impact on the final pH of the buffered reaction. Compounds 74 and 134, obtained from ChemDiv, Inc, and golgicide A (gift of Dr. David Haslam) were dissolved in 100% DMSO (Saenz et al. 2007).
ACKNOWLEDGMENTS
We thank Dr. Xiao-Ping Li for helpful comments on the manuscript, Dr. David Haslam for golgicide A, and Drs. Cheleste Thorpe and Ann Kane for Stx2. This work was supported by National Institutes of Health Grants AI072425 and AI082120.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2375411.
REFERENCES
- Applied Biosystems A. 2001. User Bulletin #2, ABI PRISM 7700 Sequence Detection System, Relative quantitation of gene expression. P/N 4303859B.
- Bai Y, Monzingo AF, Robertus JD 2009. The X-ray structure of ricin A chain with a novel inhibitor. Arch Biochem Biophys 483: 23–28 [DOI] [PubMed] [Google Scholar]
- Bai Y, Watt B, Wahome PG, Mantis NJ, Robertus JD 2010. Identification of new classes of ricin toxin inhibitors by virtual screening. Toxicon 56: 526–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri L, Valbonesi P, Bonora E, Gorini P, Bolognesi A, Stirpe F 1997. Polynucleotide:adenosine glycosidase activity of ribosome-inactivating proteins: Effect on DNA, RNA and poly(A). Nucleic Acids Res 25: 518–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri L, Ciani M, Girbes T, Liu WY, Van Damme EJ, Peumans WJ, Stirpe F 2004. Enzymatic activity of toxic and non-toxic type 2 ribosome-inactivating proteins. FEBS Lett 563: 219–222 [DOI] [PubMed] [Google Scholar]
- Baykal U, Tumer NE 2007. The C-terminus of pokeweed antiviral protein has distinct roles in transport to the cytosol, ribosome depurination and cytotoxicity. Plant J 49: 995–1007 [DOI] [PubMed] [Google Scholar]
- Brigotti M, Barbieri L, Valbonesi P, Stirpe F, Montanaro L, Sperti S 1998. A rapid and sensitive method to measure the enzymatic activity of ribosome-inactivating proteins. Nucleic Acids Res 26: 4306–4307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou JC, Li XP, Remacha M, Ballesta JP, Tumer NE 2008. The ribosomal stalk is required for ribosome binding, depurination of the rRNA and cytotoxicity of ricin A chain in Saccharomyces cerevisiae. Mol Microbiol 70: 1441–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y, Tsurugi K 1987. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J Biol Chem 262: 8128–8130 [PubMed] [Google Scholar]
- Endo Y, Tsurugi K 1988. The RNA N-glycosidase activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA. J Biol Chem 263: 8735–8739 [PubMed] [Google Scholar]
- Endo Y, Tsurugi K, Yutsudo T, Takeda Y, Ogasawara T, Igarashi K 1988. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur J Biochem 171: 45–50 [DOI] [PubMed] [Google Scholar]
- Heisler I, Keller J, Tauber R, Sutherland M, Fuchs H 2002. A colorimetric assay for the quantitation of free adenine applied to determine the enzymatic activity of ribosome-inactivating proteins. Anal Biochem 302: 114–122 [DOI] [PubMed] [Google Scholar]
- Hudak KA, Parikh BA, Di R, Baricevic M, Santana M, Seskar M, Tumer NE 2004. Generation of pokeweed antiviral protein mutations in Saccharomyces cerevisiae: Evidence that ribosome depurination is not sufficient for cytotoxicity. Nucleic Acids Res 32: 4244–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur Y, Hwang DJ, Zoubenko O, Coetzer C, Uckun FM, Tumer NE 1995. Isolation and characterization of pokeweed antiviral protein mutations in Saccharomyces cerevisiae: Identification of residues important for toxicity. Proc Natl Acad Sci 92: 8448–8452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordanov MS, Pribnow D, Magun JL, Dinh TH, Pearson JA, Chen SL, Magun BE 1997. Ribotoxic stress response: Activation of the stress-activated protein kinase JNK1 by inhibitors of the peptidyl transferase reaction and by sequence-specific RNA damage to the alpha-sarcin/ricin loop in the 28S rRNA. Mol Cell Biol 17: 3373–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XP, Baricevic M, Saidasan H, Tumer NE 2007. Ribosome depurination is not sufficient for ricin-mediated cell death in Saccharomyces cerevisiae. Infect Immun 75: 417–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior WB Jr, Tolleson WH 2010. A functional quantitative polymerase chain reaction assay for ricin, Shiga toxin, and related ribosome-inactivating proteins. Anal Biochem 396: 204–211 [DOI] [PubMed] [Google Scholar]
- Miller DJ, Ravikumar K, Shen H, Suh JK, Kerwin SM, Robertus JD 2002. Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition. J Med Chem 45: 90–98 [DOI] [PubMed] [Google Scholar]
- Parikh BA, Coetzer C, Tumer NE 2002. Pokeweed antiviral protein regulates the stability of its own mRNA by a mechanism that requires depurination but can be separated from depurination of the alpha-sarcin/ricin loop of rRNA. J Biol Chem 277: 41428–41437 [DOI] [PubMed] [Google Scholar]
- Parikh BA, Baykal U, Di R, Tumer NE 2005. Evidence for retro-translocation of pokeweed antiviral protein from endoplasmic reticulum into cytosol and separation of its activity on ribosomes from its activity on capped RNA. Biochemistry 44: 2478–2490 [DOI] [PubMed] [Google Scholar]
- Paton JC, Paton AW 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev 11: 450–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering LK, Obrig TG, Stapleton FB 1994. Hemolytic-uremic syndrome and enterohemorrhagic Escherichia coli. Pediatr Infect Dis J 13: 459–475 [DOI] [PubMed] [Google Scholar]
- Saenz JB, Doggett TA, Haslam DB 2007. Identification and characterization of small molecules that inhibit intracellular toxin transport. Infect Immun 75: 4552–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandvig K, van Deurs B 2000. Entry of ricin and Shiga toxin into cells: Molecular mechanisms and medical perspectives. EMBO J 19: 5943–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandvig K, van Deurs B 2002. Transport of protein toxins into cells: Pathways used by ricin, cholera toxin and Shiga toxin. FEBS Lett 529: 49–53 [DOI] [PubMed] [Google Scholar]
- Sandvig K, van Deurs B 2005. Delivery into cells: Lessons learned from plant and bacterial toxins. Gene Ther 12: 865–872 [DOI] [PubMed] [Google Scholar]
- Spooner RA, Smith DC, Easton AJ, Roberts LM, Lord JM 2006. Retrograde transport pathways utilised by viruses and protein toxins. Virol J 3: 26 doi: 10.1186/1743-422X-3-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stechmann B, Bai SK, Gobbo E, Lopez R, Merer G, Pinchard S, Panigai L, Tenza D, Raposo G, Beaumelle B, et al. 2010. Inhibition of retrograde transport protects mice from lethal ricin challenge. Cell 141: 231–242 [DOI] [PubMed] [Google Scholar]
- Stirpe F, Bailey S, Miller SP, Bodley JW 1988. Modification of ribosomal RNA by ribosome-inactivating proteins from plants. Nucleic Acids Res 16: 1349–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm MB, Schramm VL 2009. Detecting ricin: Sensitive luminescent assay for ricin A-chain ribosome depurination kinetics. Anal Chem 81: 2847–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita M, Chang CN, Johnson F, Will S, Grollman AP 1987. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J Biol Chem 262: 10171–10179 [PubMed] [Google Scholar]
- Taylor BE, Irvin JD 1990. Depurination of plant ribosomes by pokeweed antiviral protein. FEBS Lett 273: 144–146 [DOI] [PubMed] [Google Scholar]
- Wahome PG, Bai Y, Neal LM, Robertus JD, Mantis NJ 2010. Identification of small-molecule inhibitors of ricin and shiga toxin using a cell-based high-throughput screen. Toxicon 56: 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Hollis T, Svinth M, Day P, Monzingo AF, Milne GW, Robertus JD 1997. Structure-based identification of a ricin inhibitor. J Mol Biol 266: 1043–1049 [DOI] [PubMed] [Google Scholar]
- Zhao L, Haslam DB 2005. A quantitative and highly sensitive luciferase-based assay for bacterial toxins that inhibit protein synthesis. J Med Microbiol 54: 1023–1030 [DOI] [PubMed] [Google Scholar]