Abstract
Schizophrenia and related disorders have a major genetic component. Several large-scale studies have uncovered a number of possible candidate genes, but these have yet to be consistently replicated and their underlying biological function remains elusive. One exception is ‘Disrupted in schizophrenia 1’ (DISC1), a gene locus originally identified in a large Scottish family, showing a heavy burden of major mental illnesses associated with a balanced t(1;11)(q42.1;q14.3) chromosome translocation. Substantial genetic and biological research on DISC1 has been reported in the intervening 10 years: DISC1 is now recognized as a genetic risk factor for a spectrum of psychiatric disorders and DISC1 impacts on many aspects of central nervous system (CNS) function, including neurodevelopment, neurosignaling, and synaptic functioning. Evidence has emerged from genetic studies showing a relationship between DISC1 and quantitative traits, including working memory, cognitive aging, gray matter volume in the prefrontal cortex, and abnormalities in hippocampal structures and function. DISC1 interacts with numerous proteins also involved in neuronal migration, neurite outgrowth, cytoskeletal modulation, and signal transduction, some of which have been reported as independent genetic susceptibility factors for psychiatric morbidity. Here, we focus on the growing literature relating genetic variation in the DISC1 pathway to functional and structural studies of the brain in humans and in the mouse.
Keywords: genetics, schizophrenia, mouse models, neurodevelopment, neuroimaging
Introduction
Schizophrenia, like many other common diseases, is complex and multifactorial, with contributions from multiple susceptibility genes, epigenetic, stochastic, and environmental factors.1 Family and twin studies have shown that genetic factors play a major role in the development of schizophrenia, and a number of candidate risk genes have associated with schizophrenia2–4 of which DTNBP1, NRG1, and DISC1 are particularly relevant to this review. DISC1 has captured much attention, not only being associated with schizophrenia but also predisposing individuals to a wide range of major mental disorders. Through genetics, cell biology, animal modeling, and neuroimaging, the DISC1 pathway is emerging as a pivotal mediator of quantitative and pathological brain dysfunction.
The Scottish DISC1 Family and Further Independent Genetic Evidence
DISC1 was first identified in a large Scottish family with a high loading of major mental illness.5 Specifically, the t(1; 11)(q42;q14.3) translocation allele of the DISC1 gene associates with schizophrenia, bipolar affective disorder, and recurrent major depression6 (log of the odds ratio = 7.1) in this large family. The first independent evidence for the involvement of the DISC1 locus in psychiatric illness came from studies in the Finnish population7,8 in which evidence for linkage of schizophrenia and schizoaffective disorder to the 1q32.2–q41 region, proximal to the DISC1 gene, was reported. Further population studies of Taiwan, Scotland, and Britain/Iceland have also shown linkage of chromosome 1q32–42 to major psychiatric illness.8
DISC1 Genetic Mouse Models
Although it is difficult to model a psychiatric disorder like schizophrenia in animals (eg, hallucinations and delusions are likely to be human specific), anatomical, behavioral, and cognitive characteristics have been described in patients, and it is feasible to study these in animal models.9 There has been much debate about the functional consequence of the t(1;11) on DISC1. It may be simply the effect of haploinsufficiency (half normal levels of DISC1 protein) from conception onwards,10 but others have proposed a dominant negative effect from expressing a hypothetical truncated DISC1 protein. On this premise, several transgenic mouse models expressing an N-terminal or C-terminal truncated protein have been generated.11–13 Reduced cortical thickness and partial agenesis of the corpus callosum have been observed in transgenic mice expressing truncated DISC1 generated using a bacterial yeast artificial chromosome vector.13 Enlarged lateral and third ventricles have been shown by magnetic resonance imaging (MRI), in 2 other types of transgenic mice in which human DISC1 cDNA transgenes have been expressed postnatally in forebrain cells.11,12 These are similar to features seen in imaging studies of chronic schizophrenic patients14 (see figure 1). Furthermore, Pletnikov et al12 showed that isolated embryonic cortical neurons from these mice exhibit far less complex dendritic arborization. Both studies were also able to show that DISC1 manipulation resulted in behaviors in the transgenic mice of plausible relevance to human psychiatric illness, including hyperactivity (thought to be the equivalent of psychomotor agitation in humans), increased immobility in the forced swim test (despair), a deficit in prepulse inhibition (PPI),11 working memory deficits (females only), and decreased sociability (males only).12 Although these transgenic models showed different but overlapping phenotypes, they provide further support for altered DISC1 expression predisposing to mental illness.
Fig. 1.
DISC1 Expression, Cognitive, and Behavioral Phenotypes Reported to be Associated With Genetic Variants of DISC1 in Mice. These are representative of endophenotypes of major psychiatric illness and likely to be due to widespread DISC1 expression.
An earlier study15 looked at mice with missense mutations L100P (leucine to proline) or Q31L (glutamine to leucine) of DISC1. MRI of both strains showed the mutant mice to have reduced brain volumes compared with wild-types, especially in the cerebellum, cortex, enterohinal cortex, and thalamus, consistent with the macroscopic changes previously observed in schizophrenic patients.14 Furthermore, the L100P mutant mice showed behaviors, including profound PPI and latent inhibition (LI) deficits, akin to those seen in some with schizophrenia, and these could be reversed by antipsychotic treatment. In contrast, the Q31L mutant mice showed LI deficits, but only modest PPI deficits and exhibited a more depressive-like behavior, partially reversed by the antidepressant bupropion. Thus, as in humans, DISC1 mutations in mice can lead to 2 very different phenotypes.15 Despite the fact that none of these mouse models exactly matches all features characteristically seen in human patients, their phenotypes are consistent with human genetic studies, suggesting extensive locus and allelic genetic heterogeneity.
DISC1 Imaging Genetics in Humans
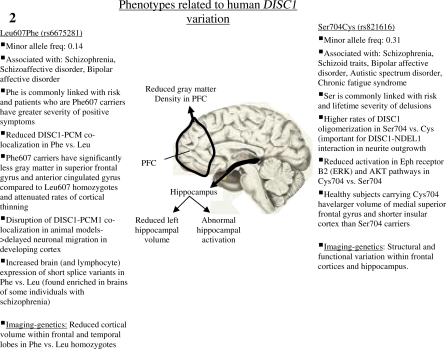
DISC1 contains 2 common nonsynonymous single nucleotide polymorphisms (SNPs)—Ser704Cys (rs821616) and Leu607Phe (rs6675281). Ser704Cys has been associated with schizophrenia and with the structure and function of the hippocampus.16 For example, the Ser allele has been associated with reduced hippocampal gray matter volume and altered engagement of the hippocampus during cognitive tasks (see figure 2). Furthermore, the Cys704 allele has been associated with major depressive disorder, a reduction in gray matter volume in the cingulate cortex, and with decreased fractional anisotropy in prefrontal white matter.
Fig. 2.
DISC1 Expression, Cognitive, and Behavioral Phenotypes Reported to be Associated With Genetic Variants of DISC1 in Humans. These are representative of endophenotypes of major psychiatric illness and likely to be due to widespread DISC1 expression.
There is robust evidence for prefrontal cortex (PFC) abnormalities in psychosis. During verbal fluency tasks, a classic test of prefrontal function, among healthy volunteers, activation of the PFC in Ser704/Ser704 subjects was greater, especially of the left hemisphere, a finding that can be interpreted as a manifestation of less efficient prefrontal function, with more activation needed to achieve the same behavioral output.17 These findings are consistent with reports of association between Ser704, schizophrenia, and impaired declarative memory in schizophrenic patients.18
A Japanese MRI study recently reported that healthy carriers of Cys704 showed significantly larger volumes of the medial superior frontal gyrus and short insular cortex than the Ser704 homozygotes, whereas schizophrenic patients who were carriers of Cys704 tended to have a smaller supramarginal gyrus than the Ser704 homozygotes.18 There was also noted to be a significant correlation between the right medial superior frontal gyrus volume and daily dose of antipsychotic medication in schizophrenic patients who were Ser704 homozygotes.18
Another study investigated the relationship between the DISC1 Leu607Phe polymorphism and prefrontal gray matter volumes using MRI.19 Among patients and healthy volunteers, carriers of Phe607 had significantly less gray matter in the superior frontal gyrus and anterior cingulate gyrus compared with Leu/Leu homozygotes. Patients who were Phe607 carriers also showed greater severity of positive symptoms suggesting that this polymorphism is clinically significant in terms of psychopathology,19 as suggested by others.20
Raznahan et al21 have recently shown that the developmental rate of cortical thinning (CT) is dependent on DISC1 genotype. Consistent with other results,19 Phe607 carriers showed attenuated rates of CT compared with Leu/Leu homozygotes in bilateral superior frontal and left angular gyri between the ages of 9 and 23 years. The Ser704 homozygotes in turn showed accelerated CT compared with Cys704 carriers in the left anterior cingulate and temporal cortices.21 Interestingly, the Ser704Cys SNP affects DISC1-NDEL1 association, which also has a well-established role in cortical maturation.22 The next section of this review focuses on DISC1 interacting partners that we have genetic data from mouse models and from imaging genetic studies.
DISC1 Interacting Partners: Structural and Anatomical Changes in Genetic Mouse Models and Imaging Genetic Studies
DISC1 interacts with numerous neuronal proteins, reflecting the diversity of potential roles that it plays in brain function. These interactors can be broadly classified into those involving: cytoskeleton, cell cycle, signal transduction, intracellular transport/exocytosis, golgi, and neurodevelopment.8 An important question remains as to which of these interactors matters most with respect to schizophrenia and as potential targets of drug development.
Neuregulin-1 and Epidermal Growth Factor Receptor B4
Neuregulin-1 (NRG1) is a large multiexon gene on chromosome 8p. It produces multiple isoforms grouped into types I–VI according to their 5′ exon, through alternate promoters and splicing.23 NRG1 has multiple roles in the central nervous system (CNS) and in processes of possible relevance to schizophrenia including: myelination, glial cell development, migration of radial glial cells during cortical development, neuronal plasticity, development of GABAergic interneurons, and expression of dopamine and serotonin receptors and monoamine transporters.23 Proteolytic cleavage of NRG1 releases the N-terminal portion, including an epidermal growth factor domain (critical for cell-cell signaling), which interacts with a membrane-associated human epidermal growth factor receptor B4 (Erb4)-type receptor tyrosine kinase. NRG1 and its receptor Erb4 are associated with schizophrenia,24 and this interaction results in receptor dimerization, tyrosine phosphorylation, and activation of downstream signaling pathways.25
Recently, it was shown that NRG1 and DISC1 link directly into a common pathway mediated by Erb (Erb 3 and Erb4) receptors and P13K/AKT1 signaling.26 This study demonstrated that NRG1 and NRG2 increased expression of a DISC1 isoform in vitro and that NRG1 knockout mice show selective reduction of DISC1 during neurodevelopment.26
Collectively, these findings suggest that NRG1, Erbs, AKT1, and DISC1 connect in a common pathway that regulates neurodevelopment, which may also contribute to schizophrenia susceptibility. Impaired AKT1 signaling has also been associated with schizophrenia.27 This study, therefore, brings together 3 prominent schizophrenia candidate genes (NRG1, AKT1, and DISC1).
The large number of NRG1 signaling mechanisms and isoforms parallels the many roles it has in the CNS, many of which could be involved in schizophrenia. A 5′ haplotype (HapICE) in the NRG1 gene has been identified that associates with schizophrenia,28 a finding supported by a number of subsequent studies. The NRG1 risk–associated variants are primarily in noncoding intronic and promoter regions, leading to the suggestion that the causative variant may be mediated by altering gene expression rather than protein levels. This hypothesis has been strengthened by a study, which demonstrated that genetic variation at SNP8NRG243177 from HapICE is associated with altered NRG1 expression.29
Mouse studies have also been informative in understanding potential roles for NRG1 in schizophrenia. Complete knockout of NRG1 expression is lethal, whereas animals with heterozygous disruption of NRG1 or of Erb4 receptor function have been shown to have reduced numbers of N-methyl-D-aspartic acid receptors30 and increased levels of dopamine receptors28 in the PFC, as well as exhibiting behavioral abnormalities consistent with schizophrenia.
Our group in Edinburgh was the first to relate NRG1 status to brain imaging measures.31 Among 79 subjects from the Edinburgh High Risk Study, none of whom were receiving treatment at the time of the study, there was a highly significant association between the risk allele at SNP8NRG243177 and psychotic symptoms, across the course of the study. During the Hayling sentence completion task (known to activate frontal and temporal brain regions), functional magnetic resonance imaging (fMRI) revealed decreased activation of right medial PFC in the subjects with the risk (T/T) genotype at SNP8NRG243177 compared with those without the risk allele in the contrast of sentence completion vs rest. We extended this study to also investigate the National Adult Reading Test, a measure of premorbid intelligence quotient (IQ), and found that the T/T group had a significantly decreased IQ compared with the C/T and C/C groups. A similar pattern of deficits was seen using the Wechsler Adult Intelligence Scale, a measure of current IQ, although this effect failed to reach statistical significance.
In a follow on study, subjects with the risk-associated SNP8NRG243177 T/T genotype showed reduced white matter density in the anterior limb of the internal capsule and evidence of reduced structural connectivity in the same region using DTI, suggesting that NRG1 may increase susceptibility to psychosis by altering connections between PFC and other brain regions.32 These findings further implicate white matter disconnectivity in psychosis. Our group also investigated DTI tractography of NRG1 and white matter integrity and showed that the 2 risk-associated variants of NRG1 (T allele at SNP8NRG243177 and C allele at SNP8NRG221533) are associated with reduced white matter integrity in the anterior thalamic radiation, which connects the dorsomedial and anterior thalamic nuclei with the PFC.33 If this connection is indeed disrupted by psychosis-related NRG1 variants, as shown here, this would provide a possible explanation of the abnormal prefrontal function and connectivity often reported in psychosis.
Collectively then, variation at the NRG1 gene has been shown to associate with the development of psychotic symptoms and abnormalities in cortical function and cognition. The fact that the association was with psychotic symptoms, whether or not subjects developed syndromal schizophrenia, suggested that NRG1 is a liability factor for psychotic symptoms rather than schizophrenia per se.
Pericentriolar Material 1
Pericentriolar material 1 (PCM1) is another putative schizophrenia susceptibility gene.34 It has also been shown to interact directly with DISC1.35 DISC1 regulates PCM1 localization to the centrosome,36 an organelle that serves as the main microtubule organizing centre. Microtubule assembly is in turn PCM1 dependent.37 Recent data show that the Leu607Phe polymorphism of DISC1 affects both centrosomal PCM1 localization and spontaneous neurotransmitter (noradrenaline) release but interestingly not microtubule dynamics.38 The assembly of other centrosomal proteins and microtubule anchorage and organization are dependent on PCM1,36 suggesting that the DISC1 Leu607Phe polymorphism could impinge on microtubule function in the absence of changes in microtubule dynamics. The fact that microtubules act as railroads along which molecular motors transport intracellular cargoes, such as precursors of synaptic vesicles to axon terminals, provides a possible explanation for why the Leu607Phe polymorphism alters neurotransmitter release via changes in PCM1-related changes in microtubule stability and organization.
The PCM1 gene has also been associated with orbitofrontal gray matter volumetric deficits,39 and there are a number of haplotypes at the PCM1 locus found to associate with schizophrenia.40 Voxel-based morphometry of cases from a UK case-control sample showed a significant relative reduction in the volume of orbitofrontal gray matter in comparison with patients with non-PCM1-associated schizophrenia, who by contrast showed gray matter volume reduction in the temporal pole, hippocampus, and interior temporal cortex. PCM1 is part of a multicomponent complex with DISC1 so it will be interesting to consider other components as putative contributors to risk and brain pathology.
Serine-threonine protein kinase AKT1
AKT1 (also known as protein kinase B), a serine/threonine kinase with multiple cellular functions including metabolism, cell stress, cell-cycle regulation, and apoptosis, also regulates actin organization and cell motility via Girdin (Girders of actin filaments), which directly binds actin at the leading edge of migrating cells.41 Girdin has been shown to be a DISC1 interactor.41 Its expression in postnatal rodent brain is localized in the dentate gyrus and pyramidal cell layers CA1 and CA3 of the hippocampus, mirroring DISC1 expression.41
There is evidence implicating the AKT pathway in schizophrenia. Genetic variants in AKT have been reported to associate with schizophrenia.9 A transgenic mouse has been generated with defective neuronal AKT1 signaling, which exhibits neurochemical and behavioral phenotypes relevant to schizophrenia, including decreases in prefrontal dopamine signaling and deficits in sensorimotor gating.42 Impaired cortical AKT1 activity in these mice significantly enhanced norepinephrine transporter function, blockade of which reversed the cortical hypodopaminergia and behavioral deficits in these mice, further supporting the potential of AKT1 and norephinephrine transporter as drug targets.
Decreased AKT1 activity and decreased AKT-dependent phosphorylation of glycogen synthase kinase-3 (GSK-3β), a protein that mediates glucose metabolism and is also thought to regulate synaptic plasticity, have been reported in postmortem schizophrenic brains.43 Furthermore, AKT1 knockout mice show impaired PPI of the startle response. Both typical and atypical antipsychotic drugs enhance AKT1 signaling by activating AKT1 or by increasing phosphorylation of GSK-3β.44 The mechanism of action is through D2 receptor activation, which results in AKT1 being inactivated by protein phosphatase 2A (PP2A), in an arrestin-dependent complex, thereby increasing GSK-3β activity. AKT1 is activated by phosphorylation, whereas GSK-3β is inactivated if phosphorylated by AKT1. Levels of the phosphorylated GSK-3 isoform, GSK-3β, have been shown to be diminished in the frontal cortex of people with schizophrenia.27 GSK-3β has recently been identified as a novel DISC1 interactor, bringing the wingless (wnt) pathway and β-catenin into the arena for DISC researchers.45 DISC1 and Girdin dimerize and bind NDEL1, which in turn binds NDE1 and LIS1, other key proteins in neurodevelopment. DISC1 therefore forms higher-order multimers, in a process that alters the binding of NDEL1 and is sensitive to polymorphic variation at the Ser704Cys position,46 which has been previously related by fMRI and working memory tasks to differential hippocampal engagement in normal human subjects and also to altered brain expression of DISC1 partners.8 Like AKT1, DISC1 signaling also inhibits GSK-3β, and interestingly, from a therapeutic viewpoint, the mood stabilizer lithium chloride also targets GSK-3β.
Dysbindin/DTNBP1
DTNBP1, located at 6p22.3, has been identified as a putative schizophrenia susceptibility gene.47 It has also been shown to interact with DISC1.48 The molecular mechanism of action of DTNBP1 is not completely understood, but it is widely distributed in the CNS and appears to be involved in a range of processes including: postsynaptic density function and presynaptic glutamatergic transmission, neuron viability via glutamate signaling resulting in modulation of dopamine and other neurotransmitter activity.25 ‘Sandy’ mice, which have a spontaneous mutation in dysbindin, show cytoarchitectural changes in the hippocampus49 and also exhibit various abnormal behaviors similar to schizophrenia.50 Genetic variation in DTNBP1 has been associated with general cognitive ability,51 while carriers of a putative schizophrenia risk haplotype have reported to have a significantly greater decline in IQ compared with noncarriers suggesting that DTNBP1 influences the severity of intellectual decline seen in schizophrenia.52 Furthermore, DTNBP1 has also been associated with imaging phenotypes in schizophrenia,53 carriers of a risk genotype exhibiting reduced brain volume and regional CT. It has been suggested from the results of these imaging phenotypes that DTNBP1 may be interacting with other schizophrenia-related risk factors to affect laminar thickness.
Conclusions
There is an abundance of genetic and biological evidence supporting a role for DISC1 in risk of psychiatric illness and in neurodevelopment and signaling. Here, we have focused on evidence from human brain imaging studies and from genetically engineered mouse models. We have extended this review to include comparable studies of DISC1 interactors to provide an overview of the role of the DISC1 pathway. Collectively, a picture is emerging of how DISC1, through a multiplicity of protein-protein interactions, including with other known risk genes, regulates 2 core etiological concepts in schizophrenia, namely neurodevelopment and neurotransmission. Further studies in both humans and rodents of defined genetic status for DISC1 and its many partners are likely to improve our understanding of schizophrenia etiology at the molecular and systems level, a prerequisite to the rational diagnosis of patients, the design of new and more effective drug therapies, and the ability to predict the course of illness and treatment responses.
Funding
The Academy of Medical Sciences/Wellcome Trust (R41455 to M.J.); The RS MacDonald Trust (D21419, together with J.H.); Research Councils UK fellowship (GR/T227983/01 to P.A.T.).
References
- 1.Ayhan Y, Sawa A, Ross CA, Pletnikov MV. Animal models of gene-environment interactions in schizophrenia. Behav Brain Res. 2009;204:274–281. doi: 10.1016/j.bbr.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh T, McClellan JM, McCarthy SE, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 5.St Clair D, Blackwood D, Muir W, et al. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- 6.Blackwood DH, Fordyce A, Walker MT, et al. Schizophrenia and affective disorders-cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hennah W, Porteous DJ. The DISC1 pathway modulates expression of neurodevelopmental, synaptogenic and sensory perception genes. PLoS One. 2009;4:e906. doi: 10.1371/journal.pone.0004906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chubb JE, Bradshaw NJ, Soares DC, et al. The DISC locus in psychiatric illness. Mol Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- 9.Arguello PA, Gogos JA. Modelling madness in mice: one piece at a time. Neuron. 2006;52:179–196. doi: 10.1016/j.neuron.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 10.Sawa A, Snyder SH. Genetics. Two genes link two distinct psychoses. Science. 2005;310:1128–1129. doi: 10.1126/science.1121114. [DOI] [PubMed] [Google Scholar]
- 11.Hikida T, Jaaro-Peled H, Seshadri S, et al. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc Natl Acad Sci U S A. 2007;104:14501–14506. doi: 10.1073/pnas.0704774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pletnikov MV, Ayhan Y, Xu Y, et al. Enlargement of the lateral ventricles in mutant DISC1 transgenic mice. Mol Psychiatry. 2008;13:115. doi: 10.1038/sj.mp.4002144. [DOI] [PubMed] [Google Scholar]
- 13.Shen S, Lang B, Nakamoto C, et al. Schizophrenia-related neural and behavioural phenotypes in transgenic mice expressing truncated Disc1. J Neurosci. 2008;28:10893–10904. doi: 10.1523/JNEUROSCI.3299-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellison-Wright I, Glahn DC, Laird AR, Thelen SM, Bullmore E. The anatomy of first-episode and chronic schizophrenia: an anatomical likelihood estimation meta-analysis. Am J Psychiatry. 2008;165:1016–1023. doi: 10.1176/appi.ajp.2008.07101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clapcote SJ, Lipina TV, Millar KJ, et al. Behavioural phenotypes of Disc1 missense mutations in mice. Neuron. 2007;54:387–402. doi: 10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 16.Callicott JH, Straub RE, Pezawas L, et al. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc Natl Acad Sci U S A. 2005;102:8627–8632. doi: 10.1073/pnas.0500515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prata DP, Mechelli A, Fu CH, et al. Effect of disrupted-in-schizophrenia-1 on pre-frontal cortical function. Mol Psychiatry. 2008;13:915–917. doi: 10.1038/mp.2008.76. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi T, Suzuki M, Tsunoda M, et al. The Disrupted-in-Schizophrenia-1 Ser704Cys polymorphism and brain morphology in schizophrenia. Psychiatry Res. 2009;172:128–135. doi: 10.1016/j.pscychresns.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Szeszko PR, Hodgkinson CA, Robinson DG, et al. DISC1 is associated with prefrontal cortical gray matter and positive symptoms in schizophrenia. Biol Psychology. 2008;79:103–110. doi: 10.1016/j.biopsycho.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeRosse P, Hodgkinson CA, Lencz T, et al. Disrupted in schizophrenia 1 genotype and positive symptoms in schizophrenia. Biol Psychiatry. 2007;61:1208–1210. doi: 10.1016/j.biopsych.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 21.Raznahan A, Lee Y, Long R, et al. Common functional polymorphisms of DISC1 and cortical maturation in typically developing children and adolescents. doi: 10.1038/mp.2010.72. [Epub ahead of print July 13, 2010]. Mol Psychiatry. doi:10.1038/mp.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burdick KE, Kamiya A, Hodgkinson CA, et al. Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia: evidence of epistasis and competitive binding. Hum Mol Genet. 2008;17:2462–2473. doi: 10.1093/hmg/ddn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9:427–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 2009;25:528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 26.Seshadri S, Kamiya A, Yokota Y, et al. Disrupted-in-Schizophrenia-1 expression is regulated by beta-site amyloid precursor protein cleaving enzyme-1-neuregulin cascade. Proc Natl Acad Sci U S A. 2010;107:5622–5627. doi: 10.1073/pnas.0909284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emamian ES, Hall D, Birnbaum MJ, Karayiorgou M, Gogos JA. Convergent evidence for impaired AKT1-GSK3beta signaling in schizophrenia. Nat Genet. 2004;36:115–116. doi: 10.1038/ng1296. [DOI] [PubMed] [Google Scholar]
- 28.Stefansson H, Sigurdsson E, Steinthorsdottir V, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Law AJ, Lipska BK, Weickert CS, et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5’ SNPs associated with the disease. Proc Natl Acad Sci U S A. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy K, Murtie JC, El-Khodor BF, et al. Loss of erb4 signaling in oligodendrocytes alters myelin and dopaminergic function, a potential mechanism for neuropsychiatric disorders. Proc Natl Acad Sci U S A. 2007;104:8131–8136. doi: 10.1073/pnas.0702157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall J, Whalley HC, Jobb DE, et al. A neuregulin 1 variant associated with abnormal cortical function and psychotic symptoms. Nat Neurosci. 2006;9:1477–1478. doi: 10.1038/nn1795. [DOI] [PubMed] [Google Scholar]
- 32.McIntosh AM, Moorhead TWJ, Job D, et al. The effects of a neuregulin 1 variant on white matter density and integrity. Mol Psychiatry. 2008;13:1054–1059. doi: 10.1038/sj.mp.4002103. [DOI] [PubMed] [Google Scholar]
- 33.Sprooten E, Lymer GKS, Munoz Maniega S, et al. The relationship of anterior thalamic radiation integrity to psychosis risk associated neuregulin-1 variants. Mol Psychiatry. 2009;14:237–238. doi: 10.1038/mp.2008.136. [DOI] [PubMed] [Google Scholar]
- 34.Gurling HM, Critchley H, Datta SR, et al. Genetic association and brain morphology studies and the chromosome 8p22 pericentriolar material 1 (PCM1) gene in susceptibility to schizophrenia. Arch Gen Psychiatry. 2006;63:844–854. doi: 10.1001/archpsyc.63.8.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabares-Seisdedos R, Rubenstein JL. Chromosome 8p as a potential hub for developmental neuropsychiatric disorders: implications for schizophrenia, autism and cancer. Mol Psychiatry. 2009;14:563–589. doi: 10.1038/mp.2009.2. [DOI] [PubMed] [Google Scholar]
- 36.Kamiya A, Tan PL, Kubo K, et al. Recruitment of PCM1 to the centrosome by the cooperative action of DISC1 and BBS4: a candidate for psychiatric illnesses. Arch Gen Psychiatry. 2008;65:996–1006. doi: 10.1001/archpsyc.65.9.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dammermann A, Mercedes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eastwood SL, Hodgkinson CA, Harrison PJ. DISC-1 Leu607Phe alleles differentially affect centrosomal PCM1 localization and neurotransmitter release. Mol Psychiatry. 2009;14:556–557. doi: 10.1038/mp.2009.13. [DOI] [PubMed] [Google Scholar]
- 39.Eastwood SL, Walker M, Hyde TM, Kleinman JE, Harrison PJ. The DISC1 Ser704Cys substitution affects centrosomal localization of its binding partner PCM1 in glia in human brain. Hum Mol Genet. 2010;19:2487–2496. doi: 10.1093/hmg/ddq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Datta SR, McQuillin A, Rizig M, et al. A threonine to isoleucine missense mutation in the pericentriolar material 1 gene is strongly associated with schizophrenia. Mol Psychiatry. 2010;15:615–628. doi: 10.1038/mp.2008.128. [DOI] [PubMed] [Google Scholar]
- 41.Enomoto A, Asai N, Namba T, et al. Roles of disrupted-in-schizophrenia 1-interacting protein girdin in postnatal development of the dentate gyrus. Neuron. 2009;63:774–787. doi: 10.1016/j.neuron.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 42.Siuta MA, Robertson SD, Kocalis H, et al. Dysregulation of te norephinephrine transporter sustains cortical hypodopaminergia and schizophrenia-like behaviours in neuronal rictor nulle mice. PLoS Biol. 2010;8:e1000393. doi: 10.1371/journal.pbio.1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arguello PA, Gogos JA. A signaling pathway AKTing up in schizophrenia. J Clin Invest. 2008;118:2018–2021. doi: 10.1172/JCI35931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freyberg Z, Ferrando SJ, Javitch JA. Roles of the Akt/GSK-3 and Wnt signaling pathways in schizophrenia and antipsychotic drug action. Am J Psychiatry. 2010;167:388–396. doi: 10.1176/appi.ajp.2009.08121873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mao Y, Ge X, Frank CL, et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3β/β-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leliveld SR, Hendrik P, Michel M, et al. Oligomer assembly of the C-terminal DISC1 domain (640-854) is controlled by self-association motifs and disease-associated polymorphism S704C. Biochemistry. 2009;48:7746–7755. doi: 10.1021/bi900901e. [DOI] [PubMed] [Google Scholar]
- 47.Duan X, Chang JH, Ge S, et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mead CL, Kuzyk MA, Moradian A, et al. Cytosolic protein interactions of the schizophrenia susceptibility gene dysbindin. J Neurochem. 2010;113:1491–1503. doi: 10.1111/j.1471-4159.2010.06690.x. [DOI] [PubMed] [Google Scholar]
- 49.Chen XW, Feng YQ, Hao CJ, et al. DTNBP1, a schizophrenia susceptibility gene, affects kinetics of transmitter release. J Cell Biol. 2008;181:791–801. doi: 10.1083/jcb.200711021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng YQ, Zhou ZY, He X, et al. Dysbindin deficiency in sandy mice causes reduction of snapin and displays behaviors related to schizophrenia. Schizophr Res. 2008;106:218–228. doi: 10.1016/j.schres.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 51.Burdick KE, Lencz T, Funke B, et al. Genetic variation in DTNBP1 influences general cognitive ability. Hum Mol Genet. 2006;15:1563–1568. doi: 10.1093/hmg/ddi481. [DOI] [PubMed] [Google Scholar]
- 52.Burdick KE, Goldberg TE, Funke B, et al. DTNBP1 genotype influences cognitive decline in schizophrenia. Schizophr Res. 2007;89:169–172. doi: 10.1016/j.schres.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narr KL, Szeszko PR, Lencz T, et al. DTNBP1 is associated with imaging phenotypes in schizophrenia. Hum Brain Mapp. 2009;30:3783–3794. doi: 10.1002/hbm.20806. [DOI] [PMC free article] [PubMed] [Google Scholar]