Abstract
Ras proteins associate with cellular membranes as a consequence of a series of posttranslational modifications of a C-terminal CAAX sequence that include prenylation and are thought to be required for biological activity. In Drosophila melanogaster, Ras1 is required for eye development. We found that Drosophila Ras1 is inefficiently prenylated as a consequence of a lysine in the A1 position of its CAAX sequence such that a significant pool remains soluble in the cytosol. We used mosaic analysis with a repressible cell marker (MARCM) to assess if various Ras1 transgenes could restore photoreceptor fate to eye disc cells that are null for Ras1. Surprisingly, we found that whereas Ras1 with an enhanced efficiency of membrane targeting could not rescue the Ras1 null phenotype, Ras1 that was not at all membrane targeted by virtue of a mutation of the CAAX cysteine was able to fully rescue eye development. In addition, constitutively active Ras112V,C186S not targeted to membranes produced a hypermorphic phenotype and stimulated mitogen-activated protein kinase (MAPK) signaling in S2 cells. We conclude that the membrane association of Drosophila Ras1 is not required for eye development.
Ras proteins regulate numerous cellular processes, including growth and differentiation. Mutations in Ras genes are associated with human cancer more frequently than those of any other oncogene. Ras is the founding member of a class of proteins known as CAAX proteins that are secondarily targeted to cellular membranes as a consequence of the posttranslational processing of a C-terminal CAAX sequence, where C is an invariant cysteine, A is usually, but not always, an aliphatic amino acid, and X is variable (30). The first step of CAAX processing is prenylation, in which a 15-carbon farnesyl or a 20-carbon geranylgeranyl polyisoprene lipid is added to the CAAX cysteine via a stable thioether linkage. Two related prenyl transferases catalyze the addition of the two polyisoprenes, farnesyltransferase (FTase) and geranylgeranyltransferase I (GGTase I). When the X amino acid of the CAAX motif is L (CAAL), the protein is a substrate for GGTase I; otherwise, FTase modifies the protein. Following prenylation the AAX amino acids are removed by Ras-converting enzyme I, and the newly C-terminal prenylcysteine is then methyl esterified by isoprenylcysteine carboxyl methyltransferase. CAAX processing thus converts the C terminus of Ras from a hydrophilic to a hydrophobic domain capable of targeting the protein to cellular membranes.
Mammalian genomes encode three Ras isoforms, N-Ras, H-Ras, and K-Ras. Following CAAX processing the C termini of N-Ras and H-Ras are further modified by the addition of one or two palmitate molecules, respectively, via a labile thioester linkage. The palmitate modifications allow efficient trafficking to the plasma membrane (PM) (3, 7). In K-Ras, a polybasic sequence immediately upstream of the CAAX motif replaces the palmitate modifications and functions by forming an electrostatic interaction with the negatively charged phospholipids of the inner leaflet of the PM (7). In mammalian systems, when the CAAX motif of a Ras protein is mutated such that it cannot be posttranslationally modified, the Ras protein loses all biological activity (29). This observation led to a quest to develop anti-Ras drugs by employing agents designed to inhibit CAAX processing, such as FTase inhibitors (30).
The Ras/mitogen-activated protein kinase (MAPK) pathway is conserved from the fission yeast Schizosaccharomyces pombe to humans, as is CAAX processing. Indeed, as a genetically tractable system, studies of Drosophila melanogaster were critical for elucidating these pathways (28). Among the numerous biological processes for which Ras is required, its role in Drosophila eye development has been most extensively characterized. The adult Drosophila eye consists of a well-ordered array of approximately 800 identical ommatidia, each of which consists of a spatially ordered group of eight photoreceptor, four cone, and eight accessory cells. Ommatidia develop posterior to a morphogenetic furrow in the eye imaginal disc of third-instar larvae. Although Ras is required for the growth and survival of all cells in the developing eye, both anterior and posterior to the morphogenetic furrow, the developing photoreceptors present a physiologically relevant and genetically tractable system wherein Ras function can be assessed. The eight photoreceptors develop in sequence and influence each other via cell-cell interactions that signal through the Ras/MAPK pathway. Loss-of-function studies have revealed that Ras is required for the development of seven of the eight photoreceptors (R1 to R7) (28). Gain-of-function studies of Ras in flies have also been reported where constitutively active Ras112V expressed from a sevenless promoter results in supernumerary R7 cells and a rough-eye phenotype (9, 27).
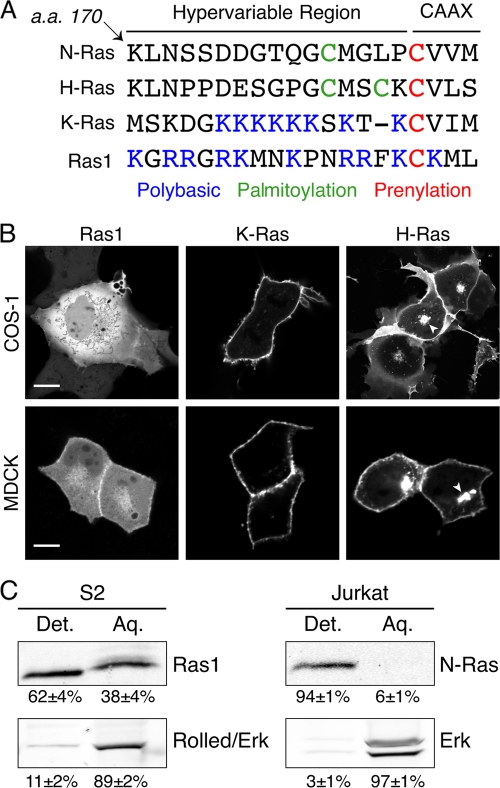
The Drosophila genome encodes one authentic Ras protein, designated Ras1 (also known as Ras85D), which is most similar to mammalian K-Ras in that it lacks palmitoylation but has a polybasic region (Fig. 1A). However, unlike mammalian K-Ras, the CAAX motif of Drosophila Ras1, like that of other insect Ras genes, specifies geranylgeranylation rather than farnesylation (16). In addition, the CAAX sequence of Drosophila Ras1, CKML, is unusual in that it includes a lysine at the A1 position (CA1A2X). Prompted by the differences in the mammalian versus fly CAAX motifs, we investigated the membrane targeting of Drosophila Ras in cultured cells and determined in vivo the requirement for Ras1 membrane targeting with respect to eye development. Surprisingly, we found that endogenous Ras1 is inefficiently prenylated and that non-membrane-targeted Ras1 can support eye development.
FIG. 1.
Drosophila Ras1 is inefficiently targeted to membranes. (A) Sequence alignment of the C terminus of human Ras isoforms and Drosophila Ras1. Features determining posttranslational modification and subcellular localization are shown in color. a.a., amino acids. (B) COS-1 and MDCK cells expressing YFP-Ras1, YFP-K-Ras, or YFP-H-Ras were imaged alive with a confocal microscope. Arrowheads indicate Golgi accumulation of YFP-H-Ras. Scale bars represent 20 μm. (C) Triton X-114 partitions of Drosophila S2 or Jurkat cell lysates were loaded as cell equivalents and immunoblotted with anti-Ras1, anti-N-Ras, or anti-Erk antibodies. Percentages of protein recovered are given as means ± SEM (n = 3). Det., detergent; Aq., aqueous.
MATERIALS AND METHODS
Plasmids.
Ras1 (wild type, Ktail, and Ktail6Q) was cloned into pEYFP-C1 (Clontech) using EcoRI and XhoI ends for expression in COS-1 and MDCK cells. Point mutants (K187V, C186S, and 12V,C186S) were generated by site-directed mutagenesis (QuikChange XL kit; Stratagene). YFP-Ras1 and mutants were then cloned into pActin5C using BamHI and XbaI ends for expression in S2 cells. Ras1 transgenes (Ktail, Ktail6Q, K187V, C186S, and 12V,C186S) were cloned into CS-UAS-6xMyc (a gift from Hyung Don Ryoo) using EcoRI and XhoI ends. Primer sequences are available upon request.
Confocal microscopy.
Confocal images of COS-1, MDCK, or S2 cells expressing the indicated yellow fluorescent protein (YFP) constructs and treated with or without 50 μM GGTI-2418 (gift of Said Sebti) (10) were obtained with a Zeiss 510 laser scanning microscope using a 63×, 1.4-numerical-aperture (NA) objective. Eye imaginal discs were stained with mouse anti-β-Gal (1:50), rat anti-Elav (1:250) (both from the Developmental Hybridoma Studies Bank, University of Iowa), and anti-c-Myc (1:200) (Santa Cruz Biotechnology), followed by fluorescent secondary antibodies (1:200) (Jackson Laboratories). Whole mounted discs were imaged with the Zeiss confocal microscope as a z series using the 63× objective.
Triton X-114 partitioning.
Equivalent numbers of S2 and Jurkat cells were lysed in ice-cold RSB (10 mM Tris [pH 7.4], 10 mM NaCl, 3 mM MgCl2) with 1% Triton X-114 (Sigma). Lysates were partitioned as previously described (6). Samples were loaded as cell equivalents on a 14% SDS-PAGE gel. Protein was immunoblotted with a 1:1,000 dilution of mouse anti-Ras1 (a gift from Marc Therrien), a 1:200 dilution of mouse anti-N-Ras (Santa Cruz Biotechnology), and a 1:5,000 dilution of rabbit anti-Erk1/2 (Santa Cruz Biotechnology) antibodies, followed by infrared-conjugated secondary antibodies (goat anti-mouse IR800 and goat anti-rabbit IR680; Li-Cor) at a 1:20,000 dilution. Membranes were scanned and bands were quantified by using a Li-Cor Odyssey scanner.
Subcellular fractionation.
Equivalent numbers of S2 cells were transfected using the indicated pActin5C-YFP-Ras1 construct using Effectene (Qiagen). Cells were resuspended in fractionation buffer (10 mM HEPES [pH 7.4], 10 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol [DTT]) and Dounce homogenized using 50 passes with a B pestle. Lysates were ultracentrifuged at 100,000 × g. Samples were loaded as cell equivalents on a 14% SDS-PAGE gel and analyzed as described above using a 1:2,000 dilution of rabbit anti-green fluorescent protein (GFP) (Invitrogen).
MAPK analysis.
Equivalent numbers of S2 cells were transfected as described above and then incubated in serum-free Schneider's S2 medium for 24 h. Cells were harvested and lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and protease inhibitor cocktail [Roche]). Lysates were sonicated to release nuclear contents, and sonicates were clarified by centrifugation. Equal amounts of protein were analyzed by SDS-PAGE and immunoblotting as described above with anti-dpErk (Sigma), anti-Erk1 (Santa Cruz), and anti-GFP (Invitrogen) antibodies. Membranes were scanned and bands were quantified by using a Li-Cor Odyssey scanner.
Fly stocks.
Ras85DΔC40B is a null allele that deletes the Ras85D gene (8), referred to in the text as Ras1. UAS-Ras1, encoding wild-type Ras1, was a kind gift of Denise Montell. UAS-Ras112V was a gift of Jessica Treisman. Fly strains used are as follows: transgenic flies were generated by w1118 embryo microinjection (CBRC Transgenic Fly Core, Charlestown, MA, and Best Gene, Inc., Chino Hills, CA) of CS-UAS-6xMyc vectors, expressing the indicated Ras1 transgenes: UAS-Ras1K187V, UAS-Ras1Ktail6Q, UAS-Ras1Ktail, UAS-Ras1C186S, and UAS-Ras112V,C186S. For mosaic analysis with a repressible cell marker (MARCM), we crossed yw ey-FLP UAS-GFP; tub-GAL4 FRT82B tub-GAL80/TM6B Tb females to males of the different UAS-Ras1 transgenes mentioned above and UAS-lacZ in a Ras85DΔC40B null background.
Clonal analysis.
Clones were generated by ey-FLP (17) using the MARCM technique (11). Eye imaginal discs were dissected and processed according to previously described methods (1) during the third larval instar and analyzed for Ras1ΔC40B clones overexpressing the different UAS-Ras1 transgenes.
RESULTS
Drosophila Ras1 is inefficiently prenylated.
The differences between the membrane-targeting regions of mammalian Ras and Drosophila Ras (Fig. 1A) led us to study, in live cells, the subcellular distribution of Ras proteins from the two phyla. We extended the Ras proteins at their N termini with yellow fluorescent protein (YFP) and expressed the fusion proteins in COS-1 fibroblasts and MDCK cells. As we reported previously (3), whereas YFP-K-Ras was observed only at the plasma membrane (PM) in both cell types, YFP-H-Ras was observed on both the PM and Golgi apparatus (Fig. 1B). In contrast, YFP-Ras1 failed to decorate any membrane in COS-1 cells and was instead seen only in the cytosol and nucleoplasm, as evidenced by negatively imaged organelles. The predominant pattern of YFP-Ras1 in MDCK cells was also cytosolic, although some YFP-Ras1 associated with the PM in the region of cell-cell contact. Thus, the membrane targeting of YFP-Ras1 is far less efficient than that of its mammalian orthologs.
To determine if the difference in the efficiency of membrane targeting holds true for endogenous Ras, we compared Triton X-114 partitioning profiles of Ras1 in Drosophila S2 cells with those of N-Ras in human lymphocytes (Fig. 1C). This method allowed the quantification of the prenylated fraction of the proteins. As expected, almost all (94% ± 1%) of N-Ras in Jurkat cells partitioned into the detergent fraction. Erk served as a cytosolic control: 97% ± 1% was recovered in the aqueous fraction. Whereas Drosophila Rolled, the ortholog of Erk, behaved in S2 cells in a manner similar to that of Erk in lymphocytes, the partitioning of Ras1 in S2 cells was dramatically different: 38% ± 4% of Ras1 was recovered in the aqueous fraction. These data indicate that endogenous Ras1 is prenylated less efficiently than its mammalian counterpart, supporting the conclusion reached with YFP-Ras1.
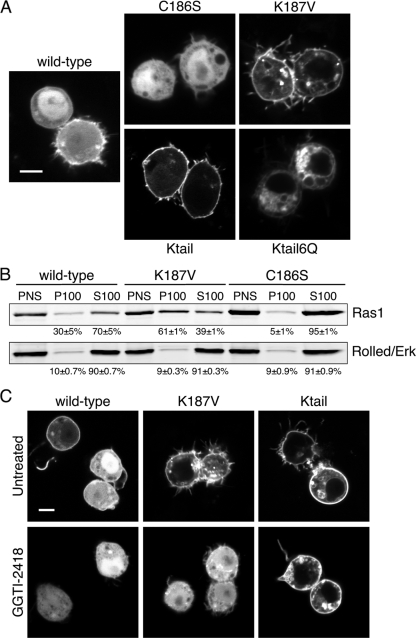
We considered that the localization of YFP-Ras1 described above may be a consequence of an inefficient modification of Drosophila Ras by mammalian GGTase I, and we therefore repeated the analysis with Drosophila S2 cells. We subcloned YFP-Ras1 into an insect expression vector in which an actin promoter drives the expression of the fusion protein (Fig. 2A). As observed for mammalian cells, YFP-Ras1 was expressed predominantly in the cytosol and nucleoplasm, with only a small fraction enriched on the PM. We mutated cysteine 186 to serine to generate a mutant version of YFP-Ras1 that could not be posttranslationally modified. As expected, YFP-Ras1C186S was expressed entirely in the cytosol and nucleoplasm and did not decorate any membrane compartment. This confirmed that the portion of YFP-Ras1 seen on the S2 cell PM was dependent on prenylation and suggested that a large pool of YFP-Ras1 behaved like YFP-Ras1C186S, consistent with a lack of prenylation. Thus, the cytosolic localization of YFP-Ras1 in mammalian cells was not a consequence of an incompatible, xenotypic GGTase I.
FIG. 2.
Subcellular localization of wild-type Ras1 and membrane-targeting mutants of Ras1 in Drosophila cells. (A) S2 cells expressing the YFP-Ras1 wild type or the indicated mutants were imaged alive with a confocal microscope. The scale bar represents 10 μm. (B) S2 cells expressing the indicated YFP-tagged Ras1 constructs were separated into membrane (P100) and cytosolic (S100) fractions by ultracentrifugation. Fractions were loaded as cell equivalents and immunoblotted with anti-GFP or anti-Erk antibodies. Percentages of protein recovered are given as means ± SEM (n = 3). (C) S2 cells expressing the indicated YFP-tagged Ras1 protein were treated with or without GGTI-2418 (50 μM for 16 h) and imaged as described above for panel A.
To determine if the membrane targeting of Ras1 could be made more efficient in Drosophila cells, we substituted the 19-amino-acid hypervariable region of human K-Ras for the analogous C-terminal amino acids of YFP-Ras1 to generate YFP-Ras1Ktail. This construct was targeted efficiently to the PM of S2 cells (Fig. 2A), demonstrating that the human sequence is a good substrate for Drosophila FTase and that Ras1 is capable of a strong membrane association when given an efficient targeting sequence. In mammalian cells, the transport of K-Ras from the endomembrane to the PM depends on a polylysine motif immediately upstream of the CAAX sequence such that the substitution of six of the lysines for glutamines (6Q) results in a K-Ras construct that associates only with the endomembrane (3). We replaced the lysines in YFP-Ras1Ktail to generate YFP-Ras1Ktail6Q and found that, like its mammalian homolog, it was restricted to the endomembrane (Fig. 2A). Thus, the trafficking of K-Ras homologs in Drosophila cells is similar to that seen for mammalian cells.
Whereas both the A1 and A2 positions of most CAAX sequences are occupied by aliphatic amino acids, the A1 position of Ras1 is occupied by lysine. Peptides with lysine in the A1 position have been shown to be relatively poor substrates for prenyltransferases (20). To determine if the lysine in the Ras1 A1 position impedes geranylgeranylation, we substituted valine for lysine in YFP-Ras1 to yield YFP-Ras1K187V. This construct was targeted efficiently to the PM and vesicles of S2 cells with clearing from the cytosol almost as complete as that seen with YFP-Ras1Ktail (Fig. 2A), suggesting that lysine 187 of Ras1 indeed inhibited modification by GGTase I. We confirmed this result with subcellular fractionation (Fig. 2B). S2 cells expressing YFP-Ras1, YFP-Ras1K187V, or YFP-Ras1C186S were disrupted by Dounce homogenization, and the postnuclear supernatants (PNS) were separated into membrane (P100) and cytosol (S100) fractions. Ninety percent of the Rolled protein was recovered in the S100 of each PNS, confirming the efficiency of the fractionation. Seventy percent of YFP-Ras1 was recovered in the S100 fraction, and this value increased to 95% ± 1% for YFP-Ras1C186S. In contrast, only 39% ± 1% of YFP-Ras1K187V was recovered in the S100 fraction, confirming that the change of the CAAX motif from CKML to CVML increased the efficiency of membrane targeting.
To confirm that Ras1 is a substrate for GGTase I and not FTase, we used a specific GGTase I inhibitor, GGTI-2418 (10). The treatment of S2 cells expressing YFP-Ras1 with GGTI-2418 eliminated all expression of the fusion protein on the plasma membrane (Fig. 2C). The same result was obtained with YFP-Ras1K187V, demonstrating that the increased efficiency of prenylation observed when the A1 lysine is converted to valine reflects increased geranylgeranylation. In contrast, YFP-Ras1Ktail was unaffected by GGTI-2418, confirming that it is a preferred substrate for Drosophila FTase. Thus, as predicted from the CAAL motif and as shown for silkworm Ras (16), Drosophila Ras1, when prenylated, is modified only by GGTase I. Moreover, our results suggest that even in the presence of a GGTase I inhibitor, alternative prenylation by FTase does not occur.
Cytosolic Ras1 rescues Drosophila eye development.
We next sought to examine the requirement for the membrane association of Drosophila Ras1 in vivo by studying eye development in larvae. We used mosaic analysis with a repressible cell marker (MARCM) (11). This exceedingly sensitive and informative approach allowed us to study the function of a variety of Ras mutants in tissue that is otherwise null for Ras1. We generated a series of transgenic flies that express Ras1 or various Myc-tagged mutations thereof under the control of a UAS promoter and crossed these with flies that are heterozygous for Ras1ΔC40B, a null allele distal to an FRT site (FRT82B). We then crossed these flies with MARCM lines that express FLP recombinase from an eyeless (ey) promoter such that mitotic recombination at FRT sites is induced in developing eye tissue. These flies also express Gal4 and its dominant repressor, Gal80, from tubulin promoters as well as GFP from a UAS promoter. If FLP is not expressed, Gal80 represses Gal4, and no UAS-linked transgenes are expressed. However, when FRT82B gal80 is placed in trans onto the FRT82B Ras1ΔC40B chromosome and a source of FLP is available, mitotic recombination is induced, and clones are generated that are homozygous for either gal80 or Ras1ΔC40B. Clones lacking gal80 are therefore null for Ras1, and Gal4 is unopposed such that both our Ras transgene and GFP are expressed.
To establish that this technique could be used to rescue the Ras1 null eye phenotype, we studied a UAS-Ras1 transgenic line along with a UAS-lacZ line as a negative control. Eye discs from UAS-lacZ larvae showed clonal defects in ommatidium development, as evidenced by gaps and partially formed rosettes within the ommatidial array. In contrast, no defects were observed for clones that expressed UAS-Ras1 (Fig. 3A). Thus, as expected, the absence of Ras1 (Ras1ΔC40B) resulted in a clear phenotype in the eye disc that could be rescued with wild-type Ras1.
FIG. 3.
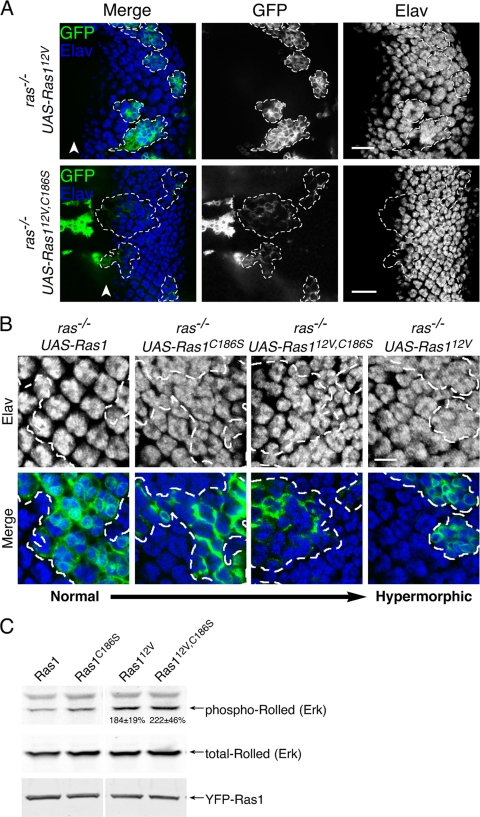
Cytosolic Ras1 rescues Ras1ΔC40B null clones in the eye imaginal disc. Confocal images of eye imaginal discs of third-instar larvae expressing LacZ or wild-type Ras1 (A) or the indicated Myc-tagged Ras1 constructs (B) in Ras1ΔC40B clones. Clones were generated by the MARCM technique and are marked by GFP (green). Dashed lines indicate the clone boundaries, which are GFP positive. Elav staining (blue) marks neuronal cells (photoreceptors). The presence of morphologically ordered, Elav-positive cells within the Ras1 clones indicates rescue. Equivalent expression of each Ras1 mutant (panel B only) is indicated by Myc staining (red). Arrowheads indicate the morphogenetic furrow. Images are representative of at least 30 clones analyzed per genotype. Scale bars represent 20 μm.
We next examined eye imaginal discs from larvae transgenic for Ras1 with an altered membrane-targeting region. We increased the efficiency of membrane targeting in two ways, both validated by the cell analysis described above: we generated a chimera with the human K-Ras-targeting sequence (UAS-Ras1Ktail), and we made a single-amino-acid substitution in Ras1 to promote efficient prenylation (UAS-Ras1K187V). Surprisingly, when analyzed by the MARCM technique, both of these membrane-targeted Ras1 alleles were, at best, able to only partially rescue the Ras1 null phenotype (Fig. 3Bi and ii). This result suggests that inefficient membrane targeting of Ras1 is required for full biological activity. Because we have shown previously that mammalian (2) and yeast (18) Ras can signal from the endomembrane, we also tested UAS-Ras1Ktail6Q, which expresses Ras1 targeted to the endoplasmic reticulum. This allele also provided only a partial rescue of ommatidium development (Fig. 3Biii). Most surprising was our result with UAS-Ras1C186S, an allele that encodes a Ras protein that is not prenylated and has no affinity for any membrane. Ras1C186S was capable of a full rescue of the Ras1 null phenotype (Fig. 3Biv). Indeed, Ras1ΔC40B MARCM clones that expressed Ras1C186S often showed an enhanced recruitment of supernumerary photoreceptor cells, suggesting that Ras1C186S is a hyperactive allele. This result suggests that the membrane targeting of Ras1 is not required for its biological activity in eye development.
To determine if constitutively active Ras1, previously shown to disrupt normal eye development through excess proliferation (9, 27), also lacked the requirement for a membrane association, we used transgenic flies with a UAS-Ras112V allele and also generated a UAS-Ras112V allele in which the CAAX sequence was nullified (UAS-Ras112V,C186S). As expected, MARCM clones null for endogenous Ras1 but instead expressing Ras112V showed a clear hyperrecruitment of photoreceptors with the attendant distortion of developing ommatidia (Fig. 4A). UAS-Ras112V,C186S, an activated allele that cannot be prenylated, efficiently rescued Ras1 null clones and showed signs of an enhanced recruitment of photoreceptors versus UAS-Ras1 and UAS-Ras1C186S although not as dramatic as was seen for UAS-Ras112V (Fig. 4B). These data reveal that untargeted, constitutively active Ras1 can support Drosophila eye development and produce a mild hypermorphic phenotype.
FIG. 4.
Constitutively active, cytosolic Ras1 produces a hypermorphic phenotype in eye imaginal discs and activates MAPK. (A) Confocal images of eye imaginal discs of third-instar larvae expressing Ras112V or Ras112V,C186S in Ras1ΔC40B null clones. Clones were generated by the MARCM technique and are marked by GFP. Dashed lines indicate clone boundaries. Elav staining (blue channel) for neuronal cells (photoreceptors) within the marked regions demonstrates that both Ras112V and Ras112V,C186S induce supernumerary photoreceptors. Arrowheads indicate the morphogenetic furrow. Images are representative of at least 30 clones analyzed per genotype. Scale bars represent 20 μm. (B) Digitally zoomed images of Elav-stained eye imaginal discs expressing Ras1, Ras1C186S, Ras112V,C186S, and Ras112V in Ras1ΔC40B null clones revealing supernumerary photoreceptors. The dashed lines indicate clone boundaries. The scale bar represents 10 μm. (C) Lysates of S2 cells expressing the indicated Ras1 allele were analyzed by immunoblotting for total and phospho-Rolled (Erk) as well as for YFP (Ras1 expression). The numbers superimposed onto the blots indicate the increase in phosphorylated/total Rolled induced by the 12V mutation, expressed as a percentage of the wild type (means ± SEM [n = 4]).
In mammalian cells, constitutively active Ras proteins without CAAX motifs not only are nontransforming but also can behave as dominant interfering proteins, presumably by sequestering Raf-1 in the cytosol, where its kinase activity cannot be enhanced (4). Our results suggest that this is not the case for insect cells, since ommatidium development requires MAPK signaling, and UAS-Ras112V,C186S led to a gain of function rather than a loss of function. To further substantiate this surprising result, we determined whether soluble, GTP-bound Ras1 could stimulate Rolled phosphorylation in S2 cells. As expected, YFP-Ras112V expression resulted in more phospho-Rolled than did the expression of YFP-Ras1 (Fig. 4C). Surprisingly, GTPase-deficient YFP-Ras112V,C186S was also more potent in stimulating Rolled phosphorylation than was its GTPase-competent counterpart, YFP-Ras1C186S, demonstrating that even in the non-membrane-targeted form, the 12V mutation is activating with regard to MAPK signaling. This suggests that in insect cells, the MAPK pathway can be driven by soluble Ras.
DISCUSSION
Our data reveal that Drosophila Ras1 is inefficiently prenylated, creating a significant pool of soluble, cytosolic protein, and that a constitutively soluble form of Ras1 is sufficient to support the development of photoreceptors. Moreover, Ras1 stringently targeted to the PM with a more efficient CAAX sequence or a mammalian K-Ras-targeting sequence cannot fully rescue a Ras1 deficiency. Thus, contrary to the current dogma, our results suggest that, at least in the context of Drosophila eye development, soluble Ras is biologically active.
Whereas at least one Ras isoform in budding and fission yeasts, worms, and vertebrates is farnesylated, as are all mammalian Ras proteins, insect genomes encode but one Ras protein that is geranylgeranylated (16). This evolutionary curiosity raises the question, What are the functional differences between a farnesyl versus a geranylgeranyl modification? The fact that two distinct prenyltransferases evolved in primitive metazoans and were conserved throughout evolution strongly suggests important biological differences between 15- and 20-carbon polyisoprene protein modifications.
One obvious difference between prenyl modifications is the affinity for membranes: a geranylgeranyl modification affords a higher degree of affinity (24). Farnesylated mammalian Ras proteins can be removed relatively easily from membranes. Studies of fluorescence recovery after photobleaching reveal that Ras recovers from a bleached region of PM not only by lateral diffusion but also via release from adjacent membranes and reassociation in the bleach zone (22). Indeed, depalmitoylated N-Ras and H-Ras are readily released from the PM and traffic, in a retrograde fashion, through the cytosol back to the Golgi apparatus, where they are again palmitoylated, creating a cycle (5, 21). K-Ras also translocates through the cytosol from organelle to organelle (23). In contrast, mammalian geranylgeranylated Ras-related proteins, such as Rho family GTPases and Rap1, are never found soluble in the cytosol except in a complex with a chaperone like RhoGDI (14, 15). Moreover, whereas AAX proteolysis and carboxyl methylation are required for the proper localization of farnesylated Ras proteins, geranylgeranylation is sufficient for the proper localization of Rho GTPases (13). Heterotrimeric G proteins perhaps best illustrate the functional differences between farnesylation and geranylgeranylation. Nine of the 12 human G protein γ subunits are geranylgeranylated. The transducin γ subunit is farnesylated, and this allows the G protein to readily dissociate from the membranes of the outer segment of photoreceptor cells (12), a process that is critical for desensitization to light (25).
Thus, it appears that the weaker membrane affinity afforded by farnesylation is important for Ras function and, with the exception of insects, has been conserved. It is therefore an attractive hypothesis that the inefficiency of CAAL processing in insects compensates for the more hydrophobic geranylgeranyl modification. Peptide inhibitor studies suggest that CAAX peptides with a lysine in the A1 position are modified by FTase only 10% as efficiently as those with aliphatic residues (20). Our observation that YFP-Ras1K187V is targeted more efficiently to membranes than the wild-type protein is consistent with this result and suggests that the lysine at the A1 position of the Drosophila CAAL motif is responsible for at least some of the inefficiency. Interestingly, whereas the mosquito Aedes aegypti has an arginine in the A1 position of its Ras CAAL sequence (16), this residue does not appear in the A1 position of the CAAX sequence of any human GTPase (19). However, a charged residue in the A1 position is not universal among insects: the CAAL motifs of the silkworm Bombyx mori and the beetle Tribolium castaneum have threonine in this position (16). Perhaps these species employ other means of rendering CAAL processing inefficient to maintain a soluble pool of Ras. Whether or not inefficient CAAL processing is universal among insects, it should be noted that insect Ras partially processed by geranylgeranylation is not equivalent to farnesylated mammalian Ras in that whereas the latter comes on and off membranes with the irreversible farnesyl modification intact, the former is soluble before processing and membrane associated afterward, and the two forms are interconverted in only one direction. Thus, it may be that unprocessed insect Ras performs a function that in other organisms is performed by a cytosolic pool of farnesylated Ras.
Two studies have suggested that the prenylation of Drosophila Ras1 is required for the gain-of-function phenotype of Ras112V in the fly eye. Therrien et al. previously performed a screen for suppressors of the Ras112V rough-eye phenotype and identified a mutation in the β-subunit of GGTase I as one such suppressor, suggesting that geranylgeranylation is required for the gain-of-function phenotype and that the loss of one GGTase I allele was sufficient to revert the rough-eye phenotype (27). In the second study, Kauffmann et al. showed that a GGTase I inhibitor injected into whole larvae could block the Ras112V rough-eye phenotype (9). However, what is striking about these studies is that neither a reduction in the genetic dose of GGTase I nor the pharmacologic inhibition of GGTase I affected normal eye development driven by endogenous Ras1. Our observation that, even in the presence of a GGTase I inhibitor, Drosophila Ras1 cannot be alternately prenylated by FTase makes the results of these two studies even more compelling. The studies suggest that whereas the GTP-bound, constitutively active Ras112V requires geranylgeranylation to produce a rough-eye phenotype, endogenous Ras1 supports normal eye development in the setting of a GGTase I deficiency. Our results using the MARCM technique are consistent with this observation in that whereas wild-type but unprocessed Ras1C186S fully rescued the Ras1 null phenotype, untargeted, constitutively active Ras112V,C186S was a weaker allele than was Ras112V in producing hypermorphic ommatidia. This suggests that whereas at least one of the pathways downstream of Ras1 required to produce a rough-eye phenotype are dependent on membrane-associated Ras1, those required for normal eye development do not require the membrane targeting of Ras1.
In mammalian systems the membrane association of Ras is believed to be required both for productive interactions with upstream guanine nucleotide exchange factors (GEFs) and for interactions with downstream effectors. Although mammalian Ras can be activated by any of several GEFs, and Ras signals through more than a dozen effectors, those comprising the Ras/MAPK pathway are best understood. In this pathway the GEF is SOS, which is brought to membranes via its PH domain and by virtue of its association with Grb2 bound to phosphotyrosines on activated receptors. Looking downstream, activated Ras recruits the MAPK kinase kinase (MAPKKK) Raf-1 to the PM, where it is activated through a poorly understood and complex process. Importantly, GTP-bound Ras binds to Raf-1 in solution but does not stimulate its kinase activity (26). It is the Ras/Raf-1/Erk pathway that is required for Drosophila eye development (28). Thus, there is a discrepancy between our current understanding of the molecular events of Ras/MAPK signaling and our unambiguous finding that nonprocessed Ras1 can rescue the Ras1 null phenotype in the developing Drosophila eye. Our observation that the canonical V12 activating mutation of Ras is a gain-of-function mutation with regard to MAPK signaling in both natively processed Ras1 and nonprocessed Ras1C186S argues that the membrane requirement for Raf-1 activation observed for mammalian cells does not apply to Draf (also known as Pole Hole) in insect cells. We conclude that, at least in the context of Drosophila eye development, soluble Ras1 is biologically active.
The question of whether Ras proteins must associate with membranes for biological activity is an important one with regard to anti-Ras drug development, because interfering with the membrane trafficking of Ras remains the most promising approach. While our results with Drosophila Ras1 do not condemn this approach to failure, they do suggest that soluble Ras may not be devoid of biological activity.
Acknowledgments
We thank Denise Montell, Jessica Treisman, Marc Therrien, the Bloomington Drosophila Stock Center, and the Developmental Hybridoma Studies Bank for flies and antibodies. We also acknowledge FlyBase for critical information. We thank Laura Ekas and Emily Olson for technical help.
This work was supported by NIH grants GM055279, CA116034, and CA118495 to M.R.P. and GM085075-01A1 to E.A.B. as well as a Basil O'Connor Starter Scholar research award (grant no. 5-FY06) from the March of Dimes Foundation and the Hirschl Charitable Trust (to E.A.B.).
Footnotes
Published ahead of print on 11 October 2010.
REFERENCES
- 1.Bach, E. A., S. Vincent, M. P. Zeidler, and N. Perrimon. 2003. A sensitized genetic screen to identify novel regulators and components of the Drosophila Janus kinase/signal transducer and activator of transcription pathway. Genetics 165:1149-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiu, V. K., T. Bivona, A. Hach, J. B. Sajous, J. Silletti, H. Wiener, R. L. Johnson, A. D. Cox, and M. R. Philips. 2002. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 4:343-350. [DOI] [PubMed] [Google Scholar]
- 3.Choy, E., V. K. Chiu, J. Silletti, M. Feoktistov, T. Morimoto, D. Michaelson, I. E. Ivanov, and M. R. Philips. 1999. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98:69-80. [DOI] [PubMed] [Google Scholar]
- 4.Fiordalisi, J. J., S. P. Holly, R. L. Johnson II, L. V. Parise, and A. D. Cox. 2002. A distinct class of dominant negative Ras mutants: cytosolic GTP-bound Ras effector domain mutants that inhibit Ras signaling and transformation and enhance cell adhesion. J. Biol. Chem. 277:10813-10823. [DOI] [PubMed] [Google Scholar]
- 5.Goodwin, J. S., K. R. Drake, C. Rogers, L. Wright, J. Lippincott-Schwartz, M. R. Philips, and A. K. Kenworthy. 2005. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J. Cell Biol. 170:261-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hancock, J. F. 1995. Prenylation and palmitoylation analysis. Methods Enzymol. 255:237-245. [DOI] [PubMed] [Google Scholar]
- 7.Hancock, J. F., H. Paterson, and C. J. Marshall. 1990. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63:133-139. [DOI] [PubMed] [Google Scholar]
- 8.Hou, X. S., T. B. Chou, M. B. Melnick, and N. Perrimon. 1995. The torso receptor tyrosine kinase can activate Raf in a Ras-independent pathway. Cell 81:63-71. [DOI] [PubMed] [Google Scholar]
- 9.Kauffmann, R. C., Y. Qian, A. Vogt, S. M. Sebti, A. D. Hamilton, and R. W. Carthew. 1995. Activated Drosophila Ras1 is selectively suppressed by isoprenyl transferase inhibitors. Proc. Natl. Acad. Sci. U. S. A. 92:10919-10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kazi, A., A. Carie, M. A. Blaskovich, C. Bucher, V. Thai, S. Moulder, H. Peng, D. Carrico, E. Pusateri, W. J. Pledger, N. Berndt, A. Hamilton, and S. M. Sebti. 2009. Blockade of protein geranylgeranylation inhibits Cdk2-dependent p27Kip1 phosphorylation on Thr187 and accumulates p27Kip1 in the nucleus: implications for breast cancer therapy. Mol. Cell. Biol. 29:2254-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, T., and L. Luo. 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22:451-461. [DOI] [PubMed] [Google Scholar]
- 12.Michaelson, D., I. Ahearn, M. Bergo, S. Young, and M. Philips. 2002. Membrane trafficking of heterotrimeric G proteins via the endoplasmic reticulum and Golgi. Mol. Biol. Cell 13:3294-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michaelson, D., W. Ali, V. K. Chiu, M. Bergo, J. Silletti, L. Wright, S. G. Young, and M. Philips. 2005. Postprenylation CAAX processing is required for proper localization of Ras but not Rho GTPases. Mol. Biol. Cell 16:1606-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michaelson, D., J. Silletti, G. Murphy, P. D'Eustachio, M. Rush, and M. R. Philips. 2001. Differential localization of Rho GTPases in live cells. Regulation by hypervariable regions and rhogdi binding. J. Cell Biol. 152:111-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mor, A., J. Wynne, I. M. Ahearn, M. Dustin, G. Du, and M. R. Philips. 2009. Phospholipase D1 regulates lymphocyte adhesion via upregulation of Rap1 at the plasma membrane. Mol. Cell. Biol. 29:3297-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moriya, K., T. Tsubota, N. Ishibashi, A. Yafune, T. Suzuki, J. Kobayashi, T. Shiotsuki, and T. Utsumi. 2009. Bombyx mori Ras proteins BmRas1, BmRas2 and BmRas3 are neither farnesylated nor palmitoylated but are geranylgeranylated. Insect Mol. Biol. 19:291-301. [DOI] [PubMed] [Google Scholar]
- 17.Newsome, T. P., B. Asling, and B. J. Dickson. 2000. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127:851-860. [DOI] [PubMed] [Google Scholar]
- 18.Onken, B., H. Wiener, M. Philips, and E. C. Chang. 2006. Compartmentalized signaling of Ras in fission yeast. Proc. Natl. Acad. Sci. U. S. A. 103:9045-9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid, T. S., K. L. Terry, P. J. Casey, and L. S. Beese. 2004. Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity. J. Mol. Biol. 343:417-433. [DOI] [PubMed] [Google Scholar]
- 20.Reiss, Y., S. J. Stradley, L. M. Gierasch, M. S. Brown, and J. L. Goldstein. 1991. Sequence requirement for peptide recognition by rat brain p21ras protein farnesyltransferase. Proc. Natl. Acad. Sci. U. S. A. 88:732-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rocks, O., A. Peyker, M. Kahms, P. J. Verveer, C. Koerner, M. Lumbierres, J. Kuhlmann, H. Waldmann, A. Wittinghofer, and P. I. Bastiaens. 2005. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307:1746-1752. [DOI] [PubMed] [Google Scholar]
- 22.Rotblat, B., I. A. Prior, C. Muncke, R. G. Parton, Y. Kloog, Y. I. Henis, and J. F. Hancock. 2004. Three separable domains regulate GTP-dependent association of H-ras with the plasma membrane. Mol. Cell. Biol. 24:6799-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silvius, J. R., P. Bhagatji, R. Leventis, and D. Terrone. 2006. K-ras4B and prenylated proteins lacking “second signals” associate dynamically with cellular membranes. Mol. Biol. Cell 17:192-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silvius, J. R., and F. l'Heureux. 1994. Fluorimetric evaluation of the affinities of isoprenylated peptides for lipid bilayers. Biochemistry 33:3014-3022. [DOI] [PubMed] [Google Scholar]
- 25.Sokolov, M., A. L. Lyubarsky, K. J. Strissel, A. B. Savchenko, V. I. Govardovskii, E. N. Pugh, Jr., and V. Y. Arshavsky. 2002. Massive light-driven translocation of transducin between the two major compartments of rod cells: a novel mechanism of light adaptation. Neuron 34:95-106. [DOI] [PubMed] [Google Scholar]
- 26.Stokoe, D., S. G. Macdonald, K. Cadwallader, M. Symons, and J. F. Hancock. 1994. Activation of raf as a result of recruitment to the plasma membrane. Science 264:1463-1467. [DOI] [PubMed] [Google Scholar]
- 27.Therrien, M., H. C. Chang, N. M. Solomon, F. D. Karim, D. A. Wassarman, and G. M. Rubin. 1995. KSR, a novel protein kinase required for RAS signal transduction. Cell 83:879-888. [DOI] [PubMed] [Google Scholar]
- 28.Wassarman, D. A., M. Therrien, and G. M. Rubin. 1995. The Ras signaling pathway in Drosophila. Curr. Opin. Genet. Dev. 5:44-50. [DOI] [PubMed] [Google Scholar]
- 29.Willumsen, B. M., A. D. Cox, P. A. Solski, C. J. Der, and J. E. Buss. 1996. Novel determinants of H-Ras plasma membrane localization and transformation. Oncogene 13:1901-1909. [PubMed] [Google Scholar]
- 30.Wright, L. P., and M. R. Philips. 2006. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J. Lipid Res. 47:883-891. [DOI] [PubMed] [Google Scholar]