Abstract
Horizontal transfer (HT), or the passage of genetic material between non-mating species, is increasingly recognized as an important force in the evolution of eukaryotic genomes1,2. Transposons, with their inherent ability to mobilize and amplify within genomes, may be especially prone to HT3–7. However, the means by which transposons can spread across widely diverged species remain elusive. Here we present evidence that host-parasite interactions have promoted the HT of four transposon families between invertebrates and vertebrates. We found that Rhodnius prolixus, a triatomine bug feeding on the blood of diverse tetrapods and vector of the Chagas disease in humans, carries in its genome four distinct transposon families that also invaded the genomes of a diverse, but overlapping, set of tetrapods. The bug transposons are ~98% identical and cluster phylogenetically with those of the opossum and squirrel monkey, two of its preferred mammalian hosts in South America. We also identified one of these transposon families in the pond snail Lymnaea stagnalis, a nearly cosmopolitan vector of trematodes infecting diverse vertebrates, whose ancestral sequence is nearly identical and clusters with those found in Old World mammals. Together these data provide evidence for a previously hypothesized role of host-parasite interactions in facilitating HT among animals3,7. Furthermore, the large amount of DNA generated by the amplification of the horizontally-transferred transposons supports the idea that the exchange of genetic material between hosts and parasites influence their genomic evolution.
In order to examine the factors underlying HT among widely diverged taxa we began our investigation with SPACE INVADERS (or SPIN), a recently described DNA transposon that has undergone repeated episodes of HT across the genomes of seven tetrapod lineages5. We first performed a series of BLASTN searches using the SPIN superconsensus sequence5 as a query against all GenBank databases (see Methods), including 102 species for which whole genome shotgun (WGS) sequences are available. In addition to the vertebrates previously known to harbor SPIN, we found highly significant hits (e-values as low as 0, corresponding here to 86% identity over >1 kb) in the triatomine bug, Rhodnius prolixus, an hemipteran insect that feeds on the blood of mammals, birds, and reptiles and serves as a vector for trypanosomes, the causal agent of Chagas disease8. Significant hits were also obtained from multiple Expressed Sequence Tag (EST) sequences generated for the freshwater snail Lymnaea stagnalis, which is an intermediate host for numerous trematodes parasitizing diverse vertebrate species9.
The discovery of SPIN in two invertebrates associated with parasitic life-cycles was intriguing, especially because triatomines are known to feed on several species in which SPIN was previously identified5. Thus, we expanded our investigation to look for evidence of HT of additional DNA transposons between R. prolixus and vertebrates by performing BLASTN searches against the WGS sequence of R. prolixus using a comprehensive collection of DNA transposons previously identified in vertebrates (see Methods). These searches yielded significant hits (e-value range: 0 – 3 × 10−108) for three families of mammalian DNA transposons: hAT110, OposCharlie1 (or OC1)11, and ExtraTerrestrial (or ET; see Methods for details on nomenclature). We confirmed the presence of all four transposon families in R. prolixus and of SPIN in L. stagnalis by PCR amplification from genomic DNA and sequencing of cloned PCR products (Supplementary Table 1). We constructed consensus (i.e. ancestral) sequences for each family of elements (SPIN, OC1, hAT1, and ET) in every species, based on a multiple alignment of copies extracted from the database. Phylogenetic analysis of consensus transposase sequences shows that like SPIN, the other families identified (OC1, hAT1, and ET) belong to the hAT superfamily, but are only distantly related to each other (Supplementary Fig. 1).
In the case of SPIN, an alignment of the L. stagnalis and R. prolixus consensus sequences with those generated previously for seven vertebrates5 revealed an extremely high level of identity between invertebrates and vertebrates across the entire length of the consensus sequences (up to 98.4% between L. stagnalis and bat and up to 95.3% between R. prolixus and opossum; Supplementary Table 2; see also Supplementary Table 3). Phylogenetic analyses of multiple individual copies revealed unrooted trees with a characteristic star topology (Supplementary Fig. 2), a pattern indicative of the accumulation of discrete substitutions in each copy and consistent with neutral evolution of transposons after their integration in the genome12 (see Supplementary Methods for additional data supporting neutral evolution). Thus, the level of sequence conservation between the invertebrate and vertebrate SPIN sequences is incompatible with vertical inheritance from their common ancestor, which occurred >500 million years ago (mya). Instead, these data indicate that SPIN was able to infiltrate the two invertebrate lineages independently, as it did in each of the seven vertebrate lineages5.
Several lines of evidence suggest that, as for SPIN, the taxonomic distribution of hAT1, OC1, and ET is the result of independent HT of these elements in each of the vertebrate lineages where they were identified (Fig.1). These include: (i) very high levels of sequence identity among species (ranging from 83.6 to 98.9% for all pairwise comparisons between consensus sequences; Supplementary Table 2; see also Supplementary Table 3), (ii) no evidence for orthologous insertions among species (Supplementary Fig. 3), and (iii) an inferred timing of transposon amplification clearly postdating the divergence of any two species harboring these transposons (Fig. 1). Remarkably, OC1 was able to infiltrate independently the germline of (at least) two prosimians and two anthropoid species, making it, to our knowledge, the most promiscuous and youngest DNA transposon ever identified in primates (~18 mya in squirrel monkey and tarsier; Fig. 1). The most recently active subfamily of OC1 was found in bat, although older subfamilies were also apparent in this species, suggesting that transposition activity persisted for a longer period in this lineage or that it was invaded recurrently (Supplementary Fig. 4 and 5). The activity of hAT1 and OC1 in different species of mammals appears to have occurred within overlapping evolutionary timeframes (9–26 mya and 12–34 mya, respectively), which also coincides with the timing of SPIN amplification (15–46 mya)5. Together, these data suggest that SPIN, hAT1, OC1, and ET have spread within the past 50 million years by means of HT among a range of animals spanning four different phyla.
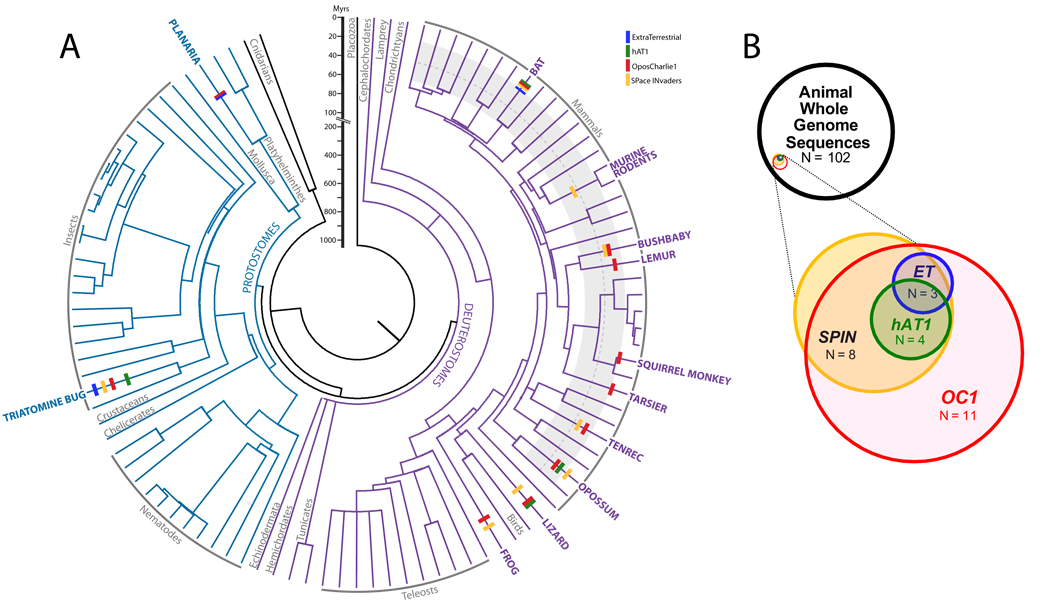
Figure 1.
Taxonomic distribution and age of SPACE INVADERS, OposCharlie1, ExtraTerrestrial, and hAT1. A. Distribution of the four elements among 102 animal genomes for which whole genome sequences are available (Supplementary Fig. 6). Branch lengths of the tree are proportional to time and tick marks illustrate the timing of amplification for each TE family. The 60 to 20 Myrs period is shaded in grey in mammals (with a dotted line at 40 Myrs) to facilitate the comparison of the different amplification times between species (see Methods). B. Venn diagram illustrating the overlap in taxonomic distribution of the four elements relative to all species for which whole genome sequence data are publicly available.
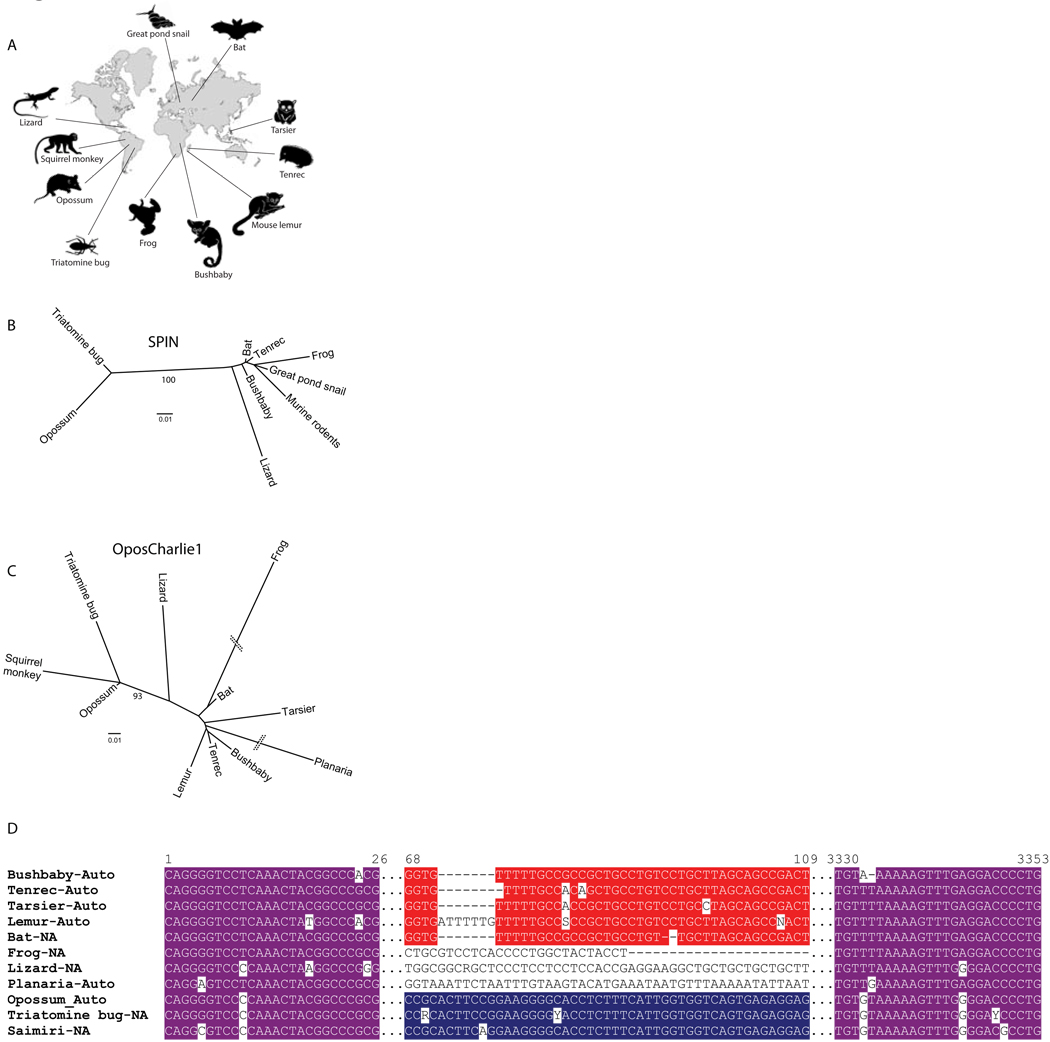
An intriguing aspect of the pattern of HT of these transposons is the widespread geographic distribution of the taxa at the presumed time of the transfers (Fig. 2A). While it is thought that tenrec, lemur, bushbaby, and frog were restricted to Africa and tarsier to Southeast Asia, there is evidence that the opossum, anole lizard, squirrel monkey, and R. prolixus were most likely confined to South America (see Methods and references therein). Bats of the genus Myotis and muroid rodents have a nearly cosmopolitan distribution, but colonized South America relatively recently (6–10 mya; see Methods), i.e. after HTs had already occurred on this continent. Thus, the biogeographical data indicate that HTs of SPIN and OC1 took place on a global scale, occurring on no less than three continents (Asia, Africa, and South America). Because South America and Africa broke apart in the Cretaceous before the HTs described here (more than 65 mya) and South America remained an island continent until the formation of the isthmus of Panama in the Pliocene (3–3.5 mya)13, the taxonomic distribution also implies at least one transoceanic movement of SPIN and OC1 transposons.
Figure 2.
Biogeographic and phylogenetic evidence supporting horizontal transfers of SPACE INVADERS and OposCharlie1 transposons on multiple continents. A. Map showing the likely geographic distribution of the taxa that contain these elements at the time of the transfers (see Methods). B., C. Maximum-likelihood phylogenies of SPIN and OC1 consensus sequences, respectively; bootstrap values above 70 are shown. D. Alignment of representatives autonomous and non-autonomous OC1 consensus sequences from species showing portions of the highly conserved 5’ and 3’ termini (purple) and the 5’ subterminal region in Old World (red) and New World (blue) species. All nucleotide positions are with reference to the bushbaby consensus (Supplementary Dataset S1).
Phylogenetic analyses using the consensus sequences of the two most widely distributed transposons (SPIN and OC1) produced trees with very similar topologies consisting of two strongly supported clusters: one grouping the elements from the South American mammals (opossum and squirrel monkey) with those found in the triatomine bug (endemic to South America) and one grouping all the other species (Fig. 2B and C). This clustering was further supported by additional molecular signatures, including a distinctive region in the sequence of OC1 elements shared by the opossum, squirrel monkey, and R. prolixus that differentiates them from all the other species (Fig. 2D). Thus, the phylogenetic relationships of the transposon sequences are discordant with those of their host species, but consistent with their continental origin. It is also noteworthy that, whereas in Old World species OC1 amplification occurred after that of SPIN, this order is reversed in New World species where OC1 appears to have amplified prior to SPIN (Fig. 1). Together, these data not only reinforce the idea that SPIN and OC1 must have traveled between the Old and New World, but also indicate that these elements have spread horizontally among New and Old World taxa within a relatively narrow timeframe (12–46 mya).
Another puzzling aspect of the horizontally-transferred transposons is the extensive overlap in their taxonomic distribution. Mapping the occurrence of these transposons onto the phylogeny of 102 animals for which whole genome assemblies are currently available, reveals a strikingly non-random pattern, with four species (bat, opossum, anole lizard, and R. prolixus) sharing all three transposon families (SPIN, OC1, hAT1; Fig. 1; Table 1; Supplementary Table 4). The probability of observing this distribution by chance alone, if the HT events were independent, is 4.9 × 10−8 (see Supplementary Methods), suggesting that some ecological factors make these species more prone to exchanging genetic material. Among New World taxa, the similarity of SPIN and OC1 consensus sequences of R. prolixus with those of opossum and squirrel monkey is striking (95.4 – 98.1%; Supplementary Table 2). We contend that this reflects HT of these transposons between triatomine bugs and one or more of their mammalian hosts. Triatomine bugs are known to feed on the blood of a variety of mammals in South America, including opossums, squirrel monkeys, and bats14,15. The exchange of large quantities of blood and saliva between the bugs and their hosts during feeding is known to facilitate the spread of trypanosomes (which cause Chagas disease in humans) and could also provide a route for the HT of transposons, possibly via these or other intracellular microparasites. Indeed, there is growing evidence for the exchange of genetic material between trypanosomes and their vertebrate hosts16.
Table 1.
Characteristics of OC1, SPIN, hAT1, and ET elements.
| TE | Copy number |
Kb of DNA |
Av distance (%) | |
|---|---|---|---|---|
| Squirrel monkey | OC1 | >344 | >64 | 4.1 |
| Tarsier | OC1 | 519 | 278 | 4.0 |
| Lemur | OC1 | 12055 | 2278 | 8.5 |
| Bushbaby | SPIN | 33123 | 3755 | 10 |
| OC1 | 14687 | 2709 | 10.6 | |
| Bat | SPIN | 50319 | 10725 | 3.4 |
| OC1 | 7190 | 1440 | 2.2 | |
| hAT1 | 19600 | 4664 | 2.5 | |
| ET | 17461 | 3908 | 4.7 | |
| Tenrec | SPIN | 99338 | 16233 | 10.5 |
| OC1 | 6837 | 1584 | 8.1 | |
| Opossum | SPIN | 4807 | 1429 | 5 |
| OC1 | 6025 | 3098 | 10.5 | |
| hAT1 | 2666 | 1377 | 8.8 | |
| Lizard | SPIN | 12138 | 373 | 9.3 |
| OC1 | 3902 | 2626 | 5.5 | |
| hAT1 | 298 | 161 | 4.7 | |
| Frog | SPIN | 3992 | 742 | 5.2 |
| OC1 | 950 | 445 | 8.9 | |
| Triatomine bug | SPIN | 211 | 75 | 3.6 |
| OC1 | 1286 | 495 | 6.4 | |
| hAT1 | 552 | 125 | 10.9 | |
| ET | 131 | 90 | 1.8 | |
| Planaria | OC1 | 46 | 267 | 8.6 |
| ET | 141 | 70 | 9.0 |
Numbers for SPIN elements are taken from Pace et al.5 except for the triatomine bug. Characteristics for each subfamily of OC1, SPIN, hAT1, and ET are given in Supplementary Table 4.
Among Old World taxa, the SPIN phylogeny (Fig. 2B) coupled to the extremely high sequence identity between L. stagnalis and the tetrapod taxa (96–98.5%; Supplementary Table 2) are suggestive of HT between snail and tetrapod(s). This transfer could be the result of another parasitic relationship because L. stagnalis is an intermediate host for diverse trematode worms that complete their life cycle in a wide range of vertebrate hosts17,18. So far, we have been unable to detect any of the horizontally-transferred transposons in the sequenced strains of Trypanosoma cruzi, one of the trypanosomes infecting R. prolixus, or in Fasciola hepatica, a mammalian trematode known to use L. stagnalis as an intermediate host. However, the streamlined and fast-evolving genomes of such microparasites might prevent the fixation or preservation of transposons in their genomes. Alternatively, HT might not require chromosomal integration in these species, but could involve extrachromosomal vector(s) such as viruses19–22.
Our findings suggest that HT of genetic material among animals has occurred on a broader scale than previously appreciated, including four families of DNA transposons and spanning four different animal phyla (Chordata, Arthropoda, Mollusca, and Platythelminthes). Although parasitism has been implicated previously to explain HT on smaller scales3,23–27, to our knowledge this is the first report of repeated HTs among invertebrates involved in host-parasite interactions with diverse vertebrate hosts. While the evolutionary consequences of the transfers described here require further investigation, the sheer amount of DNA generated by the amplification of the transposons (Table 1; Supplementary Table 4) and the myriad ways through which mobile elements can alter the structure and function of genomes28,29 supports the idea that the exchange of genetic material between host and parasite species could strongly impact genome evolution.
Methods summary
BLASTN was used to screen all GenBank databases for the presence of OC1, SPIN, hAT1, and ET transposons. A transposon family was considered to be present in a genome if the reconstructed consensus was at least 85% similar to a known transposon over 80% of its length. A total of 56 consensus sequences were constructed based on alignments of at least ten individual copies using a majority rule. Copy number and percent divergence for each TE family were determined using these consensus sequences to mask the various genomes with RepeatMasker v. 3.2.711. Estimates of the timing of amplification for each TE family in each species were derived by dividing the average percent Jukes-Cantor distance by the neutral mutation rate of the species5. Because no reliable neutral mutation rate is available for lizard, frog, triatomine bug, and planarian, we used the average mammalian neutral rate30 as an approximate estimate for the timing of amplification in these species (Fig. 1). Maximum-likelihood phylogenies were carried out with the HKY+G and HKY+I models for SPIN and for OC1 elements respectively. To verify the presence of the various transposons in all species where they were found computationally we used PCR/cloning/sequencing. To rule out DNA contamination in the two species associated with parasitic life-cycles (R. prolixus and L. stagnalis), we performed PCR using a pair of degenerate primers designed for rag-1 (a jawed vertebrate-specific gene), with human and opossum DNA as positive controls.
Methods
Identification and copy number estimation of SPIN, OC1, hAT1, and ET elements
The non-coding region of the mammalian SPIN superconsensus5 was used as a query in BLASTN (v. 2.2.1431) searches against the GenBank databases from the National Center for Biotechnology Information (NCBI), excluding the genomes of mammals, Xenopus tropicalis, and Anolis carolinensis (where SPIN had been previously identified). The following BLASTN parameters were used: gap existence penalty, 5; gap extension penalty, 2; penalty for nucleotide mismatch, −3; reward for nucleotide match, 2. SPIN was considered present in a species if the consensus was at least 85% similar at the nucleotide level to the SPIN superconsensus5 over at least 80% of its length.
In order to identify TEs other than SPIN that are shared between R. prolixus and vertebrate species, we used the Repbase library32 of vertebrate TEs as a query to perform a batch BLASTN search on the R. prolixus genome using the same parameters as above. Three TE families (OC1, hAT1, ET) were identified that are more than 85% similar to mammalian TEs over more than 80% of their length. The taxonomic distribution of these three TEs was then assessed by BLASTN searches against the animal whole genome shotgun (WGS) databases from NCBI and consensus sequences for each subfamily of OC1, hAT1, and ET were reconstructed in each species based on a multiple alignment of at least 10 individual copies (all consensus sequences are provided in Supplementary Dataset 1).
To estimate copy number and average percent divergence of each TE family, we used these respective consensus sequences to mask all genomes in which they were identified with RepeatMasker v. 3.2.711. All fragments larger than 100 bp were used to estimate copy number and calculate average percent divergence in all species except A. carolinensis where only fragments that were at least 80% of the length of the consensus were considered because of a high level of fragmentation and the presence of many chimeric elements in this species. A complete consensus sequence for OC1_NA_1_Xt, a frog-specific non-autonomous subfamily, could not be confidently reconstructed due to uncertainty of its internal region. The copy number for this subfamily was estimated based on counts of the 5’ and 3’ terminal regions, for which a reliable consensus sequence could be reconstructed. We observed that the 5’ region of hAT1_NA_1_Md (position 1–386) and that of hAT1_NA_3_Md (position 1–275) was about twice as diverged from the consensus sequence than the rest of the element copies, likely representing mutational hotspots. We therefore remasked the opossum genome without these regions in order to calculate the average percent divergence for these two non-autonomous subfamilies separately.
Given that, among the 102 species surveyed, we found OC1 in 11 species, ET in 4 species, and SPIN in 8 species, the probability of finding these three horizontally-transferred transposons in the same species, if the HTs occur by chance, is 11/102 × 4/102 × 8/102 = 3.3 × 10−4. The probability that four of the 102 species share these three transposons was then calculated using a binomial distribution B (4; 102; 3.3 × 10−4).
To test the comprehensiveness of whole genome shotgun sequences in the database, we performed TBLASTN on each of the 102 animal genomes with the ets domain from Aedes aegypti (accession: XP_00165406, region 443–529). Using this sequence as a query, we obtained 10 hits in A. aegypti and at least one hit in 93 of the other genomes (with an e-value < 1e-10). This indicates the sequencing coverage of at least 92% of genomes for which whole genome shotgun sequencing exists is confirmed to be sufficient to detect a domain from a low copy number gene. This, in turn, indicates that the sequencing coverage should be sufficient to detect the presence or absence of TEs (which are typically present in high copy number) in most, if not all, cases.
Nomenclature
Some of the OposCharlie1, hAT1, and ExtraTerrestrial subfamilies reported here correspond to subfamilies that had previously been identified in some of the species included in this study, but named differently (Supplementary Table 4). For example OposCharlie1, first described by Arian Smit in the opossum Monodelphis domestica11 was named HAT2_MD by Gentles and Jurka33 and hAT-HT2_MD by Novick et al.6. We note that an element that does not correspond to OposCharlie1 has been named hAT2_Ml in the bat Myotis lucifugus, where OposCharlie1 is also found34. To avoid confusion, we chose to use the first introduced name for this family: OposCharlie1. Also, we note that a non-autonomous subfamily of ExtraTerrestrial was identified in the bat M. lucifugus and was named Myotis_nhAT310. We have now identified the autonomous element from which this non-autonomous element derives and shown that it was not restricted to Myotis but was also present in R. prolixus and S. mediterranea. For these reasons, we decided to introduce the name ExtraTerrestrial (or ET) for this family. Lastly, hAT1 was first described in the bat, Myotis lucifugus10, and its name does not pose any particular problem.
Timing of amplification of each TE family
Estimates of the timing of amplification of each TE family in each species were calculated by dividing the average percent divergence of each TE family, to which the Jukes and Cantor correction35 was applied, by the neutral mutation rate of the different species. We used the neutral mutation rates calculated for bushbaby (2.9590 × 10−9), murine rodents (3.5411 × 10−9), tenrec (2.9173 × 10−9), opossum (3.2113 × 10−9), and bat (2.6920 × 10−9) in Pace et al.5. Because no reliable neutral mutation rate is available for lizard, triatomine bug, squirrel monkey, planaria, and African clawed frog, we used the average mammalian rate (2.2 × 10−9)30 to generate timing estimates for these species for illustrative purposes only (Fig. 1). The tree in Fig. 1 includes all 102 animals for which a complete or draft genome assembly is available in the WGS database of NCBI (http://www.ncbi.nlm.nih.gov/genomes/leuks.cgi). The tree was written in Newick format and drawn as a circular tree with branch length proportional to time in MEGA 4.036. Most phylogenetic relationships and divergence times are taken from the Timetree of life website: http://www.timetree.org/37 except for those within teleosts38,39, Drosophila40, within nematodes40,41, the genus Schistosoma42, and for the Drosophila/mosquito split43. Divergence times between Rhodnius and Acyrtosiphon and between Nasonia and Apis are not known but we placed both of them at 100 million years for illustrative purposes.
Biogeographical data
The geographical distributions of the taxa where SPIN, OC1, hAT1, and ET were identified are taken from the literature. Tenrec, bushbaby, mouse lemur, and African clawed frog are endemic to Africa and are believed to have been extant there at the time when the SPIN and OC1 invasions occurred (40–20 mya 44–47). The Philippine tarsier and all extant Tarsiidae are endemic to several islands of Southeast Asia and the fossil record indicates that the tarsiid lineage was already restricted to Southeast Asia when it was invaded by OC1 approximately 20 mya48,49. The opossum and all extant and extinct didelphoid marsupials are only known from the New World and the mouse-sized oppossums, which includes M. domestica, have been restricted to South America for at least 40 million years50. Anole lizards are endemic to Central/South America51. Bats of the genus Myotis originated in Eurasia and now, like muroid rodents, have a nearly cosmopolitan distribution but they did not disperse to South America until 6–10 mya52,53. The squirrel monkey is a platyrrhine or New World monkey, all of which diversified in South America less than 30 mya54. There is good evidence supporting South America as the sole center of origin and diversification of triatomine bugs, including R. prolixus55. Finally, although the distribution of L. stagnalis is presently holartic, the American populations are believed to be a recent introduction from a Eurasian stock56.
Phylogenetic analyses
Maximum-likelihood (ML) phylogenies of SPIN and OC1 elements were built using PHYML v. 357. Alignments were constructed manually in BioEdit58 and ambiguous regions were removed (Supplementary Dataset 2 and 3). Nucleotide substitution models were chosen using the AIC criterion in Modeltest59 (HKY+G for SPIN and HKY+I for OC1). In order to determine the phylogenetic relatedness of the four hAT transposons included in this study, we constructed an amino-acid alignment including their transposase region and that of various other hAT families taken from Repbase32. We then conducted ML analyses with PHYML v. 3 using the JTT model of amino-acid substitution60. The robustness of the nodes was evaluated for all phylogenies by performing a bootstrap analysis involving 1000 pseudoreplicates of the original matrix.
Testing for purifying selection
To examine the pattern of evolution of OC1, hAT1, and ET elements within a particular genome after horizontal transfer, dN/dS analyses were performed as follows: fifty full length OC1 copies were extracted from the opossum genome and all copies of ET (40) and hAT1 (49) that contained at least 60% of the transposase sequence were retrieved from the bat genome. A multiple alignment of the coding region of these individual copies and their respective consensus was constructed using BioEdit58 and all non-sense mutations were removed. We then tested whether the pattern of mutations observed between each copy and the consensus (an estimate of the ancestral founder element) was significantly different from what is expected if the sequence is evolving neutrally using the codon-based Z-test in MEGA 4.036 with the Nei-Gojobori method and the Jukes-Cantor correction (500 boostrap replicates). In addition, we used these multiple alignments and an alignment of fifty full length or nearly full length SPIN_NA_12_Rp (extracted from R. prolixus) to construct Neighbor-Joining phylogenies in MEGA 4.0, with the Maximum Composite Likelihood model and 1000 bootstrap replicates. See ref. 5 for examples of SPIN star-like phylogenies in other taxa.
Similarity plot (Supplementary Fig. 5A)
Similarity among copies of autonomous OC1 elements from the 6 species in which they were identified (O. garnetti, T. syrichta, M. murinus, E. telfairi, S. mediterranea, and M. domestica) was calculated using DnaSP v561. Alignments were made using ClustalW62 and corrected manually. All ambiguous sites were considered four-fold degenerate and were included in the analysis, whereas gapped sites were excluded. Polymorphism (pi) was calculated in 10 bp windows using 3 bp stepwise increments over the length of the entire element (3291 bp), including the OC1 transposase which is 1808 bp long (position 1312–3120 of the bushbaby consensus [Supplementary Dataset 1]). Values were converted to percentages and subtracted from 100 to plot similarity. Two species (S. mediterranea and M. domestica) were unalignable in an 816 bp portion of the element (position 64–880 of the opossum and 64–352 of S. mediterranea; Supplementary Dataset 1) and were excluded for this region.
PCR/cloning/sequencing
Newly identified SPIN, OC1, hAT1, and ET elements were validated in each species by PCR amplification, cloning, and sequencing. For Rhodnius prolixus, genomic DNA was extracted from insect legs in order to rule out possible contamination from ingested blood. Primers for each element are listed in Supplementary Table 5. PCR was conducted using the following temperature cycling: initial denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 sec, annealing between 48–54°C based on element-specific gradients (for 30 sec), and elongation at 72°C for 1 min, ending with a 10 min elongation step at 72°C. Fragments from the PCR were visualized on a 1–2% agarose gel, cloned, and sequenced. Cloning was performed using the Strataclone PCR cloning kit (Stratagene) following manufacturer’s protocols and successfully transformed bacterial colonies were screened by PCR (same thermocycling program as above) using M13 primers (see Supplementary Table 5) and gel electrophoresis. Amplicons from cloning products were excised from the gel and soaked in 100 ul ddH20 for 2–4 hours. PCR was used to re-amplify the products from this solution (using M13 primers) and sequencing reactions were performed using the reamplified product as template using ABI’s BigDye™ sequencing mix (1.4 ul template PCR product, 0.4 ul BigDye, 2 ul manufacturer supplied buffer, 0.3 ul reverse primer, 6 ul H2O). The thermocycler program was as follows: 2 min denaturation (96° C) followed by 30 cycles alternating between 96° C (30 sec) and 60° C (4 min), ending at 10° C for 3 minutes. Sequencing reactions were ethanol precipitated and run on an ABI 3730. Sequences were trimmed using Sequencher 4.8 (Gene Codes, Ann Arbor, Michigan) and were aligned and analyzed using MEGA 4.036. To further rule out contamination, degenerate primers were designed to amplify rag-1, a gene found only among jawed vertebrates and PCR was performed on DNA extracted from R. prolixus and L. stagnalis to ensure non-amplification (with human and opossum DNA as a positive control). The thermocycler program for this PCR amplification was the same as that described above, but using 56°C for annealing.
Supplementary Material
Acknowledgments
We thank Esther Betrán, Jeff Demuth, Trey Fondon, Britt Koskella, Jesse Meik, Ellen Pritham, Qi Wang, and members of the Feschotte lab for critical comments and helpful suggestions during the preparation of the manuscript; Mark Batzer, Ellen Dotson, Steve Goodman, Amandine Prelat, Terry Robinson, Anne Ropiquet, and the Grosell and Sánchez labs for the generous gift of tissue samples used in this study; and John Spieth and The Genome Center at Washington University School of Medicine in St. Louis for permission to use the R. prolixus assembly prior to publication. C. F. is funded by the National Institute of Health and S. S. by the National Science Foundation.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions C.G., S.S., and C.F. designed research, performed research, and analyzed data. J.K.P. contributed data and perl scripts. P.J.B. contributed reagents/material. C.G., S.S., and C.F. wrote the paper.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
Contributor Information
Clément Gilbert, Email: clement.gilbert@uta.edu.
Sarah Schaack, Email: schaack@uta.edu.
John K. Pace, II, Email: pace_john@hotmail.com.
Paul J. Brindley, Email: mtmpjb@gwumc.edu.
Cédric Feschotte, Email: cedric@uta.edu.
References
- 1.Keeling PJ, Palmer JD. Horizontal transfer in eukaryotic evolution. Nat. Rev. Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 2.Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008;320:1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- 3.Loreto EL, Carareto CM, Capy P. Revisiting horizontal transfer of transposable elements in Drosophila. Heredity. 2008;100:545–554. doi: 10.1038/sj.hdy.6801094. [DOI] [PubMed] [Google Scholar]
- 4.Diao X, Freeling M, Lisch D. Horizontal transfer of a plant transposon. PLoS Biol. 2006;4:1. doi: 10.1371/journal.pbio.0040005. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pace JK, Gilbert C, Clark MS, Feschotte C. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17023–17028. doi: 10.1073/pnas.0806548105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novick P, et al. Independent and parallel lateral transfer of DNA transposons in tetrapod genomes. Gene. 2009;449:85–94. doi: 10.1016/j.gene.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 7.Houck MA, Clark JB, Peterson KR, Kidwell MG. Possible horizontal transfer of Drosophila genes by the mite Proctolaelaps regalis. Science. 1991;6:1125–1129. doi: 10.1126/science.1653453. [DOI] [PubMed] [Google Scholar]
- 8.Lent H, Wygodzinsky P. Revision of the Triatominae (Hemiptera, Reduviidae), and their significance as vectors of Chagas disease. Bull. Amer. Mus. Nat. Hist. 1979;163:123–520. [Google Scholar]
- 9.Loy C, Haas W. Prevalence of cercariae from Lymnaea stagnalis snails in a pond system in Southern Germany. Parasitol. Res. 2001;87:878–882. doi: 10.1007/s004360100462. [DOI] [PubMed] [Google Scholar]
- 10.Ray DA, Pagan HJT, Thompson ML, Stevens RD. Bats with hATs: Evidence for recent DNA transposon activity in genus Myotis. Mol. Biol. Evol. 2007;24:632–639. doi: 10.1093/molbev/msl192. [DOI] [PubMed] [Google Scholar]
- 11.Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996–2004 http://www.repeatmasker.org.
- 12.Roberston HM. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington: ASM Press; 2002. pp. 1093–1110. [Google Scholar]
- 13.Marshall E, et al. Calibration of the great American interchange. Science. 1979;204:272–279. doi: 10.1126/science.204.4390.272. [DOI] [PubMed] [Google Scholar]
- 14.Maia Da Silva F, et al. Comparative phylogeography of Trypanosoma rangeli and Rhodnius (Hemiptera: Reduviidae) supports a long coexistence of parasite lineages and their sympatric vectors. Mol. Ecol. 2007;16:3361–3373. doi: 10.1111/j.1365-294X.2007.03371.x. [DOI] [PubMed] [Google Scholar]
- 15.Maia Da Silva F, et al. Trypanosoma rangeli isolates of bats from Central Brazil: Genotyping and phylogenetic analysis enable description of a new lineage using spliced-leader gene sequences. Acta Tropica. 2009;109:199–207. doi: 10.1016/j.actatropica.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Steglich C, Schaeffer SW. The ornithine decarboxylase gene of Trypanosoma brucei: Evidence for horizontal gene transfer from a vertebrate source. Infect. Genet. Evol. 2006;6:205–219. doi: 10.1016/j.meegid.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Rogan MT, et al. The occurence of the trematode Plagiorchis muris in the wood mouse Apodemus sylvaticus in North Yorkshire, UK. J. Helminth. 2007;81:57–62. doi: 10.1017/S0022149X07214105. [DOI] [PubMed] [Google Scholar]
- 18.Zbikowska E, et al. Infestation of Lymnaea stagnalis by digenea flukes in the Jeziorak lake. Parasitol. Res. 2006;99:434–439. doi: 10.1007/s00436-006-0178-z. [DOI] [PubMed] [Google Scholar]
- 19.Piskurek O, Okada N. Poxviruses as possible vectors of horizontal transfer of retroposons from reptiles to mammals. Proc. Natl. Acad. Sci.U.S.A. 2007;104:12046–12051. doi: 10.1073/pnas.0700531104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraser MJ, Smith GE, Summers MD. Acquisition of host cell DNA sequences by baculoviruses: Relationship between host DNA insertions and FP mutants of Autographa californica and Galleria mellonella nuclear polyhedris viruses. J. Virol. 1983;47:287–300. doi: 10.1128/jvi.47.2.287-300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friesen PD, Nissen MS. Gene organization and transcription of TED, a lepidopteran retrotransposon integrated within the baculovirus genome. Mol. Cell. Biol. 1990;10:3067–3077. doi: 10.1128/mcb.10.6.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jehle JA, Nickel A, Vlak JM, Backhaus HJ. Horizontal escape of the novel Tc1-like lepidopteran transposon TCp3.2 into Cydia pomonella granulovirus. J. Mol. Evol. 1998;46:215–224. doi: 10.1007/pl00006296. [DOI] [PubMed] [Google Scholar]
- 23.Heath BD, Butcher RDJ, Whitfield WGF, Hubbard SF. Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr. Biol. 1999;9:313–316. doi: 10.1016/s0960-9822(99)80139-0. [DOI] [PubMed] [Google Scholar]
- 24.Huigens ME, et al. Infectious parthenogenesis. Nature. 2000;405:178–179. doi: 10.1038/35012066. [DOI] [PubMed] [Google Scholar]
- 25.Yoshiyama M, et al. Possible horizontal transfer of a transposable element from host to parasitoid. Mol. Biol. Evol. 2001;18:1952–1958. doi: 10.1093/oxfordjournals.molbev.a003735. [DOI] [PubMed] [Google Scholar]
- 26.Mower JP, Stefanovic S, Young GJ, Palmer JD. Gene transfer from parasitic to host plants. Nature. 2004;432:165–166. doi: 10.1038/432165b. [DOI] [PubMed] [Google Scholar]
- 27.Davis CC, Wurdack KJ. Host to parasite gene transfer in flowering plants: Phylogenetic evidence from malpighiales. Science. 2004;305:676–678. doi: 10.1126/science.1100671. [DOI] [PubMed] [Google Scholar]
- 28.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S, Subramanian S. Mutation rates in mammalian genomes. Proc. Natl. Acad. Sci. U.S.A. 2002;99:803–808. doi: 10.1073/pnas.022629899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, et al. Basic Local Alignment Search Tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 33.Gentles AJ, Jurka J. HAT2_MD – an autonomous hAT transposon from Monodelphis domestica; consensus sequence. Repbase Report. 2005;5 [Google Scholar]
- 34.Ray DA, et al. Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus. Genome Res. 2008;18:717–728. doi: 10.1101/gr.071886.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jukes TH, Cantor CR. In: Mammalian protein metabolism. Munro HN, editor. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 36.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 37.Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 38.Yamanoue Y, et al. The mitocondrial genome of spotted green pufferfish Tetraodon nigroviridis (Teleostei: Tetraodontiformes) and divergence time estimation among model organisms in fishes. Genes Genet. Syst. 2006;81:29–39. doi: 10.1266/ggs.81.29. [DOI] [PubMed] [Google Scholar]
- 39.Steinke D, Salzburger W, Meyer A. Novel relationships among ten fish model species revealed based on a phylogenomic analysis using ESTs. J. Mol. Evol. 2006;62:772–784. doi: 10.1007/s00239-005-0170-8. [DOI] [PubMed] [Google Scholar]
- 40.Cutter AD. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol. Biol. Evol. 2008;25:778–786. doi: 10.1093/molbev/msn024. [DOI] [PubMed] [Google Scholar]
- 41.De Giorgi C, et al. Structural and evolutionary analysis of the ribosomal genes of the parasitic nematode Meloidogyne artiellia suggests its ancient origin. Mol. Biochem. Parasit. 2002;124:91–94. doi: 10.1016/s0166-6851(02)00161-5. [DOI] [PubMed] [Google Scholar]
- 42.Morgan JAT, et al. Schistosoma mansoni and Biomphalaria: past history and future trends. Parasitology. 2001;123:S211–S228. doi: 10.1017/s0031182001007703. [DOI] [PubMed] [Google Scholar]
- 43.Wiegmann BM, et al. Single-copy nuclear genes resolve the phylogeny of the holometabolous insects. BMC Biology. 2009;7:34. doi: 10.1186/1741-7007-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoder AD, et al. Ancient origin for Malagasy primates. Proc. Natl. Acad. Sci.U.S.A. 1996;93:5122–5126. doi: 10.1073/pnas.93.10.5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evans BJ, et al. A mitochondrial DNA phylogeny of African clawed frogs: phylogeography and implication for polyploidy evolution. Mol. Phylogenet. Evol. 2004;33:197–213. doi: 10.1016/j.ympev.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 46.Wilson DE, Reeder DM. Mammal species of the world: a taxonomic and geographic reference 2. Washington: Smithsonian Institution Press; 2005. [Google Scholar]
- 47.Poux C, et al. Asynchronous colonization of Madagascar by the four endemic clades of primates, tenrecs, carnivores, and rodents as inferred from nuclear genes. Syst. Biol. 2005;54:719–730. doi: 10.1080/10635150500234534. [DOI] [PubMed] [Google Scholar]
- 48.Gebo DL, et al. The oldest known anthropoid postcranial fossils and the early evolution of higher primates. Nature. 2000;404:276–278. doi: 10.1038/35005066. [DOI] [PubMed] [Google Scholar]
- 49.Rossie JB, Ni XJ, Beard KC. Cranial remains of an Eocene tarsier. Proc. Natl. Acad. Sci.U.S.A. 2006;103:4381–4385. doi: 10.1073/pnas.0509424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steiner C, Tilak M, Douzery EJP, Catzeflis FM. New DNA data from a transthyretin nuclear intron suggest an Oligocene to Miocene diversification of living South America opossums (Marsupialia: Didelphidae) Mol. Phylogenet. Evol. 2005;35:363–379. doi: 10.1016/j.ympev.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 51.Roughgarden J, editor. Anolis lizards of the Caribbean. Ecology, evolution and plate tectonics. New York: Oxford University Press; 1995. [Google Scholar]
- 52.Stadelmann B, Lin LK, Kunz TH, Ruedi M. Molecular phylogeny of New World Myotis (Chiroptera, Vespertilionidae) inferred from mitochondrial and nuclear DNA genes. Mol. Phylogenet. Evol. 2007;43:32–48. doi: 10.1016/j.ympev.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 53.Steppan SJ, Adkins RM, Anderson J. Phylogeny and divergence-date estimates of rapid radiation in muroid rodents based on multiple nuclear genes. Syst. Biol. 2004;53:533–553. doi: 10.1080/10635150490468701. [DOI] [PubMed] [Google Scholar]
- 54.Poux C, et al. Arrival and diversification of caviomorph rodents and platyrrhine primates in South America. Syst. Biol. 2006;55:228–244. doi: 10.1080/10635150500481390. [DOI] [PubMed] [Google Scholar]
- 55.Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol. Biol. Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- 56.Remigio EA. Molecular phylogenetic relationships in the aquatic snail genus Lymnaea, the intermediate host of the causative agent of fascioliasis: insights from broader taxon sampling. Parasitol. Res. 2002;88:687–696. doi: 10.1007/s00436-002-0658-8. [DOI] [PubMed] [Google Scholar]
- 57.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 58.Hall T. BioEdit version 5.0.6. 2004 Available at: < http://www.mbio.ncsu.edu/BioEdit/bioedit.html/>.
- 59.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 60.Jones DT, Jones WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. CABIOS. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 61.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 62.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids. Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hugall AF, Foster R, Lee MSY. Calibration choice, rate smoothing, and the pattern of tetrapod diversification according to the long nuclear gene RAG-1. Syst. Biol. 2007;56:543–563. doi: 10.1080/10635150701477825. [DOI] [PubMed] [Google Scholar]
- 64.Murphy WJ, et al. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.