Abstract
The TqsA (YdgG) protein of Escherichia coli has been shown to export the autoinducer-2 (AI-2) molecule, a furanosyl borate diester that bears little resemblance to previously characterized biological molecules. TqsA belongs to a large superfamily, the AI-2 exporter (AI-2E) superfamily, of putative transporters with no other functionally characterized members. These proteins derive exclusively from bacteria. Many different bacterial kingdoms contain them, although several kingdoms do not. These proteins exhibit a uniform topology with 8 putative transmembrane segments (TMSs) which we show probably arose from a 4-TMS precursor in a process that involved at least one and possibly two intragenic duplication event(s). The first halves of these proteins are more diverse in sequence than the second halves, suggesting that the first halves may serve substrate-specific functions while the second halves serve family-specific functions. Conserved residues and motifs in these proteins are identified. Some homologues include extra catalytic domains including those involved in purine nucleotide biosynthesis, ATP and GTP binding, and molecular signaling. The results presented provide guides for future functional studies on members of this superfamily of bacterial transporters.
Key Words: Transport, Metabolite exporters, Autoinducer, Phylogeny, Topology, Evolution
Bacteria use a variety of means to communicate with one another and with their eukaryotic hosts [1,2,3,4,5,6]. In some cases, social interactions in complex bacterial colonies allow bacteria to synchronize the behavior of all members of the group and thereby act like multicellular organisms [7]. By contrast, some bacterial social engagements promote individuality among members within the group and thereby foster diversity [8, 9]. Bacterial communication systems include long- and short-range chemical signaling [10]; one-way, two-way, and multi-way communication [11]; contact-mediated and contact-inhibited signaling [12, 13], and the use and spread of misinformation, even deadly information [8, 14].
Bacteria use a diversity of small molecules for extra- and intracellular signaling [8]. They scan small-molecule mixtures to access information about both their extracellular environment and their intracellular physiological status. Based on the integrated information, they continuously interpret their circumstances and react rapidly to changes [15]. They must integrate extra- and intracellular signaling information to mount appropriate responses to environmental changes.
Several small-molecule bacterial signaling pathways have been identified. These include extracellular ‘quorum-sensing’ signaling and intracellular cyclic dinucleotide signaling. Possibly, these two pathways converge to control complex processes including multicellularity, biofilm formation, and virulence [15].
The exchange of extracellular signaling molecules must be mediated by specific transporters, some involved in molecular uptake, others catalyzing efflux [16,17,18]. Unlike other autoinducers, which are specific to a particular species of bacteria, autoinducer-2 (AI-2) is produced by a large number of bacterial species. AI-2 has been proposed to serve as a ‘universal’ signal for inter-species communication [8]. The crystal structure of an AI-2 sensor protein, LuxP, in complexation with the autoinducer, revealed the bound ligand to be a furanosyl borate diester that bears no resemblance to previously characterized autoinducers [19].
Recently, the Escherichia coli YdgG (TqsA) protein was shown to control biofilm formation in E. coli K-12 by mediating AI-2 transport. YdgG had been known to be induced in E. coli biofilms. Deletion of ydgG decreased extracellular and increased intracellular concentrations of AI-2, suggesting that YdgG enhances export of AI-2 [20]. Consistent with this hypothesis, deletion of the ydgG gene increased cell motility by increasing transcription of flagellar genes that were known to be induced by AI-2. By expressing ydgG in trans, the wild-type phenotypes for extracellular AI-2 activity, motility, and biofilm formation were restored.
YdgG is a membrane-spanning protein that is conserved in many bacteria. It influences resistance to several antimicrobials, including crystal violet and streptomycin [20]. Deletion of ydgG caused 31% of the bacterial chromosome to be differentially expressed in biofilms. This fact apparently resulted because AI-2 controls the transcription of hundreds of genes. Since YdgG catalyzes export of the quorum-sensing signal, AI-2, the gene name tqsA (transporter of quorum sensing-A) was suggested [20].
In this communication, we analyze the family of putative transporters to which TqsA belongs. We name this family the AI-2 exporter (AI-2E) superfamily, based on the function of its first characterized member. We show that it is a huge superfamily (Pfam family PF01594) with members derived from a wide range of bacteria. However, no member of the family was identified from an archaeon, a eukaryote, or a member of certain bacterial kingdoms that include sequenced organisms with reduced genome sizes. Phylogenetic analyses revealed the relationships of these homologues to each other with groupings largely according to organismal type. However, this occurred in an unusual fashion suggestive of several functional types with occasional lateral gene transfer responsible for some of the anomalies. Thus, the analyses reveal groups of apparent orthologues that presumably serve the same function. Conserved motifs are identified which must play specific functional/structural roles. An evolutionary pathway for the appearance of these proteins, based on intragenic duplication of an ancestral 4-transmembrane segment (TMS) unit is established. Finally, extra domains with recognized functions in some of these proteins are identified, and these provide clues as to the associated transport functions.
Methods and Results
Initial Phylogenetic Analyses of the AI-2E Superfamily
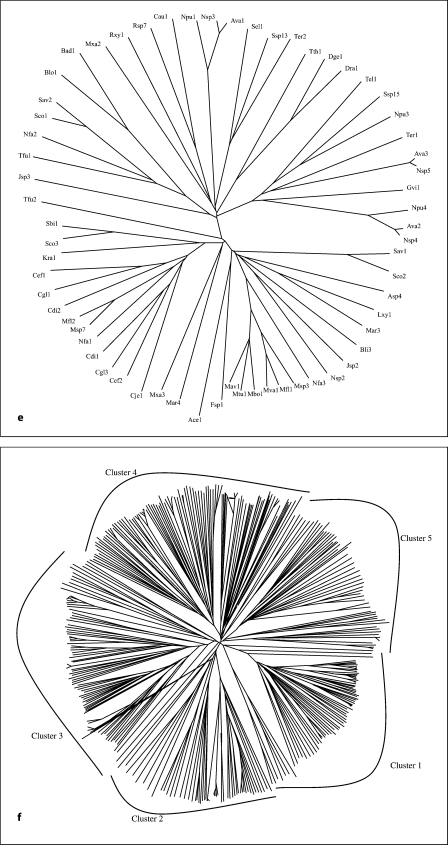
The PerM protein of E. coli (TC #2.A.86.1.1) was used as the query sequence in PSI-BLAST searches (default settings) with six iterations [21]. A modified CD hit program [22] was used to eliminate redundancies and closely similar sequences of greater than 90% identity (the default setting). This program randomly selects one protein of several that are of greater than 90% identity with each other. Membership to the AI-2E superfamily was assigned based on sequence similarity throughout the transmembrane regions. All members of the superfamily thus exhibited at least 9 SD (probability that the observed degree of similarity arose by chance is less than 10–19[23]). 391 non-redundant homologues from the NCBI protein databank remained and served as our data set for all analyses reported here (see table S1 on our website; http://www.biology.ucsd.edu/∼msaier/supmat/AI-2). A neighbor-joining phylogenetic tree (fig. 1f), based on a CLUSTAL × multiple alignment [24] (see fig. S1 on our website), and a dendogram (fig. S2), drawn using the TreeView program [25], were generated.
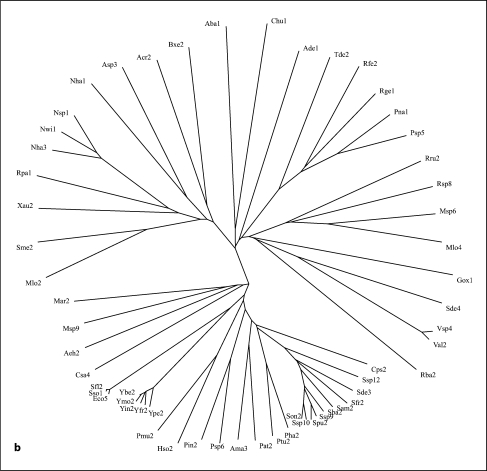
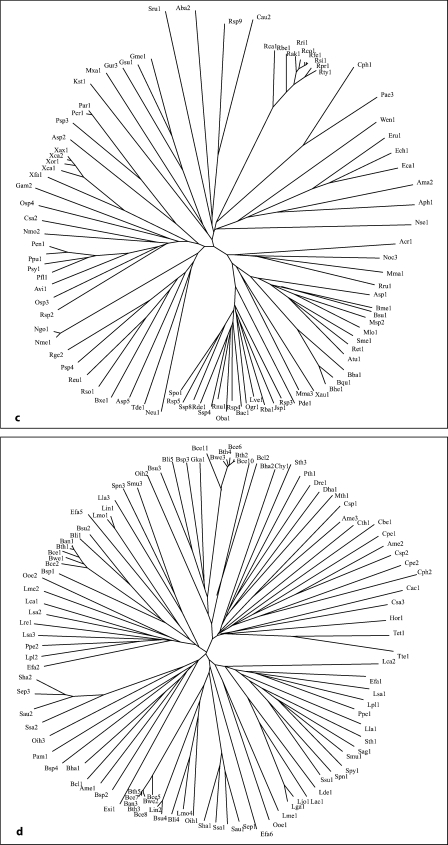
Fig. 1.
Phylogenetic trees for the 5 families of proteins in the AI-2E superfamily (a–e) as well as for the complete superfamily (f). a–e Families 1-5, respectively. f The full superfamily tree showing the positioning of families 1-5 relative to each other. The dendogram corresponding to the tree in f is presented in figure S2. It is based on the multiple alignment shown in figure S1. Programs used to derive the trees have been described (CLUSTAL × [24] and TreeView [25]). Protein abbreviations for all members of the AI-2E superfamily can be found in table S1 (alphabetically arranged) and tables S3–S7 for the 5 families (1–5, respectively). The standard abbreviations used in our laboratory and many others include three letters often followed by a number. The first capitalized letter refers to the genus name while the second two small letters are the first two letters of the species name. Thus, Eco2 refers to the second paralogue in E. coli.
The proteins generally fell into 5 distinct clusters or families of fairly similar size (fig. 1f). Cluster 1 (family 1) contains 78 proteins; family 2, 58 proteins; family 3, 89 proteins; family 4, 103 proteins, and family 5, 63 proteins. The multiple alignment of all 391 proteins (fig. S1) revealed two fully conserved prolyl residues, one at alignment position 532, and the other at alignment position 627 within hydrophobic peaks 6 and 8, respectively (see below). Several gaps were observed within the hydrophilic regions of these aligned sequences (positions 249–263, 336–384, 404–416, 441–446, 463–468, 508–515, 523–526, and 559–569 of the 1,112 residue position alignment presented in fig. S1).
Topological and Sequence Conservation Analyses
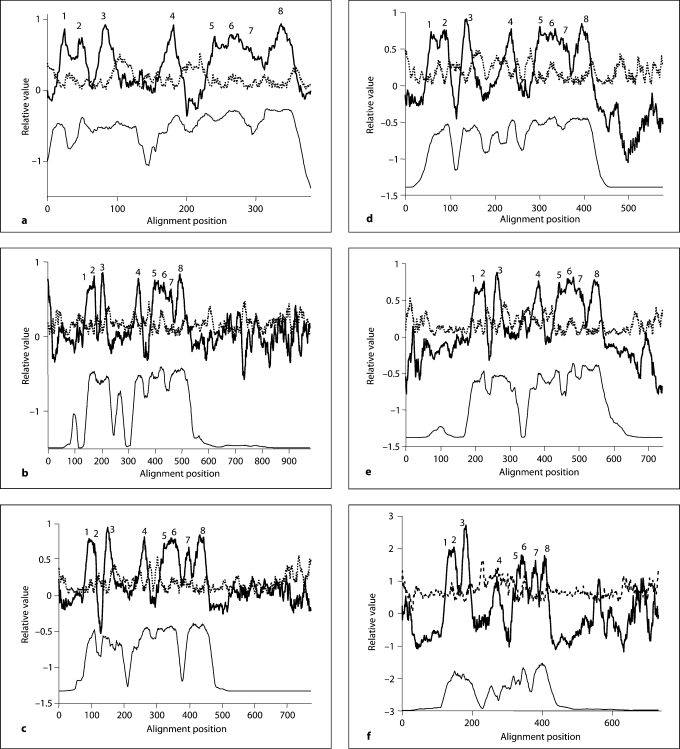
Each of the 5 families within the AI-2E superfamily was analyzed separately for sequence conservation, topology, and phylogeny. The proteins, their organismal sources, abbreviations, sizes, numbers of putative TMSs and Genbank (gi) identification numbers for the 5 clusters are provided in tables S3–S7. The 5 neighbor-joining phylogenetic trees are shown in figure 1a–e, while the average hydropathy, amphipathicity and similarity plots, based on the AveHAS program [26] are shown in figure 2a–e. The AveHAS program averages the hydropathy plots for all aligned sequences, providing much greater accuracy than would otherwise be possible. These figures provide the bases for several of the conclusions cited below. They were derived using the multiple alignments presented in figure S3–S7 on our website. Hydropathy and amphipathicity plots for individual proteins were generated using the WHAT program [26].
Fig. 2.
Relative average hydropathy (solid dark lines, top), average amphipathicity (dotted gray lines, top) and average similarity (solid but thin lines, bottom) plots for the 5 families of proteins in the AI-2E superfamily. a–e Families 1–5, respectively. The Ave-HAS program was used to derive the plots using all members of each of the 5 families included in this study [47] . Peaks of hydrophobicity that are believed to correspond to the 8 putative TMSs are numbered above the hydropathy plot. Assignments were confirmed using the WHAT [26] and TMHMM [48] programs for individual proteins. These plots are based on the multiple alignments shown in figure S3–S7.
Family 1 proteins show eight peaks of hydrophobicity (fig. 2a) and several fully conserved residues (fig. S3). These include seven prolyl residues (alignment positions 104, peak 3; 195, end of peak 4; 217, between peaks 4 and 5; 279, peak 6; 326 and 337, beginning of peak 8; and 361, end of peak 8). The second halves of these proteins proved to be better conserved than the first halves.
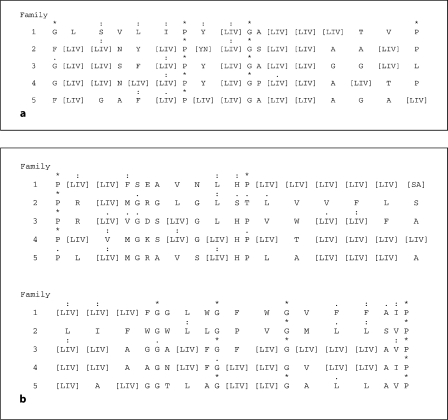
We derived consensus sequences for the two best conserved regions of these proteins for all 5 families. In peak 6, the consensus sequence was:
* : : : * : : * *
GLSVLIPY (LIV) GA (LIV)3 TVP
while in peak 8, the consensus sequence was:
* : : : : * : : : * * . * : . :. :*
P (LIV) FSEAVNLHP (LIV)4 (SA) (LIV)3 FGGLWGFWGVFFAIP
where asterisks indicate identities only, colons close similarities, and dots more distant similarities as defined by the CLUSTAL × program; alternative consensus residues at any one position are indicated in parentheses.
Figure 3 shows how these two consensus sequences differ between the proteins of the 5 phylogenetic families. Of these 5 families, family 1 appears to be the best conserved with respect to both identities and conservative substitutions. The two sets of consensus sequences show similarities in all 5 families. For example, the P at position 7 in motif 1 and the P in the terminal position of motif 2 are fully conserved in all proteins of the AI-2E superfamily as noted above, suggesting an essential structural or functional role. The G at position 10 in the first consensus sequence and those at positions 21 and 25 in the second consensus sequence are conserved in nearly all families. While the dominant residue may differ in the different families at specific positions, these are not fully conserved in any of these families. Thus, when the consensus residues differ, they are never fully conserved. We conclude that in none of these 5 families is a distinctive function, common to all members in that family, but different from those of the other four families, dictated by a specific residue or set of residues within these two conserved motifs. It appears that these motifs provide structural features or a function that is common to all members of the AI-2E superfamily.
Fig. 3.
Consensus motifs 1 (a) and 2 (b) showing degrees of relative conservation in the most conserved regions of the proteins of the 5 families within the AI-2E superfamily. Asterisks indicate identities only; colons indicate close similarities; dots indicate more distant similarities as defined by the CLUSTAL × program for each of the 5 families (1–5); alternative consensus residues at any one position are indicated in brackets. b The longer motif is presented in two lines.
The average hydropathy plots shown in figure 2a–e reveal the characteristics of each of the 5 families in the AI-2E superfamily. Family 1 is most compact, with no protein showing N- or C-terminal extensions. In this plot, peaks 1 and 2 are close to each other, peak 3 is more distant, and peak 4 is still more distant. Peaks 5–7 are all overlapping while peak 8 is distinct but close to peak 7. This pattern is observed for all 5 families (compare figure 2a–e).
The average amphipathicity plots revealed that all 5 families show similar characteristics: amphipathic peaks occur following hydrophobic peaks 3, 4 and 8 and coinciding with peak 6. Motif 1, also coinciding with peak 6, reveals a general pattern of a semipolar residue followed by three hydrophobic residues. A helical wheel depiction showed that strongly hydrophobic residues occur on one side of helix 6, while semipolar residues predominate on the other side (data not shown), thus accounting for the amphipathicity of putative TMS 6.
As noted above, families 2–5 show poorer conservation than observed for family 1 (figure 2a–e and 3). Further, families 2–4 contain proteins with short N-terminal and long C-terminal hydrophilic sequences, while family 5 contains proteins with both N- and C-terminal extensions maximally of about 160 residues each. The domains in these longer proteins will be analyzed and described below.
Phylogenetic Analyses of the 5 Families of the AI-2E Superfamily
The phylogenetic trees for the 5 AI-2E families are presented in figure 1a–e. Family 1 proteins derive exclusively from the proteobacterial phylum of the γ- and δ-orders. Family 2 is largely from α-, β-, γ- and δ-proteobacteria, but three proteins are from a single species each of the Bacteroides, Firmicute and Planctomycetes phyla, respectively. Family 3 includes proteins from the same proteobacteria as for family 2, but Chlorobi and Chloroflexi proteins are also present. Family 4 proteins are exclusively from firmicutes except for one actinobacterial protein. Finally, family 5 proteins are mostly from actinobacteria and cyanobacteria with a few proteins from Deinococcus, Chloroflexi and ε-proteobacterial species. It is therefore clear that phylogenetic clustering is predominantly according to organismal type with a few interesting exceptions. Also of interest is the absence of homologues in specific bacterial kingdoms such as the spirochetes, chlamydia, and mycoplasma. Most of the sequenced genomes of bacteria in these kingdoms are of reduced size due to their obligate parasitic lifestyles. We detected no homologues outside of the bacterial domain.
Repeat Elements in AI-2E Superfamily Proteins
The GAP and IC programs, with default settings and 500 random shuffles (to establish statistical significance) [27, 28] were used in attempts to establish homology between the first and second putative halves (TMSs 1–4 and 5–8) in proteins of the AI-2E superfamily, respectively. Figure 4a shows an alignment of putative TMSs 1–3 with putative TMSs 5–7, and figure 4b shows an alignment of putative TMS 4 with putative TMS 8. In figure 4a, the alignment gave 33% identity, 46% similarity, and a comparison score of 8.5 SD. In figure 4b, the alignment gave 30% identity, 41% similarity, and a comparison score of 9.4 SD. These values correspond to probabilities of ∼10–17 and ∼10–20, respectively, that the observed degree of sequence similarity could have arisen by chance [23]. The proteins compared are derived from family 3. Interestingly, TMS 4 and the upstream hydrophilic region of the Nmo2 protein aligned with TMSs 6–8 in the Asp2 protein, suggesting that the hydrophilic region between putative TMSs 3 and 4 may have arisen from the equivalent of TMSs 6 and 7. These results clearly suggest that these proteins arose in a process that involved intragenic duplication, but it is not clear whether part or all of the 4-TMS element was duplicated in a single event. The precise path may have involved more than one (partial) intragenic duplication event (see ‘Discussion’).
Fig. 4.
Alignments of TMSs 1–3 (top) with TMSs 5–7 (bottom; a) and TMS 4 (top) with TMS 8 (bottom; b) and their preceding regions. The alignments were generated with the GAP program [27]. Vertical lines indicate identities; colons indicate close similarities; dots indicate more distant similarities as defined by the GAP program [27]. Residue numbers in each protein are indicated at the beginning and end of each line.
Extra Domains in Large Homologues
Table S2 presents the 44 AI-2E superfamily homologues that exhibit larger than normal sizes. Twenty-two are from family 5 (actinobacteria and Chloroflexi), 14 are from family 2 (α- and β-proteobacteria and one Planctomycetes species), 5 are from family 3 (α-, β- and γ-proteobacteria), and 3 are from family 4 (firmicutes). One of the homologues, Tde1 of 665 residues, has a C-terminal domain homologous throughout its length with phosphoribosyl amino imidazole succinocarboxamide synthetase, involved in purine nucleotide biosynthesis. It is thus possible that Tde1 is an exporter specific for a purine nucleotide or one of its biosynthetic metabolites. A second protein, Bme1, also of family 3, has a C-terminal ATP/GTP binding P-loop-containing domain similar to those present in three E. coli proteins, DnaA (P03004), DNA polymerase III δ-subunit (HolA; P28630) and the ATP binding protein, IstB (AAC33916). This observation can be interpreted to suggest a function concerned with export of a nucleotide or a derivative of it.
Two large protein homologues in family 2, Nha1 and Asp3, possess C-terminal sensor GAF domains [29]. These domains can be signaling domains and are present in cyclic di-GMP-regulated cyclic nucleotide phosphodiesterases, adenylate cyclases and bacterial transcription factors [30]. They are similar in structure to PAS domains and probably bind cyclic nucleotides [30]. This observation agrees with those cited above, suggesting an involvement in cyclic nucleotide export. It should be noted that cyclic AMP efflux from E. coli is dependent on the proton motive force [31], but that no protein has yet been shown to possess such an activity. It is possible that some of these AI-2E porters are the unidentified cyclic AMP exporters [31].
Three proteins in family 2, Nwi1, Nsp1 and Nha3, appear to contain partial or full-length Tpr-like ligand-binding domains [32,33]. This suggestion is based on TC-BLAST hits when these three proteins were blasted against TCDB [34]. The searches yielded above threshold hits (e–4) to the Sec72 protein of the SRP complex of Saccharomyces cerevisiae (3.A.5.8.1). This latter protein is known to possess the Tpr domain in the region of sequence similarity. The alignment is shown in figure 5 for Nwi1 versus Sec72. This alignment, with 36% identity and 50% similarity, gave a comparison score of 13.5 SD. This value is considerably in excess of what is required to establish homology [35]. It is worth noting that the region of similarity overlaps the Tpr domain of Sec72 but includes the N-terminally adjacent region as well. This region is actually present in many of the large family 2 proteins.
Fig. 5.
Alignment of part of Nwi1 (table S1) with part of the Sec72 protein constituent of the general secretory pathway in S. cerevisiae (TC No. 3.A.5.8.1). The programs used and format of presentation are as described in the legend to figure 4.
Discussion
In this report, we have characterized a superfamily of putative secondary efflux pumps, previously named the PerM family [36, 37]. The functional characterization of the TqsA protein of E. coli (previously annotated as YdgG) by Herzberg et al. [20] revealed that this PerM family member is the AI-2 efflux pump. The present demonstration of the distribution of 391 members of this superfamily from many bacterial kingdoms has caused us to rename this family after the substrate of its only functionally characterized member. We have, consequently designated it the AI-2E superfamily.
The AI-2E superfamily exhibits some unique characteristics. First, the members fall into 5 phylogenetic clusters or families that we have analyzed separately. Second, all members seem to have a uniform 8-TMS topology, a characteristic reflected by the average hydropathy plots for all 5 families (fig. 2a–e). Two closely spaced TMSs (TMSs 1 and 2) are followed by a more distant TMS 3 and an even more distant TMS 4. These are then followed by four more TMSs in a 3 + 1 arrangement. Third, these hydrophobic domains are homologous throughout their lengths in all 391 members of the AI-2E superfamily analyzed here, and they include two fully conserved prolyl residues, one in TMS 6 and one in TMS 8. Fourth, the second 4 TMSs are better conserved than the first 4 TMSs, a generalization that appears valid for all 5 families within this superfamily. Finally, the two halves apparently arose by one or more internal duplication event(s). TMSs 1–3 are almost certainly homologous to TMSs 5–7, and TMS 4 is homologous to TMS 8.
Surprisingly, the long hydrophilic domain preceding TMS 4 (but not TMS 8) is similar in sequence to the region bearing TMSs 6 and 7 in spite of the greater hydrophilicity of the former regions. This observation suggests that the pathway by which these proteins arose may have been complex. There may have been two partial or full-length duplications of the compact C-terminal 4-TMS domain. Two partial duplications, followed by sequence divergence, or two full-length duplications, followed by partial deletion and sequence divergence, could have given rise to the current members of the superfamily. If the latter possibility is correct, the one deletion event giving rise to an 8-TMS protein from an ancestral 12-TMS protein might have occurred early during the evolution of the superfamily in order to account for their uniform topologies.
The two fully conserved prolyl residues in all members of the AI-2E superfamily are worthy of note. Full conservation in a wide range of sequence-divergent homologues suggests one or more essential functional or structural roles. No molecular genetic analyses have been reported that provide a clue as to what this role or these roles may be. However, cis/trans-prolyl isomerization has been reported to control ion gating channel activity [38]. It is possible that one or both of the conserved prolyl residues in members of the AI-2E superfamily undergo cis/trans-induced conformational changes that accompany transport. Other possibilities, such as essential structural features, can also be considered.
With only one member of the AI-2E superfamily characterized, the functions of most members remain an open question. However, there are clues. The clustering of members of this superfamily into at least 5 families, each derived from a different but often overlapping set of bacterial kingdoms, leads to the suggestion that each family has a set of characteristic properties. Usually, members of a superfamily catalyze only uptake or efflux, but occasionally large superfamilies (e.g. MFS, ABC, DMT) can catalyze both. In almost all cases where both sym- and antiporters occur within the same superfamily, the symporters cluster separately from the antiporters on a phylogenetic tree [36, 39,40,41,42,43,44,45,46]. Thus, if all AI-2E superfamily members catalyze efflux, as we tentatively predict, they are likely to transport a variety of compounds, possibly all related in structure.
What might these substrates be? For clues, we examined members of the AI-2E superfamily that are fused to other domains. We found that various hydrophilic domains of recognizable function may be fused to them, either N-terminally or C-terminally. Based on homology, these domains appeared to be involved in purine nucleotide biosynthesis, purine nucleotide binding and/or hydrolysis, and molecular signaling. On this basis, as well as the known function of TqsA, we tentatively suggest that many or all members of the AI-2E superfamily catalyze the pmf-dependent efflux of both intracellular and extracellular signaling molecules. If so, they are generally involved in transmembrane signaling. Substrates may include cyclic nucleotides such as cyclic AMP, cyclic GMP and bis-3′,5′-cyclic diguanosine monophosphate (cyclic di-GMP) [27, 32] as well as quorum sensors such as AI-2 [17]. Further studies will be required to establish the functional diversity of this large superfamily of secondary carriers.
Acknowledgements
This work was supported by NIH grant GM077402 from the National Institute of General Medical Sciences. We thank Mary Beth Hiller and Jeeni Criscenzo for assistance in the preparation of the manuscript.
References
- 1.Ansaldi M, Marolt D, Stebe T, Mandic-Mulec I, Dubnau D. Specific activation of the Bacillus quorum-sensing systems by isoprenylated pheromone variants. Mol Microbiol. 2002;44:1561–1573. doi: 10.1046/j.1365-2958.2002.02977.x. [DOI] [PubMed] [Google Scholar]
- 2.Bassler BL. Small talk. Cell-to-cell communication in bacteria. Cell. 2002;109:421–424. doi: 10.1016/s0092-8674(02)00749-3. [DOI] [PubMed] [Google Scholar]
- 3.Ben Jacob E, Becker I, Shapira Y, Levine H. Bacterial linguistic communication and social intelligence. Trends Microbiol. 2004;12:366–372. doi: 10.1016/j.tim.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Jacob E, Shmueli H, Shochet O, Tenenbaum A. Adaptive self-organization during growth of bacterial colonies. Physica A. 1992;187:378–424. [Google Scholar]
- 5.Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Visick KL. Layers of signaling in a bacterium-host association. J Bacteriol. 2005;187:3603–3606. doi: 10.1128/JB.187.11.3603-3606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taga ME, Bassler BL. Chemical communication among bacteria. Proc Natl Acad Sci USA. 2003;100(suppl 2):14549–14554. doi: 10.1073/pnas.1934514100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Bhinu VS. Insight into biofilm-associated microbial life. J Mol Microbiol Biotechnol. 2005;10:15–21. doi: 10.1159/000090344. [DOI] [PubMed] [Google Scholar]
- 10.Kolter R. Surfacing views of biofilm biology. Trends Microbiol. 2005;13:1–2. doi: 10.1016/j.tim.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Kaiser D. Signaling in myxobacteria. Annu Rev Microbiol. 2004;58:75–98. doi: 10.1146/annurev.micro.58.030603.123620. [DOI] [PubMed] [Google Scholar]
- 13.Tiller JC, Liao CJ, Lewis K, Klibanov AM. Designing surfaces that kill bacteria on contact. Proc Natl Acad Sci USA. 2001;98:5981–5985. doi: 10.1073/pnas.111143098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis K. Programmed death in bacteria. Microbiol Mol Biol Rev. 2000;64:503–514. doi: 10.1128/mmbr.64.3.503-514.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans K, Passador L, Srikumar R, Tsang E, Nezezon J, Poole K. Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1998;180:5443–5447. doi: 10.1128/jb.180.20.5443-5447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taga ME, Miller ST, Bassler BL. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol Microbiol. 2003;50:1411–1427. doi: 10.1046/j.1365-2958.2003.03781.x. [DOI] [PubMed] [Google Scholar]
- 18.Walters M, Sircili MP, Sperandio V. AI-3 synthesis is not dependent on luxS in Escherichia coli. J Bacteriol. 2006;188:5668–5681. doi: 10.1128/JB.00648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 20.Herzberg M, Kaye IK, Peti W, Wood TK. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J Bacteriol. 2006;188:587–598. doi: 10.1128/JB.188.2.587-598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 23.Dayhoff MO, Barker WC, Hunt LT. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- 24.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL × windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhai Y, Tchieu J, Saier MH., Jr A web-based Tree View (TV) program for the visualization of phylogenetic trees. J Mol Microbiol Biotechnol. 2002;4:69–70. [PubMed] [Google Scholar]
- 26.Zhai Y, Saier MH., Jr A web-based program (WHAT) for the simultaneous prediction of hydropathy, amphipathicity, secondary structure and transmembrane topology for a single protein sequence. J Mol Microbiol Biotechnol. 2001;3:501–502. [PubMed] [Google Scholar]
- 27.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhai Y, Saier MH., Jr A simple sensitive program for detecting internal repeats in sets of multiply aligned homologous proteins. J Mol Microbiol Biotechnol. 2002;4:375–377. [PubMed] [Google Scholar]
- 29.Galperin MY. Bacterial signal transduction network in a genomic perspective. Environ Microbiol. 2004;6:552–567. doi: 10.1111/j.1462-2920.2004.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho YS, Burden LM, Hurley JH. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 2000;19:5288–5299. doi: 10.1093/emboj/19.20.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saier MH, Jr, Feucht BU, McCaman MT. Regulation of intracellular adenosine cyclic 3′:5′-monophosphate levels in Escherichia coli and Salmonella typhimurium. Evidence for energy-dependent excretion of the cyclic nucleotide. J Biol Chem. 1975;250:7593–7601. [PubMed] [Google Scholar]
- 32.D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 33.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saier MH, Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saier MH., Jr Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol Rev. 1994;58:71–93. doi: 10.1128/mr.58.1.71-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Busch W, Saier MH., Jr The transporter classification (TC) system, 2002. Crit Rev Biochem Mol Biol. 2002;37:287–337. doi: 10.1080/10409230290771528. [DOI] [PubMed] [Google Scholar]
- 37.Saier MH., Jr A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev. 2000;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye S, Li Y, Chen L, Jiang Y. Crystal structures of a ligand-free MthK gating ring: insights into the ligand gating mechanism of K+ channels. Cell. 2006;126:1161–1173. doi: 10.1016/j.cell.2006.08.029. [DOI] [PubMed] [Google Scholar]
- 39.Jack DL, Paulsen IT, Saier MH. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology. 2000;146:1797–1814. doi: 10.1099/00221287-146-8-1797. [DOI] [PubMed] [Google Scholar]
- 40.Jack DL, Yang NM, Saier MH., Jr The drug/metabolite transporter superfamily. Eur J Biochem. 2001;268:3620–3639. doi: 10.1046/j.1432-1327.2001.02265.x. [DOI] [PubMed] [Google Scholar]
- 41.Kennerknecht N, Sahm H, Yen MR, Patek M, Saier MH, Jr, Eggeling L. Export of L-isoleucine from Corynebacterium glutamicum: a two-gene-encoded member of a new translocator family. J Bacteriol. 2002;184:3947–3956. doi: 10.1128/JB.184.14.3947-3956.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saier MH, Jr, Beatty JT, Goffeau A, Harley KT, Heijne WH, Huang SC, Jack DL, Jahn PS, Lew K, Liu J, Pao SS, Paulsen IT, Tseng TT, Virk PS. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- 44.Saier MH, Jr, Eng BH, Fard S, Garg J, Haggerty DA, Hutchinson WJ, Jack DL, Lai EC, Liu HJ, Nusinew DP, Omar AM, Pao SS, Paulsen IT, Quan JA, Sliwinski M, Tseng TT, Wachi S, Young GB. Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim Biophys Acta. 1999;1422:1–56. doi: 10.1016/s0304-4157(98)00023-9. [DOI] [PubMed] [Google Scholar]
- 45.Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MH., Jr The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 46.Vrljic M, Garg J, Bellmann A, Wachi S, Freudl R, Malecki MJ, Sahm H, Kozina VJ, Eggeling L, Saier MH., Jr The LysE superfamily: topology of the lysine exporter LysE of Corynebacterium glutamicum, a paradyme for a novel superfamily of transmembrane solute translocators. J Mol Microbiol Biotechnol. 1999;1:327–336. [PubMed] [Google Scholar]
- 47.Zhai Y, Saier MH., Jr A web-based program for the prediction of average hydropathy, average amphipathicity and average similarity of multiply aligned homologous proteins. J Mol Microbiol Biotechnol. 2001;3:285–286. [PubMed] [Google Scholar]
- 48.Moller S, Croning MD, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]