Abstract
The glycine riboswitch has a tandem dual aptamer configuration, where each aptamer is a separate ligand-binding domain, but the aptamers function together to bind glycine cooperatively. We sought to understand the molecular basis of glycine riboswitch cooperativity by comparing sites of tertiary contacts in a series of cooperative and noncooperative glycine riboswitch mutants using hydroxyl radical footprinting, in-line probing, and native gel-shift studies. The results illustrate the importance of a direct or indirect interaction between the P3b hairpin of aptamer 2 and the P1 helix of aptamer 1 in cooperative glycine binding. Furthermore, our data support a model in which glycine binding is sequential; where the binding of glycine to the second aptamer allows tertiary interactions to be made that facilitate binding of a second glycine molecule to the first aptamer. These results provide insight into cooperative ligand binding in RNA macromolecules.
Keywords: riboswitch, glycine, cooperativity, aptamers
INTRODUCTION
Numerous structured RNA domains capable of binding target molecules have been identified in the 5′-untranslated regions of eubacterial and eukaryotic mRNA (Roth and Breaker 2009). These RNA domains, known as riboswitches, are components of a cis-acting genetic control module, which bind to a diverse array of metabolites and control the genes involved in the biosynthesis or degradation of these metabolites (Winkler and Breaker 2005). Riboswitches have also been found that bind second-messenger molecules to regulate fundamental cellular processes (Sudarsan et al. 2008). Structural changes within the riboswitch occur upon ligand binding, allowing the riboswitch to interact with an expression platform to modulate gene expression by effects on transcriptional or translational efficiency (Yarnell and Roberts 1999; Mandal and Breaker 2004).
More than 20 distinct classes of riboswitches have been identified to date. Riboswitches respond to a variety of cofactors used by protein enzymes, purines and their derivatives, magnesium cations, and amino acids (Roth and Breaker 2009). The importance of amino acids in protein biosynthesis and their catabolic use as an alternative energy source in bacteria make it necessary to control the level of amino acids in response to environmental and cellular changes (Serganov and Patel 2009). Three riboswitches have been identified to date that control amino acid concentrations in bacteria: the lysine, glycine, and S-adenosylmethionine (SAM)-responsive riboswitches (Grundy et al. 2003; Sudarsan et al. 2003; Winkler et al. 2003; Mandal et al. 2004). Two of these, the lysine and glycine riboswitches, can directly detect amino acids. While the structure of several SAM and lysine riboswitches has been determined (Fuchs et al. 2006; Montange and Batey 2006; Gilbert et al. 2008; Serganov et al. 2008), the structural basis of glycine riboswitch function is still not known. The glycine riboswitch was identified in a bioinformatic search for conserved sequences and secondary structural elements. The search found a widespread motif termed gcvT, upstream of bacterial genes expressing enzymes involved in the metabolism of glycine (Mandal et al. 2004). The motif is represented by two similar structural types (I and II) often found in a tandem arrangement, which have a common core flanked by different termini (Mandal et al. 2004; Sudarsan et al. 2006; Welz and Breaker 2007; Tripp et al. 2009). The secondary structure of the glycine riboswitch typically comprises two glycine-binding aptamers, followed by a single expression platform (Fig. 1). In the tandem arrangement, the binding of glycine takes place cooperatively (Mandal et al. 2004). Although riboswitches with tandem architectures have been reported, most of these tandem configurations are composed of two complete riboswitches including the expression platform, and they function independently rather than cooperatively (Sudarsan et al. 2006). The glycine riboswitch is the only known example that uses a cooperative binary system to bind its ligand. Furthermore, glycine is the smallest and simplest organic ligand known to be recognized by an RNA aptamer (Cromie et al. 2006). The tandem aptamer arrangement used to bind and recognize glycine raises many molecular and cellular questions as to how the riboswitch senses a small change in glycine concentration to achieve cooperative ligand binding that results in altered gene activation and repression.
FIGURE 1.
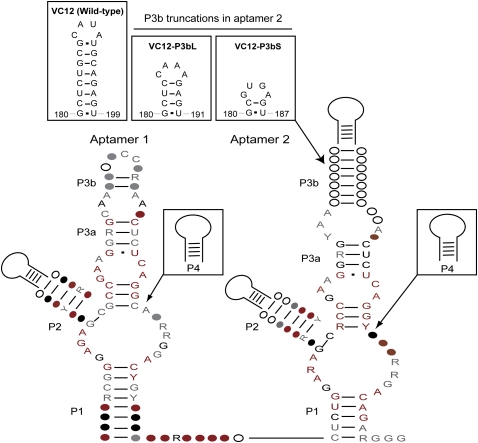
Secondary structure model of the glycine riboswitch. Consensus sequences were identified by bioinformatics. Conserved nucleotide positions in more than 50%, 75%, 90%, and 97% of the sequences are shown in white, gray, black, and red, respectively. Colored circles represent positions that are conserved and colored letters represent nucleotide identities that are conserved. Each aptamer has two conserved paired regions (P1 and P3) and two variable stem–loops (P2 and P4) whose sequence is not conserved. (Inset) Truncations of region P3b in Vibrio cholerae glycine riboswitch mutant constructs.
Cooperative binding is widely used in nature to enhance specific processes such as protein folding, polymerization, and ligand binding (Perutz 1989; Whitty 2008). Proteins that use cooperative activity have been identified in multisubunit enzymes and receptors (Bohr et al. 1904; Koshland and Hamadani 2002). Cooperativity can operate at many different levels, but in particular, allosteric cooperativity involves a “receptor” that contains two or more ligand-binding sites, in which ligand binding at one site affects the ligand affinity at the other site (Whitty 2008). The Hill coefficient provides a method for quantifying the degree of cooperativity of ligand binding to the macromolecule (Hill 1910). It describes the fraction of macromolecule saturated by ligand as a function of the ligand concentration. A Hill coefficient of 1 indicates completely independent binding, while a number greater than 1 indicates positive cooperativity (Hill 1910). Cooperativity is an economical way for the cell to sense and efficiently respond to stimuli across a narrow range of ligand concentrations (Forsen and Linse 1995).
Much of what we know about the glycine riboswitch has been obtained through biochemical approaches. Nucleotide analog interference mapping (NAIM) was used to investigate the chemical basis of glycine riboswitch cooperativity. This provided the framework for making and testing riboswitch mutants with altered cooperativity (Kwon and Strobel 2008). Hydroxyl radical footprinting and small-angle X-ray scattering (SAXS) have provided information on the structural transitions that occur upon riboswitch folding and ligand binding (Lipfert et al. 2007). In the present study we set out to understand the molecular basis for glycine riboswitch cooperativity using a series of mutants with altered cooperativity and glycine affinity. We used hydroxyl radical footprinting to compare the differences in solvent accessibility between the wild-type and mutant constructs. We further tested the ability of each individual aptamer to bind to glycine and used native gel-shift analysis to monitor the interactions between the aptamers. The results define an asymmetric tertiary interaction between the aptamers that mediates cooperative ligand binding.
RESULTS
Determining glycine riboswitch mutants with altered glycine affinity and cooperativity
NAIM studies identified several asymmetric interference sites distributed between aptamers 1 and 2 of the glycine riboswitch from Vibrio cholerae (VC) (Kwon and Strobel 2008). These interference sites provided good candidates for residues potentially involved in asymmetric cooperative interactions between the two aptamers. It is likely that each aptamer uses different functional groups to facilitate interaptamer contacts and similar functional groups to bind glycine or make contacts within the aptamer. For this study we focused on the asymmetric interference in region P1 of aptamer 1. That the P1 helix is potentially essential for riboswitch cooperativity draws parallels to the general role of the P1 helix in riboswitch function. In many different riboswitches, the modulation of the P1 helix is a key conformational switch that affects gene expression (Winkler et al. 2002, 2003; Mandal et al. 2003; Sudarsan et al. 2003; Smith et al. 2009). There are two P1 helices in the case of the tandem glycine riboswitch. Based on phylogenetic data, nucleotides on the 3′ side of the P1 helix of aptamer 2 are complementary to the sequence in the downstream expression platform (Mandal et al. 2004). Moreover, there appears to be no alternate region of complementarity to the P1 stem of aptamer 1, suggesting that this region must serve a different role in glycine riboswitch function.
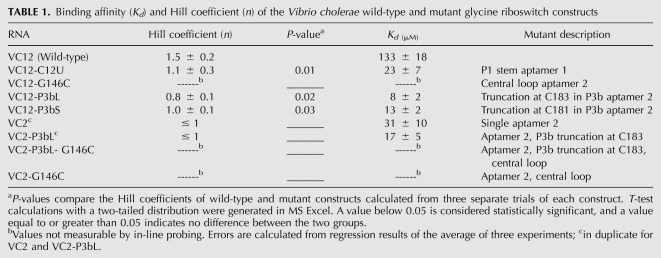
The asymmetric interference pattern suggested that the minor groove of P1 in aptamer 1 is a site of tertiary interaction facilitating cooperativity between the aptamers (Kwon and Strobel 2008). We therefore introduced a mutation to this region and measured the binding affinity and cooperativity using in-line probing to obtain the apparent Kd and Hill coefficient (n), respectively (Table 1; Regulski and Breaker 2008). NAIM experiments showed N-methyl G interference at G127, which suggested that the exocyclic amine of the nucleotide could be involved in minor groove hydrogen-bonding interactions (Rife et al. 1998; Kwon and Strobel 2008). When C12 was mutated to a U to produce a U•G wobble pair, the riboswitch also showed a loss in cooperativity (n = 1.1) compared with wild-type (n = 1.5) (Table 1). Two additional mutations were previously reported. Mutation of G127 to A, which removes the minor groove exocyclic amine, resulted in a loss of cooperativity (n = 1.1) (Kwon and Strobel 2008). To determine that the identity of the base pair was specific to aptamer 1, a C-G to U-A mutation was introduced to the P1 stem of aptamer 2. The U143-A222 mutation did not affect glycine affinity or cooperativity (Kwon and Strobel 2008). Consistent with the previous conclusion that the minor groove edge of the P1 stem of aptamer 1 is important for riboswitch cooperativity, and given that the identity of the putative tertiary interaction partner had not been determined, we set out to further characterize the C12U mutant.
TABLE 1.
Binding affinity (Kd) and Hill coefficient (n) of the Vibrio cholerae wild-type and mutant glycine riboswitch constructs
Mapping tertiary interactions involved in riboswitch cooperativity
We selected two mutants that lost cooperativity to examine whether tertiary interactions present in a fully cooperative wild-type riboswitch were missing in the mutants. To distinguish tertiary interactions involved in glycine binding from those interactions that give rise to cooperativity, we compared the wild-type riboswitch with the VC12-C12U mutant, a noncooperative riboswitch containing both aptamers, and a previously identified 105-nt RNA (VC2) that contains only the second aptamer and displays noncooperative behavior (n = 1.0 and Kd = 22 μM) (Mandal et al. 2004). Hydroxyl radical footprinting was used to monitor the folding of the riboswitch in the absence and presence of glycine. This method provides information on solvent accessibility of each nucleotide and defines regions of close packing within the RNA tertiary structure (Tullius and Greenbaum 2005). The expectation of the experiment is that differences in hydroxyl radical footprinting protection patterns between cooperative and noncooperative riboswitch RNAs would identify regions whose packing interactions change upon cooperative ligand binding.
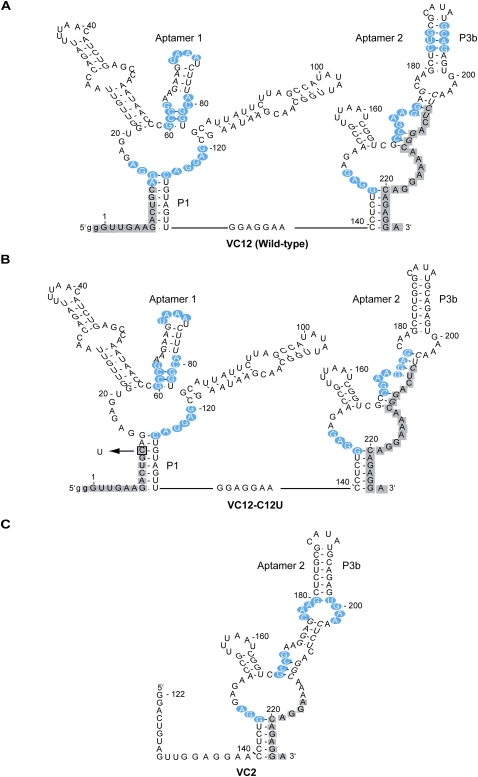
We generated protection maps of three constructs: VC12-wild-type, VC12-C12U, and VC2 (Fig. 2). Wild-type and VC12-C12U maps show the same areas of protection in the central loop and in P3/P3a of both aptamers, consistent with in-line probing (Mandal et al. 2004) and NAIM data (Kwon and Strobel 2008). VC2 also shows similar protection in the central loop and P3a hairpin. A noticeable difference in the protection pattern between the wild-type and VC12-C12U RNAs was observed in the P1 region of aptamer 1 and the P3b region of aptamer 2 (Supplemental Fig. S1). In both cases these regions were protected in the wild-type RNA (Fig. 2A), but became accessible in the C12U point mutant lacking cooperative glycine binding (Fig. 2B). Region P3b of aptamer 2 was also solvent accessible in the noncooperative construct VC2 (Fig. 2C). The loss of protection in the P1 region of aptamer 1 is potentially consistent with the loss of a tertiary contact to the minor groove edge of this helix as predicted by NAIM and mutagenesis data. That the accessibility of this region is accompanied by the loss of protection in P3b of aptamer 2 suggests that these two sites might interact with each other.
FIGURE 2.
Sites of hydroxyl radical footprinting probed from nucleotides 13–205. Depicted in blue are positions of nucleotides protected from hydroxyl radical activity. Positions that could not be quantified are depicted in gray. (A) Regions of increasing protection in double aptamer wild-type glycine riboswitch construct. (B) Protection pattern of noncooperative riboswitch mutant C12U (nucleotide change indicated by box). (C) Protection pattern of single aptamer 2 construct (VC2).
Testing cooperative interactions in the P3b stem of aptamer 2
The lack of protection in P3b of aptamer 2 in the noncooperative mutants prompted us to investigate this region further. If the P3b hairpin is important in tertiary interactions that dictate riboswitch cooperativity, eliminating or modifying this hairpin should result in a riboswitch that contains both aptamers but does not function cooperatively. We designed a series of RNAs with modifications in the P3b hairpin to test the affects of mutations on cooperativity.
We performed in-line probing on two modified riboswitch constructs with truncations in P3b of aptamer 2, capped with two GNRA tetraloop motifs. The different tetraloops were chosen to ensure effects were not dependent upon the nature of the tetraloops. VC12-P3bL is truncated at position C183 and capped with a stable GAAA loop. VC12-P3bS is truncated at position C181 and capped with a stable GUGA loop (Fig. 1, inset). Both VC12-P3bL and VC12-P3bS showed a complete loss of cooperativity (VC12-P3bL: n = 0.8, VC12-P3bS: n = 1.0) (Table 1). Thus, the same region in aptamer 2 that showed loss of solvent protection upon mutation in P1 of aptamer 1 resulted in a loss of cooperativity when mutated. These results provide evidence that the P3b hairpin in aptamer 2 participates in glycine riboswitch cooperativity.
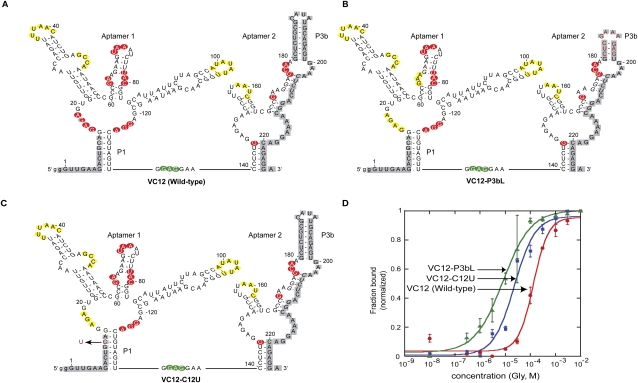
We mapped the modulation patterns obtained from in-line probing gels onto the secondary structure of the riboswitch and noticed striking differences in the cleavage patterns at the central loop of aptamer 1 between wild-type, VC12-C12U, and VC12-P3bL mutant riboswitches (Fig. 3; Supplemental Fig. S2). Mandal et al. (2004) previously showed that mutations to a conserved nucleotide in the central loop of either aptamer caused substantial loss of glycine-binding affinity to the mutated aptamer. This was determined from a decrease in cleavage upon addition of glycine in an in-line probing experiment. There was decreased cleavage in both central loops of the wild-type riboswitch, indicating changes in both aptamers when glycine is bound. However, the noncooperative mutants VC12-C12U and VC12-P3bL showed unmodulated cleavage in the central loop of aptamer 1 upon glycine addition (nt 15–18), while the central loop of aptamer 2 continued to show a pattern consistent with glycine binding. As a result, the Hill coefficient and Kd calculated for the noncooperative mutants were obtained from quantification of the modulation patterns seen in aptamer 2 (Fig. 3D). Together, these results suggest that region P3b of aptamer 2 is important for making cooperative tertiary interactions with aptamer 1, and mutation in region P3b of aptamer 2 causes a loss of glycine binding to aptamer 1.
FIGURE 3.
Sequence, secondary structure, and modulation of VC12 wild-type (A), VC12-P3bL mutant (B), and VC12-C12U mutant constructs as determined by in-line probing experiments (C) (probed from nucleotides 14–180; positions not probed indicated in gray). Cleavage modulations are indicated by yellow (unmodulated cleavage), red (decreasing cleavage), and green (increasing cleavage) residues. (D) Plots of normalized fraction of RNA bound versus glycine concentration derived from in-line probing assays. Depicted is the best fit curve as calculated from the quantified data of the bands G122–G124 for wild type and G146 for VC12-C12U and VC12-P3bL. Error bars represent standard errors from triplicate experiments. Calculated values of the binding affinity (Kd) and cooperativity (Hill coefficient, n) are shown in Table 1. The x-axis is depicted on a log scale.
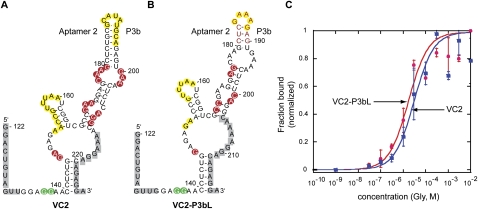
Glycine binding to isolated aptamer constructs
Several single aptamer mutants were designed to investigate whether isolated aptamers are capable of binding glycine independently. Previous studies have shown that aptamer 2 (VC2) can bind glycine and that this binding is not cooperative (Mandal et al. 2004). Along this line, we made a VC2 mutant with a truncation in P3b (VC2-P3bL) and tested whether glycine can still bind to aptamer 2 in the absence of P3b. The resulting mutant was capable of binding glycine with a Kd of 17 μM (Table 1;Fig. 4C; Supplemental Fig. S3) comparable to a Kd of 31 μM in VC2. This result indicates that while P3b is important for the cooperativity seen in the double aptamer riboswitch, it is not important for glycine binding in the context of the single aptamer riboswitch. We observed two major differences in the in-line probing patterns of constructs VC2 and VC2-P3bL, the first difference is relative to the full-length RNA. There is no modulation in P3b in both constructs. This suggests that if P3b is involved in a direct tertiary interaction with aptamer 1 that leads to cooperativity, these contacts are no longer made because the interacting partner in aptamer 1 is absent (Fig. 4A,B). The second difference is in the cleavage pattern of the internal loop in P3a (nt 177–180, 200–202) between the two constructs. Where there is decreased cleavage in the internal loop of P3a in VC2, most of the modulation is missing in VC2-P3bL. P3b may be important in the folding of the P3 stem, and if nucleotides other than those in P3b are involved in cooperative interactions with aptamer 1, a mutation in P3b would result in aptamer 2 sampling alternate conformations incapable of making correct tertiary interactions.
FIGURE 4.
Sequence, secondary structure, and modulation of VC2 (A) and VC2-P3bL constructs as determined by in-line probing (B) (probed from nucleotides 134–212 in VC2 and 134–204 in VC2-P3bL, positions not probed indicated in gray). Cleavage modulations are indicated by yellow (unmodulated cleavage), red (decreasing cleavage), and green (increasing cleavage) residues. (C) Plots of normalized fraction of RNA bound versus glycine concentration derived from in-line probing assays. Depicted is the best-fit curve calculated from the quantified data of the bands G146. Error bars represent standard deviations from duplicate experiments. Calculated values of the binding affinity (Kd) are shown in Table 1. The x-axis is depicted on a log scale.
Since we have established that aptamer 2 is capable of independent glycine binding, the question becomes what elements are necessary for isolated aptamer 1 to bind glycine? We made two aptamer 1 constructs that varied in the length of the 3′-end to test whether aptamer 1 can bind glycine independently. VC1s contained nucleotides 1–139 of the riboswitch and VC1 contained nucleotides 1–150. Interestingly, neither construct showed glycine-dependent modulation by in-line probing or hydroxyl radical footprinting experiments (Supplemental Fig. S4). This suggests that unlike aptamer 2, aptamer 1 is not capable of independent glycine binding, or glycine binding is so poor that it is not measurable by in-line probing. We assessed the importance of the linker region by placing the linker at the 5′-end to see whether this was sufficient for aptamer 1 to bind glycine. There was no modulation in the in-line probing of VC1-Lnk, indicating that placement of the linker at the 5′-end was not sufficient for glycine binding to aptamer 1 (Supplemental Fig. S4). We performed magnesium-dependent hydroxyl radical footprinting in the presence and absence of 10 mM glycine to monitor changes in folding. In the absence of glycine, footprinting gels show patterns of protection indicating that aptamer 1 is partially folded, but no additional protections were observed upon addition of glycine (Supplemental Fig. S5). We speculate that there is an element contained in aptamer 2 needed for proper folding of aptamer 1.
Native gel-shift analysis of aptamer interaction in trans
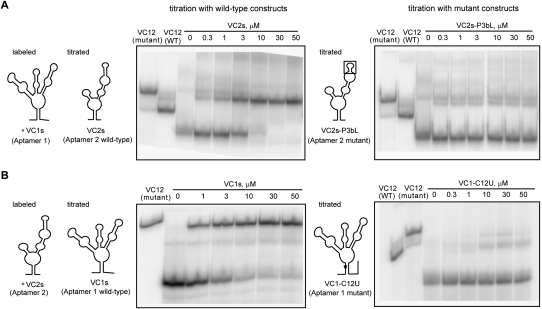
We next assessed whether the individual aptamers could interact in trans when all of the components necessary for interaction were present. If there is an element in aptamer 2 that is needed for interaction with aptamer 1, mutation to one of the aptamers would affect the interaction between them. We carried out a native gel-electrophoresis experiment based on a polyacrylamide coelectrophoresis (PACE) method (Cilley and Williamson 1999; Kwon and Strobel 2008) to measure the association between the two aptamers when they are in trans to each other. We used the wild-type riboswitch (VC12-WT) and a mutant riboswitch with a G-to-C point mutation (VC12-G146C), which abolishes glycine binding, as a size marker and measured the compactness of a full-length riboswitch. To monitor the mobility of the individual aptamers as a complex, we 5′-end labeled aptamer 1 and incubated with 5 mM glycine, and added unlabeled aptamer 2 across a range of concentrations. If aptamer 1 and aptamer 2 interact in trans, there would be an upward shift in mobility of labeled aptamer 1 such that the RNA would have approximately the same mobility as a full-length riboswitch. Moreover, the mobility of the RNA would fall in-between VC12-WT and VC12-G146C, depending on the compactness of the complex. We performed a concentration-dependent gel shift and were able to measure the affinity of association between aptamer 1 and aptamer 2. The interaction between VC1s and VC2s, which reconstitutes a full-length riboswitch, showed a complete shift in the range tested (Fig. 5A) with a Kd for aptamer interaction of 3 μM. Interestingly, the complex showed a slight degree of change in compactness as the concentration of VC2s increased and migrated closer to the VC12-WT marker.
FIGURE 5.
(A) Native gel-shift experiment of labeled VC1s titrated with VC2s (Aptamer 2 wild-type) and VC2s-P3bL (Aptamer 2 mutant) in 10 mM MgCl2 and 5 mM glycine. A picture schematic of the construct used in the titration is depicted to the left of each corresponding gel. (B) Native gel-shift experiment of labeled VC2s titrated with VC1s (Aptamer 1 wild type) and VC1-C12U (Aptamer 1 mutant).
We next investigated whether the association between the two aptamers is dependent upon the P1 region of aptamer 1 and the P3b region of aptamer 2 using this direct aptamer interaction assay. We made a P3b truncation in aptamer 2 and assayed the interaction of the mutant with aptamer 1. Results showed that the affinity of association for mutant VC2s-P3bL with VC1s could not be measured (Fig. 5A). When P1 was mutated at position C12 to a U, we saw a partial shift that was also not concentration dependent (Fig. 5B). We predict the partial shift results from other weak tertiary contacts between the aptamers. The results are consistent with the expectation that the P3b region of aptamer 2 and P1 region of aptamer 1 are in part responsible for forming a tertiary interaction that contributes to cooperative binding. As controls, we labeled VC1s and incubated with glycine and fixed concentrations of aptamer 2 variants that either contained a point mutation that abolishes glycine binding or that are truncated in P3b. A complete shift in mobility was observed only in the presence of VC2s and VC2, constructs where aptamer 2 was unaltered, while variants with a mutation in G146C or P3b showed only a partial shift (Supplemental Fig. S6).
DISCUSSION
We hypothesize that P1 and P3b are involved in a long-range tertiary interaction that facilitates communication between the two tandem aptamers. Kwon and Strobel previously reported that the P1 helix in aptamer 1 provided tertiary interactions important for glycine riboswitch cooperativity, but an attempt to identify the interacting partner in aptamer 2 using NAIM was unsuccessful. In the present study, we used hydroxyl radical footprinting to compare the wild-type riboswitch with two noncooperative mutants, and identified the P3b hairpin of aptamer 2 as the solvent-accessible region in the mutants. Mutagenesis studies confirmed that P3b of aptamer 2 is crucial for cooperativity. Additionally, footprinting data of VC12-C12U showed that P1 protection is lost when P3b is solvent accessible, whereas both regions are protected in the wild-type construct. Consistent with this prediction, we demonstrated by native gel-shift experiments that the isolated aptamers can interact in trans to form a complex with the same mobility as the tandem aptamer construct. However, when truncations or mutations are made in the P3b or P1 regions of the isolated aptamers, the in trans complex is disrupted.
The nature of the tertiary interaction between P1 and P3b may be direct or indirect. The NAIM interference pattern suggests that interaction with P1 involves contacts to the minor groove, given that interference was observed with 2′-deoxy A at A13 and N-methyl G at G127, two probes of minor groove tertiary interaction (Ortoleva-Donnelly et al. 1998; Kwon and Strobel 2008). The minor groove is also implicated in P3b, where a C194U point mutation converted the G-C to a G•U wobble pair and resulted in a loss of cooperativity between the aptamers (data not shown). Although the details of the interaction cannot be defined from these experiments, they suggest that there may be helical packing between the minor groove edges of aptamer 1 P1 and aptamer 2 P3b. Such helix–helix interactions along the minor groove have been seen in the structure of T. thermophilus 23S rRNA and characterized as ribo–base interactions and as the Along Groove Packing Motif (AGPM) (Gagnon and Steinberg 2002; Gagnon et al. 2006; Laing et al. 2009). Alternatively, an interaction that indirectly involves P3b could be facilitated through correct folding of the P3 stem of aptamer 2 such that nucleotides in the loop right below P3b make contact with P1. It is possible that the P3b mutations affect the positioning of the P3a loop of aptamer 2, resulting in a conformation that is incapable of making cooperative tertiary contacts with aptamer 1.
Given the hypothesis that region P3b in aptamer 2 is required for cooperativity, we asked whether this region was a conserved feature found in all glycine riboswitches. An examination of the phylogenetic data showed that although the nucleotide sequence is not always present in aptamer 2, P3b is found in at least one of the two tandem aptamers (Fig. 1). If P3b is not a fully conserved feature found only in aptamer 2, what does this mean for cooperativity and how do we resolve the phylogentic data? One possibility is that in those cases where P3b is located in aptamer 1, it plays the same role as it does in aptamer 2 for the V. cholerae version of the riboswitch. Another possibility is that although glycine riboswitches are found in the tandem aptamer arrangement, in the absence of P3b, the aptamers may not be cooperative. Cooperativity of four glycine riboswitches has been reported (Mandal et al. 2004; Kwon and Strobel 2008; Tripp et al. 2009). The riboswitch from V. cholerae most closely matches the consensus secondary structure with an extended P3b helix (Fig. 1, inset). It has a Hill coefficient of 1.7, comparable to a reported Hill coefficient of 1.6 (Mandal et al. 2004; Kwon and Strobel 2008). The glycine riboswitch from Fusobacterium nucleatum and Bacillus subtilis both have shorter P3b stems, and they both exhibited a slightly smaller Hill coefficient of 1.4 (Mandal et al. 2004; Kwon and Strobel 2008). The riboswitch from Candidatus Pelagibacter ubique, which lacks a P3b in aptamer 2, displayed a Hill coefficient of 1.1 (Tripp et al. 2009). Although these examples are limited in number, they do establish that not all glycine riboswitches are cooperative, and there appears to be a correlation between the degree of cooperativity and the presence of the P3b helix in one of the aptamers.
The data suggest that glycine binding is sequential. The second aptamer is the first to bind glycine, which results in a tertiary interaction that promotes glycine binding to the first aptamer. Several independent experiments support this observation. First, the isolation of aptamer 1 with various 3′ end lengths and modified placement of the linker region is not sufficient for glycine binding in aptamer 1. In the absence of glycine, protection patterns of the isolated aptamer 1 correspond to published data of aptamer 1 in the context of the wild-type riboswitch (Lipfert et al. 2007). Upon addition of glycine, the central loop and P1 helix of the wild-type tandem riboswitch become protected (Fig. 2A), whereas these regions are not protected in the isolated aptamer 1 construct (Supplemental Fig. S5). Second, point mutations in the central loop of either aptamer have differential effects on glycine binding in the unmutated aptamer. When G146 in aptamer 2 is mutated, in-line probing data show no modulation in the cleavage pattern at either central loop (Mandal et al. 2004). When G17 in aptamer 1 is mutated, modulation is seen in regions of aptamer 2, suggesting that aptamer 2 is still responsive to glycine (Mandal et al. 2004). Modulation was also seen in regions of aptamer 2 in a G17 mutant with a truncation in P3b (Supplemental Fig. S7). Taken together, these results suggest that glycine binding in aptamer 2 is independent of aptamer 1, and aptamer 2 influences glycine binding to aptamer 1. Third, an isolated aptamer 2 in the context of a full or truncated P3b hairpin is still able to bind glycine as determined by in-line probing. Hence, P3b mutations made in isolated aptamer 2 or full-length constructs have no affect on aptamer 2 glycine affinity, but effectively eliminate glycine binding to aptamer 1. We observed little modulation in the cleavage patterns of the central loop of aptamer 1 in tandem aptamer constructs that are noncooperative. However, modulation was observed and quantifiable in the central loop of aptamer 2 by in-line probing. Therefore, for the noncooperative mutants (VC12-C12U and VC12-P3bL) glycine binding only occurs in the second aptamer.
We observed structural transitions in our native gel-shift studies that also support the hypothesis that aptamer 2 binds glycine prior to glycine binding by aptamer 1. On a native gel with millimolar concentrations of Mg2+ and no amino acid present, the wild-type riboswitch is more compact, and as a result moves more quickly through the gel compared with VC12-G146C, a mutant unable to bind glycine. In the presence of glycine, there is a further downward shift in the wild-type riboswitch. Similarly, Lipfert et al. (2007) observed by SAXS analysis that in the presence of millimolar concentrations of Mg2+ and in the absence of glycine there is a significant compaction and partial folding of the riboswitch. When glycine is present there is further compaction of the riboswitch. The evidence points to the possibility that folding may occur preferentially in one aptamer to allow glycine binding. However, when glycine binding is disrupted as in the VC12-G146C mutant, folding is also interrupted, as evidenced by a slower-moving complex on a native gel. We were unable to detect a mobility shift in VC2 upon glycine binding by the PACE method because there is no further compaction in the structure of VC2 upon glycine binding. This would suggest that aptamer 2 exists in a conformation that is ready to bind glycine, or the PACE gel-shift assay is not sensitive enough to pick up small conformational changes.
A mechanism for cooperative glycine binding
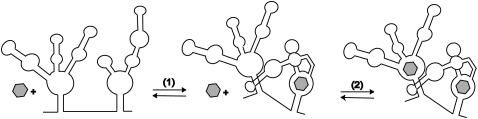
We propose a model based on these experimental observations to partially explain the cooperative nature of the glycine riboswitch. In the V. cholerae riboswitch, glycine first binds to aptamer 2, which results in a conformational change in the RNA. This allows region P3b of aptamer 2 to directly (Fig. 6) or indirectly interact with the P1 helix of aptamer 1, and promotes glycine binding to aptamer 1.
FIGURE 6.
Proposed mechanism of glycine binding and cooperativity in the glycine riboswitch. This version of the mechanism depicts a direct interaction between P3b of aptamer 2 and P1 of aptamer 1, but an indirect interaction is also consistent with the data. Glycine binding (gray hexagon) is shown schematically in the central loop, though the exact position of the binding site is unknown. Glycine binds sequentially to one aptamer of the riboswitch. This causes a subsequent change in conformation (1), leading to tertiary contacts between the two aptamers, opening up the glycine binding site (2) in the other aptamer, where a second glycine molecule can bind. Additional tertiary contacts are made allowing the riboswitch to interact with the expression platform.
Currently recognized glycine riboswitches are “on” switches for genes involved in glycine cleavage systems and symporters. It is possible that the genetic switch is made through P1 helix formation in either aptamer 1 or 2, such as observed in several other riboswitch systems (Winkler et al. 2002, 2003; Mandal et al. 2003; Sudarsan et al. 2003; Smith et al. 2009). However, unlike other examples, the P1 regions of the two aptamers do not show significant glycine-dependent structural modulation, at least in the context of the isolated riboswitch. Alternatively, the switch could involve nucleotides outside of the P1 helix, such as those in either of the central loop regions or the P3a loops, which become inaccessible upon glycine binding.
A conformational mechanism of cooperativity is plausible in comparison to the cooperative system of O2 binding to hemoglobin. In hemoglobin, the T (unbound)-to-R (bound) transition occurs when one O2 molecule binds to the heme group, causing a conformational change that displaces the F helix, inducing a shift of the intersubunit contacts that stabilize the high-affinity O2 state (Turner et al. 1992; Royer et al. 2001). The binding of a ligand at one site that causes subsequent conformational changes that affect ligand binding at another site is a fundamental mechanism inclusive of other allosteric proteins (Copeland, 2000) and in the design of allosteric ribozymes (Koizumi et al. 1999; Jose et al. 2001). Our data provide a partial view of the interactions between the aptamers that facilitate cooperativity in this novel RNA system.
MATERIALS AND METHODS
DNA oligonucleotides and chemicals
DNA oligonucleotides were synthesized by the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University and used without further purification. Glycine and other chemicals were obtained from Sigma.
DNA constructs
The Vibrio cholera VC1422 glycine riboswitch (VC12-WT) and single aptamer glycine riboswitch (VC1, VC2) constructs were made by annealing synthetic oligonucleotides and amplified by PCR. The oligonucleotides were designed to contain the T7 promoter sequence, riboswitch DNA sequence, followed by the antigenomic HDV ribozyme sequence, and flanked by restriction sites to facilitate cloning into the pUC19 (NEB) plasmid. The sequence of the cloned plasmid DNA was confirmed by DNA sequencing. The resulting plasmid DNA was used as a template for site-directed mutagenesis and cloning of modified riboswitch constructs. Mutant riboswitch constructs were made by PCR reaction using corresponding primers.
In vitro transcription
Plasmid DNA encoding the glycine riboswitch was linearized by restriction digest and used as template for transcription by T7 RNA polymerase. RNAs were transcribed in 40 mM Tris-HCL (pH 7.5), 4 mM spermidine, 10 mM DTT, 55 mM MgCl2, 0.05% Triton X-100, and 6 mM of each 5′-nucleotide triphosphate for 2.5 h at 37°C. All RNA were purified by 6% PAGE, eluted into 0.3 mM NaOAc (pH 5.2), precipitated with ethanol, and resuspended in water. RNA concentrations were determined by UV absorbance at 260 nm.
In-line probing assay and Hill plot fitting
RNA transcripts were dephosphorylated and 5′-32P labeled as previously described (Ryder and Strobel 1999). For each in-line probing reaction, ∼20 nM of end-labeled RNA was incubated at 23°C for 48 h in 50 mM Tris-HCl (pH 8.3), 20 mM MgCl2, and 100 mM KCl in the presence or absence of ligand. Spontaneously cleaved RNA products were separated by 6% denaturing PAGE and visualized by PhosphorImager (Molecular Dynamics). Individual band intensities were quantified using ImageQuaNT software. For VC12-WT and VC12 mutant constructs, four different regions in the aptamers were quantified: U70-C80 (aptamer 1), G122-A124 (aptamer 1), G134-G136 (linker), and G146 (aptamer 2). For VC2 constructs, regions G146, G170, and G176-A178 were quantified. To control for loading differences, the band intensities were normalized to nonvariable bands (U94 and G200). The Hill coefficient and dissociation constant were calculated by fitting each plot using a four-parameter cooperative Kd function (θ = m1([L]n/[L]n + KAn) + m2) in KaleidaGraph 3.51 software.
Hydroxyl radical footprinting and mapping
The 5′-end-labeled RNA (∼106 counts per minute) was dissolved in 50 mM Na-MOPS (pH 7.0) and 10 mM MgCl2 with different concentrations of glycine and equilibrated for 20 min at 50°C. The footprinting Fenton reaction (Pogozelski et al. 1995) was carried out with final concentrations of 0.1 mM Fe(NH4)2(SO4)2, 0.062 mM EDTA, and 5 mM DTT for 1 h at 23°C. The reaction was quenched with 10 mM thiourea, ethanol precipitated, and dissolved in 2x urea loading buffer (0.05% BPB, xylene cyanol, 50 mM EDTA, 8 M urea). Reaction products were separated on an 8% denaturing polyacrylamide gel for varied lengths of time to resolve the different regions of the riboswitch and visualized by PhosphorImager. Regions that showed change in cleavage were mapped onto the secondary structure of the riboswitch as areas of decreased solvent exposure and protection from hydroxyl radical cleavage. Control reactions were performed under the same conditions in the absence of Fe(NH4)2(SO4)2/EDTA and DTT.
Native gel analysis of aptamer interaction
Nondenaturing gel electrophoresis was performed as previously described (Cilley and Williamson 1999). 5′-end labeled VC1s/VC2s RNA was incubated in a TB buffer (90 mM Tris-borate at pH 8.3) containing 5 mM glycine, 10 mM MgCl2 for 20 min at 23°C. VC2/VC1 RNA constructs were then added to the reaction at increasing concentrations and incubated for an additional 20 min. One-fifth volume of glycerol loading dye (50% glycerol, 0.1% xylene cyanol) was added to the RNA mixture. Reaction mixture was loaded on to a 6% native acrylamide gel in TB buffer containing 10 mM MgCl2 and 5 mM of glycine. VC12-WT and VC12-G146C were used as size markers for shifted versus unshifted full-length riboswitches, respectively. Electrophoresis was carried out at 4°C for 2.5 h with TB buffer containing 10 mM MgCl2. The separated RNA was visualized by PhosphorImager and bands quantified using ImageQuaNT. The Kd value for the single aptamer interaction was determined by plotting the corrected fraction bound (FBcorrected = (B-FBO(T))/(T-FBO(T)), where B = bound, T = total bound + unbound and FBO = fraction bound in the label only lane) value versus the concentration of VC2/VC1 construct and fitting to a standard equation for fractional saturation (Y = 0.99([L]/([L] + Kd)).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank David Hiller, Ethan Butler, and Derek Erion for critical comments on the manuscript, Ethan Butler and Michelle Meyer for helpful discussions, and Andrew Kohlway for transcription reagents. This work was supported by NIH grant GM022778.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2271511.
REFERENCES
- Bohr C, Hasselbach KA, Krogh A 1904. Uber einen in biologischen beziehung wichtigen einfluss, den die kohlen-suer-spannung des blutes auf dessen sauerstoffbindung ubt. Skand Arch Physiol 15: 401–412 [Google Scholar]
- Cilley CD, Williamson JR 1999. PACE analysis of RNA-peptide interactions. Methods Mol Biol 118: 129–141 [DOI] [PubMed] [Google Scholar]
- Copeland RA 2000. Cooperativity in enyzme catalysis. Enzymes: A practical introduction to structure, mechanisms, and data analysis. Wiley-VCH, Inc, Berlin, Germany [Google Scholar]
- Cromie MJ, Shi Y, Latifi T, Groisman EA 2006. An RNA sensor for intracellular Mg(2+). Cell 125: 71–84 [DOI] [PubMed] [Google Scholar]
- Forsen S, Linse S 1995. Cooperativity: Over the Hill. Trends Biochem Sci 20: 495–497 [DOI] [PubMed] [Google Scholar]
- Fuchs RT, Grundy FJ, Henkin TM 2006. The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat Struct Mol Biol 13: 226–233 [DOI] [PubMed] [Google Scholar]
- Gagnon MG, Steinberg SV 2002. GU receptors of double helices mediate tRNA movement in the ribosome. RNA 8: 873–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon MG, Mukhopadhyay A, Steinberg SV 2006. Close packing of helices 3 and 12 of 16S rRNA is required for normal ribosome function. J Biol Chem 281: 39349–39357 [DOI] [PubMed] [Google Scholar]
- Gilbert SD, Rambo RP, Van Tyne D, Batey RT 2008. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol 2: 177–182 [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Lehman SC, Henkin TM 2003. The L box regulon: Lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc Natl Acad Sci 100: 12057–12062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AV 1910. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J Physiol 40: iv–vii [Google Scholar]
- Jose AM, Soukup GA, Breaker RR 2001. Cooperative binding of effectors by an allosteric ribozyme. Nucleic Acids Res 29: 1631–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi M, Soukup GA, Kerr JN, Breaker RR 1999. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nat Struct Biol 6: 1062–1071 [DOI] [PubMed] [Google Scholar]
- Koshland DE Jr, Hamadani K 2002. Proteomics and models for enzyme cooperativity. J Biol Chem 277: 46841–46844 [DOI] [PubMed] [Google Scholar]
- Kwon M, Strobel SA 2008. Chemical basis of glycine riboswitch cooperativity. RNA 14: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing C, Jung S, Iqbal A, Schlick T 2009. Tertiary motifs revealed in analyses of higher-order RNA junctions. J Mol Biol 393: 67–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert J, Das R, Chu VB, Kudaravalli M, Boyd N, Herschlag D, Doniach S 2007. Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae. J Mol Biol 365: 1393–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Breaker RR 2004. Gene regulation by riboswitches. Nat Rev Mol Cell Biol 5: 451–463 [DOI] [PubMed] [Google Scholar]
- Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR 2003. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113: 577–586 [DOI] [PubMed] [Google Scholar]
- Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR 2004. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 306: 275–279 [DOI] [PubMed] [Google Scholar]
- Montange RK, Batey RT 2006. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 441: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Ortoleva-Donnelly L, Kronman M, Strobel SA 1998. Identifying RNA minor groove tertiary contacts by nucleotide analogue interference mapping with N2-methylguanosine. Biochemistry 37: 12933–12942 [DOI] [PubMed] [Google Scholar]
- Perutz MF 1989. Mechanisms of cooperativity and allosteric regulation in proteins. Q Rev Biophys 22: 139–237 [DOI] [PubMed] [Google Scholar]
- Pogozelski WK, McNeese TJ, Tullius TD 1995. What species is responsible for strand scission in the reaction of [Fe(EDTA)]2- and H2O2 with DNA? J Am Chem Soc 117: 6428–6433 [Google Scholar]
- Regulski EE, Breaker RR 2008. In-line probing analysis of riboswitches. Methods Mol Biol 419: 53–67 [DOI] [PubMed] [Google Scholar]
- Rife JP, Cheng CS, Moore PB, Strobel SA 1998. N 2-methylguanosine is iso-energetic with guanosine in RNA duplexes and GNRA tetraloops. Nucleic Acids Res 26: 3640–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Breaker RR 2009. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem 78: 305–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer WE Jr, Knapp JE, Strand K, Heaslet HA 2001. Cooperative hemoglobins: Conserved fold, diverse quaternary assemblies and allosteric mechanisms. Trends Biochem Sci 26: 297–304 [DOI] [PubMed] [Google Scholar]
- Ryder SP, Strobel SA 1999. Nucleotide analog interference mapping. Methods 18: 38–50 [DOI] [PubMed] [Google Scholar]
- Serganov A, Patel DJ 2009. Amino acid recognition and gene regulation by riboswitches. Biochim Biophys Acta 1789: 592–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ 2008. Structure insights into amino acid binding and gene control by a lysine riboswitch. Nature 455: 1263–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA 2009. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol 16: 1218–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR 2003. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev 17: 2688–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR 2006. Tandem riboswitch architectures exhibit complex gene control functions. Science 314: 300–304 [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321: 411–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp HJ, Schwalbach MS, Meyer MM, Kitner JB, Breaker RR, Giovannoni SJ 2009. Unique glycine-activated riboswitch linked to glycine-serine auxotrophy in SAR11. Environ Microbiol 11: 230–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius TD, Greenbaum JA 2005. Mapping nucleic acid structure by hydroxyl radical cleavage. Curr Opin Chem Biol 9: 127–134 [DOI] [PubMed] [Google Scholar]
- Turner GJ, Galacteros F, Doyle ML, Hedlund B, Pettigrew DW, Turner BW, Smith FR, Moo-Penn W, Rucknagel DL, Ackers GK 1992. Mutagenic dissection of hemoglobin cooperativity: Effects of amino acid alteration on subunit assembly of oxy and deoxy tetramers. Proteins 14: 333–350 [DOI] [PubMed] [Google Scholar]
- Welz R, Breaker RR 2007. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA 13: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitty A 2008. Cooperativity and biological complexity. Nat Chem Biol 4: 435–439 [DOI] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR 2005. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol 59: 487–517 [DOI] [PubMed] [Google Scholar]
- Winkler WC, Cohen-Chalamish S, Breaker RR 2002. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci 99: 15908–15913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR 2003. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol 10: 701–707 [DOI] [PubMed] [Google Scholar]
- Yarnell WS, Roberts JW 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284: 611–615 [DOI] [PubMed] [Google Scholar]