Abstract
In flies, 22–23-nucleotide (nt) microRNA duplexes typically contain mismatches and begin with uridine, so they bind Argonaute1 (Ago1), whereas 21-nt siRNA duplexes are perfectly paired and begin with cytidine, promoting their loading into Ago2. A subset of Drosophila endogenous siRNAs—the hairpin-derived hp-esiRNAs—are born as mismatched duplexes that often begin with uridine. These would be predicted to load into Ago1, yet accumulate at steady-state bound to Ago2. In vitro, such hp-esiRNA duplexes assemble into Ago1. In vivo, they encounter complementary target mRNAs that trigger their tailing and trimming, causing Ago1-loaded hp-esiRNAs to be degraded. In contrast, Ago2-associated hp-esiRNAs are 2′-O-methyl modified at their 3′ ends, protecting them from tailing and trimming. Consequently, the steady-state distribution of esiRNAs reflects not only their initial sorting between Ago1 and Ago2 according to their duplex structure, length, and first nucleotide, but also the targeted destruction of the single-stranded small RNAs after their loading into an Argonaute protein.
Keywords: Argonaute, RNAi, miRNA, microRNA, small RNA sorting
INTRODUCTION
In Drosophila, two small silencing RNA pathways regulate gene expression in both the germ line and the soma: the microRNA (miRNA) and the small interfering RNA (siRNA) pathways. miRNAs act as post-transcriptional regulators in diverse aspects of animal development and physiology; siRNAs counteract viral infections and ensure genomic stability by targeting transposable elements (Ghildiyal and Zamore 2009). Both miRNAs and siRNAs are cleaved from longer double-stranded RNA (dsRNA) precursors by RNase III family ribonucleases. Dicer-1 liberates ∼22-nucleotide (nt)-long miRNAs from ∼65-nt stem–loop precursors (pre-miRNAs), whereas Dicer-2 cuts 21-nt exo-siRNAs off of long dsRNA derived from exogenous sources, such as viruses or 21/22-nt endo-siRNAs from endogenous dsRNA derived from convergently transcribed mRNAs, transposons, or long, partially self-complementary “hairpin” RNAs (Zamore et al. 2000; Bernstein et al. 2001; Hutvágner et al. 2001; Lee et al. 2004; Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008a,b).
Dicer-1 and Dicer-2 bind to dsRNA-binding proteins that enhance their activities: Dicer-1 partners with the PB isoform of the protein Loquacious (Loqs-PB) to process pre-miRNAs, while Dicer-2 acts with the shorter Loqs-PD isoform to generate at least some classes of endo-siRNAs (Liu et al. 2003; Förstemann et al. 2005; Jiang et al. 2005; Saito et al. 2005; Park et al. 2007; Hartig et al. 2009; Zhou et al. 2009; Marques et al. 2010).
As a consequence of their being cleaved from dsRNA precursors, both miRNAs and siRNAs are born double-stranded, and these duplexes are loaded into members of the Argonaute family of proteins, which mediate their silencing functions. In addition to its role in producing siRNAs, Dicer-2 also participates in loading siRNAs into Argonaute2 (Ago2). For this function, Dicer-2 partners with the protein R2D2, a paralog of Loqs-PB, to form a heterodimer that binds siRNA duplexes (Liu et al. 2003; Tomari et al. 2004b; Pham and Sontheimer 2005; Liu et al. 2006). Loading of small RNAs into Argonaute proteins also requires the HSP90-chaperone machinery (Iki et al. 2010; Iwasaki et al. 2010; Miyoshi et al. 2010).
Drosophila miRNAs typically assemble into Ago1, while siRNAs bind to Ago2—the Argonaute protein that mediates RNA interference—to form the RNA-induced Silencing Complex (RISC) (Okamura et al. 2004). In flies, the production and loading of small RNA duplexes are not coupled (Förstemann et al. 2007; Tomari et al. 2007). Instead, miRNAs and siRNAs are sorted between Ago1 and Ago2 according to their duplex structure and the identity of their 5′ nucleotide (Ghildiyal et al. 2008; Czech et al. 2009; Okamura et al. 2009; Ghildiyal et al. 2010). Extensive double-stranded character, as is found in siRNAs, leads to assembly into Ago2, whereas central bulges and mismatches, typically found in miRNA/miRNA* duplexes, promote assembly into Ago1 (Tomari et al. 2007; Kawamata et al. 2009).
The affinity of a small RNA duplex for the Dicer-2/R2D2 heterodimer, a component of the complex that loads small RNAs into Ago2, determines the extent to which it loads into Ago2 (Liu et al. 2003; Tomari et al. 2004a,b, 2007). R2D2 binds to the more stable 5′ end of the small RNA duplex, orienting the siRNA so that the less tightly paired 5′ end becomes the guide strand bound to Argonaute (Tomari et al. 2004b). Central mismatches reduce the affinity of the small RNA duplex for Dicer-2/R2D2, antagonizing Ago2 loading and promoting loading into Ago1 (Förstemann et al. 2007; Tomari et al. 2007; Kawamata et al. 2009). In addition to structure, the sequence of a small RNA guide influences sorting in many organisms (Lau et al. 2001; Ghildiyal et al. 2008; Mi et al. 2008; Montgomery et al. 2008; Hu et al. 2009; Ghildiyal et al. 2010). In flies, miRNAs bound to Ago1 usually start with uridine (U), whereas Ago2-bound miRNAs, miRNA* strands, and exo-siRNAs tend to start with cytidine (C) (Ghildiyal et al. 2010). The 5′ phosphate group of small RNA guides binds to a pocket in the MID (middle) domain of eubacterial and archaebacterial Argonaute proteins (Ma et al. 2005; Parker et al. 2005). The equivalent domain in eukaryotic Argonautes may “read” the identity of the first nucleotide of a small silencing RNA, as the MID domain of human Ago2, which is closely related in sequence to fly Ago1, preferentially binds to adenosine and uridine 5′ monophosphate (Frank et al. 2010). The molecular basis for the 5′ nucleotide preference of Drosophila Ago2 is not yet known and could reflect a preference for C of the Ago2-loading machinery, Ago2 itself, or both.
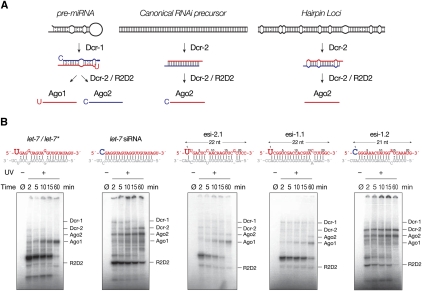
While most exo- and endo-siRNA precursors are believed to comprise long, fully complementary sense and anti-sense RNAs, the siRNA precursors transcribed from the six Drosophila “structured” loci (esi-1/hp-CG18854, esi-2/hp-CG4068, hp-CG32207, hp-CR32205, hp-pncr009/pncr009:3L, and an intergenic region adjacent to CG4770) contain mismatches, bulges, and wobble base pairs, characteristics usually associated with miRNA precursors (Fig. 1; Supplementary Figs. S1, S2; Czech et al. 2008; Kawamura et al. 2008; Okamura et al. 2008b). The duplexes of the “hp-esiRNAs” derived from these hairpin transcripts resemble miRNAs rather than canonical siRNAs (Fig. 1A). Yet hp-esiRNAs accumulate mainly in Ago2. Why?
FIGURE 1.
Hairpin-derived endo-siRNAs (hp-esiRNAs) assemble in vitro into Ago1 and Ago2 RISC. (A) Overview of the model for small silencing RNA production and sorting into Argonaute effector complexes in Drosophila. (B) The top row depicts the structures and sequences of the let-7/let-7* miRNA duplex, a canonical siRNA (let-7 siRNA), as well as the most abundant hp-esiRNAs from the esi-2 (esi-2.1) and esi-1 loci (esi-1.1 and esi-1.2). Below each small RNA duplex are representative data for a time course of assembly of the small RNA into Ago1 and Ago2 as determined by 254 nm UV crosslinking.
We recently reported that the binding of an Ago1-bound miRNA to an artificial, highly complementary target RNA triggers its tailing and 3′-to-5′ trimming, ultimately decreasing the steady-state concentration of the miRNA (Ameres et al. 2010). Hen1, the methyltransferase that adds a 2′-O-methyl group to the 3′ end of Ago2-bound small RNAs (Horwich et al. 2007; Pelisson et al. 2007), protects Ago2-bound siRNAs against such tailing and trimming (Hutvágner and Zamore 2002; Haley and Zamore 2004; Ameres et al. 2010). Here, we show that, in vitro, hp-esiRNA duplexes assemble into Ago1 or Ago2 according to their sequence, structure and length: Ago1 preferentially binds to 22 nt, bulged small RNAs starting with A or U; Ago2 associates with 21-nt, double-stranded small RNAs beginning with C or U. Like exo-siRNAs and miRNA* strands, hp-esiRNAs that begin with C bind Ago2 in vivo. In contrast, hp-esiRNAs that start with U bind mainly Ago1 in vitro yet accumulate in Ago2 in vivo. We present evidence that in vivo these hp-esiRNAs encounter complementary target mRNAs that trigger their degradation via the tailing and trimming pathway. Since Ago2-associated hp-esiRNAs bear a protective 2′-O-methyl modification at their 3′ ends, they escape tailing and trimming and accumulate. Thus, the steady-state distribution of hp-esiRNAs between Ago1 and Ago2 reflects not only their partitioning during the loading of Argonaute proteins, but also the target-directed destruction of specific hp-esiRNA species after their assembly into Ago1 complexes.
RESULTS AND DISCUSSION
Hp-esiRNAs predictably partition between Ago1 and Ago2 in vitro
Their partially double-stranded structure predicts that hp-esiRNAs should load into Ago1, yet in vivo they accumulate in Ago2 (Kawamura et al. 2008; Czech et al. 2009; Okamura et al. 2009). To test whether hp-esiRNAs violate the previously established rules for Drosophila small RNA sorting, we examined their loading into Ago1 and Ago2 in Drosophila embryo lysates. The esi-1 (hp-CG18854; Figure S1) and esi-2 (hp-CG4068; Figure S2) loci produce 98% of all mature hp-esiRNAs in fly heads and 96% in S2 cells (Supplemental Table S1; Czech et al. 2008; Kawamura et al. 2008; Okamura et al. 2008b). Among the most abundant hp-esiRNAs derived from these two loci, esi-2.1 and esi-1.1 are 22 nt long and begin with U; esi-2.1 contains a mismatch at position 8 of its predicted guide/passenger duplex (Fig. 1B, top row). These are all features of Ago1-loaded small RNAs (Ghildiyal et al. 2010). Other hp-esiRNAs resemble Ago2-associated small RNAs. For example, esi-1.2 is 21 nt long, begins with C, and has a paired central region.
We used 254-nm UV-crosslinking to measure the partitioning of synthetic, 5′ phosphorylated hp-esiRNA duplexes between Ago1 and Ago2 (Fig. 1B). Esi-2.1 and esi-1.1 crosslinked efficiently to Ago1, although at lower levels than the let-7/let-7* miRNA duplex. Crosslinking to Ago2 was detectable, but much less efficient than a canonical let-7 siRNA. In contrast, esi-1.2 crosslinked to Ago2 but not Ago1, just like a canonical siRNA (Fig. 1B). We conclude that, in vitro, hp-esiRNAs follow the established rules for small RNA sorting in flies, partitioning according to their duplex structure and first nucleotide (Czech et al. 2009; Okamura et al. 2009; Ghildiyal et al. 2010).
5′ Nucleotide identity and length influence small RNA partitioning between Ago1 and Ago2
In plants, the 5′ nucleotide of a small silencing RNA determines how it partitions among the different Argonaute proteins (Mi et al. 2008; Montgomery et al. 2008). A similar mechanism operates in some animals (Ghildiyal et al. 2008; Frank et al. 2010; Ghildiyal et al. 2010). In flies, Ago2-bound small RNAs, including exo-siRNAs and miRNA* species, typically begin with C, whereas Ago1-associated small RNAs (mostly miRNAs) begin with U (Ghildiyal et al. 2008, 2010). To disentangle the influence of structure and 5′ nucleotide identity, we examined the loading of four variants of the let-7/let-7* duplex and four variants of an siRNA in which the guide strand corresponded to let-7 (Fig. 2A). To ensure selection of let-7 as the guide strand, the first nucleotide of let-7 was unpaired in all duplexes (Schwarz et al. 2003; Tomari et al. 2007).
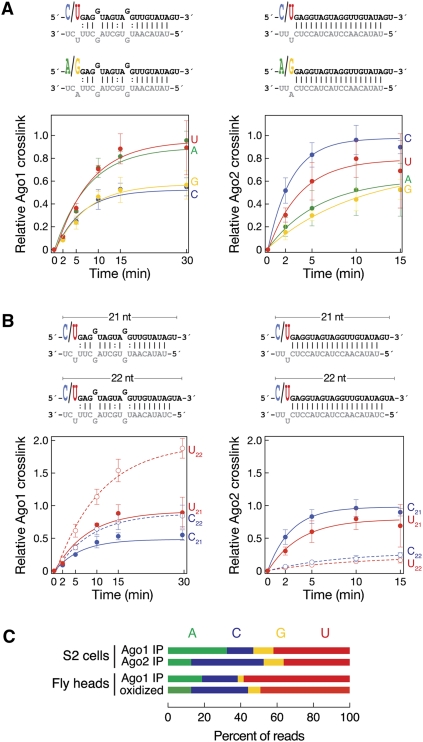
FIGURE 2.
The 5′ nucleotide of the guide strand and the length of the small RNA duplex determine sorting between Ago1 and Ago2. miRNA or siRNA duplexes that differ from each other only in their 5′ nucleotide (A) and length (B), but not their duplex structure, were assessed by UV crosslinking for their assembly into Ago1 or Ago2 RISC in vitro. Mean ± SD of three replicates is reported. (C) The 5′ nucleotide distribution for the hp-esiRNAs bound to Ago1 and Ago2 was deduced by analyzing the small RNA immunoprecipitated with Ago1 and Ago2 from S2 cell lysates or the small RNAs from fly heads immunoprecipitated with Ago1 and inferred to be bound to Ago2 by their resistance to oxidation.
As observed previously (Ghildiyal et al. 2010), the let-7/let-7* duplex more efficiently assembled into Ago1 when let-7 began with U than when it began with C (Fig. 2A, left; Supplemental Fig. S3). The let-7/let-7* duplex that began with A also loaded Ago1 efficiently, whereas loading of a let-7/let-7* duplex beginning with G was as inefficient as the duplex in which let-7 began with a C. Experiments in which each of the four let-7/let-7* variants was used as a competitor for loading a 5′ 32P-radiolabeled canonical let-7/let-7* duplex confirmed these results (Supplemental Fig. S4). Conversely, a let-7 siRNA duplex that began with a pyrimidine (C or U) was efficiently loaded into Ago2, whereas the same siRNA duplex, but starting with a purine (G or A), loaded into Ago2 less efficiently (Fig. 2A, right; Supplemental Fig. S3). Among pyrimidines, Ago2 preferred C over U.
Distinct lengths of small RNAs are bound to Ago1 and Ago2 in flies: Ago1-associated small RNAs are 22–23 nt, while nearly all small RNAs bound to Ago2 are 21 nt (Czech et al. 2008; Ghildiyal et al. 2008). The distinct RNA lengths likely reflect the intrinsic characteristics of the Dicer proteins that produce the small RNAs bound to Ago1 versus Ago2: Dicer-1 generates ∼22–23 nt miRNAs, which typically load into Ago1, while Dicer-2 produces 21 nt siRNAs, which bind to Ago2 (Zamore et al. 2000; Bernstein et al. 2001; Hutvágner et al. 2001; Lee et al. 2004). Does the propensity of Dicer-1 to cleave pre-miRNAs into 22–23 nt miRNA/miRNA* duplexes reflect a specific preference of Ago1 to load longer small RNA duplexes than Ago2? To test this idea, we compared the loading of four let-7/let-7* duplexes and four let-7 siRNA duplexes into Ago1 and Ago2. Each duplex comprised either two 21-nt or two 22-nt RNAs, with the let-7 strand beginning with either C or U. Figure 2B shows that the 22-nt miRNA/miRNA* duplexes were more efficiently loaded into Ago1, whereas the 21-nt siRNA duplexes were more efficiently loaded into Ago2.
For Ago1, the favored 22-nt length takes precedence over the disfavored 5′ C: a 22-mer miRNA starting with C loaded Ago1 as efficiently as a 21-mer miRNA starting with U (Fig. 2B, left). In contrast, Ago2 preferred a 21-nt duplex—even one in which the guide strand began with U—over a 22-nt duplex beginning with either C or U (Fig. 2B, right). These data are consistent with our finding that in Drosophila embryo lysate, esi-1.1 and esi-2.1—both 22 nt long—preferentially assemble into Ago1, whereas the 21-nt long esi-1.2 predominantly associates with Ago2 (Fig. 1B).
We analyzed high-throughput sequencing data of small RNAs that co-immunoprecipitated with Ago1 or Ago2 in S2 cells and of small RNAs from fly heads that co-immunoprecipitated with Ago1 or were inferred to be associated with Ago2 because of their resistance to oxidation (Czech et al. 2008; Ghildiyal et al. 2010). In S2 cells (Fig. 2C), Ago1-bound hp-esiRNAs tended to start with U (42%) or A (32%) rather than C (15%) or G (11%), whereas Ago2-bound hp-esiRNAs tended to start with C (40%) or U (36%) rather than A (13%) or G (11%). Similarly, in fly heads, the hp-esiRNAs bound to Ago1 tended to start with U (58%), A (19%), or C (20%), but not G (3%), whereas those bound to Ago2 began with U (49%) or C (31%), but less often with A (13%) or G (7%). Small RNA assembly in vitro (Figs. 1B, 2A) accurately recapitulated the distribution of 5′ nucleotide identities observed in vivo: Ago1 preferentially loaded small RNAs starting with U or A, but not C or G; Ago2 associated predominantly with small RNAs beginning with C or U but not G or A.
Mutations in the RNAi pathway change the hp-esiRNA repertoire
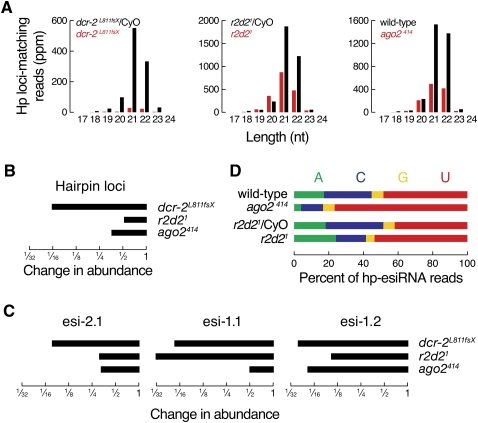
We analyzed the genetic requirements for hp-esiRNA accumulation from each of the six Drosophila hairpin loci. As reported previously, wild-type hp-esiRNA accumulation in fly heads required dicer-2, r2d2, and ago2 (Fig. 3A; Supplemental Fig. S5; Czech et al. 2008; Okamura et al. 2008b; Marques et al. 2010). In dicer-2L811fsX null mutant fly heads, total hp-esiRNAs decreased >16-fold compared to heterozygotes (Fig. 3B). In flies lacking R2D2 or Ago2, hp-esiRNAs were half (r2d21) or one-third (ago2414) as abundant as in the corresponding heterozygous siblings or in wild-type flies. (The expression of hp-esiRNAs in r2d21 heterozygotes and Oregon R was essentially identical [Supplemental Fig. S6].) These data suggest that hp-esiRNAs are produced by Dicer-2 and then loaded—at least in part—by the Dicer-2/R2D2 heterodimer into Ago2.
FIGURE 3.
Mutations in the RNAi machinery change the repertoire of hp-esiRNAs. (A) Size distribution and abundance of hp-esiRNAs in dicer-2 and r2d2 mutant fly heads compared to their heterozygous siblings or ago2414 mutants compared to Oregon R fly heads. (B) Decrease in hp-esiRNAs in dicer-2L811fsX, r2d21, and ago2414 mutant fly heads. (C) Change in full-length esi-2.1, esi-1.1, or esi-1.2 in dicer-2L811fsX, r2d21, and ago2414 mutant fly heads. (D) Relative abundance of hp-esiRNAs starting with each possible 5′ nucleotide as deduced from sequencing of small RNAs from the heads of ago2414 and r2d21 heterozygote or homozygote or Oregon R flies.
Hp-esiRNAs are generally more abundant in Ago2 than in Ago1: We analyzed high throughput sequencing data sets for the small RNAs co-immunoprecipitated with Ago1 and Ago2 in S2 cell extracts, as well as from fly heads in which Ago1-bound small RNAs were isolated by co-immunoprecipitation with Ago1 and Ago2-bound small RNAs inferred from their resistance to oxidation. In S2 cells, hp-siRNAs were twice as abundant in Ago2 than in Ago1; in fly heads, hp-siRNAs were about four times more abundant in Ago2 than in Ago1 (Figure S7).
Individual hp-esiRNA species have a common requirement for dicer-2, but distinct requirements for r2d2 and ago2 (Fig. 3C). Esi-2.1, esi-1.1, and esi-1.2 each decreased 13- to 26-fold in dicer-2L811fsX mutant fly heads, consistent with the idea that Dicer-2 cleaves hp-esiRNAs from their precursor hairpin transcripts. The concentration of esi-1.2, which in vitro was assembled into Ago2 (Fig. 1B), declined >19-fold in ago2414 and ∼10-fold in r2d21 mutant flies (Fig. 3C; Supplemental Fig. S6), yet esi-2.1 and esi-1.1, which in vitro were loaded predominantly into Ago1 (Fig. 1B), decreased only threefold (esi-2.1) or twofold (esi-1.1) in ago2414 mutants (Fig. 3C; Supplemental Fig. S6). These data suggest that esi-1.2 is destroyed when it cannot be loaded into Ago2, whereas when esi-2.1 and esi1.1 cannot be loaded into Ago2, they accumulate bound to Ago1. The >30-fold decrease of esi-1.1 in the absence of R2D2 might reflect a role for R2D2 in dicing esi-1.1 from its precursor (see below).
The hp-esiRNAs that persist in ago2414 mutants were more likely to begin with U (77%), compared to those in wild-type Oregon R fly heads (49%) (Fig. 3D). Conversely, the hp-esiRNAs that persist in ago2414 mutants were less likely to begin with C (13% in ago2414 versus 28% in wild type) or A (4% in ago2414 versus 17% in wild type). The fraction of hp-esiRNAs starting with U (54%) also increased in mutant flies homozygous for r2d21 compared to their heterozygous siblings (42%), and the fraction beginning with C decreased (17% in r2d21 homozygotes versus 33% in heterozygotes). Thus, loss of the RNAi machinery causes not only a decrease in hp-esiRNAs, but also changes the hp-esiRNA repertoire from one that bears the hallmarks of Ago2-loaded small RNAs to a population with the characteristics of Ago1-bound small RNAs.
The RISC-loading complex loads Ago2 with small RNAs that begin with C
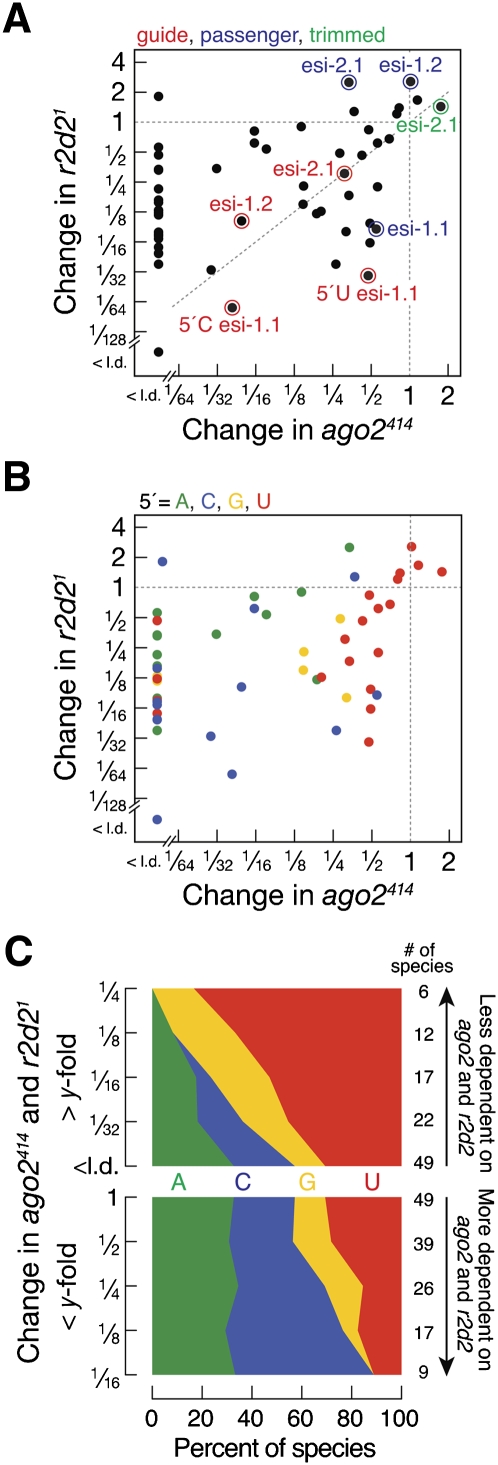
The Dicer-2/R2D2 heterodimer acts as a component of the RISC loading complex to load siRNA duplexes into Ago2. For the majority of hp-esiRNAs, R2D2 collaborates with Dicer-2 to load hp-esiRNAs into Ago2, but is dispensable for their cleavage from hairpin precursor RNAs (Marques et al. 2010). Consistent with this idea, the changes in hp-esiRNA abundance were well correlated between ago2414 and r2d21 (r = 0.47; p = 0.0002) (Fig. 4A), but not between dicer-2L811fsX and r2d21 (r = 0.12; p = 0.58) (Supplemental Fig. S8A). Consequently, abundant hp-esiRNAs such as esi-1.2 and esi-2.1, which are normally stabilized by their loading into an Argonaute protein, generally decreased in r2d21 mutants, whereas the corresponding passenger strands, which are normally destroyed during RISC maturation, accumulated (Fig. 4A). Small RNAs derived from the esi-2 locus, but not the esi-1 locus, correlated well between ago2 and r2d2 mutants (Supplemental Fig. S8B,C). For esi-1.1 both the guide and passenger strands decreased in r2d21 mutants, suggesting that R2D2 might act in its biogenesis (Fig. 4A). Nevertheless, hp-esiRNAs from the esi-1 locus decreased less in r2d2 mutants compared to ago2 mutants and their decrease in r2d2 mutants does not correlate well with dicer-2 mutants, which would be expected if R2D2 assisted Dicer-2 in the processing of esi-1 hairpins (Supplemental Fig. S8B).
FIGURE 4.
The RNAi loading complex loads hp-esiRNAs starting with C into Ago2. (A) Correlation analysis of the change in abundance of hp-esiRNAs in r2d21 mutants and ago2414 mutants (r = 0.47; P = 0.0002). The most abundant hp-esiRNA guide (red) and passenger strands (blue), as well as a trimmed species of esi-2.1 (green) are indicated. (B) Same data as in A, but with 5′ nucleotide identity indicated by color: green, 5′ A; blue, 5′ C; yellow, 5′ G; red, 5′ U. (C) The 5′ nucleotide composition of the hp-esiRNAs species in (B) ranked by the change in abundance (<y-fold, top panel; >y-fold, bottom panel) in ago2414 and r2d21. (<l.d.) Below the limit of detection.
To determine if the RISC loading complex plays an active role in selecting for specific 5′ nucleotides when loading a small RNA guide into Ago2, we analyzed the change in abundance of hp-esiRNAs in ago2414 and r2d21 mutants as a function of the 5′ nucleotide of the small RNAs (Fig. 4B,C). Since the small RNAs that increased in abundance in r2d21 mutants tend to be passenger strands (Fig. 4A), we analyzed only small RNAs that declined in r2d21. Hp-esiRNA species that decline in both ago2 and r2d2 mutants exhibited a mixed 5′ nucleotide composition (33% A, 24% C, 12% G and 31% U). Among those, the species that decreased most in r2d2 and ago2 mutants tend to start with C: 35% of hp-esiRNAs that decreased more than fourfold and 56% of those that decreased more than 16-fold in both mutants begin with C (Fig. 4C, bottom panel). These data suggest that the RISC-loading complex specifically loads into Ago2 those hp-esiRNAs that bear a 5′ C. We obtained similar results for Ago2-enriched miRNAs and miRNA* strands in ago2414, r2d21 and dicer-2L811fsX mutants (Figure S9).
In contrast, hp-esiRNAs that were less dependent on R2D2 and Ago2 tended to begin with U: 53% of species that decreased in both ago2414 and r2d21 mutants less than 16-fold and 83% of those that changed less than fourfold started with U (Fig. 4C, top panel). 5′ U small RNAs constitute 49% of Ago2-bound hp-esiRNAs in wild-type flies and 36% in S2 cells (Fig. 2C). The increase in 5′ U small RNAs in r2d21 and ago2414 mutants (Fig. 3D) is therefore likely a result of their ability to load into Ago1 (58% of small RNAs bound to Ago1 in fly heads and 42% in S2 cells start with U) (Fig. 2C), compensating partially for the loss of Ago2-bound small RNAs.
Esi-1.1 provides an example of 5′ nucleotide-dependent accumulation of small RNAs in Ago1 and Ago2: esi-1.1 was originally annotated as a 22-nt small RNA starting with a U (henceforth, 5′ U esi-1.1) (Czech et al. 2008), but also exists as an abundant 21-nt isoform, 5′ C esi-1.1, that begins with the second nucleotide of the annotated species (Supplemental Fig. S10). The two isoforms are differentially expressed in fly heads, ovaries, and S2 cells (Supplemental Fig. S10): 5′ C-esi-1.1 is more abundant in heads, whereas 5′ U esi-1.1 predominates in ovaries. In S2 cells, the two isoforms are equally abundant. Although the guide/passenger duplex structure of the two isoforms is essentially identical, they differentially partition between Ago1 and Ago2: 5′ U esi-1.1 assembled in vitro into both Ago1 and Ago2 and was bound in vivo to both Ago1 and Ago2 (Supplemental Fig. S10). In contrast, 5′ C esi-1.1 assembled in vitro almost exclusively into Ago2 and in vivo was only detected bound to Ago2 (Supplemental Fig. S10). Moreover, 5′ C esi-1.1 decreased 23-fold in ago2414 mutants, but 5′ U esi-1.1 decreased only twofold (Fig. 4A; Supplemental Fig. S11). The length of the two esi-1.1 isoforms likely contributes to their differential partitioning between Ago1 and Ago2, since in vitro, Ago1 favors 22 nt RNAs, while Ago2 favors 21 nt RNAs (Fig. 2).
Highly complementary target RNAs affect the stability of Ago1-bound hp-esiRNAs
Given the inherent preference for loading 22-nt RNAs into Ago1 and 21-nt RNAs into Ago2, we anticipated that the median hp-esiRNA length would increase in r2d21 and ago2414 mutant flies. This was not the case: 22- and 21-nt hp-esiRNAs decreased in both mutants (2.6- and 3.3-fold for 22-mers and 2.1- and 3.1-fold for 21-mers in r2d2 and ago2414 mutants, respectively) (Fig. 3A). Paradoxically, full-length hp-esiRNA isoforms (21 and 22 nt) decreased in ago2414 and r2d21 mutants, while the abundance of isoforms ≤20 nt generally was essentially unchanged (ago2414) or increased slightly (1.4-fold; r2d21), constituting >20% of all hp-esiRNAs reads in those mutants (Fig. 3A; Supplemental Fig. S12f). Such shorter species correspond to 3′ trimmed versions of full-length hp-esiRNAs.
In flies, a target mRNA that is highly complementary to a small RNA bound to Ago1 (but not Ago2) can trigger 3′-to-5′ exonucleolytic trimming and 3′ tailing of a small RNA (Ameres et al. 2010). Might the 3′ trimmed hp-esiRNAs we detected in ago2414 and r2d21 mutants be produced by 3′ tailing and trimming of Ago1-bound esiRNAs in response to complementary target mRNAs? In wild-type flies, such target-directed tailing and trimming could deplete the hp-esiRNA population of Ago1-bound small RNAs, explaining why Ago2-bound hp-esiRNAs are normally more abundant than those bound to Ago1 (Supplemental Fig. S7; Czech et al. 2008; Kawamura et al. 2008; Okamura et al. 2008b). In fact, esi-2.1, which loads into Ago1 in vitro but accumulates in Ago2 in vivo (Figs. 1C, 5C), is highly complementary to the mus308 mRNA (Supplemental Fig. S13; Czech et al. 2008; Okamura et al. 2008b). Perhaps esi-2.1 is initially loaded into both Ago1 and Ago2, but the portion of esi-2.1 loaded into Ago1 is then destroyed via the target-directed tailing and trimming pathway.
FIGURE 5.
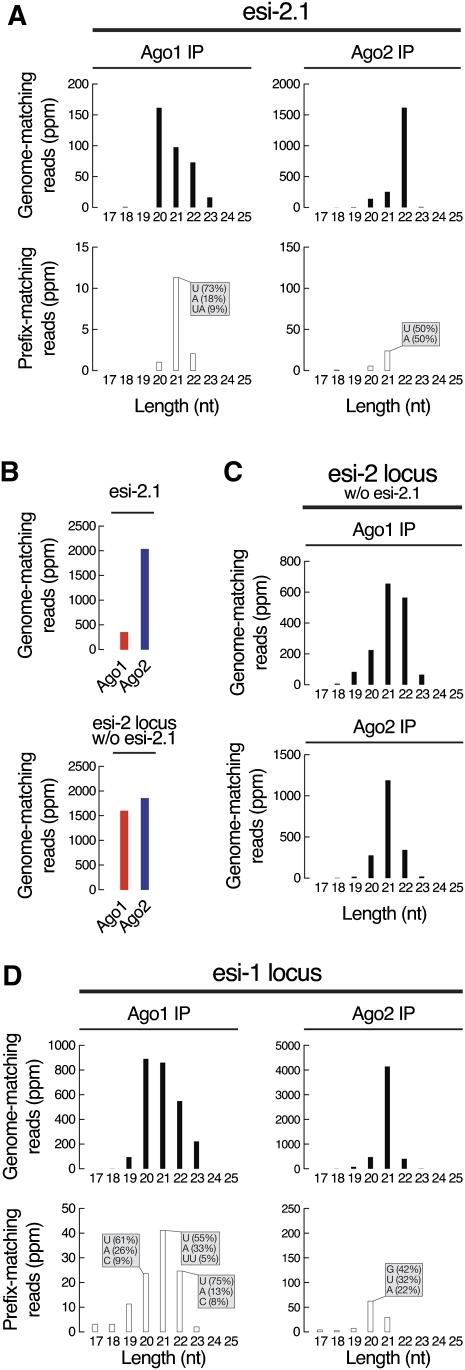
Ago1- but not Ago2-bound hp-esiRNAs are destabilized by highly complementary target RNAs. (A) Size distribution of genome-matching (top) and prefix-matching (bottom) esi-2.1 sequences associated with Ago1 (left) or Ago2 (right) in S2 cell extracts. The most abundant 3′ nontemplated, added nucleotides are noted. (B) The abundance of esi-2.1 (top) or all other small RNAs from the esi-2 locus (bottom) associated with Ago1 (red) or Ago2 (blue) in S2 cell extracts. (C) Size distribution of small RNAs, other than esi-2.1, derived from the esi-2 locus and associated with Ago1 (top) or Ago2 (bottom) in S2 cell extracts. (D) Size distribution of genome-matching (top) and prefix-matching (bottom) small RNAs from the esi-1 locus associated with Ago1 (left) or Ago2 (right) in S2 cell extracts. The most abundant 3′ nontemplated, added nucleotides are noted.
To test this idea, we compared the esi-2.1 isoforms in S2 cells that are bound to Ago1 with those that are bound to Ago2 (Fig. 5A, top row). Nearly all the Ago2-bound esi-2.1 was 22 nt, whereas the most abundant isoform of Ago1-bound ei-2.1 bound was 20 nt. In contrast, 90% of Ago1-bound miRNAs were at least 21 nt, 67% were 21 (32%) to 22 nt (35%), and <10% were 20 nt or shorter (data not shown). Target-directed trimming is accompanied by tailing, the nontemplated addition of U or A to the 3′ end of small RNAs bound to Ago1 (Ameres et al. 2010). If the shorter forms of Ago1-bound esi-2.1 reflect target-directed trimming, then we would expect to also detect tailed esi-2.1 species. Indeed, the fraction of esi-2.1 RNAs in Ago1 that had nontemplated nucleotides added to their 3′ ends was three times greater than those bound to Ago2 (Fig. 5A, bottom). The most abundant 3′ nontemplated nucleotide for Ago1-bound esi-2.1 was U (73%).
In S2 cells, esi-2.1 was approximately sixfold enriched in Ago2 versus Ago1, whereas all other esi-2-derived hp-esiRNAs accumulated equally in Ago1 and Ago2 (Fig. 5B). Unlike esi-2.1, the other Ago1-bound hp-esiRNAs derived from the esi-2 locus showed little evidence of trimming (Fig. 5C). The size distribution of these esi-2 species between Ago1 and Ago2 reflects the expected preference for 22-nt RNAs to load into Ago1 and 21-nt RNAs to load into Ago2 (Fig. 5C). We suggest that in S2 cells, the partitioning of esi-2.1 between Ago1 and Ago2 is purified by tailing and trimming directed by mus308 mRNA and possibly other highly complementary target RNAs (Supplemental Fig. S13), leading to the destruction of Ago1-bound esi-2.1. Because there are no mRNA targets with similarly high complementarity for the other hp-esiRNAs derived from the esi-2 locus, they accumulate in both Ago1 and Ago2. Fly heads express additional target mRNAs with considerable complementarity to esi-2.1: stat92E, gatA, and CG7359 (Supplemental Fig. S13B). Consistent with this observation, the amount of full-length esi-2.1 decreased in ago2414 and r2d21 mutants, while the abundance of 3′ trimmed and tailed esi-2.1 isoforms increased (Fig. 3C; Supplemental Figs. S12, S14).
Hp-esiRNAs derived from the esi-1 locus are complementary to the CG8289 mRNA (Supplemental Fig. S15; Czech et al. 2008; Okamura et al. 2008b), and, like esi-2.1, those esi-1 RNAs that were associated with Ago1, but not those bound to Ago2, were trimmed and contained 3′ nontemplated U's and A's (Fig. 5D).
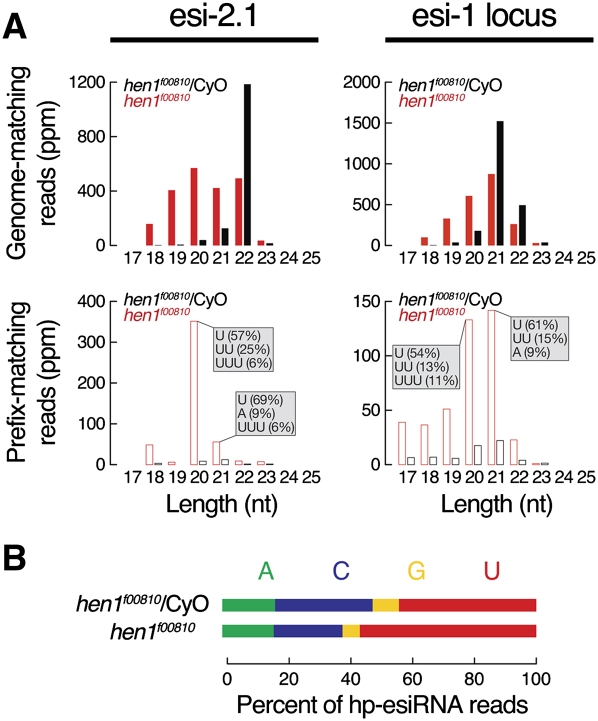
Unlike Ago1-loaded small RNAs, Ago2-loaded small RNAs are protected from tailing and trimming by a 2′-O-methyl group deposited on the 3′ end of the small RNA guide by the methyltransferase Hen1 (Horwich et al. 2007; Pelisson et al. 2007; Ameres et al. 2010). In hen1f00810 mutant fly heads, the abundance of full-length esi-2.1 and full-length (21–22 nt) hp-esiRNA species derived from the esi-1 locus each decreased approximately twofold; the abundance of tailed and trimmed hp-esiRNA 2.1 and hp-esiRNAs derived from the esi-1 locus each increased ∼17-fold and approximately fivefold, respectively, in the hen1 mutant (Fig. 6A). Additionally, loss of Hen1 changed the nature of the hp-esiRNA repertoire (Fig. 6B): the hp-esiRNAs that persisted in hen1f00810 mutants tended to start with U (56% versus 44% in hen1f00810 heterozygotes), whereas hp-esiRNAs that begin with C were reduced (22% versus 31% in hen1f00810 heterozygotes), as would be expected if Ago2-bound hp-esiRNAs are tailed and trimmed in the absence of Hen1.
FIGURE 6.
Hen1 stabilizes hp-esiRNAs. (A) The size distribution of genome-matching (top) and prefix-matching (bottom) sequences corresponding to esi-2.1 (left) or derived from the esi-1 locus (right) in the heads of hen1f00810 homozygous (red) or hen1f00810/CyO heterozygous (black) flies. The most abundant 3′ nontemplated, added nucleotides are noted. (B) The distribution of 5′ nucleotides among genome-matching hp-esiRNAs in hen1f00810 heterozygotes or homozygous mutant fly heads.
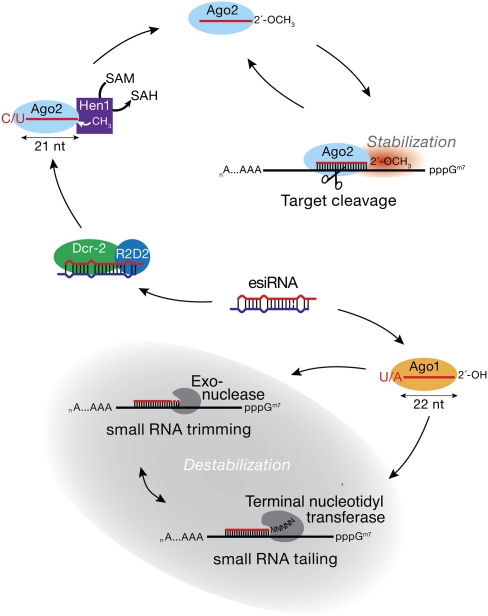
A model for target-directed purification of small RNA sorting in flies
Our data suggest that the sorting of hp-esiRNAs between Ago1 and Ago2 follows the previously established RISC assembly rules: hp-esiRNAs starting with U or A and containing mismatches in the central region of the small RNA duplex preferentially associate with Ago1, whereas those that begin with C or U and possessing a paired center are loaded into Ago2. We propose that hp-esiRNAs that begin with C are loaded into Ago2 by the RISC loading complex, but we do not yet know whether the bias toward C reflects a preference of the loading machinery, of Ago2, or of both. As the human homolog of Ago1 prefers to bind a 5′ U (Frank et al. 2010), we suspect that fly Ago1 intrinsically prefers small RNAs that start with U. Moreover, our results reinforce the view that in flies, small RNA production and Argonaute loading are not coupled. Just as miRNAs are made by Dicer-1 but can assort between Ago1 or Ago2 (Förstemann et al. 2007), so hp-esiRNA are made by Dicer-2 and can be loaded into either fly Argonaute protein (Czech et al. 2009).
The steady-state accumulation of individual small silencing RNA species in Ago1 and Ago2 reflects not only their initial sorting during the Argonaute-loading process, but also their subsequent purification by mRNA targeted tailing and trimming (Fig. 7): specific, single-stranded hp-esiRNA species, bound to Ago1, are destabilized when they bind to highly complementary targets. The result is that these hp-esiRNAs accumulate in Ago2, despite their initial loading into both Ago1 and Ago2.
FIGURE 7.
A model for the sorting of Drosophila hp-esiRNAs between Ago1 and Ago2 and their subsequent purification by target-directed tailing and trimming.
The mechanism underlying this purification is similar to that which leads to loss of specific miRNAs upon the artificial expression of a fully complementary target RNA or antisense oligonucleotide, which triggers 3′-to-5′ exonucleolytic trimming and 3′ tailing of the small RNA (Ameres et al. 2010). Ago2-bound hp-esiRNAs are protected from this process because they bear a protective 2′-O-methyl modification. Why does high complementarity between a target RNA and an Ago1-, but not an Ago2-bound, small RNA trigger small RNA destruction? Ago1 is a poor enzyme compared to Ago2 (Förstemann et al. 2007), and is therefore likely specialized to repress expression of partially complementary mRNAs through mechanisms that do not require endonucleolytic cleavage of the target (Iwasaki et al. 2009). Target-directed purification therefore might serve to ensure that highly complementary target RNAs are regulated only by Ago2, an enzyme adept at catalyzing multiple rounds of cleavage (Haley and Zamore 2004).
More puzzling still is why hp-esiRNAs are generated from partially complementary precursors that predispose them to load into Ago1 in the first place. While Ago2-bound hp-esiRNAs may generally repress high complementary target mRNAs, such as mus308, perhaps Ago1-bound hp-esiRNAs are required at a particular developmental stage or in a specific tissue to regulate less complementary target RNAs by a miRNA-like mechanism. Curiously, 5′ U esi-1.1, which is rare in S2 cells and fly heads, is more abundant than 5′ C esi-1.1 in ovaries, raising the possibility that esi-1.1, bound to Ago1 instead of Ago2, serves to regulate one or more mRNAs specific to egg production (Supplemental Fig. S10).
MATERIALS AND METHODS
General methods
Synthetic small RNAs (Dharmacon) were PAGE purified and phosphorylated using T4 polynucleotide kinase (New England Biolabs); for sequences of synthetic RNAs see Supplemental Table S2. Small RNA annealing, preparation of lysates from Drosophila melanogaster 0–2 h embryos, and RISC loading reactions were as previously described (Haley et al. 2003).
UV crosslinking analysis
UV crosslinking was as described previously (Ameres et al. 2010). Briefly, RNA duplexes (20 nM) were incubated with lysate in a standard RNAi reaction at 25°C (Haley et al. 2003). At the indicated times, the reaction was stopped by incubation on ice and then irradiated with 254 nM UV light for 5 min using a Stratalinker (Stratagene) with the sample ∼1 cm below the light bulbs. The photocrosslinked proteins were then resolved by 4%–20% gradient SDS-polyacrylamide gel electrophoresis (Criterion precast gels; Bio-Rad), dried, and exposed to storage phosphor screens (Fuji). Signal intensities were determined using ImageGauge V4.22 (Fuji). Relative crosslinking signals were determined by normalizing to the maximal crosslinking of a 5′ U guide to Ago1 for miRNAs and to a 5′ C guide to Ago2 for siRNAs. Crosslinking of 5′ U miRNA to Ago1 reached saturation after 30 min and 5′ C siRNAs to Ago2 after ∼15 min. Crosslinking of small RNAs to the respective Argonaute protein was normalized to total small RNA in the reaction as determined by 32P scintillation counting (LS6500 Multi-Purpose Scintillation Counter, Beckman Coulter).
Bioinformatic analyses
Small RNA libraries
Published small RNA libraries used in this study were Ago1 and Ago2 immunoprecipitated small RNAs (Czech et al. 2008), 19–24-nt small RNAs from S2 cells (Zhou et al. 2009), Ago1 immunoprecipitated small RNAs from Oregon R heads, oxidized 18–30-nt small RNAs from Oregon R heads, 18–30-nt small RNAs from heads of flies heterozygous or homozygous for dcr-2L811fsX, r2d21, ago2414 (Ghildiyal et al. 2010), and hen1f00810 (Ameres et al. 2010).
Extraction of sequences
Genome-matching sequences were extracted as described (Ghildiyal et al. 2010). Prefix matching reads were determined as described (Ameres et al. 2010); sequences with ambiguous base calls were excluded from the analysis.
Filtering and normalization of immunoprecipitation data
To determine the relative abundance of hp-esiRNAs in Ago1 and Ago2, we divided the hp-esiRNA reads in each immunoprecipitated or oxidized sample by a normalizing factor, computed with a subset of small RNAs that were sequenced at least once in the respective data set, as well as in the total RNA sample. To avoid ambiguity, we required these small RNAs to be detectable only in the total RNA and in the appropriate immunoprecipitated/oxidized sample and map uniquely to one location in the genome. The normalizing factor was set to the ratio of the total reads of these small RNAs in the appropriate immunoprecipitated/oxidized sample and their total reads in the total RNA sample.
5′ Nucleotide analysis
To compute the first nucleotide composition, hp-esiRNAs were weighted by their abundance.
Statistical analysis
Pearson correlation analysis was performed using Prism V5.0b (GraphPad Software Inc.).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank S. Ma and A. Boucher for help with fly husbandry and preparation of lysates, G. Farley for technical assistance, and members of the Zamore lab for advice, suggestions, and critical comments on the manuscript. This work was supported by an EMBO long-term fellowship (ALTF 522-2008) and an Erwin Schrödinger-Auslandsstipendium (J2832-B09) from the Austrian Science Fund FWF (S.L.A.) and by NIH grants GM62862 and GM65236 (P.D.Z.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2498411.
REFERENCES
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, Gordon A, Perrimon N, Hannon GJ 2009. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell 36: 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD 2005. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol 3: e236 doi: 10.1371/journal.pbio.0030236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD 2007. Drosophila microRNAs are sorted into functionally distinct Argonaute complexes after production by Dicer-1. Cell 130: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B 2010. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465: 818–822 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: an expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD 2010. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA 16: 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Zamore PD 2004. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol 11: 599–606 [DOI] [PubMed] [Google Scholar]
- Haley B, Tang G, Zamore PD 2003. In vitro analysis of RNA interference in Drosophila melanogaster. Methods 30: 330–336 [DOI] [PubMed] [Google Scholar]
- Hartig JV, Esslinger S, Bottcher R, Saito K, Förstemann K 2009. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J 28: 2932–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD 2007. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Hu HY, Yan Z, Xu Y, Hu H, Menzel C, Zhou YH, Chen W, Khaitovich P 2009. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics 10: 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, Zamore PD 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060 [DOI] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Balint É, Tuschl T, Zamore PD 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M 2010. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell 39: 282–291 [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Kawamata T, Tomari Y 2009. Drosophila Argonaute1 and Argonaute2 employ distinct mechanisms for translational repression. Mol Cell 34: 58–67 [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y 2010. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell 39: 292–299 [DOI] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q 2005. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev 19: 1674–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Seitz H, Tomari Y 2009. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol 16: 953–960 [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H 2008. Drosophila endogenous small RNAs bind to Argonaute2 in somatic cells. Nature 453: 793–797 [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81 [DOI] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925 [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang F, Kalidas S, Smith D, Liu Q 2006. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA 12: 1514–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ 2005. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434: 666–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Kim K, Wu PH, Alleyne TM, Jafari N, Carthew RW 2010. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat Struct Mol Biol 17: 24–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. 2008. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Takeuchi A, Siomi H, Siomi MC 2010. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat Struct Mol Biol 17: 1024–1026 [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC 2008. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC 2004. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev 18: 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC 2008a. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol 15: 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC 2008b. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453: 803–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Liu N, Lai EC 2009. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell 36: 431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JK, Liu X, Strauss TJ, McKearin DM, Liu Q 2007. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr Biol 17: 533–538 [DOI] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D 2005. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 434: 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A, Sarot E, Payen-Groschene G, Bucheton A 2007. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol 81: 1951–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham JW, Sontheimer EJ 2005. Molecular requirements for RNA-induced silencing complex assembly in the Drosophila RNA interference pathway. J Biol Chem 280: 39278–39283 [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi MC 2005. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol 3: e235 doi: 10.1371/journal.pbio.0030235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD 2004a. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 116: 831–841 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD 2004b. A protein sensor for siRNA asymmetry. Science 306: 1377–1380 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD 2007. Sorting of Drosophila small silencing RNAs. Cell 130: 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33 [DOI] [PubMed] [Google Scholar]
- Zhou R, Czech B, Brennecke J, Sachidanandam R, Wohlschlegel JA, Perrimon N, Hannon GJ 2009. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA 15: 1886–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]