Abstract
Insulin-like growth factor 2 (IGF-2) mRNA-binding proteins (IMPs) are a family of posttranscriptional regulatory factors with well-understood roles in embryonic development and cancer but with poorly characterized functions in normal adult cells and tissues. We now show that IMP-2, the most ubiquitously expressed member of the family, is abundant in human and mouse adult skeletal myoblasts, where it is indispensable for cell motility and for stabilization of microtubules. To explore the functions of IMP-2, we analyzed the transcripts that were differentially regulated in IMP-2-depleted myoblasts and bound to IMP-2 in normal myoblasts. Among them were the mRNAs of PINCH-2, an important mediator of cell adhesion and motility, and MURF-3, a microtubule-stabilizing protein. By gain- and loss-of-function assays and gel shift experiments, we show that IMP-2 regulates the expression of PINCH-2 and MURF-3 proteins via direct binding to their mRNAs. Upregulation of PINCH-2 in IMP-2-depleted myoblasts is the key event responsible for their decreased motility. Our data reveal how the posttranscriptional regulation of gene expression by IMP-2 contributes to the control of adhesion structures and stable microtubules and demonstrate an important function for IMP-2 in cellular motility.
Terminal differentiation of skeletal muscle leads to irreversible mitotic arrest, accompanied by a decrease in general transcriptional activity. Execution of the myogenic program and maintenance of skeletal muscle tissue depend on various posttranscriptional regulatory mechanisms. We have recently demonstrated how the mRNA-binding protein Lin-28 interacts with translation initiation complexes and enhances the translation of a crucial muscle cytokine, insulin-like growth factor 2 (IGF-2), a function that is indispensable for terminal muscle differentiation (31).
Here, we have studied the role of Lin-28 protein partners, the RNA chaperones of the IMP family (IGF-2 mRNA binding proteins), in posttranscriptional regulation of myogenesis. The IMPs (IMP-1, -2, and -3), were first discovered in rhabdomyosarcoma (RMS) cells and were characterized as RNA-binding proteins that share significant structural and functional homology with a number of other RNA-binding posttranscriptional regulators, such as Vg1 RNA binding protein (Vg1RBP), zipcode-binding protein (ZBP), coding region instability determinant binding protein (CRD-BP), and KH-domain-containing protein overexpressed in cancer (KOC) (28, 44). These proteins have been shown to bind to various regions of multiple RNA targets, such as c-myc, β-actin, IGF-2, H19, CD44, and many others, and regulate their stability, transport, and/or translation (27, 32, 36, 42). The total number of IMP-regulated transcripts can be as high as 8,400 in HEK293 cells (12). IMPs are well-characterized markers of various human cancers (13, 15, 18, 34, 38), and the molecular mechanisms underlying the function of IMP-1 and IMP-3 have been explored in multiple cancer cell lines (16, 23, 42). In contrast, next to nothing is known about IMP-2, the most ubiquitously expressed member of the IMP family (13), which is directly regulated by the HMGA2 oncogene in both mouse and human cells (2, 7). Recent studies suggested a specific role for IMP-2, but not for the other members of IMP family, in the development of type 2 diabetes (6), indicating the necessity to better characterize the specific functions and RNA targets of IMP-2.
Here we report that IMP-2 is highly expressed in mouse and human primary myoblasts, in normal myoblast cell lines, in embryonic and alveolar RMS, and during early regeneration of skeletal muscle in vivo. Interestingly, our RNA interference (RNAi) experiments showed that IMP-2 is necessary for motility, migration, and microtubule (MT) stability in mouse and human myoblasts and in RMS cells in culture. Knockdown (KD) of IMP-2 specifically decreases MT stabilization, which depends on posttranslational C-terminal detyrosination of tubulin. Stabilization of MTs is important for early myogenic differentiation (37) and also contributes to cell motility, metastatic capacity, and progression of several human cancers (17, 22, 24). Posttranslational modifications of tubulin in cells are regulated by a cycle of tyrosination/detyrosination, performed by tubulin tyrosine ligase (TTL) and tubulin carboxypeptidase (4). In addition, several proteins have been reported to directly bind to and stabilize the MTs via specific accumulation of detyrosinated tubulin (37, 40). Another mechanism of regulation of MT remodeling occurs at the level of focal adhesions (FAs), cytoplasmic and membrane structures which are indispensable for cellular adhesion, motility, and cytoskeleton integrity (11, 30).
To explore the molecular mechanisms of IMP-2-dependent regulation of cellular motility and MT stability, we performed genome-wide analysis and identified 118 transcripts that are both bound to IMP-2 complexes and regulated by IMP-2 in myoblasts. Thirty-five percent of these mRNAs code for proteins involved in adhesion, cytoskeleton remodeling, and membrane integrity. In particular, the regulation of MURF-3 (MT stabilization) and PINCH-2 (FA remodeling) by IMP-2 was found to largely contribute to the motility deficiency of IMP-2 KD myoblasts and RMS cells. Using affinity purification and mass spectrometry, we have characterized the proteins interacting with IMP-2 in myoblasts and have further identified the protein partners of IMP-2 that have functional importance in the regulation of motility and cytoskeleton stability.
(This study is part of the Ph.D. thesis work of Selim Boudoukha, conducted with the expert technical help of Sylvain Cuvellier under the direction of Anna Polesskaya.)
MATERIALS AND METHODS
Detailed experimental protocols are available on request.
Cell culture, stable cell lines, and affinity purification of IMP-2 complexes.
Cell culture media and additives were purchased from Invitrogen. C2C12 mouse myoblasts and RMS cell lines RD and Rh30 were purchased from ATCC and cultured as recommended. Human primary fetal myoblasts (HFS) were a kind gift from B. Peault and were cultured in Dulbecco's modified Eagle's medium (DMEM)/F12 medium supplemented with 20% fetal calf serum (FCS). Differentiation of C2C12 and HFS myoblasts was induced by transferring the cells to DMEM with 2% horse serum or with 0.01 mg/ml insulin or 0.1 mg/ml transferrin, respectively.
C2C12 myoblasts stably expressing IMP-2 tagged with FLAG-tagged hemagglutinin (HA) on the N terminus or on the C terminus were obtained with the retroviral vector pOZFHH (a kind gift from V. Ogryzko), as described in reference 41. C2C12 cells stably expressing FLAG-HA-CBFB and G9a were a kind gift from S. Ait-Si-Ali. Protein complexes were immunoprecipitated from whole-cell extracts with lysis buffer (10% glycerol, 20 mM Tris-HCl, pH 8, 0.2 mM EDTA, 0.1% NP-40, 0.5 M KCl, protease inhibitors [Complete; Roche]), using M2-agarose beads, eluted with FLAG peptide, treated or not with 1 mg/ml protease-free RNase A (Roche), reprecipitated with anti-HA-agarose beads, and eluted with HA peptide, as described in reference 25. (All reagents were purchased from Sigma.) Complexes were separated on a 4 to 12% polyacrylamide gel (Invitrogen) and stained with the Silver Quest kit from Invitrogen, according to the manufacturer's instructions. Mass spectrometry identification of proteins was carried out by R. Tomaino, Harvard Medical School. To separate the purified IMP-2 complexes according to their molecular weight, they were loaded onto a 25 to 49% glycerol gradient, and centrifugation was done at 49,000 rpm for 18 h with an MLS 50 rotor.
Purification of RNA from IMP-2 complexes was accomplished by digestion with proteinase K (Roche) (0.2 mg/ml in 0.5% SDS for 45 min at 37°C), followed by phenol-chloroform extraction and ethanol precipitation in the presence of 20 μg of glycogen (Invitrogen) per sample.
Wound healing and Transwell migration assays.
For the wound healing test, cells were seeded at a density of 5 × 104 in 200 μl of 2% fetal bovine serum (FBS)-DMEM in six-well plates and transfected with 20 nM small interfering RNA (siRNA), using Hiperfect (Qiagen). After 48 h, the monolayers were wounded by being scratched with a plastic pipette tip. Images of wound healing were taken every 24 h and analyzed with an inverted microscope.
For the quantitative migration assay, 48 h after siRNA transfection, cells were resuspended in 2% FBS-DMEM. Cell invasion assays were performed with an 8-μm-pore membrane (Transwell; Becton Dickinson). Ten percent FBS-DMEM was placed in the lower chamber as a chemoattractant. Cells were plated in the upper chambers of triplicate wells at a density of 5 × 104 in 200 μl of 2% FBS-DMEM and incubated for 12 to 16 h at 37°C in a humidified atmosphere with 5% CO2.After removal of the cells on the upper membrane, the cells on the lower membrane were fixed with methanol for 10 min and stained with Hoechst 33342 for 15 min. Ten randomly chosen microscopic fields were counted, and the results are shown as means ± standard deviations.
Transfections and RNAi assays.
Transient transfection of siRNA was performed using HiPerfect (Qiagen), according to the manufacturer's instructions. siRNA duplexes were transfected at 20 nM overnight. Efficiency of RNAi was evaluated by quantitative reverse transcription-PCR (qRT-PCR) and/or Western blotting (WB).
The following siRNA target sequences were used: hsIMP-2(1) [i.e., Homo sapiens IMP-2(1)], TCCGCTAGCCAAGAACCTATA; hsIMP-2(2), GTGGAGGAAAGTAGAAATTTA; mmIMP-2(1) [i.e., Mus musculus IMP-2(1)], TCGGGTAAAGTGGAATTGCAT; mmIMP-2(2), GGCATCAGTTTGAGGACTATT; mmIMP-2(3), TCAAACAGCTCGCTCGATTTT; mmIMP-2(4), CGCAAGATCAGGGAAATTGTA; hsPINCH-2(1), CAGGGACAGGAGCAAATTGCA; hsPINCH-2(2), CGAGCGCATTGTCAACAGCAA; mmPINCH-2(1), ACCCTTGGGCTTGGCTGAGAA; mmPINCH-2(2), CTGCAGTACCTTAGCACTCAT; hsMURF-3(1), GAGGCAGAAGCAGTTGTTAAA; hsMURF-3(2), CTCGAGCGTCCCAGACCCGTA; and control, irrelevant siRNA (Qiagen), AACTGCGGTGGGCTAGACCAT.
siRNAs for the RNAi screens' anti-IMP-2 partners were designed by Qiagen (see Table S3 in the supplemental material).
Skeletal muscle regeneration assay.
Skeletal muscle regeneration was induced by injecting 10 μl of 10 μM cardiotoxin (Latoxan) in phosphate-buffered saline (PBS) into the tibialis anterior (TA) muscles of 7- to 8-week-old C57BL/6 female mice. TA muscle was collected at the indicated time points and lysed in buffer containing 50 mM Tris-HCl (pH 7.4), 0.1% Triton X-100, 5 mM EDTA, 250 mM NaCl, and protease inhibitors (Complete; Roche). Total muscle lysates were analyzed by Western blotting. Expression of embryonic myosin heavy chain (MHC) was used as a positive control of regeneration efficiency. Animals received humane care in accordance with the guidelines of the Direction Départmentale des Services Vétérinaires du Val de Marne, Service de la Santé et de la Protection Animale.
Sucrose gradients.
Total lysate from C2C12 myoblasts or from primary myoblasts (PMs) (3 × 107 cells) was prepared as described in reference 28, treated or not with 10 mM puromycin or 1 mg/ml RNase A for 15 min at 37°C, and applied to a 21 to 47% sucrose gradient in a mixture of 20 mM Tris-HCl (pH 8.0), 140 mM KCl, and 5 mM MgCl2. Centrifugation was carried out at 40,000 rpm for 2 h 15 min with a Beckman SW41 rotor. Fractions of 0.8 ml were collected, adsorbance at 260 nm was measured, half of each fraction was precipitated with 10% trichloroacetic acid, and the proteins were analyzed by Western blotting. The second half of each fraction was ethanol precipitated and treated with DNase (Promega), and RNA was extracted with phenol-chloroform and used for qRT-PCR analysis.
qRT-PCR.
The qRT-PCR primer sequences are as follows: cyclophilin A, forward, GTCAACCCCACCGTGTTCTT, and reverse, CTGCTGTCTTTGGGACCTTGT; mmIMP-2, forward, AGAGGCCTTTGAGAACGACA, and reverse, TGGAGAAGTATCCGGAGTGG; mmMurf-3, forward, CAACTTGGAGAAGCAGCTCA, and reverse, GGGATTGCCACAGAGGATTA; 36b4 (Arbp), forward, ATGTGCAGCTGATAAAGACTGG, and reverse, AGGCCTTGACCTTTTCAGTAAG; mmPINCH-2, forward, CGGATTCTGTGGTGAATTTGTCA, and reverse, CTGGCAGATGAATTTGCCCAA; and hsPINCH-2, forward, TGCAGCCATGTGATTGAAGGCGAT, and reverse, CACGGGCTTCATGTCGAACT. For hsIMP-2, the Qiagen Quantitect primer assay (hs_IMP2_1_SG ref: QT00008610) was used.
RNA gel shift.
Reaction mixtures (20 μl) containing 16 μl of binding buffer (50 mM Tris [pH 8], 150 mM NaCl, 10% glycerol, 0.01% Triton X-100), 0.5 μg of tRNA (Sigma), 0.5 μg of heparin (Sigma), 1 μl of RNase out (Invitrogen), 1 μl of 32P-labeled RNA probe (600 cpm), and 0, 50, and 200 fM of recombinant glutathione S-transferase (GST)-tagged IMP-2 were incubated for 1 h on ice. Complexes were resolved by electrophoresis on a 5% polyacrylamide-1× Tris-borate-EDTA (TBE) native gel run at 180 V. The gel was dried and visualized by autoradiography.
Antibodies for Western blotting and immunofluorescence.
The following antibodies were used: IMP-2, rabbit antibodies kindly provided by F. C. Nielsen and mouse monoclonal H00010644-A01, purchased from Abnova; Ge-1, a kind gift from Dominique Weil; DDX6/p54, 9407 (Cell Signaling); nucleolin, N2662 (Sigma); MHC, clone MY-32 (Sigma); β-actin, clone AC-15 (Sigma); Glu-tubulin, δ-2 tubulin, Tyr-tubulin, a kind gift from Didier Job and Laurence Lafanechere; FAK Y397P, BD Transduction Laboratories; PINCH-2, rabbit polyclonal, a kind gift from Reinhard Fässler; MURF-3, sc-50252 (Santa Cruz); RENT-1, A300-036A (Bethyl Laboratories); MOV-10, A301-571A (Bethyl Laboratories); HSPA8, SPA-816D (Stressgene); vinculin, V9131 (Sigma); and phosphotyrosine, mouse monoclonal 4G10, a kind gift from Eric Rubinstein.
Immunofluorescence.
C2C12 mouse myoblasts and RD cells were seeded on collagen-coated glass cover slides. Cells were fixed with 4% paraformaldehyde for 15 min at room temperature and permeabilized with 0.2% Triton X-100 in PBS for 20 min. After being blocked with a solution containing 1% bovine serum albumin (BSA) and 5% serum in PBS for 1 h at room temperature, the cells were incubated with the primary antibody for 1 h at room temperature, washed three times for 5 min in PBS, and visualized with Alexa Fluor 488- or 568-conjugated anti-mouse or anti-rabbit (Invitrogen). Images were acquired using an Axiovert 2000 fluorescent microscope (Zeiss), a Hamamatsu camera, and SimplePCI software.
RESULTS
IMP-2 is expressed in primary myoblasts, in the myogenic mouse cell line C2C12, in RMS cells, and in mouse skeletal muscle during regeneration.
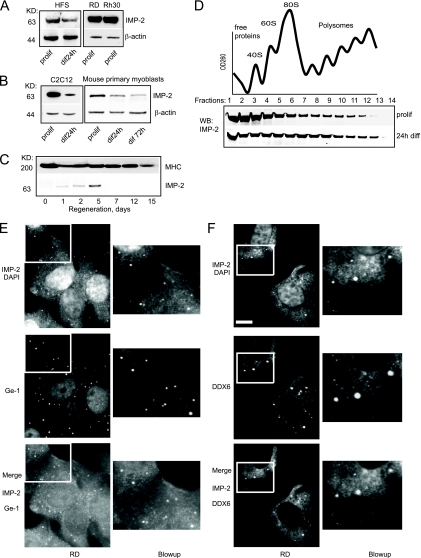
IMP-2 was originally cloned from RD cells, an embryonic RMS cell line (28), and to date, there have been no reports of IMP-2 expression in normal adult cells and tissues. Therefore, our observation that endogenous IMP-2 is associated with ectopically expressed Lin-28 in the mouse myoblast cell line C2C12 (31) attracted our attention. Using Western blotting, we detected IMP-2 in human RMS cells and human primary myoblasts, in murine primary myoblasts, in the C2C12 cell line, and in mouse skeletal muscle during cardiotoxin-induced regeneration (Fig. 1A to C). During terminal myogenic differentiation of primary myoblasts, IMP-2 is gradually downregulated, though the expression of IMP-2 is easily detected in myotubes at as late as 3 days of differentiation in culture (Fig. 1B). Sucrose gradient separation of total cell lysates showed that IMP-2 is distributed between monosomal and light polysomal fractions in the cell, with a slight shift toward heavy polysomal fractions at the onset of differentiation (Fig. 1D). This observation suggests that IMP-2 in cells can be bound by a variety of structurally and/or functionally distinct RNP complexes.
FIG. 1.
Expression levels and subcellular localization of IMP-2. (A to C) A Western blot shows that IMP-2 is expressed in human primary myoblasts (HFS) and in human RMS lines RD (embryonic) and Rh30 (alveolar) (A), in the mouse C2C12 cell line and primary myoblasts (B), and at the onset of skeletal muscle regeneration of mouse tibialis anterior muscle (C). (D) Sucrose gradient analysis of IMP-2 distribution between polysomal and monosomal fractions in C2C12 myoblasts. OD260, optical density at 260 nm. (E and F) IMP-2 forms distinct cytoplasmic granules and colocalizes with P-body markers Ge-1 and DDX6 in the RMS cell line RD. diff, differentiation; prolif, proliferation; DAPI, 4′,6-diamidino-2-phenylindole. Masses in kilodaltons are shown to the left of panels A to C. Bar, 9 μm.
Indeed, IMP-2 is diffusely distributed in the cytoplasm of myoblasts and RMS cells (Fig. 1E) but is also detected in the nucleoli of myoblasts during terminal differentiation (see Fig. S1 in the supplemental material). Interestingly, in RMS cells, a fraction of IMP-2 also forms distinct cytoplasmic granules, where it colocalizes with Ge-1 and DDX6/p54, markers of processing bodies (P-bodies), suggesting that in RMS cells certain RNA targets of IMP-2 can be transported to these RNP structures for storage and/or degradation (Fig. 1E and F).
Loss-of-function assays in myoblasts and RMS cells show that IMP-2 controls cell motility and invasive capacity, as well as MT stabilization via accumulation of Glu-tubulin.
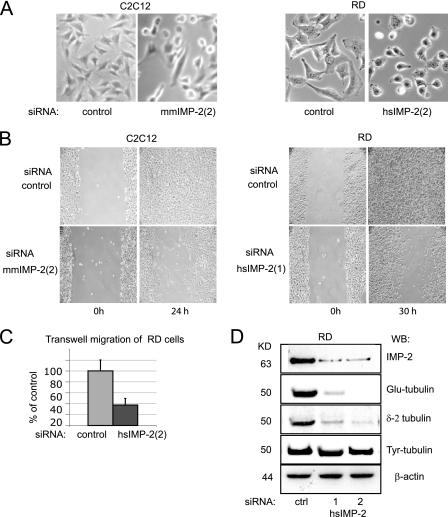
To study the function of IMP-2 in normal and cancerous cells, we transfected C2C12 myoblasts and RMS cells with siRNAs targeting IMP-2 and analyzed the phenotypes of IMP-2 KD (knocked-down) cells. In all cell lines tested, downregulation of IMP-2 led to a striking modification of cell shape (Fig. 2A) and to a dramatic decrease in cell motility and migration capacity, as measured by a wound test (Fig. 2B; see Fig. S2A and B in the supplemental material) or a quantitative Transwell migration test (Fig. 2C).
FIG. 2.
Loss-of-function assays in myoblasts and RMS cells show that IMP-2 controls cell shape and motility as well as microtubule (MT) stabilization via accumulation of detyrosinated tubulin. (A) Changes of cell shape in IMP-2 KD myoblasts and RMS cells 48 h after siRNA transfection. (B) A wound test shows deficiency in the motility of RD and C2C12 IMP-2 KD cells. (C) Transwell test quantification of RMS RD cell migration with control (ctrl) cells versus IMP-2 KD. (D) IMP-2 KD induces a specific decrease of levels of detyrosinated tubulin in RD cells.
Further analysis of IMP-2 KD cells revealed that downregulation of IMP-2 leads to a specific decrease of posttranslationally modified α-tubulin (Glu-tubulin), which is associated with stabilized, nondynamic MTs (4). The levels of nonmodified α-tubulin (Tyr-tubulin) do not change in these cells, suggesting that IMP-2 does not influence the balance between tyrosination and detyrosination of α-tubulin, but rather controls an unknown mechanism specifically responsible for the integrity of stable MTs. This hypothesis was further confirmed by our observation that another form of posttranslationally modified α-tubulin (δ-2 tubulin) also associated with stable MTs is similarly downregulated in myoblasts and RMS cells in the absence of IMP-2 (Fig. 2D).
Characterization of potential IMP-2 RNA targets in C2C12 myoblasts.
In order to explore the mechanisms of regulation of cell motility by IMP-2, our first step was to characterize IMP-2 RNA targets in myoblasts by loss-of-function assays. First, we purified total RNA from C2C12 myoblasts transfected with control siRNA or with two different siRNAs directed against IMP-2 (all samples were analyzed in duplicate) and identified the transcripts that are differentially expressed when IMP-2 is downregulated. Out of 35,029 transcripts represented on the genome-wide microarray (CodeLink; Amersham), 1,193 were found to be differentially regulated with a difference greater than 1.37-fold. Surprisingly, 91.8% of these transcripts were upregulated in the absence of IMP-2. Upregulated transcripts might be indirect targets of IMP-2; alternatively, IMP-2 might regulate its target mRNAs by different mechanisms, having opposite effects on the levels of individual transcripts.
To identify direct targets, we further analyzed the transcripts that are physically associated with IMP-2 in C2C12 myoblasts. To this end, we have stably expressed mouse IMP-2, C-terminally or N-terminally tagged with FLAG-HA, in C2C12 myoblasts and immunoprecipitated the IMP-2-containing RNP complexes, as described in reference 31. Proteins found in these IMP-2 complexes were identified by mass spectrometry, and IMP-2-bound transcripts were characterized in triplicate on genome-wide microarrays (Affymetrix). By comparing the results of the two above-described analyses, we obtained a list of 118 transcripts showing both physical association with IMP-2 complexes and significant changes in expression levels when IMP-2 was downregulated. These transcripts were likely to represent a selection of putative RNA targets of IMP-2 in myoblasts (see Table S1 in the supplemental materials). A qRT-PCR analysis of expression of a number of these RNAs in IMP-2 KD cells confirmed differential regulation for approximately 71.5% of the transcripts (15 confirmed out of 21 analyzed) (see Table S1).
The majority of putative IMP-2 target transcripts correspond to proteins involved in cell motility, adhesion, and cytoskeleton and membrane remodeling (35% of all potential IMP-2 targets). These results are consistent with the defects in cell motility and cytoskeleton remodeling that we observe when IMP-2 is downregulated in myoblasts and RMS cells. Other large groups of functionally distinct IMP-2 target transcripts are involved in protein synthesis (mostly ribosomal proteins), oxidative phosphorylation, endosome transport, and the Golgi apparatus. Interestingly, only 10% of the potential IMP-2 RNA targets in C2C12 cells were downregulated in IMP-2 KD cells, whereas the vast majority were significantly upregulated. This observation suggested that IMP-2 can have different effects on the levels of individual target transcripts. Two validated targets of IMPs, IGF-2 and the noncoding RNA H19 (28, 33), were found among the IMP-2-bound transcripts that were also differentially regulated in IMP-2 KD myoblasts, thus validating our experimental approach. IGF-2 was not found to be significantly downregulated in all IMP-2 KD cells in a high-throughput screen, but additional qRT-PCR experiments in C2C12 and RD cells have confirmed that this previously validated IMP target was decreased at least 2-fold when IMP-2 was depleted by 85% or more (data not shown).
Interestingly, the regulation of transcripts by IMP-2 did not appear to correlate with their translational status in the cell. Among the previously characterized targets of IMP regulation, the noncoding RNA H19 is monosomal in both proliferating and differentiating C2C12 myoblasts, and the mRNA of IGF-2 is associated with both monosomal and polysomal fractions. The novel IMP-2 targets characterized in this study are mostly polysomal in C2C12 cells. However, the abundant and actively translated mRNAs of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin are neither bound to nor regulated by IMP-2 in C2C12 cells, suggesting that association of a transcript with the polysomal fraction is not sufficient for its regulation by IMP-2 in myoblasts (data not shown; see Fig. S3 in the supplemental material).
A selection of putative IMP-2 targets linked to cell motility and confirmed by qRT-PCR is shown in Table 1.
TABLE 1.
Selection of transcripts demonstrating changed expression in IMP-2 KD cellsa
| Protein and group | Fold change in IMP-2 KD versus control | RefSeq no. | Function (reference) |
|---|---|---|---|
| Proteins increased in IMP-2 KD cells | |||
| PINCH-2 (Lims2) | +5.70 | NM_144862 | FA remodeling (46) |
| HoxA11 | +3.5 | NM_010450 | Migration of pre-myoblasts in embryogenesis (14) |
| Figf (VEGFD) | +1.99 | NM_010216 | Fos-induced, prometastatic integrin ligand (43) |
| CD81 | +1.84 | NM_133655 | Integrin-interacting membrane protein, migration (39) |
| Trip6 | +1.63 | NM_011639 | FA-interacting regulator of cell migration (1, 46) |
| Eps8 | +1.58 | NM_007945 | Rac activator, motility, cytoskeleton remodeling (10) |
| Map1lc3b | +1.45 | NM_026160 | Microtubule-associated protein, autophagy (20) |
| Proteins decreased in IMP-2 KD cells | |||
| Vcam1 | −1.79 | NM_011693 | Regulator of cell adhesion and migration (26) |
| Trim54 (MURF-3) | −1.88 | NM_021447 | MT stabilization during myogenesis (37) |
| Ptpn12 (PTP-PEST) | −2.45 | NM_011203 | FA-interacting tyrosine phosphatase, cytoskeleton (35) |
Shown are high-throughput data confirmed by qRT-PCR.
PINCH-2 mRNA is bound by IMP-2 complexes in myoblasts and upregulated in IMP-2 KD myoblasts and RMS cells.
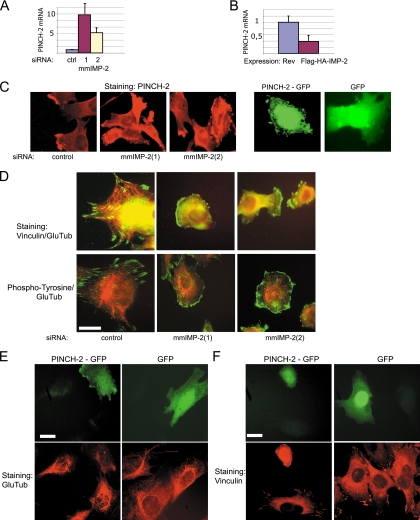
In IMP-2 KD myoblasts, the most significantly changed transcript was the mRNA of PINCH-2 (upregulated 5.7-fold in genome-wide microarrays and up to 10-fold in qRT-PCR assays) (Fig. 3A and Table 1). Conversely, in stable C2C12 cell lines expressing tagged IMP-2, the mRNA of PINCH-2 was downregulated 3.3-fold (Fig. 3B). This mRNA was easily detected within the FLAG-HA-IMP-2 complexes purified from C2C12 myoblasts, as described above (our qRT-PCR experiments; data not shown).
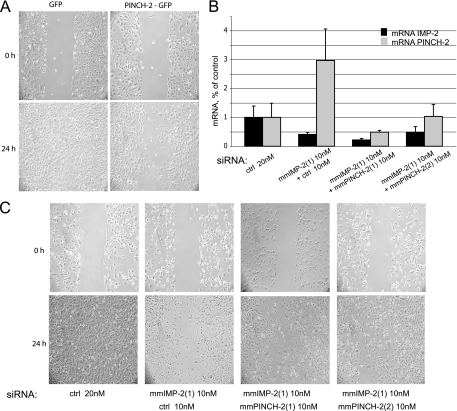
FIG. 3.
In C2C12 myoblasts, increased levels of PINCH-2 mRNA and protein lead to changes in cell shape, to clustering of FA structures, and to decrease in stable MTs. (A) KD of IMP-2 by two distinct siRNAs increases the levels of PINCH-2 mRNA. (B) Stable ectopic expression of IMP-2 decreases the levels of PINCH-2 mRNA. (C) Expression of PINCH-2 protein in IMP-2 KD C2C12 cells (left and middle panels) and in PINCH-2-GFP-transfected C2C12 cells (right panels); (D) immunofluorescent staining of FAs (green) and stable MTs (red) in control and IMP-2 KD cells; (E), decrease of Glu-tubulin staining in PINCH-2-GFP-expressing cells; (F) immunofluorescent staining of FAs in control and PINCH-2-GFP-expressing C2C12 myoblasts. Bars in panels D to F, 12 μm.
PINCH-2 is an LIM domain protein that binds to and partially dissociates the focal adhesion (FA) complexes. Depending on the cell type, PINCH-2 had been shown to localize to focal adhesions (NIH 3T3 fibroblasts) or to the nuclei and focal adhesions (human lung cell line WI-38) or to be diffused in the cytoplasm, with preferential presence in M-bands and Z-disks (primary cardiomyocytes) (3). By overexpressing PINCH-2 in C2C12 myoblasts, we obtained a population of cells with a dramatically changed shape (see Fig. S4A and B in the supplemental material). Our experiments show that in normal C2C12 myoblasts, endogenous PINCH-2 is expressed at a low level and is detectable at the periphery of 10 to 15% of the cells (Fig. 3C, left panel). In IMP-2 KD C2C12 myoblasts, endogenous PINCH-2 shows strong and aberrant expression and aggregates at the perinuclear region and at the periphery of 75 to 80% of the cells, a phenotype which can be reproduced by ectopic expression of PINCH-2-green fluorescent protein (GFP) (Fig. 3C, right panels).
FAs in the cytoplasm serve as anchoring points for the cytoskeleton, notably for F-actin filaments (stress fibers) and for microtubules. Interestingly, a number of studies have suggested that the FAs are the sites of local stabilization of microtubules and that Glu-tubulin is rapidly and specifically downregulated when the cells are maintained in suspension and lose FAs (19, 30). Based on these observations and on our data, we hypothesized that upregulation of PINCH-2 in IMP-2 KD myoblasts and RMS cells can contribute to remodeling of FAs, to decreased motility of the cells, and to the specific downregulation of Glu-tubulin observed in these cells.
Indeed, in IMP-2 KD C2C12 cells, the size and subcellular distribution of FAs were visibly modified: vinculin and phosphotyrosine staining revealed large clusters at the periphery of the cells (Fig. 3D). The stable MTs containing Glu-tubulin became restricted to the perinuclear region in these cells, which could be linked to the aberrant expression of PINCH-2 and the remodeling of FAs, but could also be an indirect consequence of IMP-2 KD. To resolve this issue, we have studied stable MTs in C2C12 cells expressing PINCH-2-GFP, or GFP as a control. Staining for stable MTs was weak in cells expressing PINCH-2-GFP, as compared to nontransfected cells (Fig. 3E), suggesting that in IMP-2 KD cells, the disappearance of Glu-tubulin-containing stable MTs can be at least in part due to increased expression of PINCH-2 and remodeling of FAs. Ectopic expression of PINCH-2-GFP in C2C12 cells leads to changes in cell shape and to peripheral clustering or FAs (visualized by vinculin and phosphotyrosine staining in Fig. 3F; see Fig. S4C in the supplemental material). Taken together, these results show that PINCH-2 upregulation in IMP-2 KD myoblasts can significantly contribute to the phenotypic changes observed in these cells.
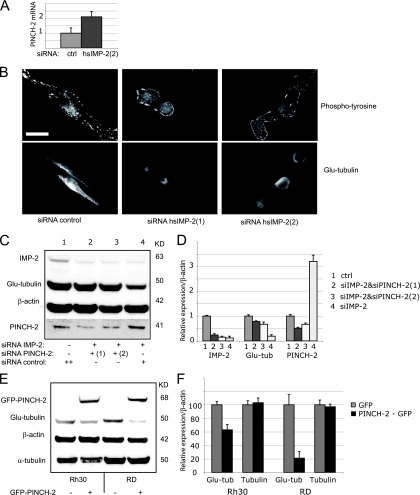
In human RMS cells, KD of IMP-2 leads to a 2-fold upregulation of PINCH-2 mRNA, showing that PINCH-2 is regulated by IMP-2 in different cell systems—mouse and human (Fig. 4A). Moreover, IMP-2 KD in an RMS cell line RD leads to changes in cell morphology similar to those observed in IMP-2 KD- or PINCH-2-overexpressing C2C12 myoblasts: cells become rounded, with barely detectable stable MTs and clustered adhesion structures (Fig. 4B). In RMS cells, Glu-tubulin is highly expressed and can be easily detected by Western blotting. We have thus attempted to quantify the PINCH-2-dependent regulation of Glu-tubulin following IMP-2 KD in these cells. IMP-2 KD led to a 2.8-fold increase of PINCH-2 protein in these cells (Fig. 4C and D, compare lanes 1 and 4), accompanied by a decrease of Glu-tubulin to 22% of the control. When IMP-2 KD RD cells were treated with two distinct anti-PINCH-2 siRNAs, the levels of Glu-tubulin in these cells could be “rescued” to 78 and 67% of the control, respectively (Fig. 4C and D, lanes 2 and 3). Conversely, ectopic expression of PINCH-2 in RMS cells leads to a decrease of endogenous Glu-tubulin by 37% in Rh30 cells and by 80% in RD cells (Fig. 4E and F).
FIG. 4.
In IMP-2 KD human RMS cells (RD), PINCH-2 regulates the remodeling of focal adhesions (FAs) and of stable microtubules (MTs). (A) KD of IMP-2 increases the levels of PINCH-2 mRNA in RD cells. (B) Immunofluorescent staining of FAs and stable MTs in IMP-2 KD RD cells. Bar, 10 μm. (C) siRNA-induced decrease of PINCH-2 in IMP-2 KD cells “rescues” the levels of Glu-tubulin; shown is one experiment out of two. (D) Quantification of two independent experiments described in panel C; error bars indicate standard deviation. (E) Ectopic expression of PINCH-2 in RD cells leads to a specific decrease of Glu-tubulin levels; shown is one experiment out of two. (F) Quantification of two independent experiments described in panel E. Error bars indicate standard deviation.
Previously published studies and our results show that overexpression of PINCH-2 leads to dramatically decreased cell motility, presumably by changing integrin-mediated signaling at FA sites (21, 46) (Fig. 5A). Therefore, upregulation of PINCH-2 in IMP-2 KD cells could be responsible for their diminished spreading and motility. Consistent with this hypothesis, the double-siRNA KD assay showed an almost fully restored motility of IMP-2 KD cells when PINCH-2 was specifically downregulated with two distinct siRNAs (Fig. 5B and C).
FIG. 5.
PINCH-2 regulates cell motility in C2C12 myoblasts. (A) Ectopic expression of PINCH-2-GFP in C2C12 myoblasts leads to their decreased motility; shown is a representative experiment out of three performed. (B) Relative levels of mRNAs of IMP-2 and PINCH-2 in C2C12 myoblasts transfected with the indicated siRNAs. (C) Decreased motility of IMP-2 KD cells is restored by a simultaneous decrease of PINCH-2; shown is a representative experiment out of two done in biological duplicates.
Taken together, these results show that upregulation of PINCH-2 in IMP-2 KD myoblasts is the key event responsible for the decreased motility of these cells, as well as a major factor contributing to the cell phenotype: depleted Glu-tubulin and aggregation of FA structures. However, the levels of Glu-tubulin in IMP-2 KD cells are not fully restored by downregulating PINCH-2 (Fig. 4C and D) (data not shown), suggesting that IMP-2 regulates other important targets in these cells.
MURF-3 mRNA is bound by IMP-2 complexes in myoblasts and downregulated in IMP-2 KD myoblasts.
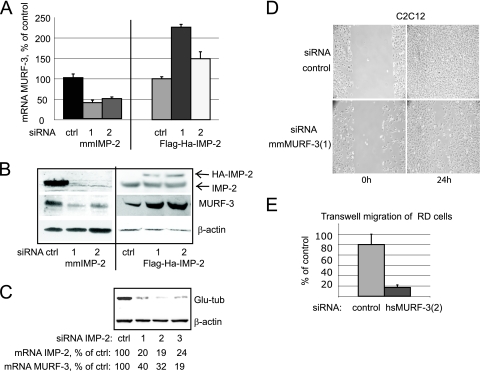
MURF-3 RNA was associated with IMP-2 in C2C12 myoblasts (qRT-PCR analysis of transcripts bound to FLAG-HA-IMP-2 complexes; data not shown). MURF-3 is a RING finger protein specifically expressed in skeletal muscle and in differentiating myoblasts in culture. Expression of MURF-3 is necessary for accumulation of Glu-tubulin and for stabilization of MTs during differentiation (37). In contrast to PINCH-2, MURF-3 mRNA and protein levels correlated with the levels of IMP-2 in IMP-2 KD myoblasts and in myoblasts stably expressing tagged IMP-2 (Fig. 6A to C). The level of unspliced precursor of MURF-3 mRNA (pre-mRNA), however, did not correlate with the expression of IMP-2 (data not shown), indicating that regulation of MURF-3 by IMP-2 in myoblasts occurs at the posttranscriptional level. Consistent with the role of MURF-3 in inducing the stabilization of MTs via accumulation of Glu-tubulin, in IMP-2 KD C2C12 myoblasts (using three different siRNAs against IMP-2) the levels of Glu-tubulin diminished in correlation with the decrease in IMP-2 and MURF-3 (Fig. 6C).
FIG. 6.
MURF-3 mRNA and protein are regulated by IMP-2 in C2C12 myoblasts. (A and B) MURF-3 mRNA (A) and protein (B) are downregulated in IMP-2 KD C2C12 myoblasts (obtained with two distinct siRNAs [1 and 2]) and upregulated in two distinct stable C2C12 lines expressing FLAG-HA-IMP-2, tagged on the N terminus (siRNA 1) or C terminus (siRNA 2). (C) In IMP-2 KD C2C12 myoblasts (obtained with three distinct siRNAs [1, 2, and 3]), downregulation of MURF-3 is followed by a specific decrease of Glu-tubulin. The indicated mRNA levels of IMP-2 and MURF-3, expressed as a percentage of the control (transfection with control siRNA), were quantified by qRT-PCR and normalized by 36b4. (D and E) MURF-3 KD myoblasts and RMS cells show decreased motility (wound test [D]) and migration capacity (Transwell test [E]). Shown are the means of at least three independent experiments; error bars indicate standard deviation.
The typical phenotype of IMP-2 KD cells included both the specific loss of Glu-tubulin and impaired motility. We thus tested the hypothesis that decreased MURF-3 in IMP-2 KD cells could contribute to both phenomena. Indeed, specific siRNAs against MURF-3 moderately decreased the motility of mouse myoblasts and human RMS cells and dramatically reduced their migration capacity in a Transwell assay (Fig. 6D and E; see Fig. S2B in the supplemental material).
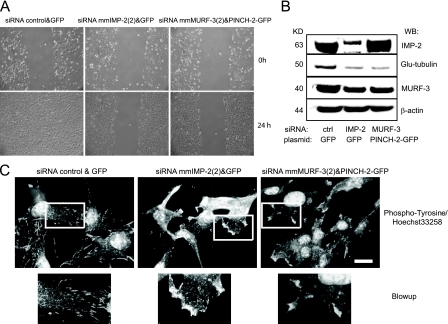
Finally, we have reconstituted the phenotype of IMP-2 KD cells by overexpressing PINCH-2 and simultaneously downregulating MURF-3 in C2C12 myoblasts. Both PINCH-2 high/MURF-3 low cells and IMP-2 KD cells demonstrate decreased motility, decreased spreading, diminished Glu-tubulin levels, and clustered adhesion structures (Fig. 7). Therefore, we conclude that IMP-2 in cells orchestrates the posttranscriptional regulation of multiple RNAs, notably the mRNAs of PINCH-2 and MURF-3, thus influencing cytoskeleton remodeling, cell adhesion, and cell motility.
FIG. 7.
The phenotype of IMP-2 KD C2C12 myoblasts can be reproduced by ectopic expression of PINCH-2 and depletion of MURF-3. (A to C) The cells were transfected with PINCH-2-GFP expression vector or with a control GFP vector, sorted by fluorescence-activate cell sorting 24 h later to select GFP-positive cells, and transfected with the indicated siRNAs. The control cells (GFP-siRNA control) were compared to IMP-2 KD cells [GFP-siRNA mmIMP-2(4)] and to cells expressing ectopic PINCH-2 and depleted of MURF-3 [siRNA mmMURF-3(2) and PINCH-2 GFP] 48 h after the transfection of siRNAs. (A) Comparison of cell motility by a wound test; (B) Western blot analysis of expression of IMP-2, Glu-tubulin, MURF-3, and actin (loading control); (C) immunostaining with phosphotyrosine antibody to analyze the adhesion structures. Bar, 14 μm.
IMP-2 directly binds the mRNA of MURF-3 and PINCH-2.
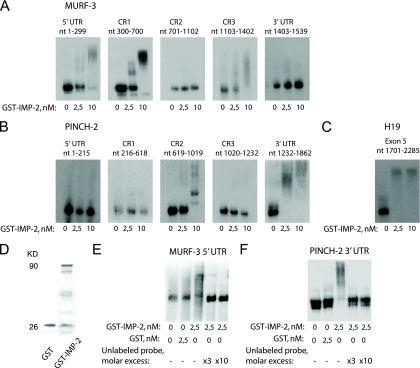
Previous studies have shown that IMPs can bind multiple sites in their target mRNAs, within 5′ or 3′ untranslated regions (UTRs), as well as within the coding region. We have mapped the IMP-2 binding sites within MURF-3 and PINCH-2 whole-length transcripts, using gel shift experiments and recombinant IMP-2. The results shown in Fig. 8A and B demonstrate that IMP-2 directly binds within the 5′ part of MURF-3 transcript (5′ UTR and 5′ part of the coding region), whereas in PINCH-2 transcript, IMP-2 strongly binds within the 3′ UTR. The efficiency of binding of these novel targets by IMP-2 was comparable to that of H19, a previously characterized IMP target (Fig. 8C).
FIG. 8.
Recombinant IMP-2 directly binds the mRNAs of MURF-3 and PINCH-2. (A and B) RNA bandshift assays using increasing concentrations of GST-IMP-2 map the binding sites of IMP-2 in MURF-3 and PINCH-2 transcripts. The results for each RNA fragment were reproduced at least twice; shown is a typical result. nt, nucleotides. (C) Binding of recombinant IMP-2 to a fragment of H19, a previously characterized RNA target of IMPs. (D) GST-IMP-2 and GST proteins used in control experiments. (E and F) Specific binding of IMP-2 to the 5′ UTR of MURF-3 and 3′ UTR of PINCH-3 is confirmed by control experiments with GST and cold-probe competition.
The mRNAs of PINCH-2 and MURF-3 are bound by distinct multiprotein complexes in myoblasts.
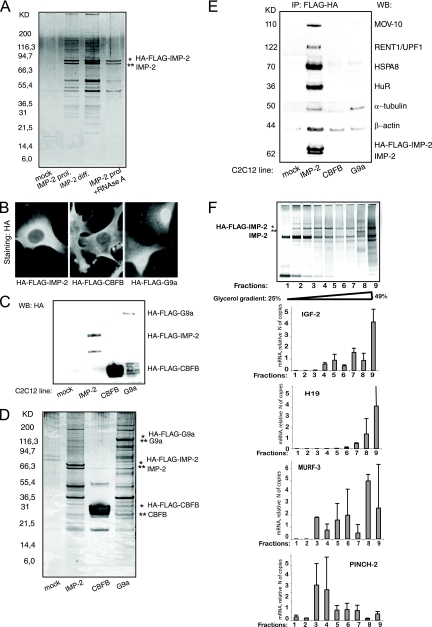
We next explored the regulation of PINCH-2 and MURF-3 mRNAs by IMP-2 in cells. IMPs are found within large and various RNP complexes in different cells and tissues. Thus, we proceeded to characterize the protein partners of IMP-2 in myoblasts. To this end, we have stably expressed mouse IMP-2, C-terminally or N-terminally tagged with FLAG-HA, and immunoprecipitated the IMP-2-containing RNP complexes, as described in reference 31. Six independent experiments were performed to purify IMP-2 complexes: two purifications of N-terminally tagged IMP-2 from proliferating cells and two from differentiating cells, followed by two purifications of C-terminally tagged IMP-2 from proliferating and differentiating cells. The protein contents of IMP-2 complexes were identified by mass spectrometry and, in part, confirmed by Western blotting. The results of these six experiments, shown in Fig. 9 A and Table S2 in the supplemental material, were found to be highly similar, suggesting that IMP-2 complexes did not vary from proliferating to differentiating myoblasts and that the position of the tag did not influence IMP-2 interaction with its partners. The specificity of IMP-2 partners was confirmed by control experiments with two irrelevant FLAG-HA-tagged proteins (Fig. 9B to E). Direct IMP-2 partners were identified by RNase treatment, as described in Materials and Methods. We then used systematic RNAi screens to identify the partners of IMP-2 that impacted the expression levels of PINCH-2 and MURF-3.
FIG. 9.
IMP-2 and its protein partners form distinct multiprotein-RNA complexes in myoblasts. (A) Silver staining of FLAG-HA-IMP-2 protein complexes purified from C2C12 myoblasts (lanes 2 and 3) proliferating (prol.) or differentiating (diff.) for 24 h. Mock, FLAG-HA immunoprecipitate (IP) from control C2C12 cells (lane 1). Lane 4, RNase A-treated IMP complexes from differentiating C2C12 cells. (B) Subcellular localization of IMP-2 and control FLAG-HA-tagged proteins stably expressed in C2C12 myoblasts; (C) expression levels of proteins shown in panel B; (D and E) FLAG-HA-IMP-2 complexes compared to irrelevant FLAG-HA complexes purified from C2C12 myoblasts shown in panels B and C; (D) silver staining; (E) Western blot against selected IMP-2 partners. (F) HA-FLAG-IMP-2 complexes were immunity purified from C2C12 myoblasts, separated on glycerol gradient, and analyzed by silver staining. RNAs were purified from each fraction, and the presence of the indicated IMP-2 RNA targets was quantified in each fraction by qRT-PCR.
Our results showed that the upregulation of PINCH-2, induced by KD of IMP-2, is also observed in KDs of MOV-10, HuR, matrin-3, and all three Hspa protein chaperones that are direct (RNA-independent) protein partners of IMP-2 (Table 2). A decrease of MURF-3 is observed in KDs of two Hspa proteins and hnRNP K. These observations strongly suggest that the transcripts of MURF-3 and PINCH-2 are regulated by distinct, but overlapping, IMP-2-containing complexes. To test this hypothesis, we immunopurified the FLAG-HA-IMP-2-bound RNP complexes from C2C12 myoblasts, as described above, and then separated them on a 25 to 49% glycerol gradient, according to their molecular weights (Fig. 9F). Subsequently, we analyzed individual fractions by qRT-PCR, to quantify the relative presence of IMP-2 RNA targets in the revealed low- and high-molecular-weight IMP-2-containing complexes. As shown in Fig. 9F, the mRNA of MURF-3 is preferentially associated with high-molecular-weight IMP-2 complexes, similarly to the known targets of the IMP family IGF-2 and H19. All of these transcripts are downregulated in IMP-2 KD cells. On the contrary, PINCH-2, the mRNA which is significantly upregulated in IMP-2 KD myoblasts and RMS cells, is preferentially associated with low-molecular-weight IMP-2 complexes. These results suggest that IMP-2 regulates its individual RNA targets by distinct molecular mechanisms that remain to be elucidated.
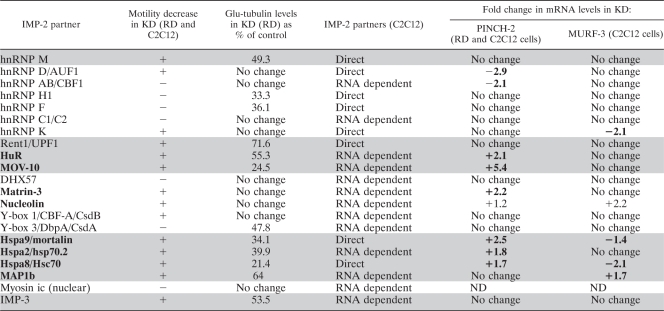
TABLE 2.
Identification of the protein partners of IMP-2 that change cell motility and levels of Glu-tubulin in RD and C2C12 cells and specifically impact the levels of PINCH-2 and/or MURF-3 in these cellsa
RNAi loss-of-function assays identify the protein partners of IMP-2 that change cell motility and levels of Glu-tubulin in RD and C2C12 cells (highlighted in gray) and specifically indicate the IMP-2 partners that impact the levels of PINCH-2 and/or MURF-3 in these cells (in boldface). Changes in cell motility were analyzed when the wound in the control cell monolayer (transfected with scrambled siRNA) was fully closed (24 h for C2C12 and 20 h for RD). Decreased motility of cells transfected with siRNAs against IMP-2 partners was considered significant if at the indicated times at least 50% of the original wound was clearly visible. Glu-tubulin levels were evaluated by Western blotting (see Fig. S5 in the supplemental material).
Our RNAi screens have also allowed us to identify the protein partners of IMP-2 that impact cellular motility and/or remodeling of MTs in C2C12 myoblasts and in the embryonic RMS cell line RD (the data are summarized in Table 2, columns 1 to 4, and illustrated in Fig. S5 in the supplemental material). Predictably, IMP-2 protein partners that regulate the mRNA levels of PINCH-2 and MURF-3 (Hspa8 and -9) are important both for cellular motility and for Glu-tubulin levels in cells. Upregulation of PINCH-2 alone (in KDs of MOV-10, HuR, matrin-3, and Hspa2) leads to impaired cellular motility, accompanied in most cases by a decrease in Glu-tubulin. Our results show that MURF-3 expression has to be decreased by at least 60% to have a clear effect on Glu-tubulin levels in the absence of upregulation of PINCH-2 (Fig. 6C). Consistent with these observations, Glu-tubulin is stable in hnRNP K KD cells, in which MURF-3 is decreased by 50% and PINCH-2 is not affected (Table 2).
KDs of IMP-2 partners hnRNP D/AUF1 and hnRNP AB/CBF1 unexpectedly lead to a decrease in PINCH-2 levels, with some or no effect on cellular motility, and stable Glu-tubulin levels. Interestingly, KDs of three RNA-binding protein partners of IMP-2 in myoblasts (hnRNP M, Rent1/UPF1, and IMP-3) do not impact expression of PINCH-2 and MURF-3 but bring on a decrease in cellular motility and Glu-tubulin levels by unknown mechanisms. Finally, a KD of the indirect IMP-2 partner, MT-binding and stabilizing protein MAP1b, also significantly changes cell motility and Glu-tubulin levels. The latter observations suggest that certain protein partners of IMP-2 could contribute to the phenotype of IMP-2 KD cells by mechanisms independent of IMP-2 targets PINCH-2 and MURF-3.
DISCUSSION
In this study, we show that the RNA-binding protein IMP-2 is strongly expressed in normal human and mouse myoblasts and in human RMS cells. The most striking phenotypes induced by the KD of IMP-2 in all these cell types include changes in cell motility, adhesion structures, and the levels of detyrosinated tubulin, the component of stable MTs. Consistent with these observations, we have found that the most significant group of IMP-2-regulated transcripts corresponds to proteins involved in cytoskeleton remodeling, adhesion, and maintenance of membrane integrity. In particular, our data indicate that IMP-2 posttranscriptionally regulates the expression of PINCH-2, a protein responsible for the remodeling of FAs, cell adhesion, and cell motility, as well as MURF-3, which is indispensable for stabilization of MTs in myoblasts. It is noteworthy, however, that the modes of action of IMP-2 are quite distinct, and in fact opposite, for these two transcripts and the corresponding proteins: the expression of MURF-3 requires the presence of IMP-2, whereas PINCH-2 is downregulated by IMP-2. Downregulation of MURF-3 and upregulation of PINCH-2 together contribute to the phenotype of IMP-2 KD cells, which are characterized by decreased motility, low levels of detyrosinated tubulin, and striking changes in cell shape. Thus, IMP-2 can be important for such processes as terminal differentiation, which involves motility/change of cellular shape as well as accumulation of Glu-tubulin and stabilization of the cytoskeleton (in particular in myogenic and neuronal differentiation). On the other hand, in numerous human tumors, cancer cells demonstrate a dramatically changed motility and invasive capacity, as well as increased levels of Glu-tubulin (17, 22, 24, 45).
It is possible that only a fraction of FAs are directly controlled by IMP-2 RNP complexes, and the involvement of the FA-remodeling PINCH-2 protein in this regulatory mechanism strongly suggests that the most “dynamic” FAs (either undergoing remodeling or newly formed) are the most IMP-2 dependent. In this respect, it is interesting that IMP-2 and a number of the IMP-2 protein partners identified in the present study have been found within the “spreading initiation centers,” pre-FA structures found in early cell spreading (9).
The pathways regulating cellular adhesion, motility, and cytoskeleton dynamics are tightly related. A number of studies have suggested that FAs are necessary and sufficient for the presence of stable, Glu-tubulin-containing MTs (19, 30). However, other cytoplasmic structures, and in particular the actin cytoskeleton, participate in the control of cellular adhesion and motility. Notably, localization of β-actin mRNA to the leading edge of multiple cell types is dependent on IMP-1/ZBP-1 protein, the founding member of the IMP family (8). In C2C12 myoblasts, the β-actin mRNA was reported to be diffused in the cytoplasm and not localized to the leading edge of the cells, suggesting that the motility of these cells is not regulated by the localized synthesis of actin and depends on other mechanisms (29). In the present study, we propose that IMP-2 influences the motility of C2C12 myoblasts and RMS cells by posttranscriptional regulation of proteins of the cytoskeleton of stable MTs and of cellular adhesion structures. All three IMPs are expressed in C2C12 myoblasts and in RMS cells, although the levels of IMP-2 in myoblasts are significantly higher than those of IMP-1 and IMP-3 (S. Cuvellier and A. Polesskaya, unpublished observation). However, specific KD of IMP-2 in myoblasts and RMS cells leads to a distinct phenotype, strongly suggesting that IMP-2 in these cells has functions which can be overlapping, but not fully redundant with, those of IMP-1/ZBP-1 and IMP-3.
We have started to characterize the molecular mechanisms of posttranscriptional regulation by IMP-2, which has an impact both on the levels of target RNAs and on the corresponding proteins. The correlation between the levels of IMP-2 target RNAs and their protein products shows that IMP-2 regulation is not likely to involve changes in splicing mechanisms and/or subsequent degradation of misspliced RNAs. The translational status of a transcript does not appear to determine its sensitivity to IMP-2. Thus, IMP-2 is likely to regulate mRNA transport and/or stability, like its family members IMP-1 and IMP-3. Our observations that IMP-2 in C2C12 cells is present both in monosomal and in light polysomal fractions and that IMP-2 RNP complexes can be separated into distinct low- and high-molecular-weight fractions strongly suggest that IMP-2 regulatory units in cells vary in size, composition, and precise function. In the case of the subset of RNAs that are downregulated in IMP-2 KD cells (such as MURF-3, IGF-2, and H19), it is tempting to suggest that the mechanisms involved include RNA transport. The decrease of these transcripts in the absence of IMP-2 could reflect mRNA degradation due to erroneous subcellular localization. MURF-3 mRNA is downregulated in the KDs of IMP-2 partners (Hspa8 and -9), suggesting that these proteins can be important for the function of IMP-2 complexes bound to this transcript.
The large subset of IMP-2 target RNAs that are upregulated in IMP-2 KD cells appears to depend on a significant number of IMP-2 protein partners. Indeed, as shown in the present study, the increase in PINCH-2 levels observed in IMP-2 KD cells can also be induced by a KD of IMP-2 partners matrin-3, HuR, and nucleolin, as well as all three Hspa proteins (Table 2). Most intriguingly, the RNA helicase MOV-10, which is involved in microRNA (miRNA)-dependent translational regulation (5), is abundant in IMP-2 RNP complexes. A KD of MOV-10 induces the increase in the levels of a number of RNAs that accumulate in IMP-2 KD cells, such as PINCH-2. In addition, IMP-2 and MOV-10, but not the Hspa proteins, colocalize in a fraction of P-bodies in RMS cells (S. Boudoukha, unpublished observation), suggesting that some IMP-2 target RNAs could be directed to these structures for storage and/or degradation. In this case, a KD of IMP-2 or MOV-10 can tilt the balance and exclude the target RNAs from the P-bodies, which should lead to their increased translation and/or stability.
In summary, our data reveal, for the first time, the specific involvement of IMP-2 in the regulation of cellular motility and MT cytoskeleton remodeling. They also identify a number of RNAs that are targeted by IMP-2 in normal myoblasts and in RMS cells, as well as protein partners of IMP-2 that are involved in these regulatory events, and can be potentially important for terminal muscle differentiation and for development of rhabdomyosarcomas.
Supplementary Material
Acknowledgments
We thank Annick Harel-Bellan and Linda L. Pritchard for critical reading of the manuscript; Jan Christianssen, Finn C. Nielsen, Eric Rubinstein, and Dominique Weil for the kind gifts of antibodies and for helpful discussions; Pascal Maire for the kind gift of RD and Rh30 cells; Slimane Ait-Si-Ali for FLAG-HA-C2C12 CBFB and FLAG-HA-G9a myoblasts; Didier Job and Laurence Lafanechere for the kind gift of anti-Tyr-tubulin, -Glu-tubulin, and -δ-2 tubulin antibodies; and Reinhard Fässler for the PINCH-2 expression construct and antibody.
This work was supported by a grant from the Association Français contre les Myopathies to A.P.
Footnotes
Published ahead of print on 18 October 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bai, C. Y., M. Ohsugi, Y. Abe, and T. Yamamoto. 2007. ZRP-1 controls Rho GTPase-mediated actin reorganization by localizing at cell-matrix and cell-cell adhesions. J. Cell Sci. 120:2828-2837. [DOI] [PubMed] [Google Scholar]
- 2.Brants, J. R., T. A. Ayoubi, K. Chada, K. Marchal, W. J. Van de Ven, and M. M. Petit. 2004. Differential regulation of the insulin-like growth factor II mRNA-binding protein genes by architectural transcription factor HMGA2. FEBS Lett. 569:277-283. [DOI] [PubMed] [Google Scholar]
- 3.Braun, A., R. Bordoy, F. Stanchi, M. Moser, G. G. Kostka, E. Ehler, O. Brandau, and R. Fassler. 2003. PINCH2 is a new five LIM domain protein, homologous to PINCH and localized to focal adhesions. Exp. Cell Res. 284:239-250. [DOI] [PubMed] [Google Scholar]
- 4.Bulinski, J. C., and G. G. Gundersen. 1991. Stabilization of post-translational modification of microtubules during cellular morphogenesis. Bioessays 13:285-293. [DOI] [PubMed] [Google Scholar]
- 5.Chendrimada, T. P., K. J. Finn, X. Ji, D. Baillat, R. I. Gregory, S. A. Liebhaber, A. E. Pasquinelli, and R. Shiekhattar. 2007. MicroRNA silencing through RISC recruitment of eIF6. Nature 447:823-828. [DOI] [PubMed] [Google Scholar]
- 6.Christiansen, J., A. M. Kolte, T. O. Hansen, and F. C. Nielsen. 2009. IGF2 mRNA-binding protein 2: biological function and putative role in type 2 diabetes. J. Mol. Endocrinol. 43:187-195. [DOI] [PubMed] [Google Scholar]
- 7.Cleynen, I., J. R. Brants, K. Peeters, R. Deckers, M. Debiec-Rychter, R. Sciot, W. J. Van de Ven, and M. M. Petit. 2007. HMGA2 regulates transcription of the Imp2 gene via an intronic regulatory element in cooperation with nuclear factor-kappaB. Mol. Cancer Res. 5:363-372. [DOI] [PubMed] [Google Scholar]
- 8.Condeelis, J., and R. H. Singer. 2005. How and why does beta-actin mRNA target? Biol. Cell 97:97-110. [DOI] [PubMed] [Google Scholar]
- 9.de Hoog, C. L., L. J. Foster, and M. Mann. 2004. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell 117:649-662. [DOI] [PubMed] [Google Scholar]
- 10.Di Fiore, P. P., and G. Scita. 2002. Eps8 in the midst of GTPases. Int. J. Biochem. Cell Biol. 34:1178-1183. [DOI] [PubMed] [Google Scholar]
- 11.Efimov, A., N. Schiefermeier, I. Grigoriev, R. Ohi, M. C. Brown, C. E. Turner, J. V. Small, and I. Kaverina. 2008. Paxillin-dependent stimulation of microtubule catastrophes at focal adhesion sites. J. Cell Sci. 121:196-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hafner, M., M. Landthaler, L. Burger, M. Khorshid, J. Hausser, P. Berninger, A. Rothballer, M. Ascano, Jr., A. C. Jungkamp, M. Munschauer, A. Ulrich, G. S. Wardle, S. Dewell, M. Zavolan, and T. Tuschl. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141:129-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammer, N. A., T. O. Hansen, A. G. Byskov, E. Rajpert-De Meyts, M. L. Grondahl, H. E. Bredkjaer, U. M. Wewer, J. Christiansen, and F. C. Nielsen. 2005. Expression of IGF-II mRNA-binding proteins (IMPs) in gonads and testicular cancer. Reproduction 130:203-212. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto, K., Y. Yokouchi, M. Yamamoto, and A. Kuroiwa. 1999. Distinct signaling molecules control Hoxa-11 and Hoxa-13 expression in the muscle precursor and mesenchyme of the chick limb bud. Development 126:2771-2783. [DOI] [PubMed] [Google Scholar]
- 15.Ioannidis, P., T. Trangas, E. Dimitriadis, M. Samiotaki, I. Kyriazoglou, C. M. Tsiapalis, C. Kittas, N. Agnantis, F. C. Nielsen, J. Nielsen, J. Christiansen, and N. Pandis. 2001. C-MYC and IGF-II mRNA-binding protein (CRD-BP/IMP-1) in benign and malignant mesenchymal tumors. Int. J. Cancer 94:480-484. [DOI] [PubMed] [Google Scholar]
- 16.Jonson, L., J. Vikesaa, A. Krogh, L. K. Nielsen, T. Hansen, R. Borup, A. H. Johnsen, J. Christiansen, and F. C. Nielsen. 2007. Molecular composition of IMP1 ribonucleoprotein granules. Mol. Cell. Proteomics 6:798-811. [DOI] [PubMed] [Google Scholar]
- 17.Kato, C., K. Miyazaki, A. Nakagawa, M. Ohira, Y. Nakamura, T. Ozaki, T. Imai, and A. Nakagawara. 2004. Low expression of human tubulin tyrosine ligase and suppressed tubulin tyrosination/detyrosination cycle are associated with impaired neuronal differentiation in neuroblastomas with poor prognosis. Int. J. Cancer 112:365-375. [DOI] [PubMed] [Google Scholar]
- 18.Kato, T., S. Hayama, T. Yamabuki, N. Ishikawa, M. Miyamoto, T. Ito, E. Tsuchiya, S. Kondo, Y. Nakamura, and Y. Daigo. 2007. Increased expression of insulin-like growth factor-II messenger RNA-binding protein 1 is associated with tumor progression in patients with lung cancer. Clin. Cancer Res. 13:434-442. [DOI] [PubMed] [Google Scholar]
- 19.Kaverina, I., K. Rottner, and J. V. Small. 1998. Targeting, capture, and stabilization of microtubules at early focal adhesions. J. Cell Biol. 142:181-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kouno, T., M. Mizuguchi, I. Tanida, T. Ueno, T. Kanematsu, Y. Mori, H. Shinoda, M. Hirata, E. Kominami, and K. Kawano. 2005. Solution structure of microtubule-associated protein light chain 3 and identification of its functional subdomains. J. Biol. Chem. 280:24610-24617. [DOI] [PubMed] [Google Scholar]
- 21.Legate, K. R., E. Montanez, O. Kudlacek, and R. Fassler. 2006. ILK, PINCH and parvin: the tIPP of integrin signalling. Nat. Rev. Mol. Cell Biol. 7:20-31. [DOI] [PubMed] [Google Scholar]
- 22.Liang, X. J., S. Mukherjee, D. W. Shen, F. R. Maxfield, and M. M. Gottesman. 2006. Endocytic recycling compartments altered in cisplatin-resistant cancer cells. Cancer Res. 66:2346-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao, B., Y. Hu, D. J. Herrick, and G. Brewer. 2005. The RNA-binding protein IMP-3 is a translational activator of insulin-like growth factor II leader-3 mRNA during proliferation of human K562 leukemia cells. J. Biol. Chem. 280:18517-18524. [DOI] [PubMed] [Google Scholar]
- 24.Mialhe, A., L. Lafanechere, I. Treilleux, N. Peloux, C. Dumontet, A. Bremond, M. H. Panh, R. Payan, J. Wehland, R. L. Margolis, and D. Job. 2001. Tubulin detyrosination is a frequent occurrence in breast cancers of poor prognosis. Cancer Res. 61:5024-5027. [PubMed] [Google Scholar]
- 25.Nakatani, Y., and V. Ogryzko. 2003. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370:430-444. [DOI] [PubMed] [Google Scholar]
- 26.Nervi, B., D. C. Link, and J. F. DiPersio. 2006. Cytokines and hematopoietic stem cell mobilization. J. Cell Biochem. 99:690-705. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen, F. C., J. Nielsen, and J. Christiansen. 2001. A family of IGF-II mRNA binding proteins (IMP) involved in RNA trafficking. Scand. J. Clin. Lab Invest. Suppl. 234:93-99. [PubMed] [Google Scholar]
- 28.Nielsen, J., J. Christiansen, J. Lykke-Andersen, A. H. Johnsen, U. M. Wewer, and F. C. Nielsen. 1999. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol. Cell. Biol. 19:1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oleynikov, Y., and R. H. Singer. 2003. Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr. Biol. 13:199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palazzo, A. F., C. H. Eng., D. D. Schlaepfer, E. E. Marcantonio, and G. G. Gundersen. 2004. Localized stabilization of microtubules by integrin- and FAK-facilitated Rho signaling. Science 303:836-839. [DOI] [PubMed] [Google Scholar]
- 31.Polesskaya, A., S. Cuvellier, I. Naguibneva, A. Duquet, E. G. Moss, and A. Harel-Bellan. 2007. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 21:1125-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross, A. F., Y. Oleynikov, E. H. Kislauskis, K. L. Taneja, and R. H. Singer. 1997. Characterization of a beta-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 17:2158-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Runge, S., F. C. Nielsen, J. Nielsen, J. Lykke-Andersen, U. M. Wewer, and J. Christiansen. 2000. H19 RNA binds four molecules of insulin-like growth factor II mRNA-binding protein. J. Biol. Chem. 275:29562-29569. [DOI] [PubMed] [Google Scholar]
- 34.Simon, R., P. A. Bourne, Q. Yang, B. O. Spaulding, P. A. di Sant'Agnese, H. L. Wang, and H. Xu. 2007. Extrapulmonary small cell carcinomas express K homology domain containing protein overexpressed in cancer, but carcinoid tumors do not. Hum. Pathol. 38:1178-1183. [DOI] [PubMed] [Google Scholar]
- 35.Sirois, J., J. F. Cote, A. Charest, N. Uetani, A. Bourdeau, S. A. Duncan, E. Daniels, and M. L. Tremblay. 2006. Essential function of PTP-PEST during mouse embryonic vascularization, mesenchyme formation, neurogenesis and early liver development. Mech. Dev. 123:869-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sparanese, D., and C. H. Lee. 2007. CRD-BP shields c-myc and MDR-1 RNA from endonucleolytic attack by a mammalian endoribonuclease. Nucleic Acids Res. 35:1209-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spencer, J. A., S. Eliazer, R. L. Ilaria, Jr., J. A. Richardson, and E. N. Olson. 2000. Regulation of microtubule dynamics and myogenic differentiation by MURF, a striated muscle RING-finger protein. J. Cell Biol. 150:771-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suda, T., T. Tsunoda, Y. Daigo, Y. Nakamura, and H. Tahara. 2007. Identification of human leukocyte antigen-A24-restricted epitope peptides derived from gene products upregulated in lung and esophageal cancers as novel targets for immunotherapy. Cancer Sci. 98:1803-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tres, L. L., and A. L. Kierszenbaum. 2005. The ADAM-integrin-tetraspanin complex in fetal and postnatal testicular cords. Birth Defects Res. C Embryo Today 75:130-141. [DOI] [PubMed] [Google Scholar]
- 40.Utreras, E., E. M. Jimenez-Mateos, E. Contreras-Vallejos, E. Tortosa, M. Perez, S. Rojas, L. Saragoni, R. B. Maccioni, J. Avila, and C. Gonzalez-Billault. 2008. Microtubule-associated protein 1B interaction with tubulin tyrosine ligase contributes to the control of microtubule tyrosination. Dev. Neurosci. 30:200-210. [DOI] [PubMed] [Google Scholar]
- 41.Viens, A., U. Mechold, F. Brouillard, C. Gilbert, P. Leclerc, and V. Ogryzko. 2006. Analysis of human histone H2AZ deposition in vivo argues against its direct role in epigenetic templating mechanisms. Mol. Cell. Biol. 26:5325-5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vikesaa, J., T. V. Hansen, L. Jonson, R. Borup, U. M. Wewer, J. Christiansen, and F. C. Nielsen. 2006. RNA-binding IMPs promote cell adhesion and invadopodia formation. EMBO J. 25:1456-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wissmann, C., and M. Detmar. 2006. Pathways targeting tumor lymphangiogenesis. Clin. Cancer Res. 12:6865-6868. [DOI] [PubMed] [Google Scholar]
- 44.Yisraeli, J. K. 2005. VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol. Cell 97:87-96. [DOI] [PubMed] [Google Scholar]
- 45.Yoon, S. O., S. Shin, and A. M. Mercurio. 2005. Hypoxia stimulates carcinoma invasion by stabilizing microtubules and promoting the Rab11 trafficking of the alpha6beta4 integrin. Cancer Res. 65:2761-2769. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, Y., K. Chen, L. Guo, and C. Wu. 2002. Characterization of PINCH-2, a new focal adhesion protein that regulates the PINCH-1-ILK interaction, cell spreading, and migration. J. Biol. Chem. 277:38328-38338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.