Abstract
Neighbor of BRCA1 (Nbr1) is a highly conserved multidomain scaffold protein with proposed roles in endocytic trafficking and selective autophagy. However, the exact function of Nbr1 in these contexts has not been studied in detail. Here we investigated the role of Nbr1 in the trafficking of receptor tyrosine kinases (RTKs). We report that ectopic Nbr1 expression inhibits the ligand-mediated lysosomal degradation of RTKs, and this is probably done via the inhibition of receptor internalization. Conversely, the depletion of endogenous NBR1 enhances RTK degradation. Analyses of truncation mutations demonstrated that the C terminus of Nbr1 is essential but not sufficient for this activity. Moreover, the C terminus of Nbr1 is essential but not sufficient for the localization of the protein to late endosomes. We demonstrate that the C terminus of Nbr1 contains a novel membrane-interacting amphipathic α-helix, which is essential for the late endocytic localization of the protein but not for its effect on RTK degradation. Finally, autophagic and late endocytic localizations of Nbr1 are independent of one another, suggesting that the roles of Nbr1 in each context might be distinct. Our results define Nbr1 as a negative regulator of ligand-mediated RTK degradation and reveal the interplay between its various regions for protein localization and function.
Receptor tyrosine kinases (RTKs) are a major superfamily of membrane-spanning growth factor receptors with intrinsic kinase activity, which regulate fundamental cellular processes such as proliferation, migration, differentiation, and survival. RTKs include, among others, insulin-like growth factor receptor (IGFR), epidermal growth factor (EGF) receptor (EGFR), fibroblast growth factor (FGF) receptor (FGFR), vascular endothelial growth factor receptor (VEGFR), and platelet-derived growth factor receptor (PDGFR) (26). Most RTKs are activated by the binding of their cognate ligands, which induces receptor dimerization and the autophosphorylation of specific tyrosine residues within the kinase domain and the C-terminal cytoplasmic tail, as well as the phosphorylation of constitutively associated scaffold proteins such as FRS2. This results in the recruitment and subsequent phosphorylation of other signaling molecules, the assembly of multiprotein signaling complexes, and the ultimate activation of downstream signal transduction pathways. Such pathways include, among others, the RAS/mitogen-activated protein kinase (MAPK) cascade also known as the extracellular signal-regulated kinase (ERK) pathway, the phosphatidylinositol-3-OH kinase (PI3K)/AKT pathway, and the phospholipase C (PLC)/protein kinase C (PKC) pathway (26). These signaling pathways in turn act on a variety of transcription factors, the activities of which define the particular cellular outcome of a given stimulation.
Signaling from RTKs is tightly regulated by a variety of extrinsic and intrinsic control mechanisms, the deregulation of which is a major contributor to most cancers (35). In particular, endocytic trafficking has emerged as a key regulator of RTK signaling (30). Internalization is often triggered by ligand binding and the activation of RTKs on the cell surface. Receptor internalization occurs through clathrin-dependent or -independent mechanisms followed by trafficking through different vesicular compartments once inside the cell (30).
Endocytic trafficking can regulate RTK signaling at least at two levels. First, it can result in selective pathway propagation or downregulation by compartmentalization. For example, the removal of activated RTKs from the plasma membrane results in the termination of those downstream signaling pathways which require plasma membrane-associated molecules. These include the PLC/PKC and the PI3K/AKT pathways, both of which require phosphatidylinositol-4,5-phosphate (PI45P2) as a substrate, which is absent in endosomes (8). In contrast, ERK1/2 signaling is not limited to plasma membrane. In fact, many studies suggested that full ERK1/2 activation depends on the endocytosis of RTKs (10, 11, 14, 34). This has been attributed to the ERK1/2 propagating scaffolding complex P14-MP1, which is localized specifically to late endosomes (19, 32, 33). Another example of specific endosomal signal propagation has been shown for AKT. The adaptor protein-containing pleckstrin homology (PH) domain, the PTB domain, and leucine zipper motif 1 (APPL1) can bind specifically to AKT on early endosomes and enhance its activity (25). APPL1 also binds to the AKT substrate glycogen synthase kinase 3β (GSK-3β) and increases its phosphorylation by AKT without affecting the phosphorylation of another AKT substrate, tuberous sclerosis complex protein 2 (TSC2), thereby providing pathway selectivity (25).
Second, it is well established that endocytosis can act to downregulate RTK signaling by trafficking receptors to lysosomes for degradation. En route, signaling receptors are removed from the limiting membrane of endosomes and enclosed within intraluminal vesicles of multivesicular bodies (MVBs), rendering them unable to interact with signaling molecules and susceptible to degradation by lysosomal hydrolyses (22, 30). MVB formation begins in early endosomes and continues in late endosomes before mature MVBs are finally fused with lysosomes (22). MVB sorting of RTKs depends on ubiquitination by specific ubiquitin ligases such as CBL, followed by the concerted action of the endosomal sorting complex required for transport (ESCRT) proteins, which direct ubiquitinated cargos to the intraluminal vesicles of MVBs (22). The importance of RTK regulation by MVB sorting and lysosomal degradation is manifested by the fact that a variety of cancer-associated genetic and epigenetic alterations specifically target this pathway (18). Once internalized, however, the lysosomal degradation route is not the only available itinerary for RTKs. Internalized receptors can undergo recycling back to the plasma membrane for reusage, and the balance between recycling and lysosomal targeting can be a crucial determinant of the signaling outcome (27, 28).
Nbr1 is a ubiquitously expressed multidomain scaffold protein of 988 amino acids with ∼90% conservation between mouse and human (4, 37). It contains an N-terminal phox/Bem1p (PB1) domain, a ZZ-type zinc finger (ZZ), a coiled-coiled (CC) region, and a C-terminal ubiquitin association (UBA) domain capable of binding to both K48- and K63-type polyubiquitin chains (9, 36). The PB1 domain of NBR1 can bind to the open catalytic domain of the giant muscle kinase titin and regulate its downstream signaling in muscle cells (12). In addition, the PB1 domain can heterodimerize with the PB1 domain of P62/sequestosome-1 (SQSTM1) (12), another scaffold protein of similar architecture with proposed roles in endosomal trafficking (7) and selective autophagy (2, 21). A role in selective autophagy has also been proposed for NBR1, as it was shown to associate with the mammalian homologue of Atg8, an essential autophagosomal protein also known as microtubule-associated protein 1 light chain 3 (LC3) (9, 36). A main LC3-interacting region (LIR) as well as a secondary LC3-interacting region (LIR2) were identified in NBR1 (9), and it was proposed previously that similarly to P62, NBR1 can act as a specific adaptor for ubiquitinated cargos destined for degradation by autophagosomes (9, 36). Recently, an in vivo role for NBR1 in the regulation of bone mass and density has been revealed by its genetic truncation in mice (38).
We recently reported that NBR1 associates with Spred2, a negative regulator of ERK1/2 signaling downstream of RTKs (15). We showed that Spred2 activity is dependent on its interaction with NBR1 and that Spred2 downregulates ERK1/2 signaling downstream of FGFR by targeting the receptor to the lysosomal degradation pathway (15). These results led us to propose NBR1 as a novel regulator of RTK trafficking, but it remained unclear whether the protein had any effect on RTK trafficking on its own. Here we investigated the role of NBR1 in the context of RTK trafficking in more detail. We show that ectopic Nbr1 expression inhibits the ligand-mediated lysosomal degradation of endogenous RTKs. Live-cell imaging revealed that this effect is probably due to the inhibition of receptor internalization from the cell surface. In contrast, small interfering RNA (siRNA) depletion of endogenous NBR1 enhances receptor degradation. The C terminus of Nbr1 is essential but not sufficient for this function. Similarly, the C terminus of Nbr1 is essential but not sufficient for the late endocytic localization of the protein. We also show that in addition to the UBA domain, the C terminus of Nbr1 contains a membrane-interacting amphipathic α-helix, which is necessary for the late endocytic localization of the protein but not for its effect and RTK degradation. Finally, we demonstrate that the late endocytic and autophagic localizations of Nbr1 are independent, suggesting that the function of Nbr1 in each context might be distinct. Our results establish Nbr1 as a novel negative regulator of RTK trafficking and reveal the relationship between its different regions for protein localization and function.
MATERIALS AND METHODS
Plasmid constructs.
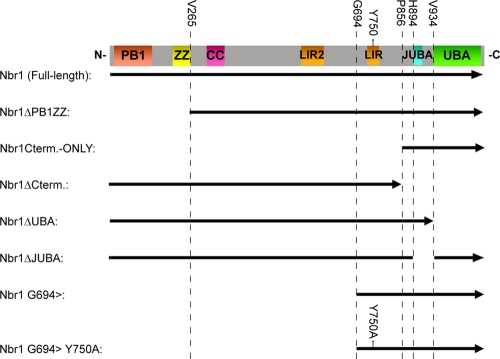
A schematic representation of Nbr1 constructs described in the study can be found in Fig. 1. N-terminally Myc- and green fluorescent protein (GFP)-tagged full-length mouse Nbr1 constructs as well as an N-terminally glutathione S-transferase (GST)-tagged C-terminal-only Nbr1 mutation (P856-Y988) construct were described previously (15). All N- and C-terminal truncation mutation constructs were made by Gateway cloning (Invitrogen) according to the manufacturer's instructions and as described previously (31). Briefly, forward and reverse primers with in-frame gateway 5′ overhangs corresponding to the beginning and end of a target sequence were used in a PCR to generate Gateway-compatible coding fragments. These fragments were then recombined into the Gateway pDONR201 entry vector (Invitrogen) using the BP clonase enzyme (Invitrogen). Myc-tagged constructs were generated by recombining entry vectors into a Myc-pRK5 gateway destination vector described previously (31) using the LR clonase enzyme (Invitrogen). GFP-tagged constructs were similarly generated by using the pcDNA DEST53 destination vector (Invitrogen). The Y750A point mutation was generated by site-directed mutagenesis using overlapping forward and reverses primers harboring the specific mutation in the middle. Finally, the Nbr1 juxta-UBA (JUBA) deletion mutation was generated from a full-length coding construct using primers with AgeI restriction digestion site 5′ overhangs. These forward and reverse primers were targeted to just after and just before the JUBA coding sequence, respectively, and were used in a PCR to amplify the whole construct minus the JUBA coding sequence. The linear product was then circularized by AgeI digestion (NEB) and self-ligation using T4 ligase (Invitrogen). The C-terminally GFP-tagged EGFR construct was described previously (3, 23).
FIG. 1.
Schematic representation of Nbr1 mutations used in this study. The arrow is in an N- to C-terminal direction. The indicated amino acids are the first or last included residues in the constructs. Nbr1 regions are represented linearly (the JUBA label applies to the cyan portion).
Reagents and antibodies.
Mouse monoclonal anti-NBR1 antibodies were purchased from Abcam and Abnova (clone 6B11). Mouse monoclonal antibody against LAMP2 (clone H4B4) was also obtained from Abcam. Mouse monoclonal anti-Myc tag antibody (clone 9E10) was obtained from Roche. Rabbit monoclonal anti-Myc tag (clone 71D10) antibody was obtained from Cell Signaling Technology. Anti-EGFR, GST, EEA1, cleaved procyclic acidic repetitive protein (PARP), and cleaved CASP-9 rabbit polyclonal antibodies were also obtained from Cell Signaling Technology. Rabbit polyclonal anti-FGFR2 (Bek), ERK1, and mouse monoclonal anti-pERK1/2 (clone E4) antibodies were obtained from Santa Cruz Biotechnology Inc. Mouse monoclonal anti-α-tubulin (clone DM 1A) was obtained from Sigma. All fluorescently labeled (Alexa Fluor 488, Alexa Fluor 594, and Texas Red) secondary antibodies were obtained from Invitrogen. Horseradish peroxidase-conjugated anti-mouse and anti-rabbit IgG secondary antibodies were obtained from Amersham Biosciences Inc. IRDye infrared anti-mouse and anti-rabbit IgG secondary antibodies were obtained from Li-COR Biosciences.
EGF was purchased from Sigma. z-VAD-FMK and bafilomycin-A1 (BafA) were also obtained from Sigma. Nontargeting control and NBR1 siRNAs were purchased from Santa Cruz Biotechnology Inc. The custom-made JUBA peptide was obtained from Alta Bioscience. The GST-tagged PH domain of PLC-δ1 (PIP2 Grip) was purchased from Echelon Inc. Membrane lipid strips, phosphatidylinositol-phosphate (PIP) arrays, and all phospholipids apart from phosphatidylcholine were also purchased from Echelon Inc. Phosphatidylcholine was purchased from Avanti Polar Lipids Inc.
Cell culture, transfection, stimulation, lysis, and Western blotting.
Cell culture and transfections were performed as described previously (15). Briefly, cells—human embryonic kidney (HEK) 293T and 293A LC3-GFP cells as well as African green monkey simian virus 40 (SV40)-transformed kidney (COS7) cells—were grown at 37°C in 5% CO2 in Dulbecco's modified Eagle medium (Invitrogen) supplemented with 10% fetal bovine serum (Labtech International). DNA transfections were performed by using Genejuice transfection reagent (Novagen) according to the manufacturer's instructions, and the cells were analyzed at 24 h posttransfection. Lipofectamine RNAiMAX (Invitrogen) was used for siRNA transfections according to the manufacturer's instructions for forward or reverse transfections, and the cells were analyzed after 24, 48, or 72 h, with the transfection being renewed every 24 h. For EGF and FGF2 stimulations, 293T cells were serum starved for 6 h, and ligand (plus 10 μg/ml heparin in the case of FGF2) was added to the cells at the indicated concentrations. Lysis was performed by the addition of 2× SDS sample buffer directly to the cells (50 μl/cm2 of cells), followed by rigorous vortexing and heating to 95°C for 10 min. Lysates were run on 4 to 12% NuPAGE Bis-Tris SDS gels (Invitrogen), and Western blotting was performed by using Immobilon PVDF-FL (Millipore) membranes. Membranes were dried and incubated with primary antibodies overnight at 4°C, followed by washing and incubation with secondary antibodies for 1 h at room temperature. After additional washes, blots were visualized by Enhanced Chemiluminescence Plus reagent (GE Healthcare) or an Odyssey infrared imaging system (Li-COR).
Live-cell imaging, immunostaining, and confocal microscopy.
Live-cell imaging with a Nikon A1R/TIRF microscope system (Nikon UK) was used to analyze receptor endocytosis and degradation. Cells transfected with GFP-tagged EGFR, with or without the cotransfection of Myc-Nbr1, were imaged by time-lapse epifluorescence microscopy employing a 60× objective and an Andor iXon 885 electron multiplier charge-coupled-device (EM-CCD) camera. Cells were imaged following stimulation with EGF, and EGFR-GFP fluorescence was analyzed to quantify endocytosis (number of intracellular endosomes positive for EGFR at each time point) and degradation (total cellular intensity at each time point). Five cells per group were analyzed, and nonstimulated cells were analyzed as a photobleaching control. Immunostaining and confocal microscopy were performed essentially as described previously (15). Briefly, cells grown on glass coverslips were fixed, permeabilized, blocked, and subjected to primary and fluorescently labeled secondary antibodies for 1 h per antibody with three washes in between before being mounted onto Mowiol. Coverslips were subsequently analyzed by laser scanning confocal microscopy using a Leica TCS SP2 confocal microscope system. All images were taken sequentially and as a single section at 200 Hz using the 63× objective lens. Final images were generated from the averages of eight consecutive scans. All presented images are representative of the majority of cells investigated.
Liposome preparation and circular dichroism.
Liposomes were prepared as described previously (20). Briefly, phosphatidylcholine and the PIP of interest were mixed in a glass vial at 95:5 molar ratio. The lipid mixture was dried under an N2 gas stream, redissolved in 1:1 CHCl3-CH3OH, and dried again under an N2 stream followed by vacuum centrifugation for 1 h to remove residual chloroform. The lipids were subsequently resuspended in ultrapure H2O and subjected to multiple rounds of freezing at −80°C and thawing in a 45°C sonicating water bath until the mixture became optically clear. Circular dichroism (CD) was performed as described previously (6). Briefly, spectra were acquired with a Jasco J715 spectropolarimeter using 0.5-mm cuvettes (Starna/Optiglass) with peptide concentrations of 1 mg/ml and liposome concentrations of 2 mg/ml where applicable. Spectra were recorded from 300 nm to 180 nm with a bandwidth of 2 nm, a data pitch of 0.2 nm, a scan speed of 100 nm/min, and a response time of 0.5. Four consecutively recorded spectra were averaged for each sample, and the relevant buffer baseline spectra were subtracted from each sample spectrum.
RESULTS
Nbr1 inhibits lysosomal degradation of RTKs.
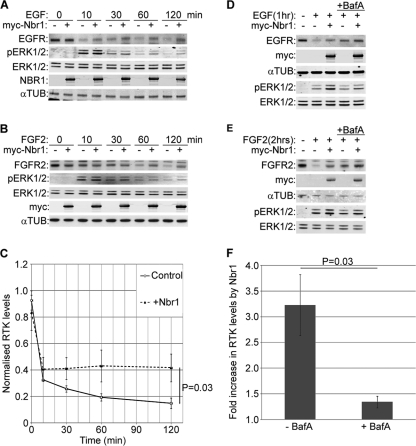
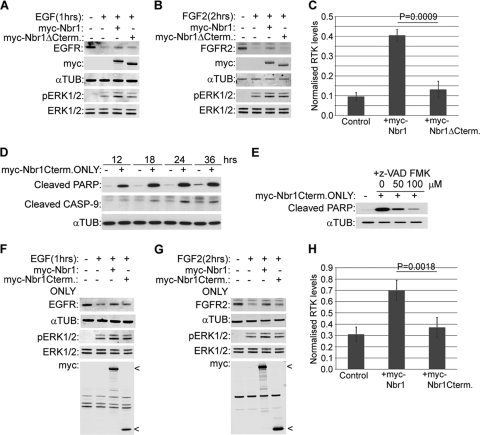
We previously reported that NBR1 is a specific late endosomal protein and that the regulation of FGFR trafficking and signaling by Spred2 depends on its interaction with NBR1 (15). These results suggested a novel role for NBR1 as a regulator of receptor trafficking, but it remained unclear whether the protein had any direct effect on RTK trafficking on its own. We therefore set out to investigate the trafficking role of NBR1 in more detail, focusing on its effect on EGFR and FGFR trafficking. We observed that ectopic Nbr1 expression inhibited the ligand-mediated degradation of endogenous EGFR (Fig. 2A and C). Similarly, the ligand-mediated degradation of endogenous FGFR2 was inhibited by ectopic Nbr1 expression (Fig. 2B and C). As a result, downstream ERK1/2 signaling was enhanced in response to both EGF and FGF2 (Fig. 2A and B). In neither case did Nbr1 have any effect on basal receptor levels as judged by immunoblotting (Fig. 2C). Moreover, the effect of ectopic Nbr1 expression on EGFR (Fig. 2D and F) or FGFR2 (Fig. 2E and F) was minimal in cells pretreated with the lysosomal inhibitor bafilomycin-A1 (BafA), supporting the notion that the observed difference in receptor levels is due to the inhibition of lysosomal degradation (Fig. 2F). These results demonstrate that Nbr1 acts to abrogate ligand-mediated RTK degradation.
FIG. 2.
Nbr1 abrogates ligand-mediated lysosomal degradation of RTKs and enhances downstream signaling. All error bars represent standard errors of the means (SEM). P values were calculated by a one-tailed paired t test (n = 3). (A) Overexpression of Nbr1 inhibits ligand-mediated EGFR degradation and enhances downstream ERK1/2 signaling. HEK293T cells transfected with Myc-Nbr1 or GFP as a control were serum starved and stimulated for the indicated time points with 50 ng/ml EGF. Cells were lysed and analyzed by Western blotting. (B) Overexpression of Nbr1 inhibits ligand-mediated FGFR2 degradation and enhances downstream ERK1/2 signaling. HEK293T cells transfected with Myc-Nbr1 or GFP as a control were serum starved and stimulated for the indicated time points with 20 ng/ml FGF2 plus 10 μg/ml heparin. Cells were lysed and analyzed by Western blotting. (C) Densitometry analysis of RTK degradation in the presence or absence of Myc-Nbr1 from panels A and B. (D) Nbr1 inhibition of ligand-mediated EGFR degradation is BafA sensitive. HEK293T cells transfected with Myc-Nbr1 or GFP as a control were starved and stimulated for the indicated times with 50 ng/ml EGF. Thirty minutes prior to stimulation, the specified cells were pretreated with 200 nM BafA. Cells were lysed and analyzed by Western blotting. (E) Nbr1 inhibition of ligand-mediated FGFR2 degradation is BafA sensitive. HEK293T cells transfected with Myc-Nbr1 or GFP as a control were starved and stimulated for the indicated times with 20 ng/ml FGF2 plus 10 μg/ml heparin. Thirty minutes prior to stimulation, the specified cells were pretreated with 200 nM BafA. Cells were lysed and analyzed by Western blotting. (F) Densitometry analysis of the effect of BafA treatment on Myc-Nbr1-mediated inhibition of RTK degradation from panels D and E. αTUB, α-tubulin.
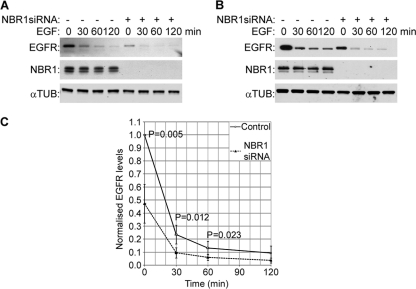
We next investigated the effect of the depletion of endogenous NBR1 on the degradation of EGFR and FGFR2. In contrast to ectopic Nbr1 expression, the depletion of endogenous NBR1 in 293T cells by siRNA enhanced EGFR degradation (Fig. 3A and C). Similar results were obtained with HeLa cells, which express considerably higher endogenous EGFR levels (Fig. 3B and C). The reduction in receptor levels could be seen even in the absence of ligand treatment, suggesting that depleting endogenous NBR1 must be sufficient to force receptors toward the degradative endocytic route (Fig. 3C). However, a similar effect could not be detected for FGFR2 (data not shown). Since the prolonged knockdown of NBR1 results in cell death (15), it is very likely that the NBR1 depletion in these experiments was not complete. Thus, a possible explanation for this variation could be that FGFR2 and EGFR simply have different sensitivity thresholds toward a partial NBR1 depletion.
FIG. 3.
Depletion of endogenous NBR1 enhances EGFR degradation. All error bars represent SEM. P values were calculated by a one-tailed paired t test (n = 5). (A) 293T cells were transfected with NBR1 or nonsilencing control siRNAs for 48 h, with the transfection being renewed after 24 h. Cells were serum starved and stimulated for the indicated times with 50 ng/ml EGF before being lysed and analyzed by Western blotting. (B) HeLa cells were transfected with NBR1 or nonsilencing control siRNAs for 48 h. Cells were starved and stimulated for the indicated times with 50 ng/ml EGF before being lysed and analyzed by Western blotting. (C) Densitometry analysis of EGFR levels in control or NBR1-depleted cells from panels A and B. Note that NBR1 depletion results in a reduction of EGFR levels even prior to stimulation.
Nbr1 inhibits ligand-mediated receptor internalization from the cell surface.
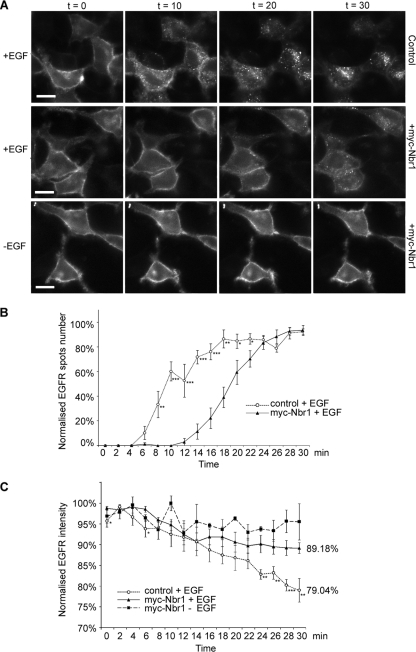
In order to generate a real-time analysis of RTK trafficking and degradation in live cells, we made use of the well-documented GFP-tagged EGFR construct (3, 23). We imaged the loss of EGFR from the plasma membrane and its appearance in cytosolic endosomes, followed by subsequent degradation, live in HeLa cells. Following stimulation, EGFR-GFP clearly redistributed to cytosolic punctae (Fig. 4A). However, the expression of Nbr1 decreased the endocytic trafficking of activated EGFR (Fig. 4A). Quantification of the number of EGFR-positive spots at each time point following stimulation revealed that endocytosis was significantly delayed in cells expressing Nbr1 (Fig. 4B). These results suggest that a likely mechanism by which Nbr1 inhibits RTK degradation is via trapping the receptor on the cell surface. Furthermore, quantification of whole-cell fluorescence demonstrated that the degradation of EGFR following EGF stimulation was significantly attenuated in cells expressing ectopic Nbr1 (Fig. 4C). Thus, the results of these imaging-based assays concur with data from the above-described biochemical studies (Fig. 2C).
FIG. 4.
Nbr1 traps EGFR on the cell surface and inhibits its degradation. All error bars represent SEM. P values were calculated by a nonpaired t test (n = 5 cells under each condition) (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (A) HeLa cells were transfected with EGFR-GFP, with or without Myc-Nbr1, and subjected to live-cell imaging by epifluorescence microscopy for 30 min following the addition of EGF (50 ng/ml). A nonstimulated control was also included (bottom). Scale bars, 10 μm. (B) Quantification of the number of EGFR-GFP-internalizing spots per area. The overexpression of Myc-Nbr1 induces a delay of 8 min in EGFR internalization. (C) Normalized total EGFR-GFP fluorescence from whole-cell areas. The overexpression of Myc-Nbr1 abrogates the degradation of EGFR.
The C terminus of Nbr1 is essential but not sufficient for its function and localization.
We next examined the contribution of different regions of Nbr1 to its inhibition of RTK degradation. Our previous study implicated the C terminus of NBR1 as being important for its function, as it was sufficient to mediate an association with SPRED2 (15). Therefore, we first compared the effect of full-length Nbr1 to that of a C-terminally deleted mutation lacking the last 133 amino acids on ligand-mediated RTK degradation. Unlike full-length Nbr1, the C-terminally deleted mutation did not inhibit EGFR degradation after EGF treatment (Fig. 5A and C), and similar results were obtained for FGFR2 (Fig. 5B and C). Accordingly, the enhancement of downstream ERK1/2 signaling was also lost when the C terminus of Nbr1 was deleted (Fig. 5A and B).
FIG. 5.
The C terminus of Nbr1 is essential but not sufficient for its function. All error bars represent SEM. P values were calculated by a one-tailed paired t test (n = 3). (A and B) A C-terminally deleted mutation of Nbr1 lacking the last 133 amino acids does not inhibit ligand-induced RTK degradation. HEK293T cells were transfected with Myc-Nbr1, Myc-Nbr1ΔCterm, or GFP as a control. Cells were serum starved and stimulated with 50 ng/ml EGF (A) or 20 ng/ml FGF2 plus 10 μg/ml heparin (B) for the indicated times before being lysed and analyzed by Western blotting. (C) Densitometry analysis of RTK degradation by Myc-Nbr1 and Myc-Nbr1ΔCterm from panels A and B. (D) Expression of a C-terminal-only Nbr1 mutation containing the last 133 amino acids (P856-Y988) induces cell death by apoptosis. HEK293T cells were transfected for the indicated times with Myc-Nbr1Cterm only or GFP as a control. Cells were lysed and analyzed by Western blotting for the apoptotic markers cleaved PARP and cleaved CASP-9. (E) Apoptosis induced by a C-terminal-only Nbr1 mutation can be inhibited by a general caspase inhibitor. HEK293T cells transfected with Myc-Nbr1Cterm only or GFP as a control were treated with increasing concentrations of z-VAD-FMK. Cells were lysed and analyzed by Western blotting for the apoptotic marker cleaved PARP. (F and G) The C-terminal-only Nbr1 mutation (P856-Y988) does not inhibit ligand-induced RTK degradation. HEK293T cells were transfected with Myc-tagged full-length Nbr1 (Myc-Nbr1), Myc-Nbr1Cterm only, or GFP as a control. A total of 50 μM z-VAD-FMK was added to all cells in order to block cell loss by apoptosis. Cells were serum starved and stimulated with 50 ng/ml EGF (F) or 20 ng/ml FGF2 plus 10 μg/ml heparin (G) for the indicated times before being lysed and analyzed by Western blotting. (H) Densitometry analysis of RTK degradation by Myc-Nbr1 and Myc-Nbr1Cterm only from panels F and G.
Next, we compared full-length Nbr1 with a C-terminal-only mutation (P856-Y988), which contains just the last 133 amino acids. We observed that similarly to the prolonged depletion of NBR1 by siRNA (15), the expression of the C-terminal-only mutation induced cell death by apoptosis (Fig. 5D), but this could be inhibited by a general caspase inhibitor, z-VAD-FMK (Fig. 5E). Unlike full-length Nbr1, the C-terminal-only Nbr1 mutation was not capable of inhibiting ligand-mediated EGFR (Fig. 5F and H) or FGFR2 (Fig. 5G and H) degradation in cells treated with z-VAD-FMK. The enhancement of downstream ERK1/2 signaling was also lost (Fig. 5F and G). Together, these results suggest that the C terminus of Nbr1 is essential but not sufficient for the inhibition of RTK degradation.
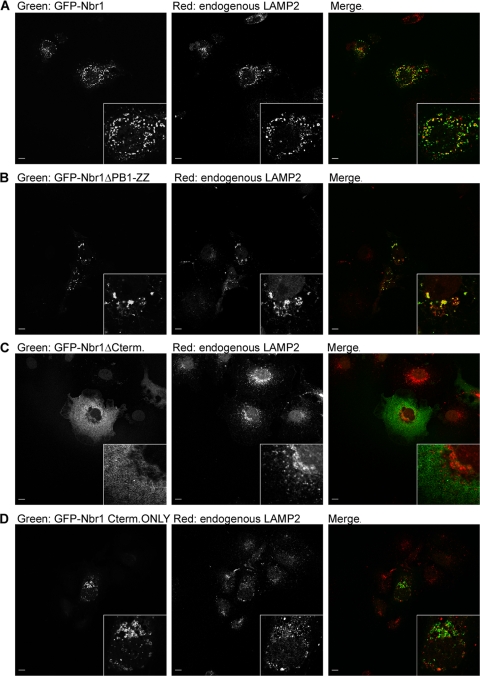
As Nbr1 specifically localizes to the limiting membrane of late endosomes (15), we asked whether this localization might be important for the effect on RTK trafficking. Using confocal microscopy, we investigated the late endocytic localization of different Nbr1 mutations. As expected, full-length Nbr1 exhibited a strong colocalization with the late endocytic marker LAMP2 (Fig. 6A). The deletion of the two N-terminal domains, the PB1 and ZZ domains, did not have an effect on protein localization (Fig. 6B). However, the loss of the C-terminal 133 amino acids (P856-Y988), which results in the loss of protein function (Fig. 5A, B, and C), had a dramatic impact on protein localization (Fig. 6C). This deletion resulted in the loss of colocalization with LAMP2, and the protein exhibited a diffuse cytosolic localization instead of a vesicular localization (Fig. 6C). Similarly, when a C-terminal-only (P856-Y988) Nbr1 mutation was investigated, it showed no colocalization with LAMP2 (Fig. 6D). However, this mutation was still localized to the limiting membrane of some intracellular vesicular structures (Fig. 6D). These vesicular structures were larger than full length (Fig. 6D) and stained positive for the early endosomal marker EEA1 (data not shown), which is suggestive of a potential defect in vesicle maturation/fission. These results indicate that the C terminus of Nbr1 is essential but not sufficient for its correct localization.
FIG. 6.
The C terminus of Nbr1 is essential but not sufficient for its correct localization. Scale bars, 10 μm. (A) Full-length Nbr1 colocalizes with the late endocytic marker LAMP2. COS7 cells transfected with GFP-Nbr1 were fixed, immunostained for LAMP2, and analyzed by confocal microscopy. (B) PB1-ZZ-deleted Nbr1 colocalizes with the late endocytic marker LAMP2. COS7 cells transfected with GFP-Nbr1ΔPB1-ZZ were fixed, immunostained for LAMP2, and analyzed by confocal microscopy. (C) C-terminally deleted Nbr1 lacking the last 133 amino acids does not colocalize with the late endocytic marker LAMP2. COS7 cells transfected with GFP-Nbr1ΔCterm were fixed, immunostained for LAMP2, and analyzed by confocal microscopy. (D)The C-terminal-only Nbr1 mutation (P856-Y988) does not colocalize with the late endocytic marker LAMP2. COS7 cells transfected with GFP-Nbr1Cterm only were fixed, immunostained for LAMP2, and analyzed by confocal microscopy.
The C terminus of Nbr1 is constituted of UBA and JUBA.
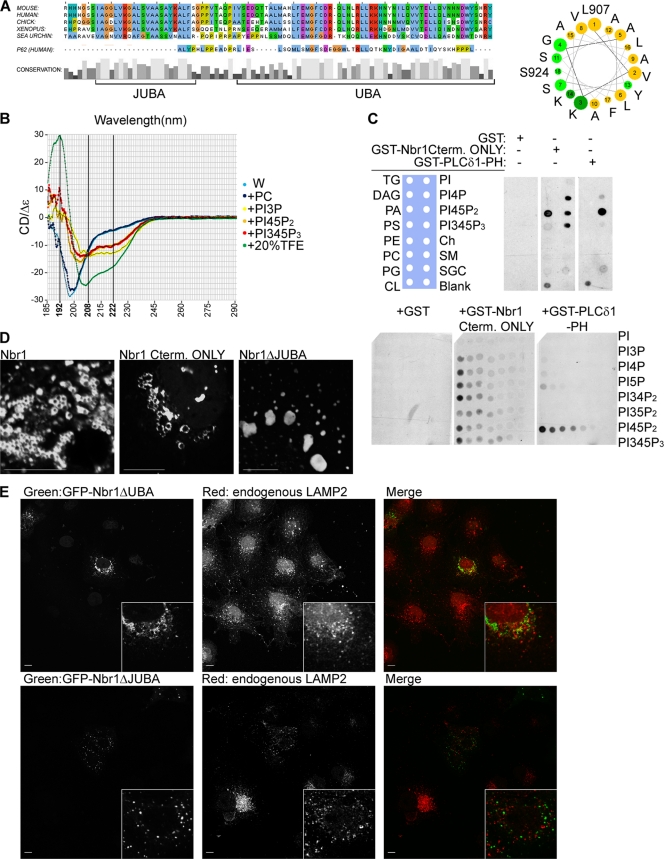
Having established the importance of the Nbr1 C terminus (P856-Y988) for protein localization and function, we sought to study this region in more detail. The C terminus of Nbr1 contains a well-conserved UBA domain (9, 36). However, a detailed study of the C terminus also revealed another region of high conservation that was located 10 amino acids N terminal to the UBA domain (Fig. 7 A). Due to its proximity to the UBA domain, we called this region juxta-UBA (JUBA). JUBA is only 22 amino acids long and does not seem to be conserved in P62 (Fig. 7A). Secondary-structure prediction by JPRED-3 (5) suggests that it predominantly has an α-helical fold. Projection along the helical axis of the predicted α-helix of JUBA showed a highly amphipathic conformation, with charged or polar amino acids arranged on one side of the helix and hydrophobic amino acids on the other (Fig. 7A). Amphipathic α-helices are found in many proteins that peripherally associate with membrane bilayers. Examples include the epsin, amphiphysin, endophilin, Arf, and Arl proteins (16). These helices are characterized by being unfolded until they come into contact with a target lipid membrane. This induces α-helical folding, and the hydrophobic side gets inserted into the bilayer, while the polar/charged side remains outside and makes ionic contacts with the negatively charged headgroups of membrane lipids such as phosphatidylinositol-phosphates (PIPs) (16).
FIG. 7.
The C terminus of Nbr1 contains UBA and JUBA, both of which are essential for its correct localization. Scale bars, 10 μm. (A, left) Cross-species multiple alignment of the C-terminal 93 amino acids of NBR1 reveals JUBA, a novel conserved stretch of amino acids next to the UBA domain, which is not present in P62. (Right) Projection along the helical axis of the predicted α-helix of JUBA (L907-S924) suggests that it would be amphipathic, with polar (light green) and charged (dark green) amino acids on one side and hydrophobic amino acids (yellow) on the other. (B) The JUBA peptide exhibits unfolded CD spectrum signatures in water or in the presence of PC and α-helical spectrum signatures in the presence of PIPs or the α-helix-inducing agent TFE. CD spectra of the JUBA peptide in water (W) and water plus phosphatidylcholine (PC) liposomes, plus phosphatidylinositol-3-phosphate (PIP3)-containing liposomes, plus phosphatidylinositol-4,5-phosphate (PI45P2)-containing liposomes, plus phosphatidylinositol-3,4,5-phosphate(PI345P3)-containing liposomes, or plus 20% TFE. (C) The C terminus of Nbr1 binds to PIPs. (Top) The C terminus of Nbr1 associates with PIPs and PA. Membranes prespotted with 15 different biologically relevant lipids were incubated with the GST-tagged C terminus of Nbr1 (GST-Nbr1Cterm only), GST alone as a negative control, or the GST-tagged pleckstrin homology (PH) domain of PLC-δ1 (GST-PLC-δ1-PH) as a positive control. Membranes were analyzed by blotting with an anti-GST antibody. (Bottom) The C terminus of Nbr1 does not show specificity to any PIP. Membranes prespotted with increasing amounts of various PIPs were incubated with GST-Nbr1Cterm only, GST, or GST-PLC-δ1-PH as described above and blotted with an anti-GST antibody. (TG, triglyceride; DAG, diacylglycerol; PA, phosphatidic acid; PS, phosphatidylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PG, phosphatidylglycerol; CL, cardiolipin; PI, phosphatidylinositol; Ch, cholesterol; SM, sphingomyelin; SGC, 3-sulfogalactosylceramide). (D) Deletion of JUBA abrogates membrane associations in vivo. COS7 cells transfected with GFP-Nbr1, GFP-Nbr1Cterm only, or GFP-Nbr1ΔJUBA were fixed and analyzed by confocal microscopy. (E) Both JUBA and UBA are necessary for late endocytic localization of Nbr1. (Top) Deletion of the UBA domain abrogates Nbr1 colocalization with LAMP2. COS7 cells transfected with GFP-Nbr1ΔUBA were fixed, immunostained for LAMP2, and analyzed by confocal microscopy. (Bottom) Deletion of JUBA also abrogates Nbr1 colocalization with LAMP2. COS7 cells transfected with GFP-Nbr1ΔJUBA were fixed, immunostained for LAMP2, and analyzed by confocal microscopy.
To find out whether JUBA is in fact one such α-helix, we used circular dichroism (CD) spectroscopy. CD spectroscopy is a powerful method to determine the secondary structure of peptides, as each secondary structure exhibits unique spectrum signatures: unfolded peptides give spectra with a single minimum at around 200 nm, whereas α-helical peptides give a maximum at around 192 nm and two minima at around 208 nm and 222 nm (6). When we subjected a peptide corresponding to the predicted α-helix of JUBA (L907-S924) to CD spectroscopy, it gave an unfolded signature spectrum (Fig. 7B). The addition of phosphatidylcholine liposomes did not affect the folding (Fig. 7B). However, when liposomes additionally containing various PIPs were added to the peptide solution, they all induced an α-helical fold, as judged by the disappearance of the ∼200-nm minimum as well as the appearance of the ∼192-nm maximum and ∼208- and ∼222-nm minima (Fig. 7B). Finally, as a positive control the addition of trifluoroethanol (TFE), an α-helix-inducing agent (6), also resulted in the induction of an α-helical fold (Fig. 7B). These results show that JUBA folds into an α-helix in the presence of PIP-containing membranes. Since every tested PIP could similarly induce an α-helical fold (Fig. 7B), the results also suggest that JUBA on its own does not display specificity toward a particular PIP.
Having shown that JUBA forms a membrane-interacting α-helix, we next tested whether the C terminus of Nbr1 exhibits a lipid binding capacity in vitro. We used membrane strips dotted with different physiologically relevant membrane lipids in a technique commonly known as fat blotting. In this method, the membrane is blotted with a GST-tagged protein of interest, and bound proteins are subsequently detected by blotting with a GST antibody. While GST on its own did not associate with any of the lipids, the GST-tagged C terminus of Nbr1 (P856-Y988) could bind to all present PIPs as well as phosphatidic acid (PA) (Fig. 7C). As a positive control we used the pleckstrin homology domain of PLC-δ1 that specifically binds PI45P2 (13), which gave the expected result (Fig. 7C). Similarly to PIPs, PA has a bivalent phosphate group, suggesting that JUBA might be interacting with bivalent phosphates of the lipid headgroups. To further confirm that the C terminus of Nbr1 does not display specificity toward a particular PIP, we performed fat blots using membranes dotted with increasing concentrations of each of seven PIPs. No significant preference was detected between different PIPs, while the pleckstrin homology domain of PLC-δ1 bound only PI45P2 (Fig. 7C). As before, GST on its own did not show any lipid binding (Fig. 7C). The results confirm that at least in vitro, the C terminus of Nbr1 binds PIPs with broad specificity.
As we reported previously, Nbr1 associates with the limiting membrane of late endosomes (15). To determine whether the vesicular membrane association of Nbr1 in vivo depends on JUBA, we compared the localization of full-length Nbr1 and the C-terminal-only Nbr1 (P856-Y988) mutation with that of a JUBA-deleted mutation. Full-length Nbr1 was specifically localized to the limiting membrane of vesicular structures in vivo (Fig. 7D). Similarly, the C-terminal-only mutation (P856-Y988) exhibited localization to the vesicular limiting membranes, although as mentioned above, the vesicles were relatively enlarged compared to that of full-length Nbr1 (Fig. 7D). However, a JUBA-deleted Nbr1 showed no localization to vesicular limiting membranes and instead exhibited an aggregate-like localization (Fig. 7D). These results reveal that JUBA is essential for the Nbr1 association with vesicular limiting membranes in vivo.
Having established that the C terminus of Nbr1 consists of UBA and JUBA and that JUBA is an amphipathic membrane-interacting α-helix that targets Nbr1 to vesicular limiting membranes, we next sought to determine the contribution of each region to the late endosomal localization of Nbr1. The deletion of either UBA or JUBA resulted in the loss of Nbr1 late endosomal localization, suggesting that both domains are crucial for the correct localization of the protein (Fig. 7E). Interestingly, the effect of the loss of both regions together (Fig. 6C) is more dramatic than that of each region individually (Fig. 7E), suggesting that the two regions cooperate to localize Nbr1.
UBA but not JUBA is necessary for Nbr1 function.
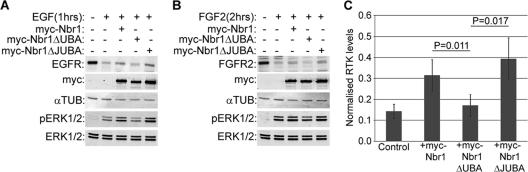
Having established the role of JUBA in Nbr1 localization, we next sought to assess its importance in the Nbr1-mediated inhibition of RTK degradation. While the deletion of the C-terminal UBA domain abrogated the effect of Nbr1 on the degradation of EGFR (Fig. 8A and C) and FGFR2 (Fig. 8B and C), the deletion of JUBA did not have any functional impact on the degradation of either receptor (Fig. 8A, B, and C). Thus, while both the UBA and JUBA regions are important for the late endocytic localization of Nbr1 (Fig. 7E), only UBA seems to be important for the Nbr1-mediated inhibition of RTK degradation.
FIG. 8.
UBA is necessary but JUBA is dispensable for Nbr1 function. All error bars represent SEM. P values were calculated by a one-tailed paired t test (n = 4). (A) Deletion of UBA but not JUBA abrogates Nbr1-mediated inhibition of EGFR degradation. HEK293T cells were transfected with Myc-Nbr1, Myc-Nbr1ΔUBA, Myc-Nbr1ΔJUBA, or GFP as a control. Cells were serum starved and stimulated with 50 ng/ml EGF for the indicated times before being lysed and analyzed by Western blotting. (B) Deletion of UBA but not JUBA abrogates Nbr1-mediated inhibition of FGFR2 degradation. HEK293T cells were transfected with Myc-Nbr1, Myc-Nbr1ΔUBA, Myc-Nbr1ΔJUBA, or GFP as a control. Cells were serum starved and stimulated with 20 ng/ml FGF2 plus 10 μg/ml heparin for the indicated times before being lysed and analyzed by Western blotting. (C) Densitometry analysis of RTK degradation by Myc-Nbr1, Myc-Nbr1ΔUBA, and Myc-Nbr1ΔJUBA from panels A and B.
Late endocytic localization of Nbr1 is LIR independent.
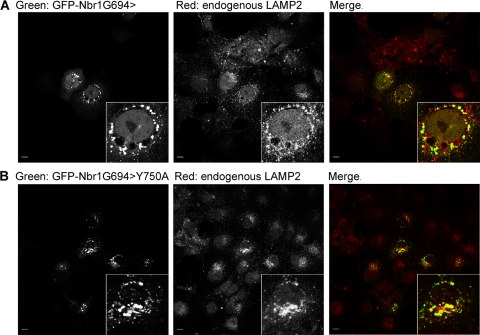
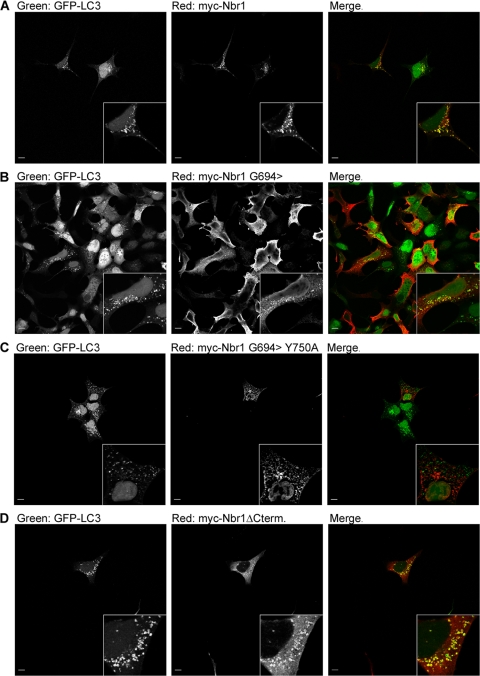
Nbr1 contains two LIRs, which can bind to LC3 and target Nbr1 to autophagosomes (9). As the C-terminal-only (P856-Y988) Nbr1 mutation that lacks both LIRs (Fig. 1) cannot correctly localize to late endosomes (Fig. 6D), we asked whether the interaction with autophagosomes via LIRs is important for the late endosomal localization of Nbr1 since autophagosomes commonly fuse with endosomes on their route toward lysosomal degradation (29). The main Nbr1 LIR is located close to the C terminus of the protein (V739-E779) and contains a pivotal tyrosine (Y750), the mutation of which results in the loss of LC3 interactions (9). The second LIR (LIR2) is located further N terminally (A543-P634) and can compensate for the loss of the main LIR (9). When we tested an N-terminal truncation mutation of Nbr1 expressing residues C terminal to G694, it could still colocalize with the late endocytic marker LAMP2 (Fig. 9A). As this mutation lacks LIR2 (Fig. 1), this result suggests that the loss of LIR2 alone does not have an effect on Nbr1 localization. However, like full-length Nbr1 (Fig. 10A), this mutation could still associate with autophagosomes (Fig. 10B). Therefore, we further mutated the pivotal tyrosine in the main LIR (Y750 to A) to assess whether the complete loss of LC3 binding affects the late endocytic localization of Nbr1. The association with autophagosomes was lost with this additional mutation (Fig. 10C), but colocalization with the late endocytic marker LAMP2 was not affected (Fig. 9B). These results reveal that the late endocytic localization of Nbr1 is independent of LC3 binding and autophagosomal association. Interestingly, we also observed that the C terminus of Nbr1 (P856-Y988), which is crucial for the late endocytic localization of the protein (Fig. 6C), is not needed for autophagosomal localization (Fig. 10D). Therefore, we conclude that the late endocytic and autophagic localizations of Nbr1 are independent of one another: the autophagic localization of Nbr1 requires the LIR but not the C terminus, while the late endocytic localization of Nbr1 requires the C terminus but not the LIRs.
FIG. 9.
Late endocytic localization of Nbr1 is independent of LIR. Scale bars, 10 μm. (A) A truncated Nbr1 mutation lacking LIR2 (Nbr1G694>) colocalizes with LAMP2. COS7 cells transfected with GFP-Nbr1G694> were fixed, immunostained for LAMP2, and analyzed by confocal microscopy. (B) This truncated mutation with an additional point mutation in the main LIR (Nbr1G694>Y750A) can still colocalize with LAMP2. COS7 cells transfected with GFP-Nbr1G694>Y750A were fixed, immunostained for LAMP2, and analyzed by confocal microscopy.
FIG. 10.
Colocalization of Nbr1 with accumulated autophagosomes depends on LIR but not the C terminus. Scale bars, 10 μm. (A) Full-length Nbr1 can colocalize with autophagosomes. 293A cells stably expressing GFP-tagged LC3 were transfected with Myc-Nbr1. Cells were treated with 200 nM BafA for 4 h to accumulate autophagosomes before being fixed, immunostained for Myc, and analyzed by confocal microscopy. (B) The Nbr1G694> mutation lacking LIR2 can colocalize with autophagosomes. 293A cells stably expressing GFP-tagged LC3 were transfected with Myc-Nbr1G694>. Cells were treated with 200 nM BafA for 4 h to accumulate autophagosomes before being fixed, immunostained for Myc, and analyzed by confocal microscopy. (C) The Nbr1G694>Y750A mutation, which lacks the LC3 binding capacity, cannot colocalize with autophagosomes. 293A cells stably expressing GFP-tagged LC3 were transfected with Myc-Nbr1G694>Y750A. Cells were treated with 200 nM BafA for 4 h to accumulate autophagosomes before being fixed, immunostained for Myc, and analyzed by confocal microscopy. (D) The C-terminally deleted Nbr1 mutation lacking the last 133 amino acids (P856-Y988) can still colocalize with autophagosomes. 293A cells stably expressing GFP-tagged LC3 were transfected with Myc-Nbr1ΔCterm. Cells were treated with 200 nM BafA for 4 h to accumulate autophagosomes before being fixed, immunostained for Myc, and analyzed by confocal microscopy.
DISCUSSION
The evidence presented in this study establishes Nbr1 as an inhibitor of ligand-mediated RTK degradation (Fig. 2 and 3). Live-cell imaging analysis suggests that the likely mechanism by which Nbr1 inhibits receptor degradation is via inhibiting receptor internalization from the cell surface (Fig. 4). The C-terminal 133 amino acids of Nbr1 are essential for its inhibition of RTK degradation (Fig. 5A, B, and C). We previously reported that the same region of Nbr1 also binds to Spred2 and that this binding is essential for the Spred2-mediated downregulation of ERK1/2 signaling by targeting the activated receptors to the lysosomal degradation pathway (15). As Nbr1 inhibits receptor degradation and consequently leads to an enhancement of downstream ERK1/2 signaling (Fig. 2A and B), it is possible that Spred2 binding to the critical C terminus of Nbr1 could be interfering with Nbr1 activity. Alternatively, Spred2 binding could be altering Nbr1 function from an inhibitor to an enhancer of RTK degradation. This is supported by the fact that while NBR1 does not have a negative impact on ERK1/2 signaling on its own, its coexpression with Spred2 results in a synergistic inhibition of ERK1/2 (15), which is suggestive of a cooperative rather than a simply antagonistic mechanism of functional interaction. Our results also reveal that further regions of functional significance other than the C terminus must exist within Nbr1, as a C-terminal-only mutation on its own is not sufficient to inhibit RTK degradation (Fig. 5F, G, and H).
The molecular interactions involved in the Nbr1-mediated inhibition of RTK internalization and degradation are not defined at the moment. A number of known binding partners of Nbr1, such as P62 (12), P14, transmembrane emp24-like trafficking protein 10 (Tmp21), and ubiquitin carboxyl-terminal hydrolase 8 (USP8/UBPY) (36), have roles in vesicular trafficking. USP8 is in particular very interesting in this regard, as its loss results in the inhibition of EGFR degradation (1, 24). USP8 deubiquitinates EGFR, and this is essential for ESCRT-dependent MVB sorting and the subsequent degradation of the receptor (1, 24). In addition, USP8 has a role in the regulation of ESCRT ubiquitin recognition machinery itself by deubiquitinating a number of its components (17). The fact that the phenotype of ectopic Nbr1 expression mimics that of the USP8 loss could be suggestive of a functional relationship. It remains to be determined whether a functional epistasis exists between the two proteins and, if so, how their interaction regulates endocytosis and receptor degradation.
We previously demonstrated that Nbr1 is a specific late endosomal protein (15). Here we reveal that the C terminus of Nbr1 is essential but not sufficient for its late endosomal localization (Fig. 6C and D). We also show that unlike its C terminus, the two LIR regions of Nbr1 are dispensable for late endocytic localization (Fig. 9). On the other hand, the autophagosomal localization of Nbr1 is dependent solely on the presence of LIRs and not the C terminus (Fig. 10). The fact that the late endocytic and autophagic localizations of Nbr1 are independent of each other indicates that the function of the protein in each context might be independent of one another as well. This is not surprising, as the association of Nbr1 with LC3 seems to result primarily in its removal from the cytoplasm and degradation via the autophagosomal pathway (9). In line with the above-described hypothesis, the inhibition of autophagy by the siRNA-mediated depletion of its key components did not affect the inhibition of RTK degradation by Nbr1 (F. K. Mardakheh and J. K. Heath, unpublished observations).
As mentioned above, the C terminus of Nbr1 contains a UBA domain, which can bind to polyubiquitin chains (9, 36). Here we show that the C terminus also contains a well-conserved membrane-interacting amphipathic α-helix, which we name JUBA. JUBA is crucial for the Nbr1 association with vesicular limiting membranes (Fig. 7D), and both UBA and JUBA are essential for the late endocytic localization of the protein (Fig. 7E). It was proposed previously that amphipathic α-helices might act as curvature sensors, being thermodynamically capable of inserting themselves into membranes with the right degree of curvature (16). In this light, JUBA could be providing further specificity with regard to Nbr1 localization by acting as a specific curvature sensor. Interestingly, despite numerous similarities at the level of domain architecture between Nbr1 and P62, JUBA seems to be a unique feature of Nbr1, as a similar region cannot be found in P62 (Fig. 7A). Thus, it is likely that the mechanisms by which these two proteins localize to late endosomes are different.
Finally, our results reveal that while JUBA is essential for the late endocytic localization of Nbr1 (Fig. 7E), it is dispensable for its effect on RTK trafficking (Fig. 8). The late endosomal localization per se, therefore, does not seem to be important for the function of Nbr1 in the context of RTK trafficking. In fact, the above-mentioned mislocalization of the C-terminal-only Nbr1 mutation to early endosomes (data not shown) suggests that it is probably trafficked through the endocytic machinery, and its late endocytic localization is therefore a steady-state phenomenon. The exact molecular mechanism by which Nbr1 associates with the endocytic machinery remains to be determined. The identification of novel Nbr1-interacting partners and determination of their mode of interaction should help shed light on this matter.
Acknowledgments
We thank Susan Brewer for the bacterial expression and purification of the GST-tagged C-terminal-only Nbr1 mutation. We also thank Sharon Tooze for 293A LC3-GFP stable cells and Steve Dove for his help with vesicle preparation. Finally, we give special thanks to all our present and past laboratory members for useful discussions and feedback.
This work was funded by Cancer Research UK (J.K.H.) and the Biotechnology and Biosciences Research Council (J.Z.R.). G.A. is funded through a Medical Research Council Ph.D. studentship, and F.K.M. is funded through a Cancer Research UK Ph.D. studentship. The Nikon A1R/TIRF microscope used in this research was obtained through the Birmingham Science City Translational Medicine Clinical Research and Infrastructure Trials Platform, with support from Advantage West Midlands (AWM).
Footnotes
Published ahead of print on 11 October 2010.
REFERENCES
- 1.Alwan, H. A., and J. E. van Leeuwen. 2006. UBPY-mediated epidermal growth factor receptor (EGFR) de-ubiquitination promotes EGFR degradation. J. Biol. Chem. 282:1658-1669. [DOI] [PubMed] [Google Scholar]
- 2.Bjørkøy, G., T. Lamark, A. Brech, H. Outzen, M. Perander, A. Øvervatn, H. Stenmark, and T. Johansen. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter, R. E., and A. Sorkin. 1998. Endocytosis of functional epidermal growth factor receptor-green fluorescent protein chimera. Biol. Chem. 273:35000-35007. [DOI] [PubMed] [Google Scholar]
- 4.Chambers, J. A., and E. Solomon. 1996. Isolation of the murine Nbr1 gene adjacent to the murine Brca1 gene. Genomics 38:305-313. [DOI] [PubMed] [Google Scholar]
- 5.Cole, C., J. D. Barber, and G. J. Barton. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36:W197-W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conner, M., M. R. Hicks, T. Dafforn, T. J. Knowles, C. Ludwig, S. Staddon, M. Overduin, U. L. Günther, J. Thome, M. Wheatley, D. R. Poyner, and A. C. Conner. 2008. Functional and biophysical analysis of the C-terminus of the CGRP-receptor; a family B GPCR. Biochemistry 47:8434-8444. [DOI] [PubMed] [Google Scholar]
- 7.Geetha, T., J. Jiang, and M. W. Wooten. 2005. Lysine 63 polyubiquitination of the nerve growth factor receptor TrkA directs internalization and signaling. Mol. Cell 20:301-312. [DOI] [PubMed] [Google Scholar]
- 8.Haugh, J. M., and T. Meyer. 2002. Active EGF receptors have limited access to PtdIns(4,5)P(2) in endosomes: implications for phospholipase C and PI 3-kinase signaling. J. Cell Sci. 115:303-310. [DOI] [PubMed] [Google Scholar]
- 9.Kirkin, V., T. Lamark, Y. S. Sou, G. Bjørkøy, J. L. Nunn, J. A. Bruun, E. Shvets, D. G. McEwan, T. H. Clausen, P. Wild, I. Bilusic, J. P. Theurillat, A. Øvervatn, T. Ishii, Z. Elazar, M. Komatsu, I. Dikic, and T. Johansen. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33:505-516. [DOI] [PubMed] [Google Scholar]
- 10.Kranenburg, O., I. Verlaan, and W. H. Moolenaar. 1999. Dynamin is required for the activation of mitogen-activated protein (MAP) kinase by MAP kinase kinase. J. Biol. Chem. 274:35301-35304. [DOI] [PubMed] [Google Scholar]
- 11.Lampugnani, M. G., F. Orsenigo, M. C. Gagliani, C. Tacchetti, and E. Dejana. 2006. Vascular endothelial cadherin controls VEGFR-2 internalization and signaling from intracellular compartments. J. Cell Biol. 174:593-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lange, S., F. Xiang, A. Yakovenko, A. Vihola, P. Hackman, E. Rostkova, J. Kristensen, B. Brandmeier, G. Franzen, B. Hedberg, L. G. Gunnarsson, S. M. Hughes, S. Marchand, T. Sejersen, I. Richard, L. Edström, E. Ehler, B. Udd, and M. Gautel. 2005. The kinase domain of titin controls muscle gene expression and protein turnover. Science 308:1599-1603. [DOI] [PubMed] [Google Scholar]
- 13.Lomasney, J. W., H. F. Cheng, L. P. Wang, Y. Kuan, S. Liu, S. W. Fesik, and K. King. 1996. Phosphatidylinositol 4,5-bisphosphate binding to the pleckstrin homology domain of phospholipase C-delta1 enhances enzyme activity. J. Biol. Chem. 271:25316-25326. [DOI] [PubMed] [Google Scholar]
- 14.MacInnis, B. L., and R. B. Campenot. 2002. Retrograde support of neuronal survival without retrograde transport of nerve growth factor. Science 295:1536-1539. [DOI] [PubMed] [Google Scholar]
- 15.Mardakheh, F. K., M. Yekezare, L. M. Machesky, and J. K. Heath. 2009. Spred2 interaction with the late endosomal protein NBR1 down-regulates fibroblast growth factor receptor signaling. J. Cell Biol. 187:265-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMahon, H. T., and J. L. Gallop. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438:590-596. [DOI] [PubMed] [Google Scholar]
- 17.Mizuno, E., K. Kobayashi, A. Yamamoto, N. Kitamura, and M. Komada. 2006. A deubiquitinating enzyme UBPY regulates the level of protein ubiquitination on endosomes. Traffic 7:1017-1031. [DOI] [PubMed] [Google Scholar]
- 18.Mosesson, Y., G. B. Mills, and Y. Yarden. 2008. Derailed endocytosis: an emerging feature of cancer. Nat. Rev. Cancer 8:835-850. [DOI] [PubMed] [Google Scholar]
- 19.Nada, S., A. Hondo, A. Kasai, M. Koike, K. Saito, Y. Uchiyama, and M. Okada. 2009. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J. 28:477-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narayan, K., and M. A. Lemmon. 2006. Determining selectivity of phosphoinositide-binding domains. Methods 39:122-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pankiv, S., T. H. Clausen, T. Lamark, A. Brech, J. A. Bruun, H. Outzen, A. Øvervatn, G. Bjørkøy, and T. Johansen. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131-24145. [DOI] [PubMed] [Google Scholar]
- 22.Piper, R. C., and D. J. Katzmann. 2007. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23:519-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rappoport, J. Z., and S. M. Simon. 2009. Endocytic trafficking of activated EGFR is AP-2 dependent and occurs through preformed clathrin spots. J. Cell Sci. 122:1301-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Row, P. E., I. A. Prior, J. McCullough, M. J. Clague, and S. Urbé. 2006. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J. Biol. Chem. 281:12618-12624. [DOI] [PubMed] [Google Scholar]
- 25.Schenck, A., L. Goto-Silva, C. Collinet, M. Rhinn, A. Giner, B. Habermann, M. Brand, and M. Zerial. 2008. The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development. Cell 133:486-497. [DOI] [PubMed] [Google Scholar]
- 26.Schlessinger, J. 2000. Cell signaling by receptor tyrosine kinases. Cell 103:211-225. [DOI] [PubMed] [Google Scholar]
- 27.Scita, G., and P. P. Di Fiore. 2010. The endocytic matrix. Nature 463:464-473. [DOI] [PubMed] [Google Scholar]
- 28.Sigismund, S., E. Argenzio, D. Tosoni, E. Cavallaro, S. Polo, and P. P. Di Fiore. 2008. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev. Cell 15:209-219. [DOI] [PubMed] [Google Scholar]
- 29.Simonsen, A., and S. A. Tooze. 2009. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J. Cell Biol. 186:773-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorkin, A., and M. von Zastrow. 2009. Endocytosis and signalling: intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 10:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sweet, S. M., F. K. Mardakheh, K. J. Ryan, A. J. Langton, J. K. Heath, and H. J. Cooper. 2008. Targeted online liquid chromatography electron capture dissociation mass spectrometry for the localization of sites of in vivo phosphorylation in human Sprouty2. Anal. Chem. 80:6650-6657. [DOI] [PubMed] [Google Scholar]
- 32.Teis, D., W. Wunderlich, and L. A. Huber. 2002. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev. Cell 3:803-814. [DOI] [PubMed] [Google Scholar]
- 33.Teis, D., N. Taub, R. Kurzbauer, D. Hilber, M. E. de Araujo, M. Erlacher, M. Offterdinger, A. Villunger, S. Geley, G. Bohn, C. Klein, M. W. Hess, and L. A. Huber. 2006. p14-MP1-MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis. J. Cell Biol. 175:861-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vieira, A. V., C. Lamaze, and S. L. Schmid. 1996. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science 274:2086-2089. [DOI] [PubMed] [Google Scholar]
- 35.Vogelstein, B., and K. W. Kinzler. 2004. Cancer genes and the pathways they control. Nat. Med. 10:789-799. [DOI] [PubMed] [Google Scholar]
- 36.Waters, S., K. Marchbank, E. Solomon, C. Whitehouse, and M. Gautel. 2009. Interactions with LC3 and polyubiquitin chains link nbr1 to autophagic protein turnover. FEBS Lett. 583:1846-1852. [DOI] [PubMed] [Google Scholar]
- 37.Whitehouse, C., J. Chambers, K. Howe, M. Cobourne, P. Sharpe, and E. Solomon. 2002. NBR1 interacts with fasciculation and elongation protein zeta-1 (FEZ1) and calcium and integrin binding protein (CIB) and shows developmentally restricted expression in the neural tube. Eur. J. Biochem. 269:538-545. [DOI] [PubMed] [Google Scholar]
- 38.Whitehouse, C. A., S. Waters, K. Marchbank, A. Horner, N. W. McGowan, J. V. Jovanovic, G. M. Xavier, T. G. Kashima, M. T. Cobourne, G. O. Richards, P. T. Sharpe, T. M. Skerry, A. E. Grigoriadis, and E. Solomon. 2010. Neighbor of Brca1 gene (Nbr1) functions as a negative regulator of postnatal osteoblastic bone formation and p38 MAPK activity. Proc. Natl. Acad. Sci. U. S. A. 107:12913-12918. [DOI] [PMC free article] [PubMed] [Google Scholar]