Abstract
The relationship between exposure to environmental chemicals during pregnancy and early childhood development is an important issue which has a spatial risk component. In this context, we have examined mental retardation and developmental delay (MRDD) outcome measures for children in a Medicaid population in South Carolina and sampled measures of soil chemistry (e.g. As, Hg, etc.) on a network of sites which are misaligned to the outcome residential addresses during pregnancy. The true chemical concentration at the residential addresses is not observed directly and must be interpolated from soil samples. In this study, we have developed a Bayesian joint model which interpolates soil chemical fields and estimates the associated MRDD risk simultaneously. Having multiple spatial fields to interpolate, we have considered a low-rank Kriging method for the interpolation which requires less computation than Bayesian Kriging. We performed a sensitivity analysis for a bivariate smoothing, changing the number of knots and the smoothing parameter. These analyses show that a low-rank Kriging method can be used as an alternative to a full-rank Kriging, reducing computational burden. However, the number of knots for the low-rank Kriging model need to be selected with caution as a bivariate surface estimation can be sensitive to the choice of the number of knots.
Keywords: environmental exposure, logistic, spatial, low-rank Kriging, Bayesian
1. Introduction
Mental retardation and development delay (MRDD) in young children has enormous public health implication as they have relatively high prevalence rates of 2-4% or more in most populations. However, the causes of approximately 50% of MRDD cases still remain unknown [1]. One possible cause for MRDD is environmental chemical exposure. Exposure to lead and mercury have been reported to be associated with MRDD in several studies. Since the cerebral neuronal development occurs in utero and in the first 2 years of postnatal life, it is necessary to investigate the association between MRDD and prenatal environmental exposure to different chemicals. This association has not been examined well since it is difficult to detect the environmental exposure.
Since it is difficult to observe environmental exposure directly, it must be inferred or interpolated from observed samples. However, the errors involved in this interpolation have been largely ignored in other studies which can result in misleading risk estimates associated with the exposures [2-4]. To overcome this problem, we combine the interpolation of soil chemicals and the estimation of the associated risk simultaneously by employing a Bayesian approach. Our method addresses the same problem as that of Fuentes et al.[5] who examined the association between the predicted fine particles and mortality. However, they used the predicted particles as ‘plug-in’ estimates and this does not account for the interpolation error associated with each spatial predictor. They also used a special form of Poisson data model with full Bayesian Kriging. Here we use a joint model which interpolates and fits the risk model simultaneously. Our approach is similar to that of Smith et al. [6] in which residential radon levels are linked to leukemia risk, although we examine a more complex multivariate interpolation problem in this study. Employing a Bayesian approach and having multiple chemicals to interpolate means that computational intensity is an important factor for choosing an interpolation method in this study.
In this paper, we will present an analysis of the effects of multiple soil chemicals on MRDD risk. In general, in environmental risk assessment studies there could potentially be a large number of spatially-referenced predictors and so the computational burden of joint modeling is an issue. Since the purpose of our paper is to develop a statistical method which can be applied to this situation, we have chosen a spline method (low-rank Kriging) instead of Kriging [7] in our Bayesian approach. To compare the predictive performance of the spline method with Kriging, a simulation study is also presented here.
The remainder of this paper is organized as follows. The motivating MRDD data are described in Section 2. The development of a Bayesian logistic spatial model relating residential chemicals to MRDD outcome is detailed in Section 3. Low-rank Kriging methods for multiple spatial fields are considered in the model development. In Section 4, a simulation study is performed to verify the prediction performance of low-rank Kriging with different number of knots in comparison with full-rank Kriging. In Section 5, results from the proposed Bayesian logistic spatial model are summarized. Finally, a discussion of our analytic approach is provided in Section 6.
2. Materials
2.1. MRDD data from Medicaid in South Carolina
This work is motivated by a study of MRDD incident cases in Medicaid population in South Carolina between 1996 and 2001. Medicaid is a health insurance for individuals with low incomes in the United States, covering low-income parents, children, seniors, and people with disabilities. During each month of pregnancy, residential addresses for mothers are recorded. Other available mom and baby characteristics include: mother's age, mother's ethnicity, mother's alcohol consumption during pregnancy, parity, birth weight, baby sex and follow-up time. We term these covariates as ‘mom and baby’ or ‘individual’ covariates. Using spatial cluster analysis, MRDD clusters were identified first and then five clusters were selected for soil sampling. There were 6535 MRDD cases and controls from the five clusters. Details of cluster analysis were described previously [8]. Data from five further clusters would be added later in the study. This will possibly increase the data set to about 13000 cases and controls. In this study, a preliminary sample from this large dataset was randomly selected and used to develop a statistical model which will be applied to the original data at a later stage, once all the data become available. This dataset comprises 141 MRDD cases and controls from month 6 of pregnancy, year 1998 in the second cluster.
2.2. Soil Samples from MRDD clusters
In each chosen cluster, soil samples were collected from a network of sample sites which has a regular grid pattern covering the whole cluster area [9]. These soil sample sites are spatially misaligned to the residential addresses of pregnant women. Thus, interpolation of soil chemical fields is required to link MRDD cases to environmental exposure risk. In this study, we used nine soil chemicals from 119 soil samples collected from the second cluster to develop our statistical model. Thus we have nine spatial fields to interpolate here but the original dataset includes nine soil chemical measures for each of ten clusters. This means we have ninety spatial fields to interpolate. The implication of the need for interpolation of a large number of predictors to a large number of outcome sites clearly supports the need for computational efficiency. Nine chemicals measured in soil samples include arsenic (As), barium (Ba), beryllium (Be), chromium (Cr), copper (Cu), lead (Pb), manganese (Mn), nickel (Ni) and mercury (Hg). We denote these observed soil chemicals at sample sites as ‘environmental’ covariates.
3. Methods
3.1. Logistic Spatial Model
Assume first that we observe a realization of a spatial process where a binary mark is observed on a set of locations. Here we assume that residential addresses of MRDD cases are the locations with binary mark yi =1 and controls as yi = 0. The controls are the residential locations of pregnant women who had a normal child and the MRDD cases are the residential locations of pregnant women who subsequently had a child with a MRDD diagnosis. This latter group is a subset of the local birth population. Thus we use a logistic regression to model this spatially-referenced outcome. We observe covariates that relate to the individual outcome such as ‘individual’ covariates and ‘environmental’ covariates which were described in Section 2. We term this model ‘logistic spatial’.
Data are in the form (xi, yi), 1 ≤ i ≤ n, where the yi is a binary outcome for xi and xi ∈ ℜ2 represents geographical locations. yi can be regarded as a Bernoulli random variable and we modeled the probability of having MRDD using a logit link function:
| (1) |
where ui = (uli,…,uqi) is a vector of individual covariates with corresponding regression parameters ; is a vector of latent soil chemicals at xi; is a vector of regression parameters of environmental covariates in the model; and εi represents a random effect term. Here, the star symbol (*) denotes the unobserved ‘true’ covariates. The inclusion of a random effect term is intended to make some allowance for confounding in the outcome and could take a variety of forms. A convolution model [10] is often assumed where an additive combination of an uncorrelated and spatially-correlated effect is employed. Often a conditional autoregressive (CAR) model is assumed for the latter effect. However, recently it has been found that CAR components can be confounded or collinear with spatially-referenced predictors (such as, in this case, soil chemicals) [11, 12] and so spatial predictor effects may be masked by this effect. We have performed a variogram analysis of residuals from a logistic model fit to non-spatial predictors (mom and baby covariates) and found negligible spatial correlation. Taking these two considerations into account, we have employed only an uncorrelated random effect term to accommodate confounding. Ma et al. [11] reported improved power in estimation of predictor effects when using such an effect with a zero mean Gaussian prior specification. We have assumed that specification here: ɛi ∼ N(0, σɛ2).
In our study, environmental covariates, z*(xi), are not observed at the outcome sites but they are observed at different geographical locations (soil sample sites). Let z(sj) = {z1(sj),…, zp(sj)}, 1 ≤ j ≤ m denote a vector of observed soil chemicals at sj where sj ∈ ℜ2 represent the soil samples sites. Then, our model must allow for both the prediction of z*(xi) from z(sj) and fitting the logistic spatial model reflecting the uncertainty in the predicted values. This can be achieved by fitting a joint model which will be developed in the following section.
3.2. Joint posterior distribution
Our logistic spatial model includes both mom and baby covariates and unobserved environmental covariates which must be predicted from the observed soil samples. For the prediction of the latent soil chemical concentrations, a low-rank Kriging [13, 14] is employed in this study. These two parts comprise our joint model and we have two likelihoods at two different sets of the locations.
The joint posterior distribution is proportional to the product of the likelihood function associated with the logistic spatial model (Equation (1) in Section 3.1.), the low-rank Kriging model (Section 3.3.2.), the predictive distribution of the latent spatial fields (Section 3.3.3.) and prior distributions for the model parameters:
| (2) |
where β0, β1 and β2 represent regression parameters for the logistic spatial model and ε represents a random effect as defined in Section 3.1.; θ represents a vector of parameters for the low-rank Kriging model; the latent soil chemical concentrations, z*(xi), are a function of θ. The models for soil chemical concentrations will be defined in the following section (Section 3.3).
We used Markov chain Monte Carlo (MCMC) methods to sample from the joint posterior distribution. It is difficult to use standard software routines, such as those found in WinBUGS to perform MCMC sampling in our case since our hierarchical model is very complex and several spatial fields need to be interpolated. Instead, we implemented a MCMC algorithm for our model within the C language. A Metropolis-Hasting algorithm [15] is used. The details of the Metropolis-Hasting algorithm used in this study are given in the Appendix.
3.3. Prediction of multiple environmental covariates
3.3.1. Kriging vs. Splines
In this study, Kriging could have been employed for the prediction of soil chemical concentrations assuming a stationary, multivariate Gaussian distribution which is often regarded as optimum interpolation in geostatistics. Quantifying spatial variability through the covariance function, Kriging can produce maps of optimal predictions from incomplete and noisy spatial data [7]. Kriging requires matrix decomposition whose complexity increases as O(n3) in the number of locations, n. If posterior sampling is employed for Kriging in a Bayesian approach, the matrix decomposition is required at every iteration of an MCMC algorithm and the computational burden increases. Note that if we assumed a prior cross-covariance structure for the spatial predictors then this would increase computation time and reduce parsimony in the model. We have not pursued that approach here. The details of Bayesian Kriging are provided by Banerjee et al. [16]. Having multiple soil chemical concentrations to interpolate, we considered an alternative spatial prediction method which is less computationally intense and decided to use a spline method: low-rank Kriging [13, 14, 17]. Low-rank Kriging requires a covariance matrix decomposition once a priori before running an MCMC algorithm fixing a spatial range parameter. This means that the computation can be reduced by 9 × O(1193) × total iteration of the MCMC algorithm for the preliminary sample dataset and by 90 × O(1193) × total iteration of the MCMC algorithm for our original dataset. Although Bayesian Kriging provides a more flexible modeling method which enables us to estimate a spatial range parameter, we chose to use a low-rank Kriging model due to this computational advantage. There is extensive literature available on spline models. A review of different spline models for univariate data is given by several authors [14, 18, 19]. For bivariate and higher-dimensional data, the details are presented by Denison et al. [20] and Ruppert et al. [14].
3.3.2. Low-rank Kriging model for multiple soil chemicals
In this section, a low-rank Kriging model for each soil chemical is developed. As a check for the assumption of prior independence of the fields we examined the empirical correlation between the observed measurements. When the sample correlations were calculated, most chemicals did not have high correlations (Table 1). This further supports our assumption for a univariate model for each soil chemical. For the p th chemical, let denotes a 1×m vector of the p th chemical concentration where 1≤ p ≤ P. Let {κ1,…,κK} be a set of K < m distinct points which is a representative subset of {sj}, 1 ≤ j ≤ m. This subset is often selected by a space filling algorithm and we have examined that approach here. These points are referred to as knots. Different strategies for the selection of knots are described by Ruppert et al. [14]. The number of knots was determined by K = max{20, min(m/4, 150)} following Ruppert et al. [14]'s recommendation. In this study, two knot location patterns were used: 1) knots were placed over the whole study area using a space filling algorithm, (this was performed by the R package, field), and 2) knots were placed close to the prediction locations generating random points based on a kernel smoothed intensity of the outcome sites. This was carried out by the R package ( spatstat).
Table 1. Correlations of different soil chemical concentrations (n=119).
| As | Ba | Be | Cr | Cu | Pb | Mn | Ni | Hg | |
|---|---|---|---|---|---|---|---|---|---|
| As | 1.000 | 0.143 | 0.080 | 0.038 | 0.031 | 0.363 | 0.281 | -0.021 | 0.133 |
| Ba | 1.000 | 0.735 | 0.396 | 0.504 | 0.128 | 0.703 | 0.651 | 0.063 | |
| Be | 1.000 | 0.635 | 0.657 | 0.101 | 0.642 | 0.723 | 0.001 | ||
| Cr | 1.000 | 0.628 | 0.178 | 0.493 | 0.733 | 0.009 | |||
| Cu | 1.000 | 0.317 | 0.347 | 0.671 | 0.174 | ||||
| Pb | 1.000 | 0.274 | 0.211 | 0.329 | |||||
| Mn | 1.000 | 0.501 | 0.098 | ||||||
| Ni | 1.000 | -0.010 | |||||||
| Hg | 1.000 |
The low-rank Kriging model for the p th chemical can be represented as follows:
| (3) |
where αp is a fixed-effect parameter vector for Wp; ṽp is a random-effect parameter vector for Z̃p; E(ṽp) = 0; ; and .
Here, the first-order trend is assumed and C(r) is an inter-point covariance function. In what follows, we have assumed a special case of Matérn family covariance model for C(r) fixing the Matérn smoothness parameter at ν =3/2:
| (4) |
The spatial range parameter ρp controls the smoothness of the fitted surface and the larger ρp, the smoother the surface. Note that if we fix ρp a priori, we can fit the model using a generalized linear model framework. Equation (3) becomes a linear mixed model:
| (5) |
where ; and .
Employing a Bayesian approach, both the spatial range parameter and the partial sill parameter can be estimated. In this study, the partial sill parameter is estimated using the joint distribution described in Section 3.2 whereas the spatial range parameter for each chemical (ρp) is estimated a priori from a variogram analysis. This provides significant savings in computation albeit at the expense of some bias. After fixing ρp, we need to invert the 119×119 dimension covariance matrix, Ωp, only once at the beginning but not at every MCMC iteration for each chemical. In several studies, the spatial range parameter has been fixed as it is difficult to obtain a reliable estimate due to its lack of identification with the partial sill, without resort to profile likelihood within 2 stage procedures (see Dietrich and Osborne [21]; Hoeting et al. [22])
After inspecting the contour plots of nine soil chemicals, a variogram analysis was performed for each chemical by the R package, geoR assuming a Matérn covariance function. The estimated spatial range parameter for each chemical is used to fix ρp a priori for the suggested logistic-spatial model. The rationale for fixing this parameter is two-fold: 1) the need to avoid estimation problems and 2) to reduce the computation time.
While French et al. [13] suggest an approach whereby ρp is fixed at 1) and 2) , using a much smaller value to assess the effect of extreme values, we have used variogram analysis which yields a different value for ρ̂p for each chemical more closely matching the spatial structure of the predictor.
3.3.3. Prediction of multiple soil chemicals using Low-rank Kriging
As explained in Section 3.1., we measured soil chemical concentrations at the sample site but not at the outcome site. Thus, we treat the latent soil chemical concentrations as missing values. The details of fitting linear mixed-effect models with missing values are presented by Schafer and Yucel [23].
For the p th chemical, let zp(obs) denote a vector of the observed chemical concentrations at the sample sites and zp(mis) denote a vector of the latent chemical concentrations at the outcome sites. In our MCMC algorithm, let and zp(mis)(t) represent current versions of the unknown parameters and missing data respectively. Then, θ(t) and are updated as follows:
| (6) |
| (7) |
In our low-rank Kriging model, the posterior predictive distribution of zp(mis) is as follows:
| (8) |
where C(r), κK, and Ωp are as defined in Equation (3).
At the t th iteration of the MCMC algorithm, both zp(obs) and are used as the complete data to update θp for the low-rank Kriging model described in Section 3.3.2. Then, we update using the joint posterior distribution in Equation (2) which comprises the posterior predictive distribution above (Equation (8)) and the logistic spatial model likelihood. In this way, the risk estimate can reflect the uncertainty involved in the prediction procedure properly which cannot be achieved by using plug-in estimates. For the details of the Metropolis-Hasting algorithm for the prediction, please refer to the Appendix.
4. Simulation Study
Using the low-rank approximation helps to improve computational speed [24]. For one-dimensional data, it has been found that low-rank Kriging has a similar prediction mean square error compared to full-rank Kriging [25]. It has also been found that low-rank smoothing performs as well as full-rank smoothing in terms of mean square error for one-dimensional data [26]. For a penalized spline model, it has been shown that sensitivity to the knot locations in one dimension is quite low [26, 27]. However, for higher-dimensional data, the effects of the number of knots and their locations on low-rank Kriging have not been tested previously to our knowledge. Thus, as part of our analysis we performed a simulation study to evaluate the effect of the number of knots on the performance of low-rank Kriging for bivariate data prior to fitting the multiple field models.
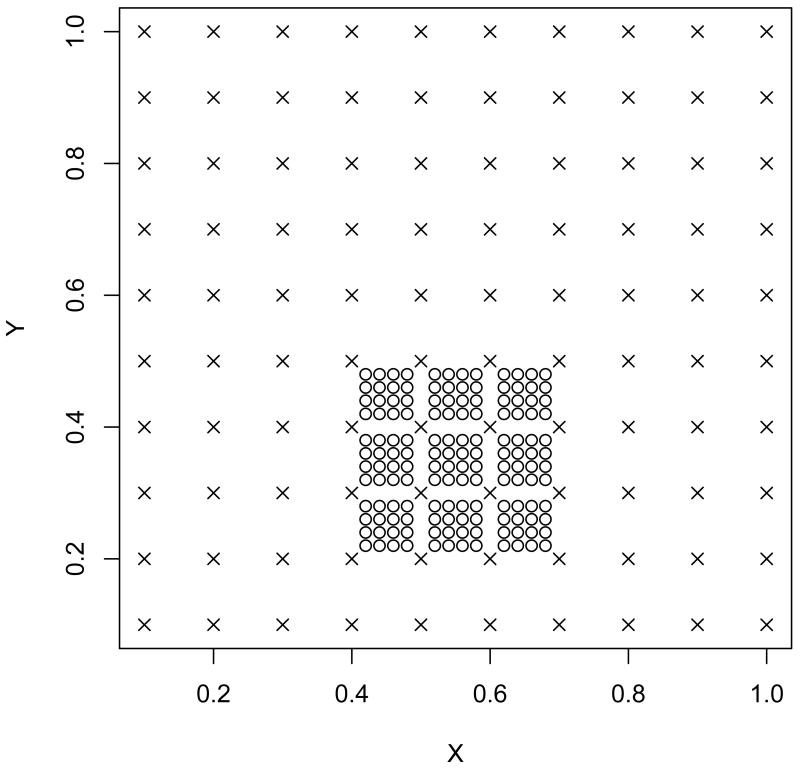
Firstly, a 10 by 10 regular grid was created within a unit square {0.1,…,1}×{0.1,…,1}. This first grid was assumed to be soil sample sites. Let sj denote the first grid where sj ∈ ℜ2 and 1 ≤ j ≤ 100. Imitating our MRDD outcome which was observed on a fine grid, the second grid was created from all combination of x2 = {0.42, 0.44, 0.46, 0.48, 0.52, 0.54, 0.56, 0.58, 0.62, 0.64, 0.66, 0.68} and y2 = {0.22, 0.24, 0.26, 0.28, 0.32, 0.34, 0.36, 0.38, 0.42, 0.44, 0.46, 0.48}. Let xi denote the second grid where xi ∈ ℜ2 and 1 ≤ i ≤ 144. The graphical representation of these two grids is shown in Figure 1.
Figure 1.
Two sets of grids for simulation. × represents the first grid and ○ represents the second grid.
We generated ytrue(sj) and ytrue(xi) from a stationary spatial Gaussian random field on both sj and xi This simulation was performed in R using the GaussRF package assuming zero mean and Matérn covariance function. Let WT = {sj xi} be the grid which includes both the first grid (sj) and the second grid (xi), then the Gaussian random field can be represented as follows:
We used ytrue(xi) as a prediction set and ytrue(sj) as a modeling set here. Bayesian Kriging was performed assuming the correct form of the model (zero mean, Matérn covariance function). This model was fitted using the SpBayes package in R. Low-rank Kriging models were fitted with different number of knots (K =5, 10, …,95) using the nlme package in R. For each K, knots were selected as a representative subset of xi via a space-filling algorithm using the field package in R. We tested the performance of low-rank Kriging for each number of knots using a different set of values for σ2 and ρ. All the combinations of (1, 0.5, 0.25) for each parameter were used.
The procedure for our simulation study is as follows:
Generate 500 samples of ytrue(sj) and ytrue(xi)
For each sample, leave out ytrue(xi) and use only ytrue(sj) to fit Bayesian Kriging and low-rank Kriging.
Predict ŷfull(xi) using the fitted Bayesian Kriging model
Predict ŷlow_K(xi) where K =5, 10, …,95 using the fitted low-rank Kriging model with K knots.
-
Calculated mean squared error of prediction (MSEP) for 500 samples as follows:
Find the 2.5th and 97.5th quantiles of MSEPfull. We call this ‘95% MSEP Range of Bayesian Kriging’. Then, count how many times MSEPlow_K falls in this range
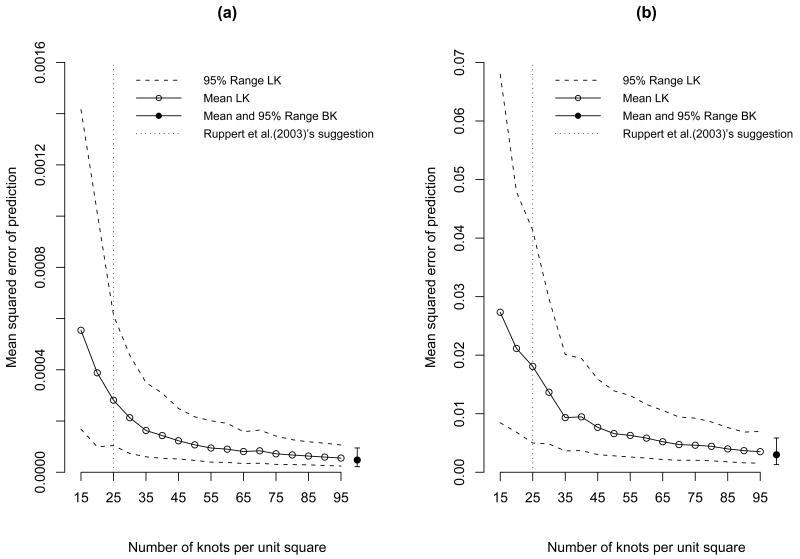
Figure 2 shows the mean squared error of prediction (MSEP) for the low-rank Kriging with each number of knots and the full-rank Bayesian Kriging with different σ2 and ρ. The MSEP is the average squared difference between the estimate and the true value. The separate error-bar plot represents the mean and the 2.5th and 97.5th quantiles of MSEP of Bayesian Kriging estimates. These plots indicate that the MSEP is smaller in general when the spatial surface is smoother, i.e. ρ is larger. For both plots, the 95% MSEP range of Bayesian Kriging is narrow indicating that the model recovers the simulated data well. For both cases, as the number of knots increases, the MSEP decreases; it decreased rapidly at the beginning and then improvement became smaller. Ruppert et al. [14] recommend K=25 knots which is depicted by the vertical line. We can see that the 95% range of low-rank Kriging with 25 knots is much wider than that of Bayesian Kriging for both cases. When we used different sets of σ2 and ρ, similar results were observed.
Figure 2.
Mean-squared error of prediction of low-rank Kriging (LK) and Bayesian Kriging (BK) with (a) σ2=1 and ρ=1 and (b) σ2=1 and ρ=0.25.
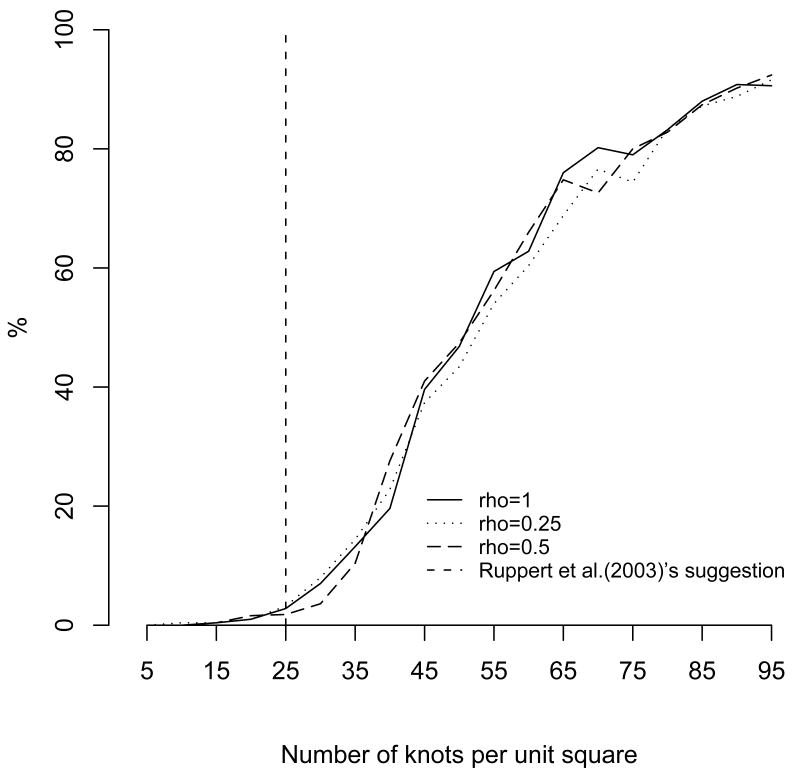
To compare the prediction performance of low-rank Kriging with different number of knots to Bayesian Kriging, the percentage of the MSEP of low-rank Kriging falling in the 95% MSEP range of Bayesian Kriging is calculated varying the smoothing parameter (Figure 3). We can see that if we followed Ruppert et al.'s suggestion, about 3% of the MSEP of low-rank Kriging fell in the 95% MSEP range of Bayesian Kriging. To achieve about 80%, at least 65 knots are required when the surface is smooth (ρ =1) and 80 knots when the surface is less smooth (ρ = 0.25). Of course, the above results suggest that Ruppert et al's suggestion is highly conservative. It should be noted that the results assume that Bayesian Kriging is the ‘gold-standard’ for comparison.
Figure 3.
Percentage of mean-squared error of prediction (MSEP) of low-rank Kriging falling in the 95% MSEP Range of Bayesian Kriging with σ2=1 different ρs.
5. Application: analysis of the MRDD outcome in a Medicaid population in South Carolina
In this section we use the MRDD data collected by Medicaid in South Carolina to fit our Bayesian joint model. The response variable is the diagnosis of MRDD (yes (yi=1) or no (yi=0)). During each month of pregnancy, residential addresses for mothers were recorded. In addition to the geographical location, mother's age, mother's ethnicity, mother's alcohol consumption during pregnancy, parity, birth weight, baby's sex and follow-up time were also available. After MRDD clusters were identified, five areas were selected for soil sampling. In this study, data from the second cluster in month 6 of pregnancy, year 1998 were randomly selected for our analysis (n=141). In the area, 119 soil samples were taken and 9 chemical concentrations were measured: arsenic (As), barium (Ba), beryllium (Be), chromium (Cr), copper (Cu), manganese (Mn), nickel (Ni), lead (Pb) and mercury (Hg). Table 1 shows the sample correlations between the nine soil chemicals. About 50% of chemical combinations had the correlation less than 0.3 and about 11% of chemical combinations had the correlation greater than 0.7. The lowest and highest correlation was observed between Be and Hg (r=0.001) and Ba and Be (r=0.735), respectively.
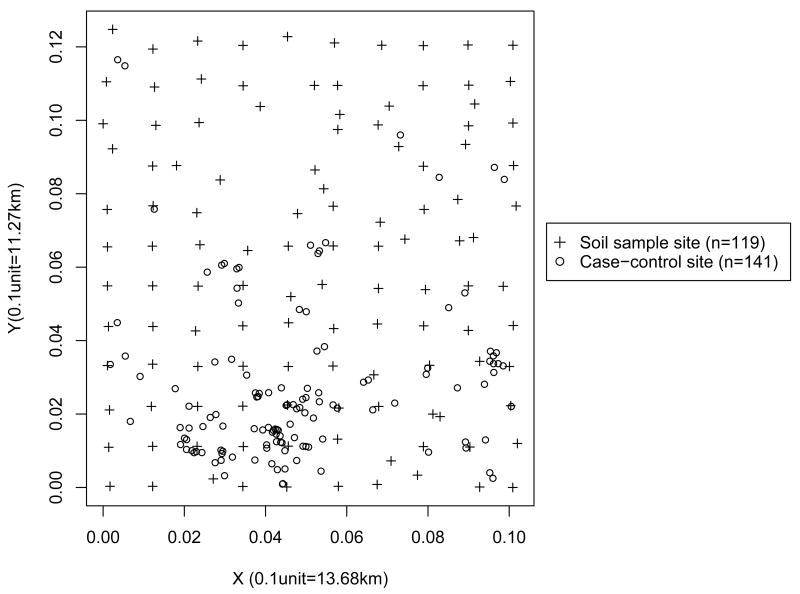
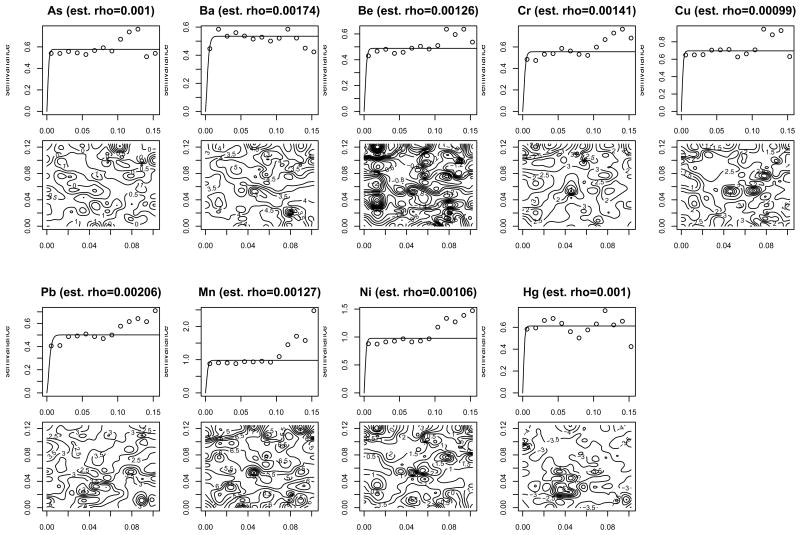
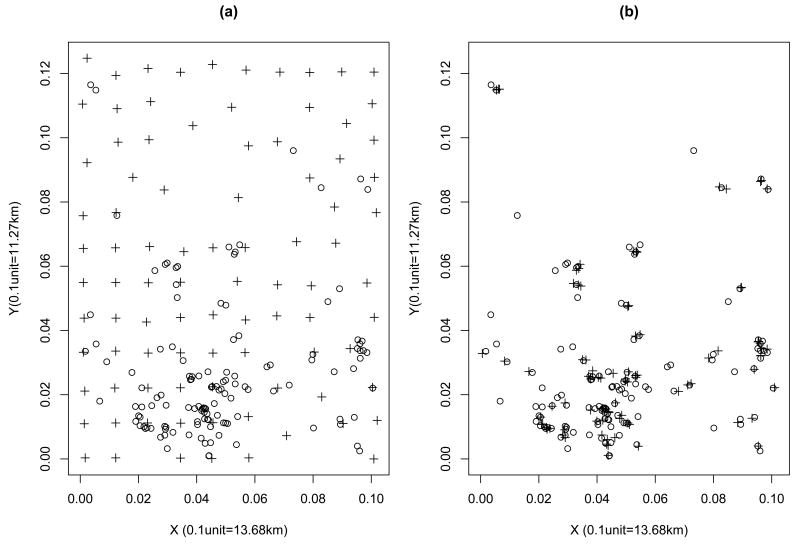
Figure 4 shows the MRDD outcome sites and the soil sample sites in the selected cluster. It shows that the soil sample sites are misaligned to the locations of pregnant women's residential addresses. It also illustrates the grid for the soil sample sites are coarser than that for the outcome sites. Figure 5 shows empirical semivariogram and contour plots of nine chemical concentrations from 119 soil samples. The empirical ssemivariogram and contour plots were drawn using the geoR and the MBA package in R, respectively. It also shows the estimated spatial range parameter. For all chemicals, we can see some peaks in certain regions and smooth variations in other regions. Spatially-varying chemical concentrations within the area suggest that potential hotspots exist. It appears that soil chemicals have different spatial patterns and thus a variogram analysis, was performed separately, for each chemical to estimate a spatial range parameter ρp. The estimated values found for ρ̂p were 0.001, 0.0017, 0.0013, 0.0014, 0.001, 0.0021, 0.0013, 0.0011, 0.001 for As, Ba, Be, Cr, Cu, Pb, Mn, Ni and Hg, respectively.
Figure 4.
Plot of case-control sites and soil sample sites
Figure 5.
Empirical semivariogram (circle) with fitted variogram model (line) and associated contour plots of soil chemical measures (n=119).
5.1. Results of the logistic-spatial model
5.1.1 Plug-in Model vs. Joint Model
Using the estimated spatial range parameter from the variogram analysis, we fitted a Bayesian joint model with K=90 knots. These knots were selected from the soil sample sites using a space filling algorithm. Two chains with different initial values were run and thinning=10 was used to reduce autocorrelation. The details of the MCMC algorithm are provided in the Appendix. For comparison, a logistic spatial model was fitted in WinBUGS using the full-rank Bayesian Kriging estimates as plug-in which is a common modeling strategy. The Bayesian full-rank Kriging estimates were obtained using the R package SpBayes. For the plug-in model, it took about 2.8 hour to obtain Kriging estimates for soil chemicals and 5 minutes to run the logistic spatial model. For the joint model, it took about 15 hours. All models were run on a laptop with Intel Core 2 Duo T9500 processor and 3.5 GB RAM.
Table 2 shows the summary statistics for the two models. The logistic spatial model parameters are reported as a posterior mean and the 95% credible interval for both models. The first model is a logistic model with plug-in estimates of soil chemicals obtained from the full-rank Kriging (Model with FK plug-in). The second model is our suggested model, the joint model with a low-rank Kriging with 90 knots (Joint model with LK_90) developed in Equation (1). 90 Knots (75% of data points) were selected as our simulation study suggests the use of at least 65% of data points to achieve about 80% prediction performance comparable to the full-rank Kriging.
Table 2. Summary of a logistic spatial model with plug-in estimates from a full-rank Kriging (Model with FK plug-in) and a joint model with a low-rank Kriging with 90 knots placed on soil sample sites (Joint Model with LK_90).
| Model with FK plug-in | Joint Model with LK_90 | |||||
|---|---|---|---|---|---|---|
| Mean | 95% Cred. Int. | Mean | 95% Cred. Int. | |||
| Intercept | 2.036 | -3.911 | 11.141 | 0.536 | -9.557 | 12.356 |
| Mother's age (year) | -0.549 | -0.009 | -1.051 | 0.041 | ||
| Mother_Black (ref. Mother_non_Black) | -0.523 | -3.221 | 1.871 | -1.724 | -7.706 | 3.046 |
| Birth weight (Kg) | -0.129 | -2.014 | 1.558 | -0.849 | -4.493 | 2.414 |
| Female Baby (ref. Male Baby) | -5.392 | 0.093 | -8.762 | 0.945 | ||
| Mother's alcohol consumption (ref. No) | -1.725 | -8.380 | 3.245 | -1.435 | -12.019 | 6.509 |
| Parity (birth) | 0.173 | -0.820 | 1.274 | 0.396 | -1.628 | 2.808 |
| Follow-up time (year) | 0.303 | -0.087 | 0.799 | 0.634 | -0.159 | 1.641 |
| As (mg/Kg) | 0.903 | -0.671 | 2.941 | 5.987 | -7.564 | 21.024 |
| Ba (mg/Kg) | -0.321 | -0.026 | -0.794 | -0.319 | ||
| Be (mg/Kg) | -3.652 | -15.455 | 4.356 | -4.062 | -19.612 | 7.684 |
| Cr (mg/Kg) | 0.016 | 0.507 | -0.123 | 1.711 | ||
| Cu (mg/Kg) | 0.015 | -0.219 | 0.239 | -0.194 | -0.997 | 0.261 |
| Pb (mg/Kg) | -0.006 | -0.061 | 0.045 | 0.066 | -0.091 | 0.217 |
| Mn (10 mg/Kg) | -0.001 | 0.011 | 0.025 | 0.050 | ||
| Ni (mg/Kg) | 0.052 | -0.132 | 0.255 | -1.317 | -4.434 | 1.018 |
| Hg (mg/Kg) | -0.062 | -11.972 | 11.610 | -0.373 | -13.014 | 11.824 |
 Significant at the 5% level
Significant at the 5% level
 Close to significant
Close to significant
When the full-rank Bayesian Kriging estimates for soil chemicals were used as plug-in values, mother's age (mean [95% credible interval]: -0.259 [-0.549, -0.009]), Ba (-0.148 [-0.321, -0.026]) and Cr (0.257 [0.016, 0.507]) were significant at the 5% level. Baby sex (-2.521 [-5.392, 0.093]), follow-up time (0.303 [-0.087, 0.799]) and Mn (0.005 [-0.001, 0.011]) were relatively close to significant, having more than 90% of posterior samples of the risk estimates lying below/above zero. In our suggested model, Ba (-0.505 [-0.794, -0.319]) and Mn (0.035 [0.025, 0.050]) were significant at the 5% level and mother's age (-0.424 [-1.051, 0.041]), baby sex (-3.304 [-8.762, 0.945]), follow-up time (0.634 [-0.159, 1.641]) and Cr (0.703 [-0.123, 1.711]) were relatively close to significant compared to other covariates which were consistent with the model with plug-in values. In general, for the significant covariates, the joint model has a larger magnitude of the logistic regression parameter and a wider 95% credible interval than the model with the plug-in values.
Model comparisons are performed with respect to the deviance information criterion (DIC). DIC is defined as the posterior mean of the deviance (-2*log likelihood) plus pD, the estimated effective number of parameters in the posterior distribution. pD is defined as half the posterior variance of the deviance [28]. DIC for the model with plug-in estimates and our suggested model was 156.221 and 2440.391, respectively. However, these DIC values cannot be compared directly since the model with plug-in values has a logistic likelihood only whereas the joint model has two likelihoods (normal and logistic likelihoods). To compare these two models, DIC for the joint model was calculated using logistic likelihoods only. The deviance for the joint model was smaller than that of the model with plug-in values (53.543 vs. 28.111) in part because it contains chemical values that are estimated parameters. However, the joint model also has greater variability in the converged sampler and to account for this variability we calculated DIC as explained above. The calculated DIC for the joint model was DIC=69.566 which was less than the half of the DIC of the model with plug-in values (DIC=156.221).
5.1.2 Joint model with different knot locations and numbers of knots
To investigate whether placing knots close to the prediction locations can help the prediction of soil chemicals and thus the joint model, our suggested model was fitted again with 90 knots placed close to the outcome sites. As explained in 3.3.2, knots were generated based on a kernel smoothed intensity of the outcome site locations by the R package, spatstat. Figure 6 shows these two sets of knots selected based on soil sample sites and outcome sites. To determine whether similar results are obtained following Ruppert et al. [14]'s suggestion, a joint model with K=30 knots (25% of data point) was also fitted.
Figure 6.
Knots selected based on: (a) soil sample sites using a space filling algorithm and (b) outcome sites using a kernel-smoothed intensity of the outcome sites (+: knots, o: outcome sites).
Table 3 shows the comparison between three joint models. First model is the originally suggested joint model presented in Table 2 (90 knots placed on sample sites). Second model is the joint model with low-rank Kriging with 90 knots placed close to the outcome sites (90 knots placed on outcome sites). Third model is the joint model with low-rank Kriging with 30 knots placed on soil sample sites (30 knots place on samples sites). As all these models are joint models which have two likelihoods, we can compare DIC to find a better model. The first model has the smallest DIC of 2440.39 followed by the second model (DIC=2848.52) and the third model (DIC=2851.308). This result suggests that the low-rank Kriging can be affected by the knot location for two-dimensional data which is in contrast to one-dimensional data where the prediction result is rather insensitive to the knot location [14, 25]. As seen in the simulation study, using reduced number of knots affected the model fit, increasing DIC by 411 compared to the originally suggested model. DIC can also be calculated using the health model likelihood only. When we compared two joint models with different knot placements, the model with knots placed on the outcome sites had a smaller DIC (DIC=45.190) than the model with knots placed on sample sites (DIC=69.566).
Table 3. Comparison of three joint models with different knot locations (sample sites vs. outcome sites) and knot numbers (90 vs. 30).
| 90 knots placed on sample sites | 90 knots placed on outcome sites | 30 knots placed on sample sites | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | 95% Cred. Int. | Mean | 95% Cred. Int. | Mean | 95% Cred. Int. | ||||
| Intercept | 0.536 | -9.557 | 12.356 | -1.342 | -14.063 | 11.339 | 1.424 | -11.344 | 19.738 |
| Mother's age (year) | -1.051 | 0.041 | -1.41 | -0.004 | -0.276 | -1.041 | 0.311 | ||
| Mother_Black (ref. Mother_non_Black) | -1.724 | -7.706 | 3.046 | -1.686 | -8.694 | 3.643 | -1.823 | -9.185 | 3.912 |
| Birth weight (Kg) | -0.849 | -4.493 | 2.414 | -2.449 | -7.13 | 1.034 | -0.555 | -4.326 | 2.678 |
| Female Baby (ref. Male Baby) | -8.762 | 0.945 | -2.998 | -9.816 | 2.294 | -0.896 | -7.674 | 4.909 | |
| Mother's alcohol consumption (ref. No) | -1.435 | -12.019 | 6.509 | 0.312 | -10.055 | 11.142 | 0.179 | -9.972 | 10.189 |
| Parity (birth) | 0.396 | -1.628 | 2.808 | -0.334 | -3.671 | 2.317 | -1.571 | -5.733 | 1.381 |
| Follow-up time (year) | -0.159 | 1.641 | 0.512 | -0.333 | 1.772 | 0.523 | -0.402 | 1.887 | |
| As (mg/Kg) | 5.987 | -7.564 | 21.024 | -1.323 | -10.599 | 6.741 | 1.376 | -6.579 | 10.227 |
| Ba (mg/Kg) | -0.794 | -0.319 | -0.478 | -0.029 | 0.121 | 0.601 | |||
| Be (mg/Kg) | -4.062 | -19.612 | 7.684 | -1.059 | -15.111 | 10.839 | 0.446 | -12.817 | 13.146 |
| Cr (mg/Kg) | -0.123 | 1.711 | 0.721 | -0.505 | 1.642 | -0.337 | -1.619 | 1.034 | |
| Cu (mg/Kg) | -0.194 | -0.997 | 0.261 | -0.293 | -3.861 | 2.117 | -1.473 | -4.677 | 0.583 |
| Pb (mg/Kg) | 0.066 | -0.091 | 0.217 | -0.033 | -0.555 | 0.467 | 0.478 | 1.300 | |
| Mn (10 mg/Kg) | 0.025 | 0.050 | 0.055 | 0.074 | -0.144 | -0.112 | |||
| Ni (mg/Kg) | -1.317 | -4.434 | 1.018 | -1.37 | -7.702 | 2.12 | 0.848 | -1.442 | 5.603 |
| Hg (mg/Kg) | -0.373 | -13.014 | 11.824 | 0.137 | -10.46 | 13.313 | 0.243 | -10.818 | 11.541 |
| DIC=2440.391 | DIC=2848.521 | DIC=2851.308 | |||||||
 Significant at the 5% level
Significant at the 5% level
 Close to significant
Close to significant
When knots are placed close to the outcome sites, mother's age (-0.680 [-1.41, -0.004]), Ba (-0.255 [-0.478,-0.029]) and Mn (0.066 [0.055, 0.074]) were significant at the 5% level. This result is similar to the originally suggested joint model. When 30 knots placed on the sample sites were used, Ba (0.330 [0.121, 0.601]), Pb (0.768 [0.478, 1.300]) and Mn (-0.122[-0.144, -0.112]) were significant at the 5% level. This results are quite different from the suggested joint model and the model with plug-in estimates since the sign of the logistic regression parameter is opposite for Ba and Mn.
5.1.3 Joint model with different spatial range parameters
In the suggested model, we estimated the spatial range parameter ρp using a variogram analysis for each soil chemical. To assess the sensitivity of the results to the choice of the spatial range parameter, a sensitivity analysis was performed. Two joint models were fitted separately using 2*ρ̂p and 1/2*ρ̂p where ρ̂p was obtained from a variogram analysis for each chemical. Table 4 shows the results of comparing the suggested model with these two modes with different ρp values. Compared to the suggested model, some different results were observed. When 2*ρ̂p was used, Ba (0.021[0.021, 0.022]), Cr (-5.385[-6.245, -4.682]), Pb (5.701[4.185, 7.794]) and Hg (0.065[0.053, 0.077]) were significant at the 5% level. When 1/2*ρ̂p was used, Ba (0.022[0.022, 0.024]), Cr(-6.628[-8.862, -4.296]) and Hg (0.271[0.236, 0.296]) were significant at the 5% level. No mom and baby covariates were significant at the 5% level. When we compared DIC, our suggested model has the smallest DIC suggesting the best model fit (DIC=2440.391 vs. 3003.621 vs. 2995.552).
Table 4. Comparison of joint models with different spatial range parameters (sensitivity analysis).
| Model with estimated rho | Model with 0.5*Estimated rho | Model with 2*Estimated rho | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | 95% Cred. Int. | Mean | 95% Cred. Int. | Mean | 95% Cred. Int. | ||||
| Intercept | 0.536 | -9.557 | 12.356 | -0.786 | -16.257 | 11.748 | -0.719 | -14.916 | 11.518 |
| Mother's age (year) | -1.051 | 0.041 | -1.999 | 0.220 | -0.650 | -2.055 | 0.319 | ||
| Mother_Black (ref. Mother_non_Black) | -1.724 | -7.706 | 3.046 | -5.021 | -18.930 | 3.603 | -22.718 | 2.116 | |
| Birth weight (Kg) | -0.849 | -4.493 | 2.414 | -1.204 | -6.342 | 3.074 | -0.770 | -6.592 | 4.031 |
| Female Baby (ref. Male Baby) | -8.762 | 0.945 | 2.429 | -4.905 | 13.104 | 1.814 | -4.522 | 10.144 | |
| Mother's alcohol consumption (ref. No) | -1.435 | -12.019 | 6.509 | 0.038 | -11.146 | 12.707 | -0.435 | -12.376 | 10.825 |
| Parity (birth) | 0.396 | -1.628 | 2.808 | -11.259 | 0.346 | -10.698 | 0.739 | ||
| Child's age last seen Medicaid (year) | -0.159 | 1.641 | 0.538 | -0.450 | 2.128 | 0.523 | -0.583 | 2.193 | |
| As (mg/Kg) | 5.987 | -7.564 | 21.024 | -0.081 | -12.565 | 13.386 | -0.100 | -11.33 | 10.075 |
| Ba (mg/Kg) | -0.794 | -0.319 | 0.022 | 0.024 | 0.021 | 0.022 | |||
| Be (mg/Kg) | -4.062 | -19.612 | 7.684 | 0.224 | -10.444 | 9.911 | -2.875 | -15.025 | 5.746 |
| Cr (mg/Kg) | -0.123 | 1.711 | -8.862 | -4.296 | -6.245 | -4.682 | |||
| Cu (mg/Kg) | -0.194 | -0.997 | 0.261 | 0.219 | -12.452 | 14.042 | -0.102 | -12.627 | 12.573 |
| Pb (mg/Kg) | 0.066 | -0.091 | 0.217 | 0.168 | -2.233 | 2.775 | 4.185 | 7.794 | |
| Mn (10 mg/Kg) | 0.025 | 0.050 | -2.538 | -10.777 | 2.637 | -10.021 | 0.766 | ||
| Ni (mg/Kg) | -1.317 | -4.434 | 1.018 | 0.637 | -0.299 | 1.312 | 0.073 | -0.517 | 0.757 |
| Hg (mg/Kg) | -0.373 | -13.014 | 11.824 | 0.236 | 0.296 | 0.053 | 0.077 | ||
| DIC=2440.391 | DIC=2995.552 | DIC=3003.621 | |||||||
 Significant at the 5% level
Significant at the 5% level
 Close to significant
Close to significant
Although some similar results were obtained such as Ba which was significant at the 5% level for all three models, different results were also observed. The sign of the effect for Ba was different in the models with different ρp values, Cr was close to significant in the suggested model but became significant at the 5% in the models with different ρp values. Other chemicals such as Pb and Hg also became significant in the model with different ρp values.
5.2. Chemical prediction by low-rank Kriging with different spatial range parameters and numbers of knots in MRDD data
In this section, four different low-rank Kriging models were considered for chemical prediction using a different set of knot numbers K, and different spatial range parameters ρp to investigate how the number of knots and the spatial range parameter affects low-rank Kriging prediction. All the combinations of ρ̂p = (0.16, 0.008) and K = (30, 90) were used. ρ̂p =0.16 was chosen as the maximum distance between sample sites within the study area as recommended by French and Wand [13]. ρ̂p=0.008 is selected as a much smaller value . K=30 (25% of data) was chosen following the suggestion of Ruppert et al. [14]. To compare the prediction performance of low-rank Kriging with different number of knots, K=90 (75% of data) was also selected since our simulation study found that about 65-80% of data points are needed to achieve similar prediction results to the full-rank Kriging. We also fitted a Bayesian full-rank Kriging model (FK) using the R package spBayes assuming a Matérn covariance function. We assume that Bayesian Kriging is the ‘gold-standard’ for comparison. The prediction performance of each low-rank Kriging was measured by the mean squared error of prediction (MSEP) which was calculated as follows:
Table 5 shows the prediction performance of different low-rank Kriging models measured by mean squared error of prediction (MSEP). When the smoothing parameter was fixed at ρ̂p=0.16, the low-rank Kriging with 30 knots and that with 90 knots had similar MSEP for all chemicals. It was because the estimated bivariate surface was too smooth for both models. In contrast, when we used ρ̂p=0.008, the low-rank Kriging with 90 knots had much smaller MSEP for all chemicals than the low-rank Kriging with 30 knots by a factor varying from 1.9 (Ba) to 14.7 (Pb). When 30 knots are used, MSEP for most chemicals are similar between ρ̂p=0.16 and ρ̂p=0.008 whereas when 90 knots are used, most chemicals have smaller MSEP when ρ̂p=0.008 is used.
Table 5. Mean squared error of prediction (MSEP) of the low-rank Kriging with 30 knots (LK_30) and that with 90 knots (LK_90).
| rho=0.16 | rho=0.008 | |||
|---|---|---|---|---|
| LK_30 | LK_90 | LK_30 | LK_90 | |
| As | 1.08 | 1.06 | 1.06 | 0.46 |
| Ba | 691.89 | 688.54 | 883.94 | 458.31 |
| Be | 0.020 | 0.020 | 0.025 | 0.0072 |
| Cr | 42.19 | 42.58 | 61.58 | 14.31 |
| Cu | 96.66 | 96.35 | 86.61 | 21.95 |
| Pb | 1402.84 | 1400.02 | 1138.83 | 77.28 |
| Mn | 75494.82 | 74572.78 | 74909.58 | 31379.03 |
| Ni | 16.90 | 16.53 | 17.03 | 2.25 |
| Hg | 0.0019 | 0.0019 | 0.0019 | 0.00083 |
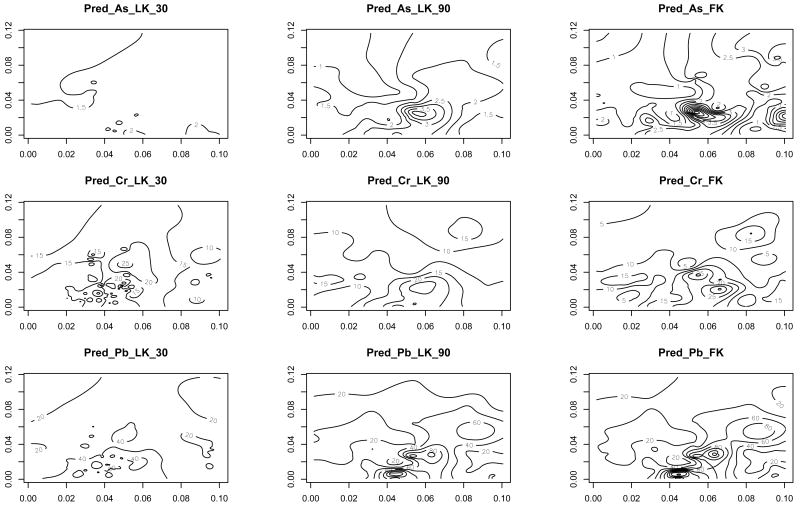
Figure 7 shows contour plots of predicted As, Cr and Pb at the outcome sites by the low-rank Kriging with 30 knots, the low-rank Kriging with 90 knots and full-rank Kriging. For both low-rank Kriging models, ρ̂p = 0.008 was used. To make the contour plots comparable, contours are drawn at the same level in three plots for each chemical. The general patterns across the region seem similar between the full-rank Kriging and the low-rank Kriging with 90 knots whereas the chemical estimates from the low-rank Kriging with 30 knots are too smooth as compared to the full rank Kriging. Similar results were also observed for other chemicals (not shown here).
Figure 7.
Contour plots of predicted As, Cr and Pb at mothers' residential addresses by low-rank Kriging with 30 knots (LK_30), low-rank Kriging with 90 knots (LK_90) and full-rank Kriging (FK).
6. Discussion
We present a Bayesian joint model which interpolates several spatial fields and estimates the associated risk simultaneously for a binary outcome. Low-rank Kriging imposes less computational burden compared to a full-rank Kriging. Employing a Bayesian approach, low-rank Kriging can reduce computational complexity significantly since it does not invert the spatial covariance matrix at each MCMC iteration as the full-rank Kriging model does. This is achieved by fixing the spatial range parameter a priori which makes low-rank Kriging have a generalized linear model specification.
Different approaches are possible for choosing the spatial range parameter. French and Wand [13] suggested fixing the spatial range parameter at the maximum distance between two sites within the study area. Another way to choose the spatial range parameter is to use a variogram analysis as we adopted in this study. The spatial range need to be selected with caution as these affect low-rank Kriging prediction performance compared to the full rank Kriging. Our sensitivity analysis suggests that the results can be sensitive to the choice of the spatial range parameter. This implies that the interpretation for those soil chemicals can be different depending on what ρp values were used to fit the joint model. Thus, it seems necessary to estimate range parameters properly. Based on our findings, it appears that employing variogram analysis is a better approach than Ruppert et al.'s recommendation.
It was found that the knot specification has little effect on low-rank smoothing splines performance for one-dimensional data [26, 27]. However, the effect of knot specification on two-dimensional data has not been investigated in depth. We investigated the effect of different number of knots and knot locations for two-dimensional data in this study. Our simulation study and data example suggest that if a surface is smooth (large spatial range parameter), less knots are required but for less smooth surface (smaller spatial range parameter) more knots are required to achieve similar prediction with the full-rank Kriging. Although Ruppert et al. [14] provide a recommendation based on their experience, our simulation study shows that it is a bit conservative. With a reasonable number of knots and spatial range parameter, low-rank Kriging performs as well as full-rank Bayesian Kriging. It is obvious that there is tradeoff between computational speed and the performance of approximation. In our data example, it was found that knots covering the study area had a better model fit supported by smaller overall DIC. This area can be an interesting area where further research is required. As our joint model has both the health model likelihood and the exposure model likelihood, DIC was calculated using both likelihoods and compared between different joint models. It can be argued that only the health model likelihood need be considered for DIC as that is the main focus of the analysis. However, the prediction of the unobserved soil chemicals is affected by the exposure model fitting and thus it seems necessary to find the best exposure model to obtain accurate prediction of those soil chemicals. Thus, we decided to use overall DIC as our criteria which consider both exposure and health model likelihoods for the DIC calculation.
There are some issues which need to be addressed with regard to our modeling strategy. First, the choice of a spatial range parameter is important as it controls the smoothness of the bivariate surface. The spatial range parameter could have been estimated in our joint model but we did not use this approach for computational expediency. If this parameter were estimated, a spatial covariance matrix needs to be inverted at each MCMC iteration which requires a computation similar to Full-rank Kriging. Secondly, we used an unstructured random effect rather than spatial random effect for our binary outcome as it has been reported that using a conditionally autoregressive (CAR) model to spatially referenced data can mask spatial predictor effects [11, 12]. Thirdly, our joint model was fitted as a full model using all the available mom and baby covariates and soil chemical concentrations. DIC is a reliable measure to compare different Bayesian model fits. We used DIC to compare different joint models in this study. However, we did not perform variable selection to find the best submodel since we have sixteen covariates and the number of possible submodels is large (216). A different variable selection approach is needed for our analysis and we will use a MCMC method for variable selection in the future work. Finally, in our joint model, each chemical concentration was modeled separately as a univariate model though we observed multiple chemical concentrations in the soil samples. A multivariate model which captures the possible correlations between chemical measures might provide an appropriate prediction although this was not investigated in this study. In the future, we will develop a method which introduces these possible correlations between chemical concentrations into a joint model.
Our joint model found significant association between MRDD outcome and Ba, Mn and possibly Cr concentrations which was consistent with the model with plug-in estimates. However, our joint model can find an association which is close to the unobserved truth reflecting the uncertainty involved in the chemical prediction which the model with plug-in estimates cannot. It should be also noted that the true association between MRDD and soil chemical concentrations could be more subtle as we did not perform variable selection.
In this paper, we showed that a low-rank Kriging can have similar prediction performance to the full-rank Kriging when the number of knots and the smoothing parameter are selected with caution. This enables us to employ a Bayesian approach and obtain the risk estimates of soil chemical concentrations reflecting the uncertainty involved in the spatial field prediction. Thus, it is recommended to use the joint model approach so that the risk associated with soil chemical concentrations can be properly assessed.
Acknowledgments
Funding for this research was provided by the National Institutes of Health, National Institute of Environmental Health Sciences, RO1 Grant No. ES012895-01A1.
Appendix
The MCMC algorithm
Markov chain Monte Carlo (MCMC) sampling is an iterative numerical method which simulates complex and non-standard multivariate distribution [29]. It generates correlated draws from the joint posterior distribution of model parameters. For our MRDD data analysis, a Metropolis-Hastings algorithm [15, 29] was employed to sample for all the parameters. This Metropolis-Hasting algorithm can draw samples from any probability distribution and does not involve knowledge of the conditional posteriors to update the parameters.
General description for Metropolis-Hastings algorithm is as follows: let y denote the observed data and θ = (θ1,…,θn) represent a vector of all the parameters. Values for θ are to be sampled sequentially at each iteration of the MCMC sampler.
Metropolis-Hastings Sampler
Assign starting values to θ(0)
Set t = 0
For 1 ≤ i ≤ n, draw from the proposal density
Compute the ratio of the densities where p is the full conditional distribution of θi
Set
Any distribution from which samples are readily obtainable can be used as the proposal distribution.
Now we present the specific use of Metropolis-Hastings in our MCMC algorithm. From Equation (2), (5) and (8), our joint posterior distribution becomes:
| (9) |
For priors, normal distributions priors are used for β0,β1,β2,ε and αp as follows:
For hyperpriors, uniform distributions are assumed for standard deviation parameters [30]:
At each iteration of our Metropolis-Hastings sampler, we
Update parameters of the low-rank Kriging model for each chemical concentrations using observed and unobserved measures
Use the low-rank Kriging parameters, predictive distribution and the logistic spatial model likelihood to update unobserved chemical concentrations at the outcome sites
Use the predicted soil chemical concentrations to update the regression parameters in the logistic spatial model
To run the Metropolis-Hastings sampler,
Select K knots as described in Section 3.3.2.
Initialize all the parameters
For the p th chemical, calculate , Zp and Zxp fixing ρ̂p using the estimated spatial range parameter from a univariate variogram analysis as described in Section 3.3.2 and 3.3.3.
At t iteration, update all the parameters for the low-rank Kriging for each p th chemical where 1 ≤ p ≤ P calculating the density ratio r using the joint distribution in (9)
At t iteration, update all the unobserved chemical concentrations for each p th chemical where 1 ≤ p ≤ P calculating the density ratio r using the joint distribution in (9)
At t iteration, update β = (β0,β1,β2), ε, σβ = (σβ0, σβ1, σβ2), and σε for the logistic spatial model calculating the density ratio r using the joint distribution in (9)
Two chains with different initial values were run for 100,000 times and the convergence was checked by Gelman-Rubin statistics and a visual inspection of trace plots of the deviance (-2 log likelihood). After convergence was checked, additional 50,000 samples were drawn for summary statistics and thinning=10 was used to reduce autocorrelation.
References
- 1.McDermott S, Durkin M, Schupf N, Stein Z. Epidemiology and Etiology of Mental Retardation. In: Jacobson JW, Mulick JA, Rojahn J, editors. Handbook of Intellectual and Developmental Disabilities. Springer Press; New York: 2007. [Google Scholar]
- 2.Fung KY, Krewski D. On measurement error adjustment methods in Poisson regression. Environmetrics. 1999;10:213–224. doi: 10.1002/(SICI)1099-095X(199903/04)10:2<213∷AID-ENV349>3.0.CO;2-B. [DOI] [Google Scholar]
- 3.Heid IM, Küchenhoff H, Wellmann J, Gerken M, Kreienbrock L, Wichmann HE. On the potential of measurement error to induce differential bias on odds ratio estimates: an example from radon epidemiology. Statistics in Medicine. 2002;21:3261–3278. doi: 10.1002/sim.1252. [DOI] [PubMed] [Google Scholar]
- 4.Reeves GK, Cox DR, Whitley SCDE. Some aspects of measurement error in explanatory variables for continuous and binary regression models. Statistics in Medicine. 1998;17:2157–2177. doi: 10.1002/(SICI)1097-0258(19981015)17:19<2157∷AID-SIM916>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 5.Fuentes M, Song HR, Ghosh SK, Holland DM, Davis JM. Spatial Association between Speciated Fine Particles and Mortality. Biometrics. 2006;62:855–863. doi: 10.1111/j.1541-0420.2006.00526.x. [DOI] [PubMed] [Google Scholar]
- 6.Smith BJ, Zhang LR, Field W. Iowa radon leukaemia study: a hierarchical population risk model for spatially correlated exposure measured with error. Statistics in Medicine. 2007;26:4619–4642. doi: 10.1002/sim.2884. [DOI] [PubMed] [Google Scholar]
- 7.Cressie N. Statistics for Spatial Data. Revised Edition. Wiley; 1993. [Google Scholar]
- 8.Zhen H, Lawson AB, McDermott S, Lamichhane AP, Aelion CM. A spatial analysis of mental retardation of unknown cause and maternal residence during pregnancy. Geospatial Health. 2008;2:173–182. doi: 10.4081/gh.2008.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aelion CM, Davis HT, McDermott S, Lawson AB. Metal concentrations in rural soils in South Carolina; Potential for human health impact. Science of the Total Environment. 2008;402:149–156. doi: 10.1016/j.scitotenv.2008.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Besag J, York J, Mollié A. Bayesian image restoration, with two applications in spatial statistics. Annals of the Institute of Statistical Mathematics. 1991;43:1–59. [Google Scholar]
- 11.Ma B, Lawson AB, Liu Y. Evaluation of Bayesian models for focused clustering in health data. Environmetrics. 2007;18:871–887. [Google Scholar]
- 12.Reich BJ, Hodges JS, Zadnik V. Effects of Residual Smoothing on the Posterior of the Fixed Effects in Disease-Mapping Models. Biometrics. 2006;62:1197–1206. doi: 10.1111/j.1541-0420.2006.00617.x. [DOI] [PubMed] [Google Scholar]
- 13.French JL, Wand MP. Generalized additive models for cancer mapping with incomplete covariates. Biostat. 2004;5:177–191. doi: 10.1093/biostatistics/5.2.177. [DOI] [PubMed] [Google Scholar]
- 14.Ruppert D, Wand MP, Carroll RJ. Semiparametric Regression. Cambridge University Press; Cambridge: 2003. [Google Scholar]
- 15.Chib S, Greenberg E. Understanding the Metropolis–Hastings Algorithm. The American Statistician. 1995;49:327–335. [Google Scholar]
- 16.Banerjee S, Carlin BP, Gelfrand AE, Carlin BP. Hierarchical Modeling and Analysis for Spatial Data. CRC Press; Boca Raton, Florida: 2003. [Google Scholar]
- 17.Kammann EE, Wand MP. Geoadditive models. Journal of the Royal Statistical Society: Series C (Applied Statistics) 2003;52:1–18. doi: 10.1111/1467-9876.00385. [DOI] [Google Scholar]
- 18.MacNab YC. Spline smoothing in Bayesian disease mapping. Environmetrics. 2007;18:727–744. doi: 10.1002/env.876. [DOI] [PubMed] [Google Scholar]
- 19.Wood S. Generalized Additive Models: An Introduction with R. CRC Press; 2006. [Google Scholar]
- 20.Denison DGT, Holmes CC, Mallick BK, Smith AFM. Bayesian Methods for Nonlinear Classification and Regression. John Wiley and Sons; Chichester: 2002. [Google Scholar]
- 21.Dietrich CR, Osborne MR. Estimation of covariance parameters in kriging via restricted maximum likelihood. Mathematical Geology. 1991;23:119–135. doi: 10.1007/BF02065971. [DOI] [Google Scholar]
- 22.Hoeting JA, Davis RA, Merton AA, Thompson SE. Model Selection For Geostatistical Models. Ecological Applications. 2006;16:87–98. doi: 10.1890/04-0576. [DOI] [PubMed] [Google Scholar]
- 23.Schafer JL, Yucel RM. Computational Strategies for Multivariate Linear Mixed-Effects Models With Missing Values. Journal of Computational & Graphical Statistics. 2002;11:437–457. doi: 10.1198/106186002760180608. [DOI] [Google Scholar]
- 24.Cressie N, Johannesson G. Proc Australian Academy of Science Elizabeth and Frederick White Conf. Canberra: Australian Academy of Science; 2006. Spatial prediction of massive datasets; pp. 1–11. [Google Scholar]
- 25.Laslett GM. Kriging and Splines: An Empirical Comparison of Their Predictive Performance in Some Applications. Journal of the American Statistical Association. 1994;89:391–400. [Google Scholar]
- 26.French JL, Kammann EE, Wand MP. Semiparametric Nonlinear Mixed-Effects Models and Their Applications. Journal of the American Statistical Association. 2001;96:1285–1288. [Google Scholar]
- 27.Ruppert D. Selecting the Number of Knots for Penalized Splines. Journal of Computational & Graphical Statistics. 2002;11:735–757. doi: 10.1198/106186002853. [DOI] [Google Scholar]
- 28.Gelman A, Carlin JB, Stern HS, Rubin DB. Bayesian Data Analysis. Chapman and Hall; London: 2003. [Google Scholar]
- 29.Hastings WK. Monte Carlo sampling methods using Markov chains and their applications. Biometrika. 1970;57:97–109. doi: 10.1093/biomet/57.1.97. [DOI] [Google Scholar]
- 30.Gelman A. Prior distributions for variance parameters in hierarchical models(Comment on Article by Browne and Draper) Bayesian Analysis. 2006;1:515–534. doi: 10.1214/06-BA117A. [DOI] [Google Scholar]