Abstract
The BCA2 protein contains a RING H2 finger and a Zn finger near the N-terminus and has E3 ligase activity. RING finger proteins play critical roles in mediating the transfer of ubiquitin and ubiquitin like modifiers to heterologous substrates as well as to the RING finger proteins themselves. Protein modification by ubiquitin and small ubiquitin-related modifier (SUMO) plays a pivotal role in protein homeostasis and is critical to regulating basic cellular processes such as proliferation, differentiation, apoptosis, intracellular signaling, and gene-transcriptional regulation. The addition of ubiquitin or SUMO can modulate the ability of proteins to interact with their partners, alter their patterns of sub-cellular localization and control their stability. It is clear that SUMO influences many different biological processes however recent data suggest that it is specifically important in the regulation of transcription. BCA2 is an E3 ligase that interacts with the SUMO conjugating enzyme Ubc9. It could therefore function as an E3 in the sumoylation of various transcription factors. We have found that the BCA2 is co-expressed with the estrogen receptor in 74% of ER-positive invasive ductal carcinomas from a 635 member breast cancer cohort (p = 0.004). At the cellular level, BCA2 co-localizes with ER and it appears that at the transcriptional level BCA2 mRNA expression is regulated by estrogen. Bioinformatic analysis of the BCA2 promoter region revealed ER and PR binding sites as well as that of other more general transcription factors. The data presented here provides an overview of the potential involvement of the BCA2 in hormone responsive breast cancer and opens up avenues that should be exploited to better understand the regulation of ER expression, growth of breast cancer cells, and the importance of BCA2.
Introduction
At the cellular level, transcription factors are tightly controlled by their rates of synthesis and degradation. Many transcription factors are maintained at an appropriate level by targeted addition of polyubiquitin chains and subsequent degradation in the proteasome [1]. While polyubiquitination targets proteins for degradation, monoubiquitination or their modification by small ubiquitin-like modifiers such as SUMO, alters subcellular localization and can change their activity [1]. Important transcription factors known to be regulated by ubiquitination or sumoylation are HIF1-α, c-Myb, c-Jun, Oct4, ETS1, and the ER among others [2-4]. Each of these transcription factors regulates the expression of a large number of target genes. Alterations of these transcription factors are frequently involved in tumorigenesis [1,5,6].
In response to circulating estrogen, the estrogen receptor (ER) regulates the genetic programs of cell cycle progression and growth in normal mammary gland and breast cancer epithelial cells. This critical transcription factor has two receptor forms, ERα and ERβ. ERβ demonstrates lower hormone-dependent transcriptional activity [7]; therefore ERα is considered the primary receptor for mammary gland development and function [8]. However, little is known of how the stability and expression of ER is regulated. Recent studies indicate that the ER is monoubiquitinated and sumoylated when interacting with BRCA1, which might lead to repression of ER transcriptional activation [9,10]. Furthermore, cancer-predisposing mutations in BRCA1 were observed to abrogate ER ubiquitination [9], implicating ubiquitin E3 ligases as playing a major role in ER regulation and hormone responsive breast cancer.
We had previously identified Breast Cancer Associated gene 2, BCA2 (synonymous with T3A12/ZNF364/Rabring7/RNF115), a novel RING-finger ubiquitin E3 ligase, by subtractive hybridization cloning in breast carcinoma cell lines [11]. Subsequently we found that BCA2 is expressed in primary invasive breast cancers and is associated with a positive estrogen receptor status and outcome [12,13]. Here we describe the interaction of the BCA2 protein with the SUMO conjugating enzyme Ubc9, its regulation by ER, and its potential involvement in transcriptional regulation of hormone responsive breast cancers.
BCA2 and ER co-expression in invasive breast cancer
Estrogen regulates the proliferation and development of tissues expressing estrogen receptors and is a risk factor for breast cancer development. Ligand binding activates both ERα-dependent transcription and ERα ubiquitination [14]. ERα ubiquitination and proteasome activity are intimately linked to ERα-dependent transcriptional activation [14,15]. Proteasome inhibitors and mutations that inhibit co-activator binding both abrogate ligand-mediated ERα proteolysis and ERE transcriptional activity [15]. Different ligands stimulate ERα proteolysis to different degrees [16], and the ubiquitin ligases BRCA1 [17], MDM2 [18], and E6AP [19] can all stimulate estrogen-induced transcriptional activity.
The BCA2 protein is an ubiquitin ligase co-expressed with the ER in breast cancers [12]. We have studied BCA2 protein expression in a large collection of over 1,000 invasive mammary carcinomas, termed the Henrietta Banting Breast Cancer Collection (HBBCC) [12,13]. This breast cancer tissue resource located at Sunnybrook Health Sciences Centre, Toronto, has been well characterized for established molecular markers such as ER, PR (progesterone receptor) and HER2/Neu, along with clinico-pathological variables and outcomes [12,13]. By performing comparative BCA2 protein expression analysis on tissue microarrays of the HBBCC and by using the HBBCC database, we obtained clues about a potential function of the E3 ligase in invasive breast cancer. BCA2 is expressed in the cytoplasm and nucleus of breast cancer tissues [12,13]. Nuclear BCA2 expression however, was the predominant site of protein location in breast cancer cells (Fig.1). Nine hundred and forty-five invasive breast carcinomas of the HBBCC were evaluable for nuclear BCA2 expression, representative tissue sections are shown in figure 1A. Four hundred and fifteen cases or 43.9 % of the invasive breast cancer cohort had low nuclear BCA2 levels (BCA2 intensity score <=1), 530 patient tissues or 56.1% of the study cohort were found to overexpress BCA2 (BCA2 score >1) (Tab. 1). Some normal breast tissues also expressed low amounts of nuclear BCA2 protein (score<=1) [12]. Sixty-seven percent (635) of the HBBCC breast cancers were estrogen receptor positive. The statistical analysis of the HBBCC nuclear BCA2 protein expression study revealed that three variables yield significant results: first, patients with low BCA2 (57.7% of 213), were more likely to have lymph node metastasis at presentation than those with higher BCA2 levels (49.9% of 229, p=0.02). Second, women with invasive breast cancers with low BCA2 (8.6 % of 415) were more likely to experience regional re-occurrence than those with higher BCA2 (5.3 % of 530) (p = 0.05) (Tab. 1). The third significant correlation was very strong: nuclear BCA2 expression was correlated with a positive estrogen receptor status (Fig. 1A). 67.1 % of all 945 invasive HBBCC cases stained positive for ER expression. 74% of breast cancers that were ER-positive had also nuclear BCA2>1 (p = 0.004) [12]. Because of the significant statistical correlation found between invasive breast cancers expressing ER and BCA2, we compared the pattern of expression of these two proteins. Core by core analysis of consecutive tissue sections indicated that BCA2 and ER are mostly co-expressed (odds ratio = 1.5) (Fig. 1A), cells strongly positive for BCA2 were positive for ER. It is important to note that high expression of BCA2 is not beneficial at the cellular level for inhibiting cancer progression and may be a contributing factor in metastasis. However the high correlation of co-expression with ER makes higher expression of BCA2 a marker for good prognosis in breast cancer due to the availability of highly effective treatment options.
Fig. 1.
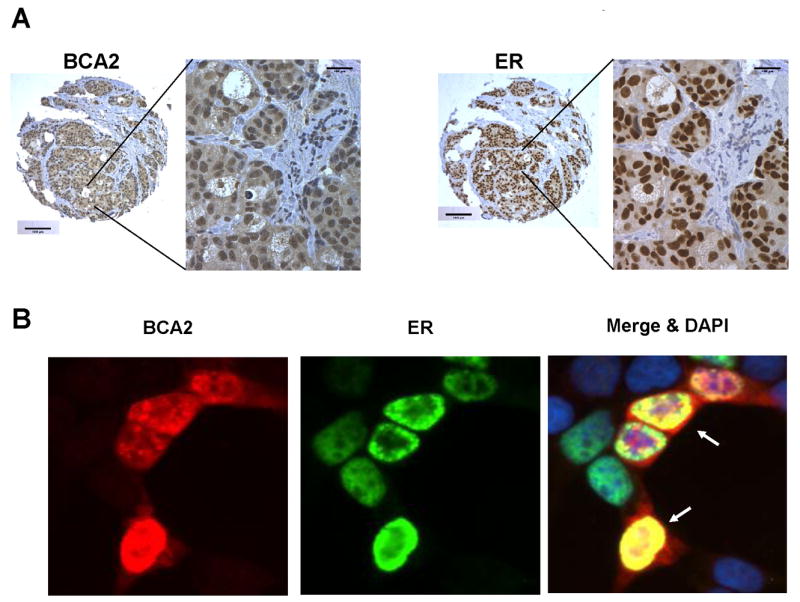
A. Comparison of BCA2 and ER expression by immunoperoxidase staining in a representative case of invasive breast cancer from the HBBCC tissue microarrays (method described in detail in [12]). Consecutive tissue microarray sections were stained for BCA2 and ER and viewed at 4X magnification, size bar = 100μm. Enlargements at 40x magnification were captured from the 4X core areas as outlined by a box. The data reveal co-expression of ER and BCA2 in the nuclei of cells staining positive for the antigens (brown color: diaminobenzidine). Overall, 67% of the 945 invasive HBBCC cases were positive for ER and of those 74% were positive for both, ER and BCA2 [12]. B. Co-localization of ER (green, FITC-labeled anti-mouse antibody) and BCA2 (red, TRITC-labeled anti-rabbit antibody) in the nuclei (merge: yellow, white arrows; nuclei were counterstained with DAPI = blue) of HEK293T cells. 5 μg of a wild type estrogen receptor vector construct [43] was co-transfected with 5 μg wild type BCA2 in pCMV-tag2B (FLAG tag) into HEK293T cells and after 48 hrs fixed in methanol:acetone (1:1) and stained with monoclonal anti-estrogen receptor α antibody clone 6F11 (Invitrogen, 08-1214) and a rabbit anti-FLAG antibody (Sigma, St. Louis, MO) following procedures as described previously [12,44]. A Leica DM5500 microscope and Improvision software with a Retiga camera were used to capture and merge fluorescence signals. Size bar = 15 μm.
Table 1.
Summary of BCA2 and ER expression pattern in invasive breast cancers.
| Total Cases | LN positive | Regional Recurrence | ||||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| BCA2 ≤ 1 | 415 | 43.9 | 213 | 51.3 | 35 | 8.6 |
| BCA2 >1 | 530 | 56.1 | 229 | 42.2 | 28 | 5.3 |
| ER+ | 635 | 67.1 | ND | ND | ||
| ER- | 310 | 32.9 | ND | ND | ||
| ER+/BCA2>1 | 392 | 74.0 | ND | ND | ||
ND, not determined; n, number; %, percent;
ER+, estrogen receptor positive; ER-, estrogen receptor negative
BCA2 may be transcriptionally regulated by the estrogen receptor
ERα is a 66-kDa nuclear hormone receptor transcription factor [20]. Upon ligand binding, ERα dimerizes and associates with co-activators and chromatin remodeling factors to activate transcription of genes containing estrogen response elements [21]. The ERα phosphorylation state affects co-activator binding and ERα-DNA binding affinity [22].
BCA2 is expressed in 74% of ER positive invasive breast cancers. Co-localization of BCA2 and ER protein can also be demonstrated in cell lines that express neither endogenous BCA2 nor ER (Fig. 1B). The nuclear co-localization of BCA2 and ER indicates that BCA2 could function as an E3 ligase for ER and regulate its stability as shown for other RING domain ubiquitin ligases.
BCA2 may also be an estrogen (17-beta-estradiol) responsive gene. T47D cells are estrogen responsive when grown in charcoal-stripped fetal calf serum (cFCS) [23]. The ER-positive, BCA2-positive T47D human breast cancer cell line was used to study estrogen effects on BCA2 RNA and protein expression in cells that were grown in medium cFCS, which is growth factor deprived (Fig. 2). For the experiment depicted in figure 2 A and B, T47D cells were grown in cFCS for an extended period of time (72 hrs.), and further cultured in RPMI 1640 medium supplemented with 5% cFCS or 5% cFCS that contained 10 or 100 nM estrogen, for 4, 24, 48 and 72 hours. Northern blotting results show that when cells were grown in 5% cFCS transcription of BCA2 mRNA is similar over a time course of 4-48 hours. Cells treated with estrogen showed increased induction of BCA2 mRNA after 48 hours. Moreover, the induction of BCA2 was concentration dependent; 100 nM estrogen strongly induced BCA2 after 24 and 48 hrs, whereas 10 nM estrogen induced BCA2 to 50% of the levels seen for 100 nM estrogen (Fig. 2A). Induction of BCA2 by estrogen at the RNA levels translated into expression of BCA2 in T47D cells at the protein level. Protein levels are shown after 72 hours in 5%cFCS or cFCS supplemented with 10 nM estrogen (Fig. 2B) indicating that estrogen induces BCA2 RNA and protein in cFCS-cultured T47D cells and that BCA2 is an estrogen responsive gene.
Fig. 2.
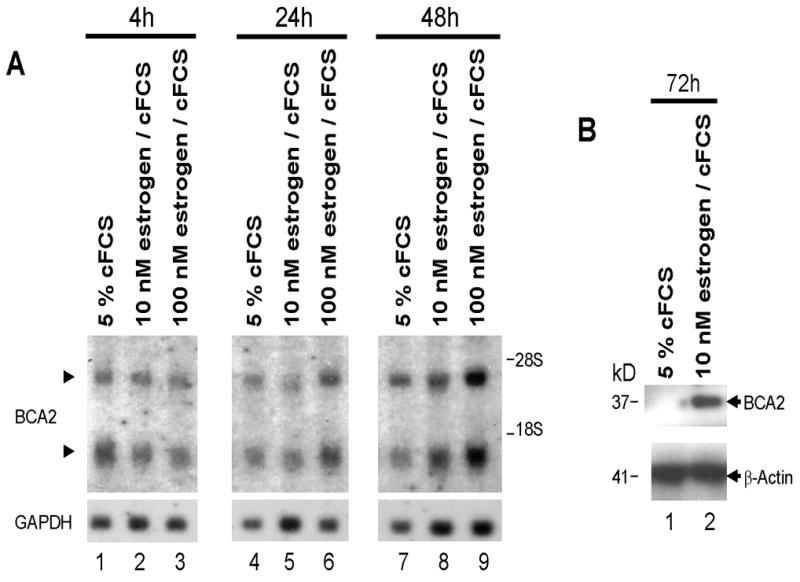
A. Northern blot probed with BCA2 cDNA showing BCA2 expression in RNA extracted from ER-positive T47D cells treated with either 5% cFCS (charcoal-stripped fetal calf serum), or cFCS treated with 10nM or 100nM of estrogen, over a time-course of 48hrs. The same membrane was re-probed for the GAPDH housekeeping gene as loading control. RNA extraction and Northern blots were performed as reported by us before [11,12]. A time and concentration dependent response of BCA2 mRNA to estrogen is seen with the most prominent effects observed at 48 hrs in estrogen-supplemented cFCS. B. Western blot probed with anti-BCA2 antibodies showing BCA2 protein expression in whole cell lysates from T47D cells that were grown in 5% cFCS or cFCS with 10nM estrogen after 72 hours [12]. Beta-actin was as a loading control.
We searched for transcription factor binding sites in the BCA2 promoter region (Fig. 3) using a bioinformatics approach. To predict the potential transcription factors binding to the BCA2 promoter region, an 1100 bp sequence upstream of exon 1 was analyzed. We preformed in silico analysis of the genomic region from 1.1 kb upstream and 235 bp downstream (-1101 to +235), with the Alibaba 2.1 (www.gene-regulation.com) software using the TRANSFAC transcription factor binding sites collection. This analysis revealed a canonical CAAT box (-737 to -734) and reverse TATA box (-656 to -653) and several putative binding sites for general transcription factors; nuclear hormone receptors were also identified and are listed in Table 2 and Figure 3.
Fig. 3.
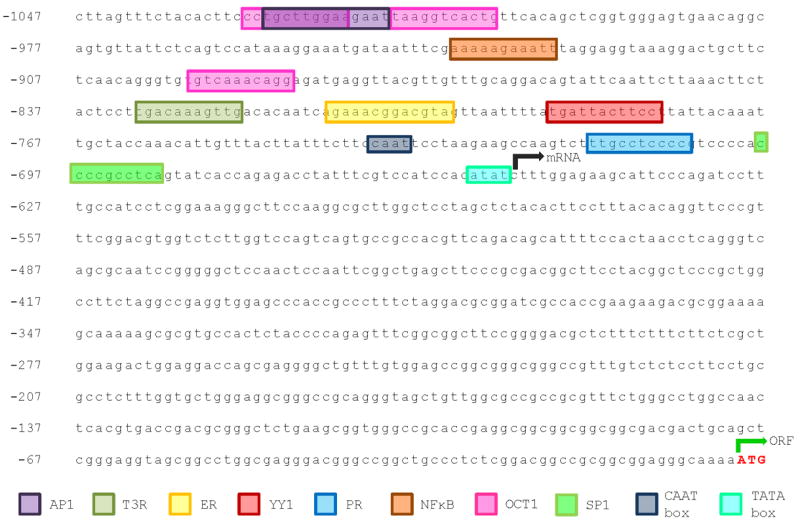
BCA2 promoter. Annotation of transcription factor binding sites (indicated as colored boxes) from bioinformatics analysis of the BCA2 promoter, accession NT_030171.4|Hs19_30426. BCA2 transcriptional targets and their position in the promoter region are also listed in table 2.
Table 2.
Potential regulatory element binding sites in the BCA2 promoter region.
| Regulatory element binding sites | Start | Stop |
|---|---|---|
| AP1 | -1027 | -1018 |
| NF-kappa B | -939 | -930 |
| T3R | -828 | -819 |
| ER | -811 | -800 |
| YY1 | -790 | -780 |
| PR | -716 | -707 |
| SP1 | -698 | -689 |
| OCT1 | -1029 | -1020 |
| -1017 | -1008 | |
| -894 | -885 | |
| TATA Box | -656 | -653 |
| CAAT Box | -737 | -734 |
AP1: Activator Protein 1, NF-kappa B: Nuclear factor kappa-light-chain-enhancer of activated B cells, T3R: Triiodothyronine Receptor, ER: Estrogen Receptor, YY1: Ying Yang 1, PR: Progesteron Receptor, OCT1: Octamer-binding transcription factor 2, SP1: Specificity Protein 1
Up-stream of the TATA box we found a half site for ER, multiple sites for Oct-1, and SP1 (Tab. 2). Other transcriptional factor binding sites include AP-1, NF-κB, T3R, YY1, and PR (Tab. 2, Fig. 3). SP1, AP-1, Oct-1 and YY1 are ubiquitous transcription factors that have a fundamental role in normal biologic processes including differentiation, replication, cellular proliferation and apoptosis [24-27].
T3R and NF-κB have more defined functions in cells [28-30]. Thyroid hormone receptor (T3R) generally requires the binding of its high affinity ligand. However mutants of the ligand-binding domain have been implicated in interactions with other proteins, suggesting participation of other transcription or accessory factors in the hormone-independent activity of T3R.
The estrogen receptor and progesterone receptor (PR) receptor binding sites are most noteworthy in view of the induction of BCA2 RNA and protein expression seen in response to estrogen (Fig. 2).
Sumoylation and transcriptional activation of ER and its target genes
Sumoylation is a dynamic process and its outcomes are extremely diverse, ranging from changes in localization to altered activity and stability of some of the modified proteins. [31]. Ubc9, a SUMO-conjugating enzyme, is essential for the sumoylation pathway as currently understood [32]. The number of known E3 ligases for SUMO modification is relatively low compared to the large number of known sumoylated targets, suggesting that many ligases await identification [33]. Our data presented in Figure 4 suggest that BCA2 may be involved in the conjugation of SUMO to targets such as ER and other transcription factors.
Fig. 4.
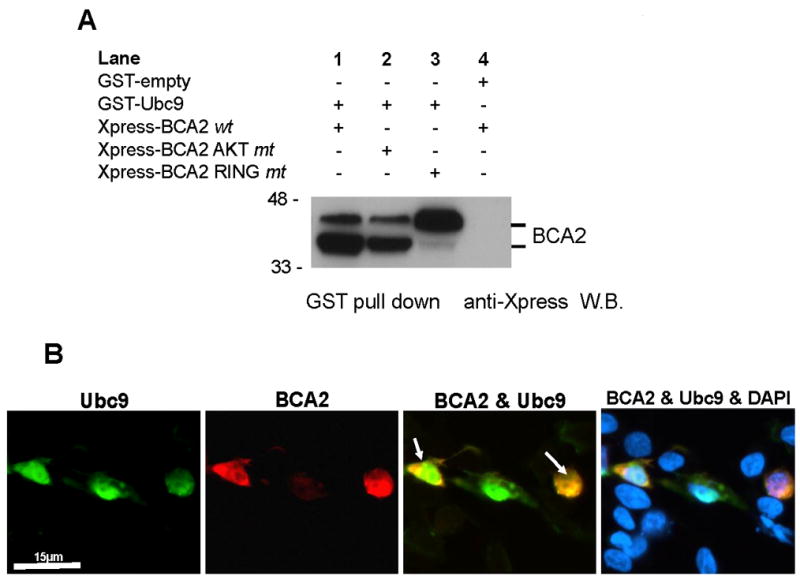
A. Bacterially expressed His/Xpress-tag vectors of BCA2 and GST vectors of Ubc9 were used for GST-pulldown assay as described in [37] and indicated to the left, lanes 1-4. Pull-down assays were performed from bacteriall cell lysates with GST-beads and Western blots (W.B.) probed with anti-Xpress tag antibodies detecting BCA2 and its mutants. B. Co-localization of Ubc9 and BCA2 in HEK293T cells. Ubc9 was cloned into pCMV-tag2B and BCA2 into a GST expression vector as described in [12] and [39]. Cells were co-transfected and processed for immunofluorescence staining as outlined in figure legend 1B. Ubc9 expression was detected by using FITC-labeled (green, against monoclonal anti-FLAG antibodies) anti-mouse antibodies and BCA2 by using TRITC-labeled (red, against our polyclonal BCA2 antibody) anti-rabbit antibodies. Co-expression of Ubc9 and BCA2 (yellow) is indicated by white arrows. Co-localization is seen in both the nucleus and the cytoplasm. Nuclei are counterstained with DAPI (blue). Size bar = 15 μm
Sumoylation has been implicated in regulation of transcriptional activity either by modification of factors which induce or repress the transcription of genes, or through modification of histones [31]. Sumoylation of histones results in the recruitment of factors which promote heterochromatin formation, decreasing the amount of active gene transcription.
In the context of hormone-receptors, androgen receptor (AR), ER and PR, all have SUMO-conjugation sites; however the consequence of modification differs between the different receptors. ER is predominantly activated upon sumoylation. In contrast, when PR and AR are sumoylated, an inhibitory effect is observed [34].
ER sumoylation occurs strictly in the presence of estradiol and it appears that SUMO-1 regulates ER-dependent transcription (Fig. 4, 5) [10]. The SUMO E3 ligases PIAS1 and PIAS3, as well as Ubc9 were found to modulate ER-dependent transcription independently from their SUMO-1 conjugation activity and provide a link between the SUMO and estrogen pathways [10].
Fig. 5.
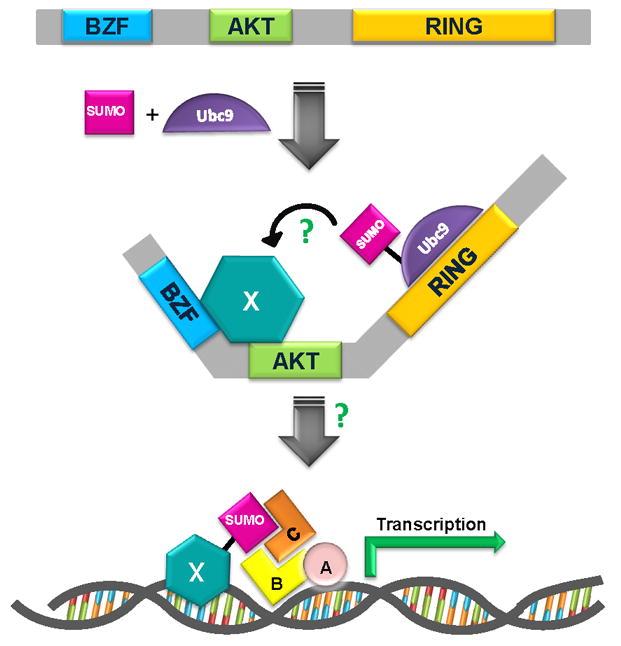
Proposed model for BCA2 involvement in sumoylation. BCA2 with its known domains: BZF, BCA2 zinc finger domain; AKT, BCA2 AKT phosphorylation site; RING-finger domain, catalytic E3 ligase and probable E2 binding site. Ubc9, the SUMO-conjugating enzyme for the sumoylation pathway binds BCA2 likely in the RING domain, similar to the binding ubiquitin-conjugated E2s. This brings SUMO-conjugated Ubc9 into close proximity with the substrate (indicated by X) and allows the transfer of SUMO. Following sumolyation, the substrate may act as a factor in gene transcription though protein-protein interaction (with A, B, C) or DNA binding.
Several of the transcription factors that bind in the BCA2 promoter region (Fig. 3, Tab.1) have been shown to be modified by sumoylation including YY1, SP1 and IκB [32,35-37].
YY1 protein is sumoylated at lysine 288 as the major sumoylation site [35]. Many post-translational modifications have been implicated in the regulation of Sp1 activity including SUMO-1. SUMO-1 is covalently conjugated to Sp1 within the N-terminal negative regulatory domain of Sp1. Compared with Sp1, sumoylation-deficient Sp1 mutants exhibit enhanced cleavage, increasing transcriptional activation. This is in contrast to the constitutively SUMO-1-modified Sp1, which is deficient in proteolytic processing and is associated with inhibition of Sp1 transcriptional activity [36].
The NF-kB inhibitory protein IκB is modified by SUMO-1 on lysine 21, which is also used for ubiquitin conjugation. Importantly, SUMO-1-modified IκB cannot be ubiquitinated and is resistant to proteasome mediated degradation, increasing the stability of the protein. As a result, signal-induced activation of NF-kB dependent transcription is inhibited [32]. While Ubc9 is the SUMO conjugating enzyme of IκB, the cooperating E3 ligase remains unknown.
BCA2 interacts with the SUMO-conjugating enzyme UBC9
RING-E3 ligases are major players in post-translational modification and protein degradation. Known to be imbalanced in cancer, many E3s have emerged as mutated, lost or overexpressed in breast cancer. E3 ligases together with kinases make-up approximately 15% of all cancer genes, suggested to be major regulators in cell growth and death pathways [38]. More recently RING-E3 ligases have been known to play a role in the SUMO pathway, which is thought to be an antagonist of the ubiquitination pathway, by stabilizing proteins and sterically competing for ubiquitin modification sites [2].
To identify potential substrates for the BCA2 E3 ligase, we had previously performed bacterial and yeast-two hybrid screens [39, unpublished data]. Several binding partners were isolated, including, Rab7, ubiquitin, 14-3-3, and the SUMO conjugating enzyme Ubc9 (Tab. 2) [40]. Ubc9 was confirmed by pull down assays using GST-tagged Ubc9 expressed from the pGEX4T3 bacterial expression vector. BCA2 variations were constructed in pET100 vectors containing N-terminal 6xHis and Xpress tags. Purified bacterial recombinant wild-type BCA2, as well as RING (C228A, C231A) and AKT (S132A, S133A) BCA2 mutants, were incubated with GST-Ubc9 from bacterial lysates. Following incubation, mixtures were subjected to SDS-page and Western blotting. Membranes were probed with anti-Xpress antibody (Invitrogen). We found that Ubc9 binds to all BCA2 variants (Fig. 4A). Moreover, when we expressed both BCA2 and Ubc9 in HEK293T cells, we saw co-localization with BCA2 in the cytoplasm and the nucleus suggesting that Ubc9 mediated sumoylation might be important to its nuclear as well as cytoplasmic function (Fig. 4B).
Conclusion
Endocrine therapy for women with metastatic breast cancer is one of the options for systemic treatment in the battle against breast cancer. The “best” strategy depends in large part on the molecular biology of particular cancers, e.g., the expression of hormone receptors and HER2/neu.
Although, several agents that interfere with ER signaling are clinically available, the understanding of their mechanisms is more complex than expected. Tissue- and cell-specific estrogen mechanisms depend upon the formation of a wide variety of co-regulatory complexes as well as variable ER subtypes and extra-nuclear signaling events.
It is well known that post-translational modifications, such as sumoylation, affect breast cancer by altering important regulatory proteins, transcription factors, growth factors and oncoproteins [41]. Innumerate cellular pathways are impacted through sumoylation, affecting transcriptional activity, protein stability and protein sub-cellular localization. Examples of sumoylated oncogenes and tumor suppressor genes include Mdm2, c-Myb, Rb, ER, and p53, all of which undergo sumoylation [41,42]. Here we showed that BCA2 is upregulated by ER and that it in turn could mediate the sumoylation of ER and other proteins through its interaction with Ubc9.
Genomic and proteomic approaches are often used to determine targets differentially regulated by sumoylation. Such work has broadened our understanding of how sumoylation affects ER associated signaling molecules function and localization in breast cancer cells. An example of this is our work with BCA2, where we identified its interactions with the ER and SUMO conjugating enzyme Ubc9. Ubc9 is the only E2 currently known to conjugate SUMO to substrates. It is possible that BCA2 may act as an E3 and catalyze the addition of SUMO to substrates, or BCA2 may auto-sumoylate to regulate its own function, similar to the POU domain transcription factor OCT4 which has been shown to bind Ubc9 [2]. We believe the interplay of SUMO, BCA2, and ER in breast cancer warrants further examination for potential clinical interventions that may exploit their molecular characteristics.
Acknowledgments
This work was supported by Award Number R01CA127258 from the National Cancer Institute, Developmental Therapeutics Program and by the Canadian Breast Cancer Research Alliance special program grant on metastasis.
List of abbreviations
- BCA2
Breast cancer associated gene 2
- SUMO
small ubiquitin-like modifier
- AP1
Activator Protein 1
- NF-kappa B
Nuclear factor kappa-light-chain-enhancer of activated B cells
- T3R
Triiodothyronine Receptor
- ER
Estrogen Receptor
- ERE
Estrogen responsive element
- YY1
Ying Yang 1
- PR
Progesteron Receptor
- OCT1
Octamer-binding transcription factor 2
- HBBCC
Henrietta Banting Breast Cancer Collection
- FITC
Fluorescein isothiocyanate
- TRITC
Tetramethylrhodamine isothiocyanate
- DAPI
4’,6-diamidino-2-phenylindole
- His
histidine
- GST
glutathione S transferase
- SP1
Specificity Protein 1
- OCT4
Octamer-binding transcription factor 4
- AR
Androgen Receptor
Footnotes
Conflict of Interest: None
Reference List
- 1.Desterro JMP, Rodriguez MS, Hay RT. Regulation of transcription factors by protein degradation. Cell Mol Life Sci. 2007;57:1207–19. doi: 10.1007/PL00000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Z, Liao B, Xu M, Jin Y. Post-translational modification of POU domain transcription factor Oct-4 by SUMO-1. FASEB J. 2007;21:3042–51. doi: 10.1096/fj.06-6914com. [DOI] [PubMed] [Google Scholar]
- 3.Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 2003;4:137–142. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishida T, Terashima M, Fukami K, Yamada Y. PIASy controls ubiquitination-dependent proteasomal degradation of Ets-1. Biochem J. 2007;405:481–88. doi: 10.1042/BJ20070026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C, Seth AK, Aplin AE. Genetic and expression aberrations of E3 ubiquitin ligases in human breast cancer. Mol Cancer Res. 2006;4:695–707. doi: 10.1158/1541-7786.MCR-06-0182. [DOI] [PubMed] [Google Scholar]
- 6.von, Mikecz A. The nuclear ubiquitin-proteasome system. J Cell Sci. 2006;119:1977–84. doi: 10.1242/jcs.03008. [DOI] [PubMed] [Google Scholar]
- 7.Huang J, Li X, Maguire CA, Hilf R, Bambara RA, Muyan M. Binding of estrogen receptor beta to estrogen response element in situ is independent of estradiol and impaired by its amino terminus. Mol Endocrinol. 2005;19:2696–2712. doi: 10.1210/me.2005-0120. [DOI] [PubMed] [Google Scholar]
- 8.Feng Y, Manka D, Wagner KU, Khan SA. Estrogen receptor-alpha expression in the mammary epithelium is required for ductal and alveolar morphogenesis in mice. Proc Natl Acad Sci USA. 2007;104:14718–23. doi: 10.1073/pnas.0706933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eakin CM, Maccoss MJ, Finney GL, Klevit RE. Estrogen receptor alpha is a putative substrate for the BRCA1 ubiquitin ligase. Proc Natl Acad Sci USA. 2007;104:5794–99. doi: 10.1073/pnas.0610887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sentis S, Le Romancer M, Bianchin C, Rostan MC, Corbo L. Sumoylation of the estrogen receptor alpha hinge region regulates its transcriptional activity. Mol Endocrinol. 2005;19:2671–84. doi: 10.1210/me.2005-0042. [DOI] [PubMed] [Google Scholar]
- 11.Burger A, Li H, Zhang XK, Pienkowska M, Venanzoni M, Vournakis J, et al. Breast cancer genome anatomy: correlation of morphological changes in breast carcinomas with expression of the novel gene product Di12. Oncogene. 1998;16:327–33. doi: 10.1038/sj.onc.1201517. [DOI] [PubMed] [Google Scholar]
- 12.Burger AM, Gao Y, Amemiya Y, Kahn HJ, Kitching R, Yang Y, et al. A novel RING-type ubiquitin ligase breast cancer-associated gene 2 correlates with outcome in invasive breast cancer. Cancer Res. 2005;65:10401–12. doi: 10.1158/0008-5472.CAN-05-2103. [DOI] [PubMed] [Google Scholar]
- 13.Burger A, Amemiya Y, Kitching R, Seth AK. Novel RING E3 ubiquitin ligases in breast cancer. Neoplasia. 2006;8:689–95. doi: 10.1593/neo.06469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reid G, Hubner MR, Metivier R, Brand H, Denger S, Manu D, et al. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 15.Lonard DM, Nawaz Z, Smith CL, O’Malley BW. The 26S proteasome is required for estrogen receptor-alpha and coactivator turnover and for efficient estrogen receptor-alpha transactivation. Mol Cell. 2000;5:939–48. doi: 10.1016/s1097-2765(00)80259-2. [DOI] [PubMed] [Google Scholar]
- 16.Wijayaratne AL, McDonnell DP. The human estrogen receptor-alpha is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J Biol Chem. 2001;276:35684–92. doi: 10.1074/jbc.M101097200. [DOI] [PubMed] [Google Scholar]
- 17.Fan S, Wang J, Yuan R, Ma Y, Meng Q, Erdos MR, et al. BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science. 1999;284:1354–6. doi: 10.1126/science.284.5418.1354. [DOI] [PubMed] [Google Scholar]
- 18.Saji S, Okumura N, Eguchi H, Nakashima S, Suzuki A, Toi M, et al. MDM2 enhances the function of estrogen receptor alpha in human breast cancer cells. Biochem Biophys Res Commun. 2001;281:259–65. doi: 10.1006/bbrc.2001.4339. [DOI] [PubMed] [Google Scholar]
- 19.Nawaz Z, Lonard DM, Smith CL, Lev-Lehman E, Tsai SY, Tsai MJ, O’Malley BW. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol Cell Biol. 1999;19:1182–9. doi: 10.1128/mcb.19.2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nilsson S, Gustafsson J. Estrogen receptor transcription and transactivation: Basic aspects of estrogen action. Breast Cancer Res. 2000;2:360–6. doi: 10.1186/bcr81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klinge CM. Estrogen receptor interaction with co-activators and co-repressors. Steroids. 2000;65:227–51. doi: 10.1016/s0039-128x(99)00107-5. [DOI] [PubMed] [Google Scholar]
- 22.Chu I, Arnaout A, Loiseau S, Sun J, Seth A, McMahon C, et al. Src promotes estrogen-dependent estrogen receptor alpha proteolysis in human breast cancer. J Clin Invest. 2007;117:2205–15. doi: 10.1172/JCI21739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddel RR, Murphy LC, Sutherland RL. Factors affecting the sensitivity of T-47D human breast cancer cells to tamoxifen. Cancer Res. 1984;44:2398–405. [PubMed] [Google Scholar]
- 24.Feng D, Kan YW. The binding of the ubiquitous transcription factor Sp1 at the locus control region represses the expression of beta-like globin genes. Proc Natl Acad Sci USA. 2005;102:9896–900. doi: 10.1073/pnas.0502041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–6. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 26.Bing D, Feng-Qi Z. Involvement of the ubiquitous Oct-1 transcription factor in hormonal induction of ß-casein gene expression. Biochem J. 2007;401:57–64. doi: 10.1042/BJ20060570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–42. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 28.Helmer EB, Raaka BM, Samuels HH. Hormone-dependent and -independent transcriptional activation by thyroid hormone receptors are mediated by different mechanisms. Endocrinology. 1996;137:390–9. doi: 10.1210/endo.137.2.8593781. [DOI] [PubMed] [Google Scholar]
- 29.Xu J, Thompson KL, Shephard LB, Hudson LG, Gill GN. T3 receptor suppression of Sp1-dependent transcription from the epidermal growth factor receptor promoter via overlapping DNA-binding sites. J Biol Chem. 1993;268:16065–73. [PubMed] [Google Scholar]
- 30.Nakshatri H, Bhat-Nakshatri P, Martin DA, Goulet RJ, Jr, Sledge GW., Jr Constitutive activation of NF-kappaB during progression of breast cancer to hormone-independent growth. Mol Cell Biol. 1997;17:3629–39. doi: 10.1128/mcb.17.7.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia-Dominguez M, Reyes JC. SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim Biophys Acta. 2009;1789:451–9. doi: 10.1016/j.bbagrm.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Mo YY, Yu Y, Theodosiou E, Ee PL, Beck WT. A role for Ubc9 in tumorigenesis. Oncogene. 2005;24:2677–83. doi: 10.1038/sj.onc.1208210. [DOI] [PubMed] [Google Scholar]
- 33.Knipscheer P, Flotho A, Klug H, Olsen JV, van Dijk WJ, Fish A, et al. Ubc9 sumoylation regulates SUMO target discrimination. Mol Cell. 2008;31:371–82. doi: 10.1016/j.molcel.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Kaikkonen S, Jaaskelainen T, Karvonen U, Rytinki MM, Makkonen H, Gioeli D, Paschal BM, Palvimo JJ. SUMO-specific protease 1 (SENP1) reverses the hormone-augmented SUMOylation of androgen receptor and modulates gene responses in prostate cancer cells. Mol Endocrinol. 2009;23(3):292–307. doi: 10.1210/me.2008-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng Z, Wan M, Sui G. PIASy-mediated sumoylation of Yin Yang 1 depends on their interaction but not the RING finger. Mol Cell Biol. 2007;27:3780–92. doi: 10.1128/MCB.01761-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spengler ML, Brattain MG. Sumoylation inhibits cleavage of Sp1 N-terminal negative regulatory domain and inhibits Sp1-dependent transcription. J Biol Chem. 2006;281:5567–74. doi: 10.1074/jbc.M600035200. [DOI] [PubMed] [Google Scholar]
- 37.Xu J, Watkins T, Reddy A, Reddy ES, Rao VN. A novel mechanism whereby BRCA1/1a/1b fine tunes the dynamic complex interplay between SUMO-dependent/independent activities of Ubc9 on E2-induced ERalpha activation/repression and degradation in breast cancer cells. Int J Oncol. 2009;34:939–49. doi: 10.3892/ijo_00000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burger AM, Seth AK. The ubiquitin-mediated protein degradation pathway in cancer: therapeutic implications. Eur J Cancer. 2004;40:2217–29. doi: 10.1016/j.ejca.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 39.Amemiya Y, Azmi P, Seth A. Autoubiquitination of BCA2 RING E3 ligase regulates its own stability and affects cell migration. Mol Cancer Res. 2008;6:1385–96. doi: 10.1158/1541-7786.MCR-08-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Connor MK, Azmi PB, Subramaniam V, Li H, Seth A. Molecular characterization of ring finger protein 11. Mol Cancer Res. 2005;3:453–61. doi: 10.1158/1541-7786.MCR-04-0166. [DOI] [PubMed] [Google Scholar]
- 41.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6:776–88. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 42.Baek SH. A novel link between SUMO modification and cancer metastasis. Cell Cycle. 2006;5:1492–5. doi: 10.4161/cc.5.14.3008. [DOI] [PubMed] [Google Scholar]
- 43.Qi X, Borowicz S, Pramanik R, Schultz RM, Han J, Chen G. Estrogen receptor inhibits c-Jun-dependent stress-induced cell death by binding and modifying c-Jun activity in human breast cancer cells. J Biol Chem. 2004;279:6769–77. doi: 10.1074/jbc.M311492200. [DOI] [PubMed] [Google Scholar]
- 44.Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJ, Double JA, et al. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005;65:1489–96. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]


