Abstract
Methyltransferase enzymes that use S-adenosylmethionine as a cofactor to catalyze 5-methyl uridine (m5U) formation in tRNAs and rRNAs are widespread in Bacteria and Eukaryota, but are restricted to the Thermococcales and Nanoarchaeota groups amongst the Archaea. The RNA m5U methyltransferases appear to have arisen in Bacteria and were then dispersed by horizontal transfer of an rlmD-type gene to the Archaea and Eukaryota. The bacterium Escherichia coli has three gene paralogs and these encode the methyltransferases TrmA that targets m5U54 in tRNAs, RlmC (formerly RumB) that modifies m5U747 in 23S rRNA, and RlmD (formerly RumA) the archetypical enzyme that is specific for m5U1939 in 23S rRNA. The thermococcale archaeon Pyrococcus abyssi possesses two m5U methyltransferase paralogs, PAB0719 and PAB0760, with sequences most closely related to the bacterial RlmD. Surprisingly, however, neither of the two P. abyssi enzymes displays RlmD-like activity in vitro. PAB0719 acts in a TrmA-like manner to catalyze m5U54 methylation in P. abyssi tRNAs, and here we show that PAB0760 possesses RlmC-like activity and specifically methylates the nucleotide equivalent to U747 in P. abyssi 23S rRNA. The findings indicate that PAB0719 and PAB0760 originated as RlmD-type m5U methyltransferases and underwent changes in target specificity after their acquisition by a Thermococcales ancestor from a bacterial source.
Keywords: 5-methyluridine, rRNA modification, MALDI-MS, Archaea, horizontal gene transfer
INTRODUCTION
RNA molecules in each of the three domains of life are subject to post-transcriptional modification. The most heavily modified RNAs are the tRNAs and rRNAs, and their nucleotide modifications play pivotal roles in protein synthesis (for reviews, see Agris 2004; Björk and Hagervall 2005; Ofengand and Del Campo 2005; Grosjean 2009). In tRNAs, modified nucleotides are clustered in the T- and D-arms where they help to fold the molecule into its L-shaped conformation, and in the anticodon loop where they facilitate mRNA decoding (Jühling et al. 2009). In rRNA, modifications are located in regions that are essential for ribosomal subunit associations, tRNA binding, mRNA decoding, and peptide bond formation (Decatur and Fournier 2002; Chow et al. 2007; Purta et al. 2009).
One RNA modification that is universally present in both the rRNA and tRNAs of bacteria is 5-methyluridine (m5U, alternatively termed ribothymidine rT) (Björk and Hagervall 2005; Ofengand and Del Campo 2005). Escherichia coli RNAs contain three m5Us, two of which are in 23S rRNA and one in tRNA (Fig. 1). In 23S rRNA, m5U747 is located in the loop of hairpin 35 and protrudes into the large subunit tunnel and possibly contacts the nascent peptide (Ban et al. 2000; Harms et al. 2001); and m5U1939 is situated at the ribosomal subunit interface where the acceptor stem of the A-site tRNA interacts (Schuwirth et al. 2005; Korostelev et al. 2006; Selmer et al. 2006). The third m5U is at position 54 in the T-loop of all the E. coli tRNAs where the modification stabilizes their tertiary folding (Davanloo et al. 1979). Each of the three m5U modifications requires its own specific methyltransferase enzyme: RlmC (previously called RumB) methylates U747 in 23S rRNA (Madsen et al. 2003; Andersen and Douthwaite 2006), while RlmD (previously RumA) is specific for m5U1939 (Agarwalla et al. 2002; Madsen et al. 2003); TrmA (previously RumT) catalyzes the m5U54 modification in tRNAs (Ny et al. 1988; Kealey et al. 1994; Gu et al. 1996). The sequence similarity between these three E. coli m5U methyltransferases indicates that they share a common evolutionary origin (Urbonavičius et al. 2008). Homologs are present in other bacteria as well as in eukaryotes where, for example, the yeast Saccharomyces cerevisiae Trm2p is specific for tRNA m5U54. All of these methyltransferases use S-adenosyl-L-methionine (AdoMet) as a donor of the methyl group, and cluster within the family of orthologous genes COG2265 (Tatusov et al. 2003).
FIGURE 1.
P. abyssi RNA transcripts and the corresponding E. coli structures (A) Pa-II is a 77-nt transcript corresponding to part of helix 33 and hairpins 34 and 35 from domain II of P. abyssi 23S rRNA; point mutations were introduced at the sites indicated by arrows. E. coli nucleotide numbering is used to facilitate a direct comparison of structures (P. abyssi numbers in italics). The P. abyssi rRNA is two nucleotides longer than (B) the corresponding structure in E. coli 23S rRNA. The RlmC target at nucleotide U747 and two other modifications are indicated in the E. coli rRNA. (C) The Pa-IV-80 transcript corresponds to hairpins 69–71 from domain IV of P. abyssi 23S rRNA; the methyltransferases responsible for adding the modifications in this region (light shading) still await identification. The 5′- and 3′-ends of a shorter transcript (Pa-IV-56) used in a previous study (Urbonavičius et al. 2008) are indicated by arrows. (D) The homologous region from E. coli 23S rRNA containing the RlmD target at U1939; the enzymes responsible for the neighboring modifications have been identified (for review, see Purta et al. 2009). (E) Structure of P. abyssi tRNAAsp showing the site of PAB0719 methylation at U54 (Urbonavičius et al. 2008). (F) A schematic consensus of tRNA structures indicating the U54 target of the E. coli TrmA (Ny et al. 1988) and S. cerevisiae Trm2p homolog (Nordlund et al. 2000), as well as the structurally unrelated enzyme TrmFO that modifies the same nucleotide in Bacillus subtilis using a tetrahydrofolate cofactor as the methyl donor (Urbonavičius et al. 2005, 2007).
The genome of the hyperthermophilic archaeon Pyrococcus abyssi contains two open reading frames, PAB0719 and PAB0760, which have been shown by comparative phylogenetic analyses to belong to the COG2265 m5U methyltransferases. PAB0719 and PAB0760 are 44% identical (62% similar) and both are more closely related to RlmD than to either RlmC or TrmA (Urbonavičius et al. 2008). Surprisingly, however, neither PAB0719 nor PAB0760 targets the RlmD site at the equivalent of E. coli 23S rRNA nucleotide U1939 (U2040 in the P. abyssi numbering system) (Fig. 1). Recombinant PAB0719 was shown to function like TrmA and catalyze m5U formation at position 54 in P. abyssi tRNAs, while the function of PAB0760 remained undetermined (Urbonavičius et al. 2008).
In this study, we used reverse transcriptase primer extension and matrix-assisted laser desorption/ionization (MALDI) mass spectrometry (MS) to establish whether the 747 and 1939 regions of P. abyssi 23S rRNA contain nucleotide modifications that could be 5-methyluridines. We then determined the type and position of any modification added by recombinant PAB0760 enzyme to RNA transcripts in vitro. It was shown that PAB0760 is indeed an m5U methyltransferase and recognizes a specific uridine within the loop of 23S rRNA hairpin 35 at the same location as the E. coli RlmC target U747 (U859 in the P. abyssi numbering) (Fig. 1A). Mutagenesis of this rRNA region, as well as attempts to substitute a bacterial rRNA substrate, showed that PAB0760 is highly specific for the P. abyssi hairpin 35 structure.
RESULTS
Modifications in P. abyssi 23S rRNA
Total rRNA from P. abyssi was analyzed by primer extension and mass spectrometry to identify possible sites of modification within the 747 and 1939 regions of the 23S rRNA (the E. coli rRNA nucleotide numbering system is used to aid direct comparison of structures). Primer extension with reverse transcriptase (RT) from 23S rRNA-specific primers confirmed that these regions matched the sequences in the databases (Fig. 2A,B). No complete RT stops were observed, and therefore there are presumably no nucleotide modifications that block Watson-Crick base pairing; however, several bands might represent labile phosphodiester bonds, or RT pausing at 2′-O-methylated nucleotides (Fig. 2A,B, K lanes).
FIGURE 2.
Analyses of authentic P. abyssi 23S rRNA. (A) Primer extension sequencing (lanes C,U,A,G) through the 747 region and (B) the 1939 region of P. abyssi 23S rRNA. The K lanes are extension reactions without dideoxynucleotide triphosphates; sites of possible (A744) and confirmed (C1934) 2′-O-methylation are indicated by RT pauses immediately before the modified nucleotides. (C,D) MALDI mass spectra of RNase T1 oligos from the 747 region. All RNase T1 digestions yielded a mixture of products with 2′-3′-cyclic and 3′-linear phosphates; these differ by 18 Da in mass and are, respectively, designated with “>p” and “p.” (E) Relevant RNase A oligos from the 747 region. From the 1939 region: (F) trimer and (G) decamer RNase T1 oligos; and (H) RNase A fragment containing C1934 and U1939. Only the spectral regions where methylation was detected are shown; oligos containing methylated nucleotides are shaded.
Sequence stretches of about 50 nucleotides (nt) containing U747 and U1939 were isolated from the P. abyssi 23S rRNA and digested with RNase A or T1 to yield oligos that were suitable for MALDI-MS analysis. The MS spectra showed several anomalous masses, indicating methylation on the 747 region oligos GUp and UGp (Fig. 2C), AUGp (Fig. 2D) and AGUp or GAUp (Fig. 2E); and within the 1939 region, methylation was detected on GCp, CCGp (Fig. 2F), and AAAUUCCUUGp (Fig. 2G). The decamer AAAUUCCUUGp is unique in P. abyssi rRNA and could be unambiguously identified as the sequence containing U1939; there is no peak at m/z 3185 (or m/z 3167 in the cyclic phosphate form) indicating stoichiometric addition of a single methyl group to this sequence. The overlapping RNase A fragment GAAAUp (nucleotides 1935–1939) was missing from the spectra, both in unmethylated (m/z 1657) and singly methylated forms (m/z 1671). However, a new unexpected peak was evident at m/z 2664, which fits with the mass of the sequence AGCGAAAUp (1932–1939) plus two methyl groups (Fig. 2H). One of these would be a 2′-O-methyl on C1934, which would block RNase A cutting and is also consistent with an RT pause (Fig. 2B), and the second methyl group has tentatively been assigned to U1939 (although formally this could be on any of the three adenosines, 1936–1938). Unfortunately, the quality of the fragments containing nucleotide U1939 was inadequate for tandem MS analysis to ascertain the position of this second methyl group.
In the smaller oligos, the combination of RNase A and T1 digestions helped to locate some of the methylations. For instance, the increases in the masses of the GCp and CCGp peaks fit with methylation at C1957; and the lack of an RT stops here (Fig. 2B) suggests that this is m5C1957. In the 747 region, only U747 and U762 can be cleaved into both GUp and UGp oligos, and the occurrence of methylated peaks together with the lack of any RT stop are consistent with the presence of either m5U747 or m5U762 modification. Finally, the increase in mass of the AUGp and AGUp/GAUp oligos indicates methylation at either U745 or A744, and an RT pause is consistent with a 2′-O-methylation on A744 (Fig. 2A).
The role of the putative m5U ortholog PAB0760 in adding any of the above modifications was investigated in vitro using a recombinant version of the enzyme and RNA transcripts.
In vitro RNA substrates
RNA transcripts (Fig. 1) used to test the activity of recombinant PAB0760 protein correspond to regions of domain II and IV of P. abyssi 23S rRNA that contain nucleotides U747 and U1939 (P. abyssi rRNA numbering is given in italics, where expedient). The P. abyssi domain II transcript is a 77-nt RNA (Pa-II) (Fig. 1A) corresponding to the region around U747 in E. coli 23S rRNA (Fig. 1B).
Two domain IV transcripts were used to test for methylation at U1939: a 56-nt transcript (Pa-IV-56), which had been used in previous studies (Urbonavičius et al. 2008), and a longer 80-nt transcript (Pa-IV-80) (Fig. 1C); both transcripts span the equivalent structure in E. coli 23S rRNA that contains U1939 (Fig. 1D).
Nucleotides U747 and U1939 are located within single-stranded regions of highly conserved secondary structures (Noller 1984; Cannone et al. 2002). A transcript of the E. coli 23S rRNA sequence equivalent to Pa-IV-56 was previously shown to be an effective substrate for in vitro methylation at 37°C by the E. coli RlmD methyltransferase (Madsen et al. 2003). Probing of Pa-IV-56 using N-methylisatoic anhydride (Ying et al. 2007) indicated that its secondary structure was maintained at 37°C (data not shown). This structure would be less stable at the temperatures required for PAB0760 methylation, and therefore the longer version of the transcript, Pa-IV-80 with a higher thermostability, was also used in methylation assays.
PAB0760 enzyme catalyzes formation of m5U in the Pa-II transcript
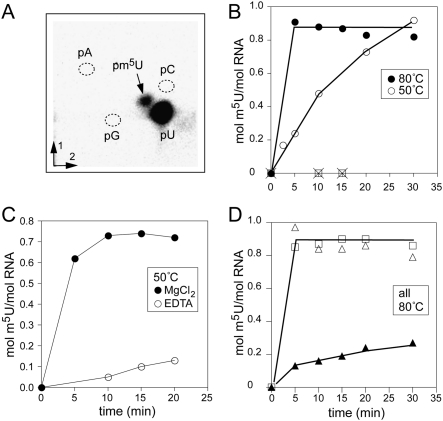
RNAs corresponding to the pyrococcal sequences (Fig. 1A,C) were transcribed containing 5′-[32P]-uridines. Each RNA was incubated with recombinant PAB0760 enzyme in the presence of AdoMet as the methyl donor. The RNAs were digested with nuclease P1, and the nucleotide products were separated by two-dimensional thin-layer chromatography. Incubation of PAB0760 with the Pa-II RNA resulted in a new radiolabeled spot corresponding to m5UMP (Fig. 3A) that was absent in control samples lacking PAB0760 or AdoMet. This indicated that PAB0760 directed m5U-methylation on the Pa-II RNA, and that the reaction was dependent on AdoMet.
FIGURE 3.
Ribothymidine formation in vitro by PAB0760 on RNA substrates (A) Autoradiogram of a two-dimensional, thin-layer plate showing the nuclease P1 hydrolysate of [α-32P]UTP-labeled Pa-II RNA after modification with PAB0760 for 20 min at 50°C. (B) Methylation of the Pa-II RNA at 50°C (○) and 80°C (•) in buffer with 10 mM MgCl2. Same reactions with the Pa-IV-80 RNA (□ and × on the base line). (C) Pa-II RNA incubated with PAB0760 at 50°C with 10 mM MgCl2 (•) or 0.5 mM EDTA (○). (D) Comparative kinetics of m5U formation for the mutants G746A (▲), G761A (△) and U762C (□) at 80°C under the same assay conditions as in panel B.
As would be expected for a hyperthermophilic enzyme, the kinetics of m5U formation by PAB0760 proceeded faster at 80°C than at 50°C (Fig. 3B). Magnesium ions stimulated methylation of the Pa-II RNA (Fig. 3C) similar to the effect seen for PAB0719 methylation at nucleotide U54 in full-length tRNA (Urbonavičius et al. 2008). The bivalent cation probably helps to fold and stabilize the structure of the RNA at high temperature (Misra and Draper 2002; Grilley et al. 2006), promoting an RNA conformation that is recognized and modified by PAB0760.
PAB0760 failed to catalyze m5U formation on the Pa-IV-56 and the tRNAAsp transcript, consistent with previous studies on this enzyme (Urbonavičius et al. 2008). Likewise, there was no methylation on the more stable Pa-IV-80 transcript (Fig. 3B) confirming that nucleotide U1939 is not a target for PAB0760 in vitro.
Identification of the uridine methylated by PAB0760
The location of the PAB0760 target nucleotide was pinpointed using MALDI mass spectrometry. The Pa-II RNA transcript was methylated by PAB0760 at 80°C and was then digested with RNase A or RNase T1 to produce smaller oligonucleotides of an optimal size for MALDI analysis. The hairpin 35 region of P. abyssi rRNA is 2 nt longer than the E. coli sequence, leaving some doubt as to whether the E. coli nucleotide U747 was equivalent to U857 or U859 in P. abyssi (Fig. 1A). No mass increase in the RNase T1 and RNase A digestion products AUGp and GAUp was observed, ruling out U857 (P. abyssi numbering) as the PAB0760 target; nor was there a mass increase in any of the other trinucleotides or larger oligonucleotides (Fig. 4). However, in the lower mass regions of both RNase A and T1 spectra, a new peak appeared at m/z 684 that was absent in unmethylated control samples. The m/z 684 peaks fit with a 14-Da mass increase in one of the UGp RNase T1 digestion products (Fig. 4D) and one of the GUp RNase A products. As in the previous case, when we used these two RNases on authentic 23S rRNA (Fig. 2), we could narrow down the methylation target to either U747 or U762, but we could not distinguish between them.
FIGURE 4.
MALDI-MS analyses of PAB0760 methylation in vitro on Pa-II RNAs. (A) Spectrum of RNase T1 oligonucleotide from the unmethylated wild-type Pa-II RNA. The empirical masses are given above the peaks; all digestion products are shown with the exception of Gp, and the terminal 5′-pppG and CCOH-3′ fragments (these were evident in the lower m/z region). The unlabeled peaks in the lower mass region are fragments from the matrix. (B) Upper m/z region after digestion of the Pa-II G761A mutant, where loss of the G761 cleavage site increases the length of the m/z 1890 fragment by 2 nt to include nucleotide U762 in a new peak at m/z 2525. (C) Enlargement of the m/z 2525 region from the Pa-II G761A mutant after incubation with PAB0760, showing the absence of methylation product here (+14 Da). (D) Enlargement of the dinucleotides m/z region to show peak distribution before PAB0760 methylation. (E) Enlargement of the same m/z region from wild-type Pa-II RNA after incubation with PAB0760. (F) Theoretical monoisotopic masses of the RNase T1 digestion products; the theoretical masses match the empirically derived values to within 0.1 Da. The m/z 2525 fragment is seen only in the RNA with the G761A mutation (encircled).
To differentiate the two possible uridine targets, several variants of Pa-II (Fig. 1A) were synthesized in which G746 or G761 was mutated to an adenosine (mutants G746A and G761A) or U762 was changed to a cytidine (U762C). In the in vitro methylation assays, both the G761A and U762C mutants were equally good substrates for PAB0760 as the wild-type RNA (Fig. 3D). The G761A mutation moves U762 into a unique RNase T1 octanucleotide at m/z 2525 in the spectrum (Fig. 4B), and the mass of this oligo remained unaltered after PAB0760 treatment (Fig. 4F) ruling out U762 as the methylation target. This result was confirmed by changing U762 to a cytosine with no resultant loss in PAB0760 methylation activity (Fig. 3D). The m5UGp peak at m/z 684 in the wild-type RNA was also evident in both the G761A and U762C mutants, and could only correspond to methylation at nucleotide U747 (U859 in P. abyssi numbering).
After mutation of the adjacent guanosine in the G746A RNA, nucleotide U747 becomes situated within a unique RNase T1 pentanucleotide (AUAUGp-748) and a unique RNase A dinucleotide (AUp-747), which can be assayed unambiguously by mass spectrometry. Although a minor amount of radiolabeled m5U (0.2 mol) was incorporated into G746A RNA (Fig. 3D), no methylation was detected by MALDI-MS. The MS signal-to-noise ratio for the peak containing U747 was favorable enough to identify 0.2 mol of modified fragment, so possibly the methylation detected with the radiolabel was distributed at more than one uridine in the RNA. It can be concluded that the identity of the base at 746 is important (and possibly crucial) for methylation by PAB0760 at the neighboring nucleotide U747.
DISCUSSION
The RNA m5U methyltransferase family
The superfamily of AdoMet-dependent RNA methyltransferases responsible for m5U and m5C modifications probably evolved from a common enzyme ancestor. This idea is supported by structural studies showing clear similarity between the folding of the E. coli m5U methyltransferase RlmD (Lee et al. 2005) and the m5C methyltransferase YccW (renamed RlmI) (Sunita et al. 2008); in fact, before YccW was shown to introduce the m5C1962 modification in 23S rRNA (Purta et al. 2008), phylogenetic analyses had misleadingly indicated that it was an m5U methyltransferase. The common ancestor of present-day m5U and m5C RNA methyltransferases most likely evolved from an RlmD-type enzyme that originated amongst the Bacteria (Urbonavičius et al. 2008). This ancestral enzyme is conserved in most bacterial lineages, although it has generally been lost in Mycoplasma species and other bacteria with minimalistic genomes (de Crécy-Lagard et al. 2007; Urbonavičius et al. 2008).
The two other E. coli m5U methyltransferases, RlmC and TrmA, are restricted to Proteobacteria and probably originated via two duplication events followed by specificity shifts to their respective targets at U747 in 23S rRNA and U54 in tRNA. It cannot be excluded that the Proteobacteria acquired these enzymes by horizontal gene transfer (HGT) subsequent to the specificity shifts, although in either case it seems that an rlmD ancestral gene was first duplicated and then paralogs evolved independently, resulting in methylation at different targets. From the available data on the structures and function of these methyltransferases, it is not possible to deduce the target of the ancestral RlmD-type enzyme and, as discussed below, it is quite probable that this particular question is unanswerable.
Archaeal RNA m5U methyltransferases
In Archaea, an AdoMet-dependent rlmD-type gene was apparently acquired by the common ancestor of the Thermococcales and Nanoarchaeota lineages (Brochier et al. 2005) via a single HGT from a bacterial donor (Urbonavičius et al. 2008). Following the separation of the lineages, the rlmD-type gene was duplicated within the Thermococcales lineage giving rise to PAB0719 and PAB0760 in P. abyssi. Since both of these genes are more closely related to RlmD than to other types of methyltransferase enzyme, it might be expected that at least one of the pyrococcal enzymes was specific for the nucleotide corresponding to the RlmD target at U1939 in E. coli 23S rRNA. However, according to the in vitro methylation data, this seems not to be the case: PAB0719 catalyzes the formation of m5U54 in tRNA (Urbonavičius et al. 2008) and the target for PAB0760 is shown here to correspond to the E. coli RlmC site at U747 in 23S rRNA.
The presence of the m5U747 modification within authentic P. abyssi 23S rRNA was supported by the primer extension and MS analyses. In addition, four other methylations were observed in the 747 and 1939 regions of P. abyssi 23S rRNA. Nucleotide C1934 is 2′-O-methylated, C1957 appears to be methylated at the five-position, and a methyl group is present on either nucleotide A744 or U745 (Fig. 2D). The exact position of the fourth methyl group could not be pinpointed, and while rRNA modifications in other organisms indicate that m5U1939 is the most likely location, it remains possible that one of the neighboring nucleotides (A1936-38) carries the methylation.
The putative m5U1939 modification is presently difficult to explain seeing that neither PAB0719 nor PAB0760 modifies this nucleotide in vitro, and no other COG2265 candidates are evident in the P. abyssi genome. It seems unlikely that PAB0719 or PAB0760 would extend their target specificity in vivo to include U1939. There have indeed been examples of methyltransferases becoming more promiscuous in vitro (e.g., Auxilien et al. 2007), but despite this, all m5U and m5C RNA methyltransferase that have so far been purified in active form still retain a preference for their cognate nucleotide targets.
Target specificity shifts in archaeal RNA m5U methyltransferases
The activities of the PAB0719 and PAB0760 enzymes are consistent with the occurrence of two specificity shifts in the Thermococcales lineage. As discussed above, the specificity for one distinct target (such as U1939) could have shifted to the present targets or, alternatively, the stringency of a relaxed RlmD-type ancestral enzyme could have increased so that it became specific for single nucleotide targets. Although we cannot determine the extent to which one or both of these mechanisms occurred, the structural and genetic data do indicate that the specificity of m5U methyltransferases is pliable.
Pertinent to this point are the crystal structures for the E. coli RlmD and TrmA AdoMet methyltransferases bound to RNA fragments that encompass their respective 23S rRNA U1939 (Lee et al. 2005) and tRNA U54 nucleotide targets (Alian et al. 2008). In these two structures there are striking similarities in the RNA conformation at the target nucleotides and also in the complementary enzyme folds that recognize the targets. Specificity is achieved by interaction of a few enzyme residues with nucleotides immediately adjacent to the target uridine (Hur et al. 2006; Alian et al. 2008). Conversely, it can be imagined that activity or specificity might be lost by changing key residues in either the enzyme or the substrate. In keeping with this, we note that changing G746 to an adenosine markedly reduces the ability of PAB0760 to recognize its methylation target at U747 and, not surprisingly, that PAB0760 was incapable of methylating the equivalent Streptococcus pneumoniae RNA transcript (Lebars et al. 2003), even though the secondary structure with an accessible U747 is similar (data not shown).
Common for all the characterized RNA m5U AdoMet methyltransferases (Fig. 1) is that they recognize and methylate naked RNA substrates. Each of the methylated uridines occurs in a single-stranded loop, and the inherent flexibility of these structures is exploited by RlmD (Lee et al. 2005) and TrmA (Alian et al. 2008) to achieve binding and target recognition. Similar refolding of RNA substrates might represent a common theme for how other m5U AdoMet methyltransferases function, although additional RNA–enzyme structures need to be solved to establish whether this is generally the case.
Based on the presently available data on the similarities in the molecular folds at the sites of RNA–enzyme contact, taken together with the flexibility of the RNA substrate and the lack of requirement for other components such as r-proteins, it seems that relatively minor sequence changes in either the enzyme or the RNA substrate could have been sufficient to promote specificity shifts during m5U AdoMet methyltransferase evolution. Feasibly, ancestral enzymes had a broad specificity range, and a series of later adaptations produced the contemporary orthologs that target single nucleotides. A consequence of this would be that the identity of the nucleotide(s) modified by the primordial enzyme will remain an open question.
MATERIALS AND METHODS
Preparation of rRNA from P. abyssi cells
P. abyssi strain GE5 was cultivated in a Trypticase-salt-yeast extract medium (Balch and Wolfe 1976) at 30–45 bar of pressure and 95°C; cells were collected by centrifugation. The cell paste (5 g) was resuspended in 25 mL lysis buffer (50 mM Tris-Cl, pH7.6, 6 mM MgCl2, 100 mM NH4Cl, and 6 mM β-mercaptoethanol) containing 600 U DNase 1 (Sigma). After incubation for 1 h at 4°C, the cell suspension was disrupted by two passages through a Constant Cell Disruption system (BazicZ, CellD) at 1.5 kbars. Cell debris was removed by centrifugation at 5000 g, and ribosomes were then pelleted from the supernatant by centrifuging at 100,000 g for 4 h. Ribosomes were resuspended in 2 mL lysis buffer and extracted with 2 mL phenol equilibrated with 0.1 M ammonium citrate, pH 4.5. Total rRNA was recovered from the aqueous phase by precipitation with 2 vol ethanol, and was purified further by guanidine thiocyanate/acidic phenol treatment (Chomczynski and Sacchi 2006). The purified rRNA was precipitated with ethanol in 0.5 M ammonium acetate pH 5.0, and after centrifugation the pellet was washed with 70% ethanol, air dried, and dissolved in water.
Analysis of P. abyssi rRNA
P. abyssi total rRNA was analyzed first by primer extension with reverse transcriptase (Stern et al. 1988). The 23S rRNA was sequenced using the 5′-end 32P-labeled primers 5′-TTCGCCGGGAACCAGCTA (hybridizing to nucleotides 803–820 to view the 747 region) and 5′-GGACCTCGTTACGCCATTC (hybridizing to nucleotides 1972–1990 to view the 1939 region).
The 747 and 1939 regions of P. abyssi 23S rRNA were then analyzed by MALDI-MS. The sequences from G725-C767 and U1917-C1962 were isolated by hybridization to the complementary deoxyoligonucleotides 5′-GCCCGGGGTCACGGGAGCGAACTGCACATCAGCACCCCTAGCGGGCCTCC and 5′-GCAGGTCGGCATTTAACCGACAGGAATTTCGCTACCTTAAGAGGGTTA, respectively (Andersen et al. 2004; Auxilien et al. 2007). Briefly, 100 pmol of total rRNA was heated with 400 pmol of deoxyoligonucleotide at 85°C for 1 min, followed by slow cooling to 45°C over 2 h. Regions of the rRNAs that were not protected by hybridization were digested away with mung bean nuclease (NE Biolabs) and RNase A (Sigma), and the rRNA sequences paired to the deoxyoligonucleotide were separated by gel electrophoresis and were extracted. Each of the rRNA sequences of ∼50 nt was digested with 20 units of RNase T1 (USB) or 0.5 μg RNase A at 37°C for 3 h in 2 μL aqueous solution of 60 mM 3-hydroxypicolinic acid. Samples were dried and resuspended in 1 μL H2O prior to analysis by MALDI-MS (Voyager Elite, Perseptive Biosystems) recording in reflector and positive ion mode (Kirpekar et al. 2000). Spectra were analyzed using the program m/z (Proteometrics Inc) and were left unsmoothed.
Purification of recombinant PAB0760 enzyme
A recombinant version of the PAB0760 gene was expressed from plasmid pET28b in E. coli. The recombinant PAB0760 enzyme has six histidine residues at its N-terminus and could be purified to near homogeneity by Ni2+-affinity chromatography (Urbonavičius et al. 2008). Further purification of the enzyme was achieved by gel filtration on a Superdex 75 HR 10/30 column (GE Healthcare) equilibrated with 50 mM sodium phosphate pH 8.0 and 300 mM NaCl. Enzyme fractions were aliquoted and stored at −20°C.
Production of RNA substrates
The structures of the RNA substrates Pa-II, Pa-IV-56, and Pa-IV-80 were based on the sequence of the P. abyssi 23S rRNA. The Pa-IV RNAs (Fig. 1) were transcribed from complementary single-stranded DNA templates containing the T7 promoter sequence at 3′-end, and 2′-O-methyl G at the first two positions to prevent nontemplated addition of nucleotides at the 3′-end of the nascent RNA (Kao et al. 1999). A 17-mer top-strand DNA was annealed to the 5′-end of the template forming a double-stranded promoter for the T7 RNA polymerase (Promega). The wild-type and mutant Pa-II sequences were transcribed in a slightly different manner using double-stranded DNA templates generated by PCR from two overlapping oligodeoxynucleotides; the T7-promoter was encoded at the 5′-end of the plus strand, and the minus-strand oligonucleotides contained 2′-O-meG at the first two positions. In vitro transcription of DNA templates in the presence of [α-32P]-UTP (GE Healthcare) using T7 RNA polymerase and purification of the resulting radiolabeled transcripts on denaturing gels have been described previously (Grosjean et al. 2007). RNAs for MALDI-MS analyses were prepared without radiolabeled nucleotides.
RNA methylation assays
Unlabeled RNA (480 nM) was incubated with 200–400 nM recombinant PAB0760 enzyme in 25 μL of reaction mix containing 25 mM Tris-Cl, pH 7.5, 50 mM ammonium acetate, 2 mM dithiotreitol, 10 mM MgCl2, 0.1 mg/mL RNase-free bovine serum albumin (Roche Applied Science), and 80 μM AdoMet (Sigma) for 15 min at either 50°C or 80°C. The [α-32P]-UTP-labeled samples were methylated by incubating 2 nM RNA with recombinant PAB0760 in the same buffer (substituting ammonium acetate for KCl) for up to 30 min at 50°C or 80°C. In some experiments, MgCl2 was substituted with 0.5 mM EDTA. Reactions were stopped by extracting with phenol/chloroform and RNAs were recovered by ethanol precipitation after addition of 0.3 M ammonium acetate pH 5.5. The analysis of radiolabeled nucleotide products following nuclease P1 digestion (Roche Applied Science) was carried out by two-dimensional, thin-layer chromatography (Grosjean et al. 2007).
MALDI-MS analyses of transcripts were carried out as described about using 10 pmol RNA and digesting with either RNase A or RNase T1.
ACKNOWLEDGMENTS
We thank Jean-Pierre Rousset (IGM, University Paris-Sud) and Beatrice Golinelli-Pimpaneau (CNRS, Laboratory of Enzymology, Gif-sur-Yvette) for encouragement and financial support. We also thank Nicolas Soler (CNRS, Laboratory of Structural Chemistry and Biology, Gif-sur-Yvette) for assistance with RNA purification and Christina Sørensen and Lene Jakobsen (SDU, Odense) for help with the MALDI-MS analyses. This work was supported in part by the Agence Nationale de la Recherche (to Beatrice Golinelli-Pimpaneau), by the CNRS (through the program GEOMEX to H.G and D.F. and the ATIP program to C.B.-A.), by grants of University of Paris-Sud (to J.-P. Rousset), and by grants from the Danish Research Agency (FNU-rammebevilling 272-07-0613) plus the Nucleic Acid Center of the Danish Grundforskningsfond (to S.D.). C.H. was supported by a fellowship from Institut de Chimie des Substances Naturelles.
Footnotes
Abbreviations: AdoMet, S-adenosyl-L-methionine; COG, cluster of orthologous genes; HGT, horizontal gene transfer; MALDI, matrix assisted laser desorption/ionization; MS, mass spectrometry; m5U, 5-methyluridine, ribothymidine; nt, nucleotide; RT, reverse transcriptase.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2323411.
REFERENCES
- Agarwalla S, Kealey JT, Santi DV, Stroud RM 2002. Characterization of the 23S ribosomal RNA m5U1939 methyltransferase from Escherichia coli. J Biol Chem 277: 8835–8840 [DOI] [PubMed] [Google Scholar]
- Agris PF 2004. Decoding the genome: a modified view. Nucleic Acids Res 32: 223–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alian A, Lee TT, Griner SL, Stroud RM, Finer-Moore J 2008. Structure of a TrmA-RNA complex: A consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc Natl Acad Sci 105: 6876–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen NM, Douthwaite S 2006. YebU is a m5C methyltransferase specific for 16S rRNA nucleotide 1407. J Mol Biol 359: 777–786 [DOI] [PubMed] [Google Scholar]
- Andersen TE, Porse BT, Kirpekar F 2004. A novel partial modification at 2501 in Escherichia coli 23S ribosomal RNA. RNA 10: 907–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auxilien S, El Khadali F, Rasmussen A, Douthwaite S, Grosjean H 2007. Archease from Pyrococcus abyssi improves substrate specificity and solubility of a tRNA m5C methyltransferase. J Biol Chem 282: 18711–18721 [DOI] [PubMed] [Google Scholar]
- Balch WE, Wolfe RS 1976. New approach to the cultivation of methanogenic bacteria: 2-Mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl Environ Microbiol 32: 781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Björk GR, Hagervall TG 2005. Transfer RNA modification. In Escherichia coli and Salmonella: Cellular and molecular biology (ed. Böck A, et al. ), Chap. 4.6.2. ASM, Washington, DC [Google Scholar]
- Brochier C, Gribaldo S, Zivanovic Y, Confalonieri F, Forterre P 2005. Nanoarchaea: representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales? Genome Biol 6: R42 doi: 10.1186/gb-2005-6-5-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2 doi: 10.1186/1471-2105-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N 2006. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc 1: 581–585 [DOI] [PubMed] [Google Scholar]
- Chow CS, Lamichhane TN, Mahto SK 2007. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem Biol 2: 610–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanloo P, Sprinzl M, Watanabe K, Albani M, Kersten H 1979. Role of ribothymidine in the thermal stability of transfer RNA as monitored by proton magnetic resonance. Nucleic Acids Res 6: 1571–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crécy-Lagard V, Marck C, Brochier-Armanet C, Grosjean H 2007. Comparative RNomics and modomics in Mollicutes: Prediction of gene function and evolutionary implications. IUBMB Life 59: 634–658 [DOI] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ 2002. rRNA modifications and ribosome function. Trends Biochem Sci 27: 344–351 [DOI] [PubMed] [Google Scholar]
- Grilley D, Soto AM, Draper DE 2006. Mg2+-RNA interaction free energies and their relationship to the folding of RNA tertiary structures. Proc Natl Acad Sci 103: 14003–14008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H 2009. DNA and RNA modification enzymes: Structure, mechanism, function and evolution. Landes Biosciences, Austin TX [Google Scholar]
- Grosjean H, Droogmans L, Roovers M, Keith G 2007. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabelled tRNA substrates. In RNA modification (ed. Gott JM), pp. 57–101 Elsevier, New York: [DOI] [PubMed] [Google Scholar]
- Gu X, Ivanetich KM, Santi DV 1996. Recognition of the T-arm of tRNA by tRNA m5U54 -methyltransferase is not sequence specific. Biochemistry 35: 11652–11659 [DOI] [PubMed] [Google Scholar]
- Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107: 679–688 [DOI] [PubMed] [Google Scholar]
- Hur S, Stroud RM, Finer-Moore J 2006. Substrate recognition by RNA 5-methyluridine methyltransferases and pseudouridine synthases: a structural perspective. J Biol Chem 281: 38969–38973 [DOI] [PubMed] [Google Scholar]
- Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J 2009. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37: D159–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C, Zheng M, Rudisser S 1999. A simple and efficient method to reduce nontemplated nucleotide addition at the 3′-terminus of RNAs transcribed by T7 RNA polymerase. RNA 5: 1268–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kealey JT, Gu X, Santi DV 1994. Enzymatic mechanism of tRNA m5U54 methyltransferase. Biochimie 76: 1133–1142 [DOI] [PubMed] [Google Scholar]
- Kirpekar F, Douthwaite S, Roepstorff P 2000. Mapping posttranscriptional modifications in 5S ribosomal RNA by MALDI mass spectrometry. RNA 6: 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Trakhanov S, Laurberg M, Noller HF 2006. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 126: 1065–1077 [DOI] [PubMed] [Google Scholar]
- Lebars I, Yoshizawa S, Stenholm AR, Guittet E, Douthwaite S, Fourmy D 2003. Structure of 23S rRNA hairpin 35 and its interaction with the tylosin-resistance methyltransferase RlmAII. EMBO J 22: 183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TT, Agarwalla S, Stroud RM 2005. A unique RNA fold in the RumA-RNA-cofactor ternary complex contributes to substrate selectivity and enzymatic function. Cell 120: 599–611 [DOI] [PubMed] [Google Scholar]
- Madsen CT, Mengel-Jorgensen J, Kirpekar F, Douthwaite S 2003. Identifying the methyltransferases for m5U747 and m5U1939 in 23S rRNA using MALDI mass spectrometry. Nucleic Acids Res 31: 4738–4746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra VK, Draper DE 2002. The linkage between magnesium binding and RNA folding. J Mol Biol 317: 507–521 [DOI] [PubMed] [Google Scholar]
- Noller HF 1984. Structure of ribosomal RNA. Annu Rev Biochem 53: 119–162 [DOI] [PubMed] [Google Scholar]
- Nordlund ME, Johansson JO, von Pawel-Rammingen U, Byström AS 2000. Identification of the TRM2 gene encoding the tRNA(m5U54)methyltransferase of Saccharomyces cerevisiae. RNA 6: 844–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ny T, Lindström HR, Hagervall TG, Björk GR 1988. Purification of transfer RNA m5U54-methyltransferase from Escherichia coli. association with RNA. Eur J Biochem 177: 467–475 [DOI] [PubMed] [Google Scholar]
- Ofengand J, Del Campo M 2005. Modified nucleotides of E. coli ribosomal RNA. In Escherichia coli and Salmonella: Cellular and molecular biology (ed. Böck A, et al. ), Chap. 4.6.1. ASM, Washington, DC [Google Scholar]
- Purta E, O'Connor M, Bujnicki JM, Douthwaite S 2008. YccW is the m5C methyltransferase specific for 23S rRNA nucleotide 1962. J Mol Biol 383: 641–651 [DOI] [PubMed] [Google Scholar]
- Purta E, O'Connor M, Bujnicki JM, Douthwaite S 2009. YgdE is the 2′-O-ribose methyltransferase RlmM specific for nucleotide C2498 in bacterial 23S rRNA. Mol Microbiol 72: 1147–1158 [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Stern S, Moazed D, Noller HF 1988. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol 164: 481–489 [DOI] [PubMed] [Google Scholar]
- Sunita S, Tkaczuk KL, Purta E, Kasprzak JM, Douthwaite S, Bujnicki JM, Sivaraman J 2008. Crystal structure of the Escherichia coli 23S rRNA:m5C methyltransferase RlmI (YccW) reveals evolutionary links between RNA modification enzymes. J Mol Biol 383: 652–666 [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4: 41 doi: 10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavičius J, Skouloubris S, Myllykallio H, Grosjean H 2005. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria-evolutionary implications. Nucleic Acids Res 33: 3955–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavičius J, Brochier-Armanet C, Skouloubris S, Myllykallio H, Grosjean H 2007. In vitro detection of the enzyme activity of folate-dependent tRNA(U54, C5)-methyltransferase. In RNA modification (ed. Gott JM), Vol 425, pp. 103–119 Elsevier, New York: [DOI] [PubMed] [Google Scholar]
- Urbonavičius J, Auxilien S, Walbott H, Trachana K, Golinelli-Pimpaneau B, Brochier-Armanet C, Grosjean H 2008. Acquisition of a bacterial RumA-type tRNA(uracil-54, C5)-methyltransferase by Archaea through an ancient horizontal gene transfer. Mol Microbiol 67: 323–335 [DOI] [PubMed] [Google Scholar]
- Ying BW, Fourmy D, Yoshizawa S 2007. Substitution of the use of radioactivity by fluorescence for biochemical studies of RNA. RNA 13: 2042–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]