Abstract
Inhibition of eukaryotic DNA replication leads to the rapid suppression of histone synthesis, via 3′ uridylation of cytoplasmic histone mRNAs followed by their Lsm1–7-mediated decapping and degradation. Here we show that the human cytoplasmic RNA terminal U-transferase ZCCHC11, recently implicated in microRNA metabolism, associates with replication-dependent histone mRNAs. Knockdown of ZCCHC11 selectively blocked histone mRNA degradation following inhibition of DNA replication, whereas knockdown of PAPD1 or PAPD5, previously proposed as candidate histone mRNA U-transferases, had no such effect. Furthermore, a reduction in the proportion of histone transcripts that were uridylated was observed following ZCCHC11 knockdown. Our data indicate that ZCCHC11 is the terminal U-transferase responsible for targeting human histone mRNAs for degradation following inhibition or completion of DNA replication.
Keywords: RNA terminal nucleotidyl transferase, hydroxyurea, inhibition of DNA replication, mRNA turnover
INTRODUCTION
Histones have the capacity to interfere nonspecifically with a variety of cellular processes, and hence to be toxic if they accumulate to levels that exceed the capacity of the cell to incorporate them into nucleosomes and chromatin (Osley 1991). Synthesis of most core histones is therefore tightly linked to ongoing DNA replication during S phase; this is achieved in large part by cell cycle-dependent changes in histone mRNA levels.
The mRNAs encoding mammalian “replication-dependent” histones lack poly(A) tails but instead share a characteristic 3′ stem–loop structure. Through its interaction with a stem–loop binding protein (SLBP/HBP), this structure mediates processing of precursor histone mRNAs in the nucleus, the nuclear export, stability, and translational competence of the mRNAs, as well as their specific regulation in the cytoplasm (Dominski and Marzluff 1999). Inhibition of DNA replication in mammalian cells, for example on exposure to the ribonucleotide reductase inhibitor hydroxyurea (HU), leads to the rapid and selective degradation of replication-dependent histone mRNAs. A number of cellular factors or processes have been implicated in HU-induced histone mRNA degradation, including SLBP/HBP, ongoing translation, the signaling kinases ATR and DNA-PK, a 3′–5′ exonuclease termed 3′hExo, and Upf1, a protein also involved in nonsense-mediated mRNA decay (Dominski et al. 2003; Kaygun and Marzluff 2005; Muller et al. 2007).
A recent study further suggested that 3′ oligouridylation of replication-dependent histone mRNAs is a rate-limiting step in their degradation both by 5′–3′ and by 3′–5′ exonuclease activities, and tentatively identified two enzymes (PAPD1/hmtPAP/TUTase1 and PAPD5/TUTase3) as candidates for performing the oligouridylation step (Mullen and Marzluff 2008). Here we describe data that implicate a different enzyme, the cytoplasmic RNA terminal U-transferase ZCCHC11, as a rate-limiting mediator of histone mRNA degradation.
RESULTS AND DISCUSSION
The reported involvement of PAPD1 and PAPD5 in cytoplasmic histone uridylation (Mullen and Marzluff 2008) was unexpected for two reasons. Firstly, PAPD1 has been identified as the enzyme (hmtPAP) responsible for polyadenylation of mitochondrial mRNAs, and immunofluorescence microscopy data suggest that it is an exclusively mitochondrial protein (Tomecki et al. 2004; Nagaike et al. 2005). Secondly, the amino acid sequence of PAPD5 indicates that it is one of two human orthologs of the yeast Trf4/Trf5 poly(A) polymerases, which are involved in adenylation-mediated turnover of nuclear RNAs; furthermore, PAPD5 was recently shown to be responsible for the adenylation of incomplete ribosomal RNA transcripts in the nucleus (Shcherbik et al. 2010). We therefore considered the possibility that other RNA nucleotidyl transferase(s) might be responsible for uridylation of cytoplasmic replication-dependent histone mRNAs. PAPD1 and PAPD5 are members of a family of seven related human proteins that have been described as noncanonical poly(A) polymerases or as ribonucleotidyl transferases that can incorporate residues other than AMP (Schmidt and Norbury 2010). Three members of this family have been found to have uridyl transferase activity and, of these three, the nuclear enzyme RBM21 (U6TUTase) was identified as a highly specific terminal U-transferase that uridylates only the U6 snRNA (Trippe et al. 2006). The nucleotide preference of the two remaining uridyl transferases, ZCCHC6 and ZCCHC11, was initially identified by an RNA tethering approach in Xenopus oocytes (Kwak and Wickens 2007); the in vitro activity of ZCCHC6 as a terminal U-transferase was also demonstrated using tandem affinity purified enzyme from human cells (Rissland et al. 2007). ZCCHC11 (with its orthologs in mouse and C. elegans) has been the subject of much recent attention due to its identification as the enzyme responsible for 3′ uridylation of microRNAs (miRNAs) and their cytoplasmic precursors (Hagan et al. 2009; Heo et al. 2009; Jones et al. 2009; Lehrbach et al. 2009).
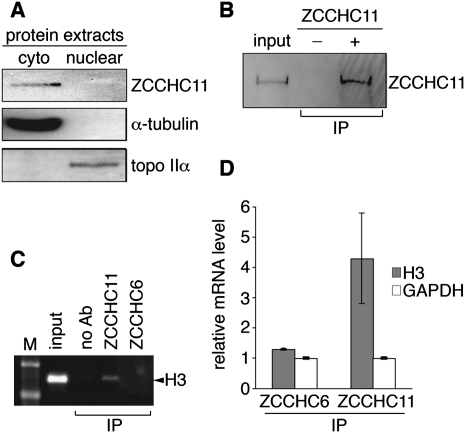
Cytoplasmic ZCCHC11 associates with histone H3 mRNA
We raised and affinity-purified a polyclonal antibody against recombinant ZCCHC11. Western blotting of fractionated human HEK293T cell extracts with this antibody indicated that ZCCHC11 is an exclusively cytoplasmic protein (Fig. 1A), in agreement with recently described immunofluorescence microscopy of endogenous ZCCHC11 (Heo et al. 2009). Our antibody was also able to immunoprecipitate ZCCHC11 from nondenaturing cell lysates (Fig. 1B), which allowed us to test directly the possibility that ZCCHC11 might be associated with replication-dependent histone mRNAs. After formaldehyde cross-linking, immunoprecipitation, and RT-PCR, we were able to detect replication-dependent histone H3 (HIST2H3) mRNA specifically in association with ZCCHC11, but not with ZCCHC6 (Fig. 1C,D). Both proteins were readily detectable by Western blotting (data not shown), suggesting that failure to recover HIST2H3 in association with ZCCHC6 is not simply due to a lack of ZCCHC6 protein in these cells. In contrast, GAPDH mRNA, a control mRNA representative of those not subject to S phase arrest-specific degradation, was not specifically immunoprecipitated by either antibody. These data indicate that ZCCHC11 can bind to histone mRNAs in vivo.
FIGURE 1.
ZCCHC11 is a cytoplasmic protein and specifically associates with replication-dependent histone mRNAs. (A) Cytoplasmic (cyto) and nuclear HEK293T cell extracts were analyzed by Western blotting with a ZCCHC11-specific antibody. α-Tubulin and topoisomerase IIα served as controls for cytoplasmic and nuclear localization, respectively. (B) ZCCHC11 was immunoprecipitated from cytoplasmic extracts in the presence (+) or absence (−) of polyclonal anti-ZCCHC11 antibody. (C) Following formaldehyde cross-linking RNPs were immunoprecipitated (IP) using specific antibody or no antibody (ab). Precipitates were subjected to RT-PCR using primers specific for the ORF of replication-dependent histone HIST2H3 (H3). Products were separated by agarose gel electrophoresis. M, molecular weight marker. (D) As in C, but H3 mRNA (and GAPDH mRNA as a control) were analyzed using real time RT-PCR. Data (mean of three biological replicates, ±SD) are expressed relative to the signal obtained in the absence of specific antibody.
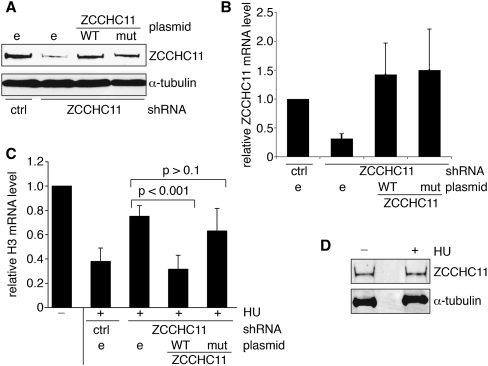
ZCCHC11 is required for efficient histone mRNA turnover
Its association with HIST2H3 mRNA suggested a potential role for ZCCHC11 in histone mRNA degradation. To test this possibility, we expressed a short hairpin RNA (shRNA) corresponding to a sequence in the ZCCHC11 3′ UTR to knock down ZCCHC11 expression in HEK293T cells (Fig. 2A,B). Cells expressing the ZCCHC11-specific or control nonspecific shRNAs were then treated with HU, and replication-dependent histone H3 mRNA levels were determined by quantitative RT-PCR (Fig. 2C). As expected, H3 mRNA was rapidly degraded in the control shRNA-expressing cells within 30 min of HU treatment. Expression of the ZCCHC11-specific shRNA largely prevented the HU-induced decrease in histone mRNA level, and this effect was attributable to knockdown of ZCCHC11, as it was completely reversed by co-expression from a plasmid of a ZCCHC11 cDNA lacking the 3′ UTR sequence targeted by the shRNA (Fig. 2A–C). A site-directed mutant version of ZCCHC11 lacking two aspartate residues essential for catalysis was unable to support HU-induced histone mRNA turnover in this assay, indicating that ZCCHC11 catalytic activity is required for its mRNA destabilizing function. This function of ZCCHC11 was not associated with any change in its abundance, as judged by Western blotting (Fig. 2D).
FIGURE 2.
ZCCHC11 is required for efficient degradation of replication-dependent histone mRNAs upon inhibition of DNA replication. (A,B) ZCCHC11 was knocked down using shRNAs directed against its 3′ UTR. A scrambled shRNA control (ctrl) was used in parallel. Forty-eight hours following shRNA transfection, HEK293T cells were transfected with either empty (e), WT, or catalytically inactive mutant (mut) ZCCHC11 expression vector as indicated. The expression of ZCCHC11 was analyzed by Western blot using anti-ZCCHC11 (and anti-tubulin as a control; A) and by quantitative RT-PCR (B) 72 h after shRNA transfection. (C) As in B, but cells were treated with (+) or without (−) 5 mM hydroxyurea (HU) for 30 min before being harvested. Total RNA was isolated and analyzed by real time RT-PCR. H3 mRNA values (mean of five biological replicates ±SD) were normalized to GAPDH mRNA and expressed relative to the RNA amount in untreated samples, which was arbitrarily set to 1.0. (D) ZCCHC11 levels (and α-tubulin as a loading control) were monitored by Western blotting extracts of cells treated (+) or untreated (−) with 5 mM HU for 30 min.
Importantly, no such restoration of histone mRNA degradation was seen on over-expression either of the other known cytoplasmic terminal U-transferase ZCCHC6 or of mitochondrial PAPD1, previously implicated in histone mRNA turnover (data not shown). Equivalent results were obtained for HIST2H2A mRNA (data not shown), indicating a general requirement for ZCCHC11 in turnover of replication-dependent histone mRNAs following exposure to HU.
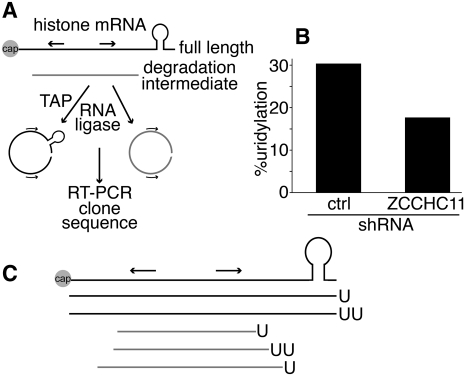
Reduced histone mRNA uridylation on ZCCHC11 knockdown
The requirement for the catalytic activity of this RNA terminal uridyl transferase in histone mRNA destabilization suggested that ZCCHC11 might be directly responsible for the previously observed uridylation of histone mRNAs (Mullen and Marzluff 2008). To address this possibility, we used a circularized rapid amplification of cDNA ends (cRACE) approach to detect terminal uridylation of histone mRNAs and to determine the effect of ZCCHC11 knockdown on these sequences (Fig. 3A). Our initial experiments recapitulated those described in an earlier study (Mullen and Marzluff 2008) and used RNA prepared from asynchronous HeLa cells. In our hands the frequency of histone mRNA uridylation under these circumstances was too low to allow a statistically robust investigation of its dependence on ZCCHC11 activity. This low frequency suggests that uridylated mRNAs are turned over very rapidly in vivo, consistent with the documented roles of 3′ UMP residues in RNA turnover pathways. We therefore used instead RNA from cells synchronized in late S phase by double thymidine blockade and release. Under these circumstances, the variant of the cRACE protocol using untreated RNA at the ligation step (to selectively monitor de-capped degradation intermediates; Fig. 3A) yielded insufficient material for quantitative analysis, although a few clones were isolated corresponding to RNAs that had undergone extensive 3′–5′ degradation and terminated in nontemplated uridyl residues. This observation suggests that 3′ uridylation continues during histone mRNA decay, and might serve, for example, to reinitiate stalled exonucleolysis. Nonetheless, pretreatment of the RNA with tobacco acid pyrophosphatase (TAP) to remove 5′ caps allowed the cloning and sequencing of substantial numbers of HIST2H2AC cDNAs (Fig. 3B,C; Supplemental Table 1). Approximately 30% of the 46 sequences included one or two terminal nontemplated uridyl residues. It should be noted that the UMP tails detected in our study would be too short to allow their detection by oligo(dA)-primed reverse transcription as used by Mullen and Marzluff (2008); the fact that we did not observe longer oligo(U) tails suggests that such tails are comparatively rare and/or unstable.
FIGURE 3.
The impact of ZCCHC11 knockdown on histone mRNA uridylation. (A) Overview of the cRACE procedure used to capture capped HIST2H2AC transcripts (black) and degradation intermediates (gray). Arrows indicate the position of the PCR primers used. (B) cRACE was performed on TAP-treated RNA isolated from HeLa cells harvested at the end of S phase following shRNA plasmid transfection. The percentage of cRACE products containing terminal uridyl residues is shown for control and ZCCHC11 knockdown cells (n = 14/46, 8/48, respectively). (C) Diagram indicating the positions of uridyl residues detected by cRACE.
When the cRACE protocol was carried out using TAP-treated RNA from cells in which ZCCHC11 had been knocked down, there was an almost twofold reduction in the proportion of uridylated sequences (P = 0.15, χ2 test). These data are consistent with the notion that HIST2H2AC, and by inference other replication-dependent histone genes, are direct targets of the ZCCHC11 uridyl transferase. The lack of a larger effect may be due to incomplete ZCCHC11 knockdown, or functional redundancy between ZCCHC11 and other uridyl transferases, even though ZCCHC11 knockdown alone was sufficient to stabilize histone mRNAs significantly.
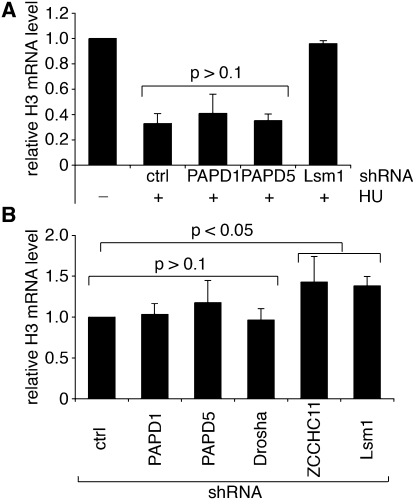
Knockdown of PAPD1 or PAPD5 has no effect of histone mRNA turnover
Given their reported function in histone mRNA turnover (Mullen and Marzluff 2008), we performed assays analogous to those described above using shRNAs specific for PAPD1 or PAPD5 (Fig. 4A). Although the extent of knockdown in each case was comparable to that achieved with ZCCHC11 (Supplemental Table 2), and PAPD1 knockdown resulted in decreased abundance of mitochondrial cytochrome oxidase mRNA (Supplemental Fig. 1), as reported previously (Nagaike et al. 2005), neither shRNA had any significant impact on HU-induced histone mRNA destabilization. Our data are therefore at odds with those previously described, also using HeLa cells, which implicated PAPD1 and PAPD5, but not ZCCHC11, in this process (Mullen and Marzluff 2008). The reason for this discrepancy is currently unclear, but given the involvement of 3′ uridylation in the histone mRNA turnover pathway, it is easier to envisage a role in this pathway for the cytoplasmic terminal U-transferase ZCCHC11 than for the poly(A) polymerases PAPD1 or PAPD5, especially as the former is a mitochondrial enzyme.
FIGURE 4.
Effect of PAPD1 and PAPD5 knockdown on the degradation of replication-dependent histone mRNAs upon S phase arrest. (A) HEK293T cells were transfected with control (ctrl) or specific shRNAs directed against PAPD1, PAPD5, or Lsm1. Seventy-two hours after transfection, cells were treated with (+) or without (−) 5 mM hydroxyurea (HU) for 30 min. RNA was analyzed as in Figure 2C. The mean of at least three biological replicates ±SD is indicated. (B) Steady-state H3 mRNA levels in asynchronous cells transfected with the shRNA expression vectors indicated were compared to those in cells expressing the scrambled control shRNA (ctrl).
In line with earlier studies that identified roles for the Lsm1 protein, and by extension the cytoplasmic Lsm1–7 heptameric complex, in uridylation-dependent mRNA degradation in human cells and fission yeast (Mullen and Marzluff 2008; Rissland and Norbury 2009), we found that expression of an Lsm1-specific shRNA in HEK293T cells abolished the HU-induced histone mRNA destabilization (Fig. 4A). This RNA turnover pathway does not appear to operate only under the nonphysiological condition of HU exposure, as steady-state histone H3 mRNA levels were significantly elevated in asynchronous cell populations expressing shRNAs targeting ZCCHC11 or Lsm1 (Fig. 4B). Again, knockdown of PAPD1 or PAPD5 had no such effect.
ZCCHC11 expression declines during S phase progression
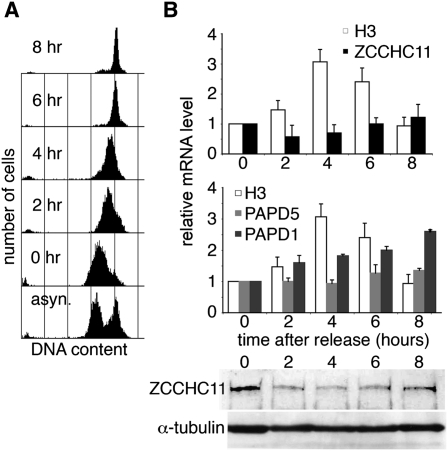
As ZCCHC11 is required for histone mRNA regulation, we investigated the possibility that it might itself be regulated in some way during S phase progression. HeLa cells were synchronized by double thymidine block and release (Fig. 5A), and mRNA levels were monitored during the subsequent progression through S phase. As expected, histone H3 mRNA levels rose to a peak by four hours after release, when cells were in mid-S phase. In contrast, ZCCHC11 mRNA and protein levels declined slightly by this time point, but subsequently recovered as cells completed S phase (Fig. 5B). This pattern of expression would be consistent with suppression of ZCCHC11 activity during S phase progression and a physiological role for this activity in the degradation of histone mRNAs on completion of DNA replication, as suggested by our observation of histone mRNA accumulation on ZCCHC11 knockdown in asynchronous cells (Fig. 4B). Comparable S phase-specific regulation was not seen in the case of PAPD1, expression of which increased steadily on release from the thymidine block, or of PAPD5, which was expressed at a constant level (Fig. 5B).
FIGURE 5.
ZCCHC11 expression negatively correlates with H3 expression during S phase. HeLa cells were blocked in early S phase by a double thymidine block and then released. (A) The cell cycle distribution of propidium iodide stained cells was analyzed by flow cytometry at the indicated times after release from the block, and in asynchronous cells. (B) Upon removal of the block, total RNA was isolated at time points indicated and analyzed by real time RT-PCR using specific oligonucleotides. H3 and ZCCHC11 (upper panel) and H3, PAPD1, and PAPD5 (middle panel) mRNA levels were normalized to GAPDH mRNA and expressed in relation to RNA obtained from cells at the time when the block was removed (0 h). ZCCHC11 and α-tubulin protein levels were measured by Western blotting at the same time points (lower panel).
ZCCHC11 was previously found to be responsible for the uridylation of cytoplasmic pre-miRNAs on recruitment by its RNA-binding partner Lin28, and for the uridylation of mature miRNAs in such a way as to influence their biological activity. To exclude the possibility that the ZCCHC11-dependent effect on histone mRNA turnover is mediated indirectly via miRNAs, we knocked down the Drosha ribonuclease in order to inhibit miRNA maturation. We did not observe any changes in the rate of histone mRNA degradation upon S phase arrest under these conditions (Supplemental Fig. 2) or any change in steady-state histone mRNA levels (Fig. 4B). In addition to the role of ZCCHC11 in miRNA regulation, our data clearly implicate the same enzyme in a uridylation-dependent pathway of histone mRNA turnover. The biological significance of the combination of these two important regulatory activities in a single RNA uridyl transferase awaits further clarification. ZCCHC11 does not display inherent substrate specificity (Jones et al. 2009), suggesting the involvement of other factors in targeting specific RNAs. It will be interesting to determine whether there is a functional analog of Lin28 that recruits ZCCHC11 to histone mRNAs. Further studies may also help to explain the significance of the comparatively large size of ZCCHC11, and its possession of zinc finger motifs and a duplication of the catalytic domain, while only one of the domains retains amino acid residues critical for activity.
Amino acid sequence comparisons and biochemical assays suggest that the ZCCHC6 protein might perform ZCCHC11-like roles, but these do not seem to relate to turnover of the histone mRNAs examined here. It will be interesting to determine whether either of these enzymes is also involved in the uridylation and turnover of polyadenylated mRNAs, as is the related uridyl transferase Cid1 in fission yeast (Rissland and Norbury 2009).
MATERIALS AND METHODS
Oligonucleotides
Sequences of oligonucleotides used for cloning, site-directed mutagenesis, and shRNA expression are given in Supplemental Table 3.
Plasmids
All shRNA expression plasmids were generated by inserting respective annealed oligonucleotides into the pre-linearized siSTRIKE vector (Promega). To create pcDNA-ZCCHC11-TAP, ZCCHC11 was amplified from clone KIAA0191 (Kazusa Research Institute) using oligonucleotides SacIIHC11f and NotIHC11r and the procedure described previously (Rissland et al. 2007). Site-directed mutagenesis using oligonucleotides HC11DADAf and HC11DADAr with plasmid pcDNA-ZCCHC11-TAP was used to create a derivative encoding catalytically inactive ZCCHC11.
RNAi
HEK293T cells were transfected with 5 μg of siSTRIKE plasmid (Promega) expressing specific shRNA or a scrambled shRNA control using Lipofectamine 2000 (Invitrogen). The efficiency of the RNAi was monitored by real time RT-PCR using specific oligonucleotides. Two micrograms of pcDNA-ZCCHC11-TAP was used for subsequent transfection when needed. Seventy-two hours after siSTRIKE transfection, cells were treated with 5 mM HU for 30 min and harvested at 4°C.
RNA isolation and RT-PCR
Total RNA was isolated using Tri-reagent (Sigma-Aldrich). Contaminating DNA was removed by DNase treatment (Promega). Reverse transcription was performed using Superscript III and 10 pmol random hexamers (Invitrogen) according to the manufacturer's guidelines. Five microliters of SYBR Green mix (Bioline) and one twentieth of the cDNA were used for subsequent real time PCR reactions. For each sample, a control reaction was performed in the absence of reverse transcriptase. All RNA levels were normalized to GAPDH mRNA.
Circularized rapid amplification of cDNA ends analysis
cRACE was performed as described previously (Mullen and Marzluff 2008; Rissland and Norbury 2009). In brief, capped transcripts were captured by treating RNA with tobacco acid pyrophosphatase prior to RNA circularization. Half of the ligation was subjected to cDNA synthesis using primer H2cRACE-r. One twentieth of the cDNA was amplified by PCR using oligonucleotides H2-f and H2cRACE-r for 30 cycles. PCR products were cloned and sequenced.
ZCCHC11 immunoprecipitation
ZCCHC11 was immunoprecipitated as described (Dostie and Dreyfuss 2002) using anti-ZCCHC11. RNA immunoprecipitation experiments were performed as described (West and Proudfoot 2008); immunoprecipitation was performed for two hours using anti-ZCCHC6 or anti-ZCCHC11.
Human cell synchronization
HeLa cells were synchronized using a double thymidine block as described (Whitfield et al. 2000). Upon release of the block, total RNA was isolated at time points indicated. The DNA content of the cells was determined by flow cytometry (FACScan, Becton Dickinson) of propidium iodide stained cells.
Cell fractionation and Western blot analysis
Total protein extracts were obtained by resuspension of cell pellets in RIPA buffer (15 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and 10% glycerol [v/v]) including protease inhibitors. Cell fractionation was performed as described (West et al. 2008). After centrifugation, cytoplasmic extracts were obtained from the top 0.2 mL while nuclear pellets were resuspended in 0.2 mL RIPA buffer.
Western blotting was performed using ECL (Santa Cruz Biotechnology). Antibodies against α-tubulin (TAT-1) and topoisomerase IIα were generous gifts from Keith Gull and Ian Hickson, respectively.
Preparation of ZCCHC6 and ZCCHC11 antibodies
Polyclonal antibodies were raised against recombinant peptides of ZCCHC6 and ZCCHC11 encoded by nucleotides 1920–2480 and 160–720 of the open reading frame, respectively. Immunoglobulins were purified from crude serum by preparative Western blotting. In brief, the recombinant GST-tagged peptide was expressed and purified essentially as described previously (Rissland et al. 2007) but eluted from glutathione-Sepharose using 25 mM glutathione. Eluted protein was transferred to a PVDF membrane, which was incubated overnight with 2 mL of crude serum. The bound antibody was eluted using 1 mL of 0.1 M glycine (pH 2.5) and subsequently neutralized with 120 μL of 1 M Tris pH 8.0.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Andrea Mikulasova for generating antibodies against ZCCHC6 and ZCCHC11, Keith Gull and Ian Hickson for providing other antibodies, and members of the laboratory for their comments on the manuscript. This work was supported by Cancer Research UK, the Biotechnology and Biological Sciences Research Council, The EP Abraham Research Fund, and the Wellcome Trust.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2252511.
REFERENCES
- Dominski Z, Marzluff WF 1999. Formation of the 3′ end of histone mRNA. Gene 239: 1–14 [DOI] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Kaygun H, Dadlez M, Marzluff WF 2003. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol Cell 12: 295–305 [DOI] [PubMed] [Google Scholar]
- Dostie J, Dreyfuss G 2002. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol 12: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI 2009. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol 16: 1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708 [DOI] [PubMed] [Google Scholar]
- Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu S, Ivanov AR, Wolf DA, Mizgerd JP 2009. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol 11: 1157–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaygun H, Marzluff WF 2005. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol 12: 794–800 [DOI] [PubMed] [Google Scholar]
- Kwak JE, Wickens M 2007. A family of poly(U) polymerases. RNA 13: 860–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA 2009. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol 16: 1016–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller B, Blackburn J, Feijoo C, Zhao X, Smythe C 2007. DNA-activated protein kinase functions in a newly observed S phase checkpoint that links histone mRNA abundance with DNA replication. J Cell Biol 179: 1385–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaike T, Suzuki T, Katoh T, Ueda T 2005. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J Biol Chem 280: 19721–19727 [DOI] [PubMed] [Google Scholar]
- Osley MA 1991. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem 60: 827–861 [DOI] [PubMed] [Google Scholar]
- Rissland OS, Norbury CJ 2009. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol 16: 616–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Mikulasova A, Norbury CJ 2007. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol Cell Biol 27: 3612–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MJ, Norbury CJ 2010. Polyadenylation and beyond: Emerging roles for noncanonical poly(A) polymerases. WIREs RNA 1: 142–151 [DOI] [PubMed] [Google Scholar]
- Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG 2010. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep 11: 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP 2004. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res 32: 6001–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ 2006. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 12: 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Proudfoot NJ 2008. Human Pcf11 enhances degradation of RNA polymerase II-associated nascent RNA and transcriptional termination. Nucleic Acids Res 36: 905–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Proudfoot NJ, Dye MJ 2008. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol Cell 29: 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, Zheng LX, Baldwin A, Ohta T, Hurt MM, Marzluff WF 2000. Stem–loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol 20: 4188–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]