Abstract
The E26 transformation-specific (Ets) proteins are a family of transcription factors with important roles in a variety of cellular processes ranging from proliferation and differentiation to transformation and metastasis. Tissue-specific expression of Ets proteins and their ability to interact with other families of transcription factors contribute to their versatility. In this study, we investigated the regulation of Ets factors in primary human monocytes and macrophages, and their role in matrix metalloprotease (MMP) and cytokine production. The macrophage-activating Toll-like receptor ligand, lipopolysaccharide (LPS), induced the expression of Ets family members epithelium-specific Ets factor 3 (ESE-3) and TEL-2 but rapidly suppressed Friend leukemia virus integration 1 (FLI-1) expression. Modulation of FLI-1 expression using either RNA interference or forced expression identified a positive role for FLI-1 in contributing to LPS-induced expression of MMP-1, MMP-3, MMP-10, and interleukin-10 (IL-10). Thus, the rapid downregulation of FLI-1 expression after LPS stimulation attenuates the induction of various MMPs and IL-10 under inflammatory conditions. In contrast, the expression of IL-6 and TNFα and the effects of interferon (IFN)γ on LPS responses were not dependent on FLI-1. Our results define a novel FLI-1-mediated self-regulatory feedback loop that limits MMP expression and thus may attenuate extent of tissue destruction associated with inflammatory responses.
Introduction
Since Nunn and others (1983) and Leprince and others (1983) discovered its first member in 1983, the E26 transformation-specific (Ets) family of transcription factors have become increasingly recognized as key regulators of different cellular functions. As of today, ∼30 members of the Ets family proteins have been identified in mammals, which share a conserved DNA-binding domain (ETS domain) that recognizes a consensus GGA(A/T) motif (Ets binding site, or EBS) in promoter elements. In addition, the ETS domain allows protein–protein interactions among members of the Ets family and between Ets and a variety of other transcription factors, including AP-1 proteins (Li and others 2000). This adds another level in their orchestrated regulation of a wide variety of biological processes, which include cellular proliferation, differentiation, development, apoptosis, angiogenesis, hematopoiesis, tissue remodeling, and malignant transformation of cells and metastasis (Oikawa Yamada 2003; Cardone and others 2005; Jung and others 2005; Seth and Watson 2005; Carella and others 2006).
The ability of multiple Ets proteins to recognize similar or even identical EBS response elements requires that the function and specificity of each Ets transcription factor be regulated differentially at multiple levels. Not surprisingly, the expression of certain Ets proteins was found to be tissue selective, e.g., epithelium-specific Ets factor (ESE)-1, 2, and 3 are preferentially expressed in epithelium and induced in hematopoietic cells (Kas and others 2000; Tugores and others 2001); others such as TEL can change their subcellular localization through interaction with other nuclear proteins (Chakrabarti and others 2000). Moreover, posttranslational modification of Ets factors by specific signaling pathways (eg, Ets-1 phophorylation by mitogen-activated protein kinase or calcium signaling pathways) and their interaction with other unrelated transcription factors (eg, AP-1 proteins) on composite DNA elements also contribute to the context-specific nature (ie, either as activators or repressors) of gene regulation by Ets proteins (Li and others 2000; Oikawa and Yamada 2003; Seth and Watson 2005). For instance, Ets related gene (ERG) has been found to activate matrix metalloprotease 1 (MMP-1) gene expression by interacting with c-Fos/c-Jun, but it represses MMP-3 promoter by preventing its activation by ETS-2 (Basuyaux and others 1997). On the other hand, the same Ets transcription factor can also behave differently in different cell types, eg, overexpression of Friend leukemia virus integration 1 (FLI-1) led to induction of the chicken GATA-1 promoter in COS and HeLa cells but not in chicken bone marrow erythroblasts (Seth and others 1993; Pereira and others 1999), whereas in human (K562) and murine (HB60) erythroleukemia cell lines, expression of FLI-1 negatively regulates GATA-1 gene expression (Tamir and others 1999; Athanasiou and others 2000).
In this study, we investigated the expression profile of Ets factors in primary human monocytes and macrophages under inflammatory conditions and explored the role of specific Ets proteins in the induction of gene expression by the Toll-like receptor 4 (TLR4) ligand lipopolysaccharide (LPS). We found that activation of TLRs led to a strong induction of ESE-3 and TEL-2, but a rapid reduction in the level of FLI-1 gene expression in primary human macrophages. Forced expression and siRNA knockdown experiments involving FLI-1 further revealed an important role of this Ets protein in the LPS-induced expression of multiple MMPs and of the anti-inflammatory cytokine interleukin-10 (IL-10).
Materials and Methods
Biological reagents and cell culture
Peripheral blood mononuclear cells (PBMCs) were obtained from whole blood from disease-free volunteers by density gradient centrifugation with Ficoll (Invitrogen, Carlsbad, CA). CD14+ monocytes were purified from fresh PBMCs with anti-CD14 magnetic beads (Miltenyi Biotec, Auburn, CA), as recommended by the manufacturer. Purity of monocytes was greater than 97% as verified by FACS, and freshly isolated monocytes were used in many experiments, as noted in the text and figure legends. Some experiments were performed using macrophages, which were derived by culturing monocytes in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS (Hyclone, Logan, UT) in the presence of 10 ng/mL of human macrophage-colony stimulating factor (M-CSF) (Peprotech, Rocky Hill, NJ). THP-1 human monocytic cells were obtained from ATCC (Manassas, VA) and cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS (Hyclone, Logan, UT). LPS (100 ng/mL) and actinomycin D (5 μg/mL) were purchased from Sigma-Aldrich (Milwaukee, WI).
Lentiviral gene transduction and RNA interference
A lentivirus-based vector expressing the human ESE-3 or FLI-1 cDNA driven by a human phospho-glycerol kinase (hPGK) promoter was used to generate recombinant lentiviral particles as described (Rubinson and others 2003). A construct that contained a transcription cassette encoding enhanced green fluorescent protein (eGFP) driven by the hPGK promoter was used to generate control viral particles for ESE-3 or FLI-1 expression experiments. THP-1 cells were incubated overnight with recombinant lentiviral particles at a ratio of 1:50 in the presence of 4 μg/mL polybrene. The efficiency of transduction was evaluated using flow cytometry and fluorescence microscopy to monitor eGFP expression and was typically >90%. For RNA interference (RNAi) experiments with primary monocytes (FLI-1 and TEL-2), prevalidated siRNAs and nontargeting control siRNAs were purchased from Dharmacon (Lafayette, CO). siRNAs were transfected into primary human macrophages using the Amaxa Nucleofector device set to program Y-001 with the Human Monocyte Nucleofector kit (Amaxa, Cologne, Germany).
Real-time, quantitative RT-PCR
For real-time PCR, total RNA was extracted using an RNeasy Mini kit and 1 μg of total RNA was reverse transcribed using a First Strand cDNA Synthesis kit (Fermentas, Hanover, MD). Real-time, quantitative PCR (qPCR) was performed using iQ™ SYBR-Green Supermix and iCycler iQ™ thermal cycler (Biorad, Hercules, CA), following the manufacturer's protocols. Triplicate reactions were run for each sample and mRNA levels were normalized relative to β-actin. The generation of the correct-size amplification products was confirmed using agarose gel electrophoresis.
Results
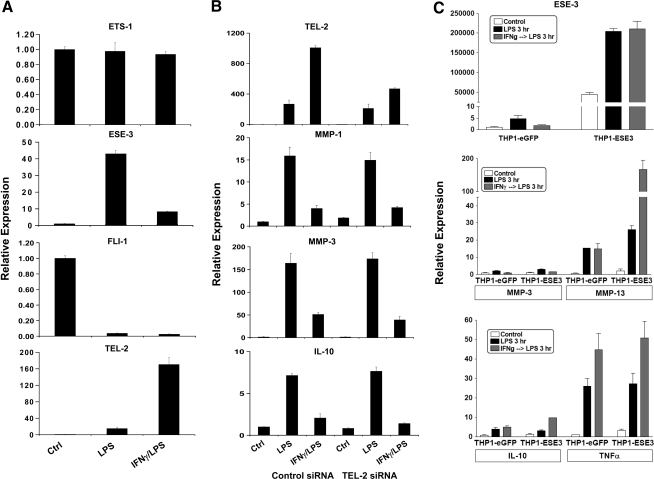
We first determined which Ets factors were induced or inhibited in monocytes and macrophages by inflammatory stimuli. Primary monocytes were treated with inflammatory factors (LPS and/or IFNγ) and various Ets mRNA levels (that included ETS-1, ETS-2, ELK-1, ERF, ERG, FLI-1, NET, PE-1, SAP-1, TEL, TEL-2) were measured using real-time RT-qPCR. Upon exposure of monocytes to the TLR4 ligand LPS, changes in expression of most Ets mRNAs were modest, except for strong >10-fold induction of ESE-3 and TEL-2 expression and marked 80% suppression of FLI-1 expression (Fig. 1A and data not shown). Similar results were obtained using the TLR2 ligand Pam3Cys and IL-1β. Changes in ESE-3, TEL-2, and FLI-1 mRNA expression were validated using different sets of amplification primers and were also reflected by hybridization signals on microarray experiments (data not shown). Preincubation of monocytes with IFNγ did not alter the effect of LPS on FLI-1 expression but suppressed ESE-3 induction. Interestingly, IFNγ and LPS synergistically induced TEL-2 expression (Fig. 1A, B).
FIG. 1.
Effects of lipopolysaccharide (LPS) and interferon (IFN)γ on Ets mRNA expression in primary monocytes. (A) Primary human monocytes were stimulated with 100 ng/mL of LPS with or without IFNγ pretreatment (100 U/mL, added 3 h before LPS) for 3 h. (B) Gene expression in primary human monocytes transfected with control or Tel-2-specific short interfering RNAs. (C) THP-1 cells were transduced with lentiviral particles expressing enhanced green fluorescent protein (eGFP) or epithelium-specific Ets factor 3 (ESE-3) and stimulated with 100 ng/mL of LPS with or without IFNγ pretreatment (100 U/mL, added 3 h before LPS) for 3 h. mRNA levels were analyzed using real-time PCR and normalized relative to β-actin; results are expressed as mean ± SD of triplicate determinants. Ets, E26 transformation-specific; MMP, matrix metalloprotease; IL, interleukin; FLI-1, Friend leukemia virus integration 1.
To assess the role of these Ets factors in LPS/IFNγ signaling, we modulated the cellular levels of ESE-3, FLI-1, and TEL-2 through either forced expression or RNAi in monocytic cells. Subsequently, we screened for changes in downstream gene expression profile for several known LPS/IFNγ-inducible genes in our system that included various matrix metalloproteinases (e.g., MMP-1, MMP-3, and MMP-10) and cytokines such as IL-6, IL-10, and TNFα. TEL-2 siRNA knockdown experiments to attenuate its induction by LPS/IFNγ (by ∼50%–70%) did not result in any noticeable difference in the expression of any of the analyzed downstream genes (Fig. 1B). On the other hand, high-level overexpression of ESE-3 only led to increased MMP-13 expression basally and upon LPS stimulation, without affecting the expression of other MMPs and cytokines (Fig. 1C). Because of the limited results obtained with TEL-2 and ESE-3, we focused on the analysis of FLI-1 function in mediating macrophage TLR responses.
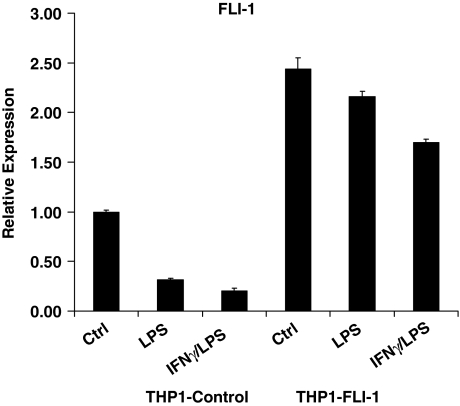
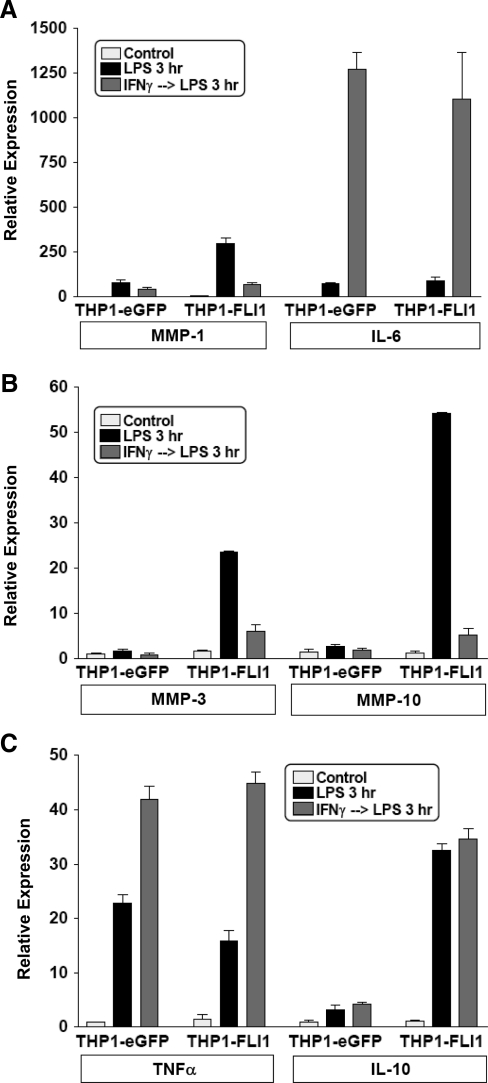
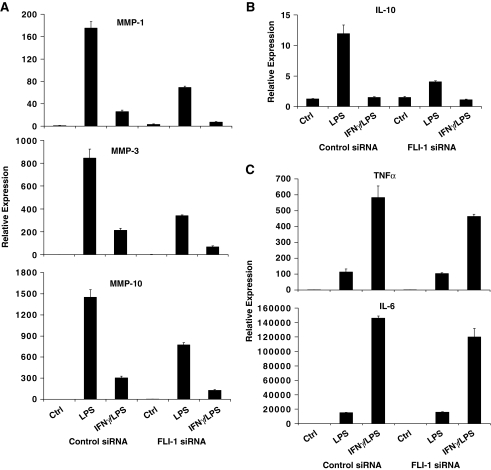
Using lentivirus-mediated transduction of monocytic cells, we were able to increase the basal FLI-1 expression by 2-fold, a level that is comparable to physiological expression. More importantly, transgene-encoded FLI-1 transcripts (which do not contain 3′ untranslated region sequences) were resistant to LPS-induced downregulation. Thus, in these transduced monocytes, FLI-1 levels were maintained at levels comparable to baseline physiological expression even after LPS stimulation (Fig. 2). As shown in Fig. 3, when downregulation of FLI-1 upon LPS stimulation was prevented, we observed a marked increase in LPS-induced expression of MMP-1, MMP-3, and MMP-10 compared with the control cells that were transduced with eGFP-encoding lentivirus (Fig. 3A, B). In terms of cytokine expression, increased FLI-1 levels specifically enhanced IL-10 expression (Fig. 3C), without affecting LPS induction of IL-6 and TNFα (Fig. 3A, C). Interestingly, neither the inhibitory effect of IFNγ preincubation on LPS-mediated induction of MMPs nor the synergistic effect of IFNγ and LPS on IL-6 and TNFα production was affected by the increased FLI-1 level (Fig. 3). These results suggested a role for FLI-1 in LPS regulation of MMP and cytokine expression, and this was further investigated using RNAi.
FIG. 2.
Modulation of FLI-1 expression in THP-1 monocytic cells. THP-1 cells were transduced with lentiviral particles expressing eGFP or FLI-1 and stimulated with LPS with or without IFNγ pretreatment, and FLI-1 mRNA was measured using real-time PCR. mRNA levels were normalized relative to β-actin, and real-time PCR results are expressed as mean ± SD of triplicate determinants.
FIG. 3.
FLI-1 overexpression enhances LPS-induced MMP expression but variably affects cytokine production. (A–C) THP-1 cells were transduced with lentiviral particles expressing eGFP or FLI-1 and stimulated with 100 ng/mL of LPS with or without IFNγ pretreatment (100 U/mL, added 3 h before LPS) for 3 h. mRNA levels were analyzed using real-time PCR and normalized relative to β-actin; results are expressed as mean ± SD of triplicate determinants. TNF, tumor necrosis factor.
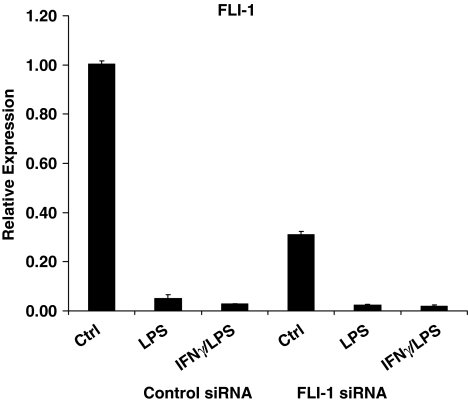
Using siRNAs against FLI-1, we were able to knockdown the basal level of FLI-1 in primary monocytes to about 30% of control (Fig. 4). When FLI-1 expression was reduced, we found that LPS-mediated induction of MMP-1, MMP-3, and MMP-10 was dramatically diminished (Fig. 5A), as was the LPS induction of IL-10 (Fig. 5B). In contrast, expression of TNFα and IL-6 was once again not affected by FLI-1 knockdown (Fig. 5C), and there was no effect on IFNγ modulation of LPS-induced gene expression. Overall, our findings support a role for FLI-1 in the positive and negative regulation of MMPs and IL-10 expression under inflammatory conditions and suggest that the downregulation of FLI-1 levels by LPS may represent a self-regulatory feedback loop in TLR signaling in monocytic cells.
FIG. 4.
Modulation of FLI-1 expression in primary monocytes. Primary monocytes transfected with control or FLI-1 siRNAs were stimulated with LPS with or without IFNγ pretreatment, and FLI-1 mRNA was measured using real-time PCR. mRNA levels were normalized relative to β-actin, and real-time PCR results are expressed as mean ± SD of triplicate determinants.
FIG. 5.
FLI-1 knockdown suppresses LPS-induced MMP expression but variably affects cytokine production. (A–C) Primary monocytes transfected with control or FLI-1 siRNAs were stimulated with LPS with or without IFNγ pretreatment. mRNA levels were analyzed using real-time PCR and normalized relative to β-actin; results are expressed as mean ± SD of triplicate determinants.
Discussion
Macrophage-derived MMP production during chronic inflammatory conditions has been shown to be involved in pathological conditions such as rheumatoid arthritis (RA) and atherosclerosis (Takahashi and others 2002; Stoll and Bendszus 2006; Szekanecz and Koch 2007). For instance, the secretion of MMPs by atheromatous plaque-associated macrophages can cause plaque destabilization, leading to acute coronary events with high morbidity and mortality in people with cardiovascular disease (Libby 2000; Takahashi and others 2002; Stoll and Bendszus 2006). Although Ets proteins have been implicated in the regulation of various MMPs expression in different cell types (Buttice and Kurkinen 1993; Basuyaux and others 1997; Bidder and others 2000; Fenrick and others 2000; Oikawa 2004; Jinnin and others 2005; Seth and Watson 2005; Baillat and others 2006), little is known about their role in MMPs production in primary cells involved in inflammation, such as monocytes and macrophages.
The Ets factor FLI-1 has generated considerable interest in recent years secondary to its association with malignancy (e.g., Ewing's sarcoma and erythroleukemia) and its role in malignant transformation and tumor invasion (Ben David and others 1991; Truong and Ben David 2000; Fuchs and others 2003; Oikawa and Yamada 2003; Seth and Watson 2005). However, FLI-1 also plays a role in the regulation of hematopoiesis, vasculogenesis, lymphoid cell function, and embryogenesis (Zhang and others 1995; Brown and others 2000; Spyropoulos and others 2000) and is highly expressed in hematopoietic tissues (Ben David and others 1991). The downregulation of FLI-1 in response to inflammatory mediators such as LPS has previously been reported by Klemsz and others (1993), but there was no follow-up on its significance. We now demonstrate here that FLI-1 downregulation plays an important role in limiting the production of MMPs during inflammation and thus restraining the associated tissue damage during prolonged immune activation.
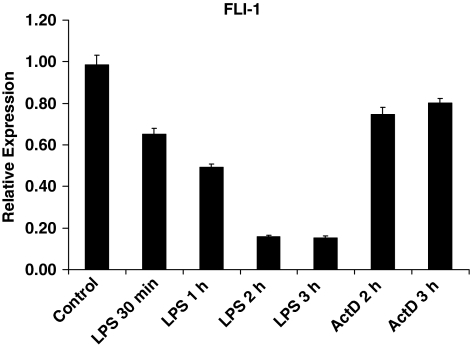
Further analysis of the magnitude and kinetics of FLI-1 downregulation revealed that the FLI-1 mRNA level rapidly diminished upon LPS stimulation in primary monocytes. As shown in Fig. 6, as little as 30 min into LPS challenge, FLI-1 mRNA was already down by as much as 35% and reached a trough in about 2 h. Addition of actinomycin D to prevent new FLI-1 mRNA synthesis showed that FLI-1 RNA transcripts are relatively stable in control cells (Fig. 6, last 2 bars to the right), indicating that LPS actively induces rapid FLI-1 mRNA degradation. In the continuous presence of LPS, FLI-1 level remained near its trough for at least 24 h (data not shown). Interestingly, under a similar condition (with prolonged LPS stimulation), mRNAs of MMP-1, MMP-3, and MMP-10 reached a peak at about 3–6 h and gradually declined back to basal level after 12–24 h (unpublished observation), suggesting that FLI-1 may be necessary for sustained expression of MMPs. Moreover, the fact that only certain LPS-inducible genes [e.g., MMPs and IL-10 (Fig. 5A, B), but not IL-6 and TNFα (Fig. 5C)], are affected by FLI-1 level points to the more distal effect of this Ets factor on LPS-signaling pathways—findings consistent with a general role of Ets proteins as activators and/or repressors at the individual promoter level (i.e., without altering the expression levels of the TLR4/CD14/MD-2 receptor complex and/or positive/negative regulators of TLR4 signaling).
FIG. 6.
LPS stimulation rapidly downregulates FLI-1 expression in primary monocytes. Primary monocytes were stimulated with LPS from 30 min to 3 h, and FLI-1 mRNA was measured using real-time PCR. mRNA levels were analyzed using real-time PCR and normalized relative to β-actin; results are expressed as mean ± SD of triplicate determinants. ActD, actinomycin D.
The genes that were regulated by FLI-1, MMPs 1, 3, and 10, and IL-10 share a common feature of being induced by AP-1 family transcription factors (Unemori and others 1991; Benbow and Brinckerhoff 1997; Westermarck and others 1997; Chakraborti and others 2003; Hu and others 2006). Indeed, the MMP-1 promoter contains a composite Ets/AP-1 site that is critically important for induction of MMP-1 expression. As Ets and AP-1 proteins can interact, it is possible that FLI-1 contributes to MMP-1 activation via this promoter element; we were unable to address this possibility because of limitations of currently available antibodies, which did not work in supershift electrophoretic mobility shift assay (EMSA) or immunoblotting experiments. We have previously reported that LPS-mediated induction of MMPs and IL-10 is suppressed by IFNγ by a mechanism that involves inhibition of AP-1 proteins (Hu and others 2006; Ho and others 2008). We were interested in testing whether IFNγ can regulate FLI-1 expression and/or function, and whether such regulation could contribute to IFNγ-mediated suppression of gene expression. However, IFNγ did not regulate FLI-1 expression, and changes in FLI-1 expression did not noticeably alter the inhibition of MMP-1, 3, and 10 and IL-10 expression by IFNγ. Thus, IFNγ inhibits the expression of these genes by a mechanism independent of FLI-1.
In summary, our results demonstrate that FLI-1 may be necessary for the initial expression of MMPs by activated macrophages (e.g., for tissue remodeling); and that the ensuing rapid downregulation of FLI-1 prevents excessive MMPs production—a presumably self-protective mechanism to minimize nonspecific tissue damage in prolonged inflammatory conditions. Moreover, the fact that reduction in FLI-1 levels specifically leads to suppression of IL-10 (an anti-inflammatory cytokine) but not IL-6 and TNFα (pro-inflammatory cytokines) suggests another role of FLI-1 in immunoregulation, in which downregulation of FLI-1 may be required for sustained immune activation. Taken together, FLI-1 downregulation may represent a novel mechanism by which macrophages limit undesirable damage to tissues without compromising their ability to eradicate infectious pathogens during sustained immune and inflammatory reactions.
Acknowledgment
This work was supported by grants from the NIH (to L.B.I.).
Author Disclosure Statement
No competing financial interests exist.
References
- Athanasiou M. Mavrothalassitis G. Sun-Hoffman L. Blair DG. FLI-1 is a suppressor of erythroid differentiation in human hematopoietic cells. Leukemia. 2000;14(3):439–445. doi: 10.1038/sj.leu.2401689. [DOI] [PubMed] [Google Scholar]
- Baillat D. Leprivier G. Regnier D. Vintonenko N. Begue A. Stehelin D. Aumercier M. Stromelysin-1 expression is activated in vivo by Ets-1 through palindromic head-to-head Ets binding sites present in the promoter. Oncogene. 2006;25(42):5764–5776. doi: 10.1038/sj.onc.1209583. [DOI] [PubMed] [Google Scholar]
- Basuyaux JP. Ferreira E. Stehelin D. Buttice G. The Ets transcription factors interact with each other and with the c-Fos/c-Jun complex via distinct protein domains in a DNA-dependent and -independent manner. J Biol Chem. 1997;272(42):26188–26195. doi: 10.1074/jbc.272.42.26188. [DOI] [PubMed] [Google Scholar]
- Ben David Y. Giddens EB. Letwin K. Bernstein A. Erythroleukemia induction by Friend murine leukemia virus: Insertional activation of a new member of the ets gene family, Fli-1, closely linked to c-ets-1. Genes Dev. 1991;5(6):908–918. doi: 10.1101/gad.5.6.908. [DOI] [PubMed] [Google Scholar]
- Benbow U. Brinckerhoff CE. The AP-1 site and MMP gene regulation: What is all the fuss about? Matrix Biol. 1997;15(8–9):519–526. doi: 10.1016/s0945-053x(97)90026-3. [DOI] [PubMed] [Google Scholar]
- Bidder M. Loewy AP. Latifi T. Newberry EP. Ferguson G. Willis DM. Towler DA. Ets domain transcription factor PE1 suppresses human interstitial collagenase promoter activity by antagonizing protein-DNA interactions at a critical AP1 element. Biochemistry. 2000;39(30):8917–8928. doi: 10.1021/bi000343+. [DOI] [PubMed] [Google Scholar]
- Brown LA. Rodaway AR. Schilling TF. Jowett T. Ingham PW. Patient RK. Sharrocks AD. Insights into early vasculogenesis revealed by expression of the ETS-domain transcription factor Fli-1 in wild-type and mutant zebrafish embryos. Mech Dev. 2000;90(2):237–252. doi: 10.1016/s0925-4773(99)00256-7. [DOI] [PubMed] [Google Scholar]
- Buttice G. Kurkinen M. A polyomavirus enhancer A-binding protein-3 site and Ets-2 protein have a major role in the 12-O-tetradecanoylphorbol-13-acetate response of the human stromelysin gene. J Biol Chem. 1993;268(10):7196–7204. [PubMed] [Google Scholar]
- Cardone M. Kandilci A. Carella C. Nilsson JA. Brennan JA. Sirma S. Ozbek U. Boyd K. Cleveland JL. Grosveld GC. The novel ETS factor TEL2 cooperates with Myc in B lymphomagenesis. Mol Cell Biol. 2005;25(6):2395–2405. doi: 10.1128/MCB.25.6.2395-2405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carella C. Potter M. Bonten J. Rehg JE. Neale G. Grosveld GC. The ETS factor TEL2 is a hematopoietic oncoprotein. Blood. 2006;107(3):1124–1132. doi: 10.1182/blood-2005-03-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti SR. Sood R. Nandi S. Nucifora G. Posttranslational modification of TEL and TEL/AML1 by SUMO-1 and cell-cycle-dependent assembly into nuclear bodies. Proc Natl Acad Sci USA. 2000;97(24):13281–13285. doi: 10.1073/pnas.240315897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborti S. Mandal M. Das S. Mandal A. Chakraborti T. Regulation of matrix metalloproteinases: An overview. Mol Cell Biochem. 2003;253(1–2):269–285. doi: 10.1023/a:1026028303196. [DOI] [PubMed] [Google Scholar]
- Fenrick R. Wang L. Nip J. Amann JM. Rooney RJ. Walker-Daniels J. Crawford HC. Hulboy DL. Kinch MS. Matrisian LM. Hiebert SW. TEL, a putative tumor suppressor, modulates cell growth and cell morphology of ras-transformed cells while repressing the transcription of stromelysin-1. Mol Cell Biol. 2000;20(16):5828–5839. doi: 10.1128/mcb.20.16.5828-5839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs B. Inwards CY. Janknecht R. Upregulation of the matrix metalloproteinase-1 gene by the Ewing's sarcoma associated EWS-ER81 and EWS-Fli-1 oncoproteins, c-Jun and p300. FEBS Lett. 2003;553(1–2):104–108. doi: 10.1016/s0014-5793(03)00984-0. [DOI] [PubMed] [Google Scholar]
- Ho HH. Antoniv TT. Ji JD. Ivashkiv LB. Lipopolysaccharide-induced expression of matrix metalloproteinases in human monocytes is suppressed by IFN-gamma via superinduction of ATF-3 and suppression of AP-1. J Immunol. 2008;181(7):5089–5097. doi: 10.4049/jimmunol.181.7.5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X. Paik PK. Chen J. Yarilina A. Kockeritz L. Lu TT. Woodgett JR. Ivashkiv LB. IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity. 2006;24(5):563–574. doi: 10.1016/j.immuni.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Jinnin M. Ihn H. Mimura Y. Asano Y. Yamane K. Tamaki K. Matrix metalloproteinase-1 up-regulation by hepatocyte growth factor in human dermal fibroblasts via ERK signaling pathway involves Ets1 and Fli1. Nucleic Acids Res. 2005;33(11):3540–3549. doi: 10.1093/nar/gki648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HH. Lee J. Kim JH. Ryu KJ. Kang SA. Park C. Sung K. Nam DH. Kang WK. Park K. Im YH. STAT1 and Nmi are downstream targets of Ets-1 transcription factor in MCF-7 human breast cancer cell. FEBS Lett. 2005;579(18):3941–3946. doi: 10.1016/j.febslet.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Kas K. Finger E. Grall F. Gu X. Akbarali Y. Boltax J. Weiss A. Oettgen P. Kapeller R. Libermann TA. ESE-3, a novel member of an epithelium-specific ets transcription factor subfamily, demonstrates different target gene specificity from ESE-1. J Biol Chem. 2000;275(4):2986–2998. doi: 10.1074/jbc.275.4.2986. [DOI] [PubMed] [Google Scholar]
- Klemsz MJ. Maki RA. Papayannopoulou T. Moore J. Hromas R. Characterization of the ets oncogene family member, fli-1. J Biol Chem. 1993;268(8):5769–5773. [PubMed] [Google Scholar]
- Leprince D. Gegonne A. Coll J. de Taisne C. Schneeberger A. Lagrou C. Stehelin D. A putative second cell-derived oncogene of the avian leukaemia retrovirus E26. Nature. 1983;306(5941):395–397. doi: 10.1038/306395a0. [DOI] [PubMed] [Google Scholar]
- Li R. Pei H. Watson DK. Regulation of Ets function by protein–protein interactions. Oncogene. 2000;19(55):6514–6523. doi: 10.1038/sj.onc.1204035. [DOI] [PubMed] [Google Scholar]
- Libby P. Changing concepts of atherogenesis. J Intern Med. 2000;247(3):349–358. doi: 10.1046/j.1365-2796.2000.00654.x. [DOI] [PubMed] [Google Scholar]
- Nunn MF. Seeburg PH. Moscovici C. Duesberg PH. Tripartite structure of the avian erythroblastosis virus E26 transforming gene. Nature. 1983;306(5941):391–395. doi: 10.1038/306391a0. [DOI] [PubMed] [Google Scholar]
- Oikawa T. ETS transcription factors: Possible targets for cancer therapy. Cancer Sci. 2004;95(8):626–633. doi: 10.1111/j.1349-7006.2004.tb03320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T. Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- Pereira R. Quang CT. Lesault I. Dolznig H. Beug H. Ghysdael J. FLI-1 inhibits differentiation and induces proliferation of primary erythroblasts. Oncogene. 1999;18(8):1597–1608. doi: 10.1038/sj.onc.1202534. [DOI] [PubMed] [Google Scholar]
- Rubinson DA. Dillon CP. Kwiatkowski AV. Sievers C. Yang L. Kopinja J. Zhang M. McManus MT. Gertler FB. Scott ML. Van Parijs L. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33(3):401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Seth A. Robinson L. Thompson DM. Watson DK. Papas TS. Transactivation of GATA-1 promoter with ETS1, ETS2 and ERGB/Hu-FLI-1 proteins: Stabilization of the ETS1 protein binding on GATA-1 promoter sequences by monoclonal antibody. Oncogene. 1993;8(7):1783–1790. [PubMed] [Google Scholar]
- Seth A. Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41(16):2462–2478. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Spyropoulos DD. Pharr PN. Lavenburg KR. Jackers P. Papas TS. Ogawa M. Watson DK. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol Cell Biol. 2000;20(15):5643–5652. doi: 10.1128/mcb.20.15.5643-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll G. Bendszus M. Inflammation and atherosclerosis: Novel insights into plaque formation and destabilization. Stroke. 2006;37(7):1923–1932. doi: 10.1161/01.STR.0000226901.34927.10. [DOI] [PubMed] [Google Scholar]
- Szekanecz Z. Koch AE. Macrophages and their products in rheumatoid arthritis. Curr Opin Rheumatol. 2007;19(3):289–295. doi: 10.1097/BOR.0b013e32805e87ae. [DOI] [PubMed] [Google Scholar]
- Takahashi K. Takeya M. Sakashita N. Multifunctional roles of macrophages in the development and progression of atherosclerosis in humans and experimental animals. Med Electron Microsc. 2002;35(4):179–203. doi: 10.1007/s007950200023. [DOI] [PubMed] [Google Scholar]
- Tamir A. Howard J. Higgins RR. Li YJ. Berger L. Zacksenhaus E. Reis M. Ben David Y. Fli-1, an Ets-related transcription factor, regulates erythropoietin-induced erythroid proliferation and differentiation: Evidence for direct transcriptional repression of the Rb gene during differentiation. Mol Cell Biol. 1999;19(6):4452–4464. doi: 10.1128/mcb.19.6.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AH. Ben David Y. The role of Fli-1 in normal cell function and malignant transformation. Oncogene. 2000;19(55):6482–6489. doi: 10.1038/sj.onc.1204042. [DOI] [PubMed] [Google Scholar]
- Tugores A. Le J. Sorokina I. Snijders AJ. Duyao M. Reddy PS. Carlee L. Ronshaugen M. Mushegian A. Watanaskul T. Chu S. Buckler A. Emtage S. McCormick MK. The epithelium-specific ETS protein EHF/ESE-3 is a context-dependent transcriptional repressor downstream of MAPK signaling cascades. J Biol Chem. 2001;276(23):20397–20406. doi: 10.1074/jbc.M010930200. [DOI] [PubMed] [Google Scholar]
- Unemori EN. Bair MJ. Bauer EA. Amento EP. Stromelysin expression regulates collagenase activation in human fibroblasts. Dissociable control of two metalloproteinases by interferon-gamma. J Biol Chem. 1991;266(34):23477–23482. [PubMed] [Google Scholar]
- Westermarck J. Seth A. Kahari VM. Differential regulation of interstitial collagenase (MMP-1) gene expression by ETS transcription factors. Oncogene. 1997;14(22):2651–2660. doi: 10.1038/sj.onc.1201111. [DOI] [PubMed] [Google Scholar]
- Zhang L. Eddy A. Teng YT. Fritzler M. Kluppel M. Melet F. Bernstein A. An immunological renal disease in transgenic mice that overexpress Fli-1, a member of the ets family of transcription factor genes. Mol Cell Biol. 1995;15(12):6961–6970. doi: 10.1128/mcb.15.12.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]