Abstract
Protein splicing is a naturally-occurring process in which a protein editor, called an intein, performs a molecular disappearing act by cutting itself out of a host protein in a traceless manner. In the two decades since its discovery, protein splicing has been harnessed for the development of several protein-engineering methods. Collectively, these technologies help bridge the fields of chemistry and biology, allowing hitherto impossible manipulations of protein covalent structure. These tools and their application are the subject of this Primer.
Introduction
Molecular biologists have developed powerful methods to study the details of protein function. Approaches such as X-ray crystallography and site-directed mutagenesis have furnished countless insights, highlighting how even the most byzantine of problems can yield to the right tools. Nonetheless, there is always demand for more tools. This is perhaps best illustrated by considering protein post-translational modifications (PTMs). Most, if not all, proteins are modified at some point; it is nature‘s way of imposing functional diversity on a single polypeptide chain (Walsh et al., 2005). Moreover, many proteins are modified in manifold ways as exemplified by the histones, where dozens of discrete PTMs have been identified. Existing tools based on site-directed mutagenesis offer limited opportunities for determining what all these PTMs are doing. Although it is straightforward to mutate a protein in such a way as to prevent a PTM from being installed, the reverse strategy whereby a mutation is introduced that mimics a PTM is a haphazard business at best. To fill this and other voids, protein chemists have come up with an array of approaches for the introduction of countless chemical modifications into proteins, including all of the major types of PTM.
The chemical modification of proteins can be accomplished through a variety of means, including bioconjugation techniques (Hermanson, 2008), total chemical synthesis (Kent, 2009), enzyme mediated reactions (Lin and Wang, 2008), nonsense suppression mutagenesis (Wang et al., 2006), and a variety of protein ligation methods (Hackenberger and Schwarzer, 2008). The latter group of strategies include the protein semi-synthesis methods (defined herein as a protein manufactured from pre-made fragments) expressed protein ligation (EPL) and protein trans-splicing (PTS) (Muir, 2003; Muralidharan and Muir, 2006; Mootz, 2009). These are unique technologies in that they combine the power of biotechnology, which provides accessibility to significant amounts of large proteins, with the versatility of chemical synthesis, which allows the site-specific incorporation of almost any chemical modification into the target protein. In the following sections we provide an overview of EPL and PTS and illustrate how these technologies have been used to tackle problems in molecular biology that have proven refractory to other methods.
Expressed Protein Ligation
Expressed protein ligation (EPL) allows a recombinant protein and a synthetic peptide to be linked together under mild aqueous conditions (Muir et al., 1998; Evans et al., 1998). The process involves a chemo-selective reaction that yields a final protein product with a native peptide bond between its two building blocks. The synthetic nature of one of the fragments enables the site-specific introduction of almost any chemical modification in the protein of interest, including fluorophores, caging groups, crosslinkers, PTMs and their analogues, as well as almost any imaginable combination of modifications. At the same time, the recombinant nature of the other fragment conveniently gives access to large proteins, thereby overcoming the size restriction associated with total chemical synthesis.
EPL is based on the well-known reaction between a polypeptide bearing a C-terminal thioester (α-thioester) and a peptide possessing an N-terminal cysteine residue. This reaction, termed native chemical ligation (NCL), originated in the field of peptide chemistry and has proven extraordinarily powerful for the total synthesis of small proteins and their analogues (Kent, 2009). However, the generation of large proteins using total synthesis is still a daunting task for the non-specialist, largely due to the technical issues associated with performing the multiple ligation reactions needed to access polypeptides greater than ~100 amino acids. One solution to this size problem is to employ recombinant polypeptide building blocks in the process; indeed, this semi-synthetic NCL approach was demonstrated early on by using a recombinant protein fragment containing an N-terminal cysteine (Erlanson et al., 1996). Nonetheless, the full integration of NCL and semi-synthesis awaited the development of a general approach to install an α-thioester moiety into recombinantly-derived proteins. The solution to this problem came from the discovery of a most unusual PTM, termed protein splicing (Paulus, 2000).
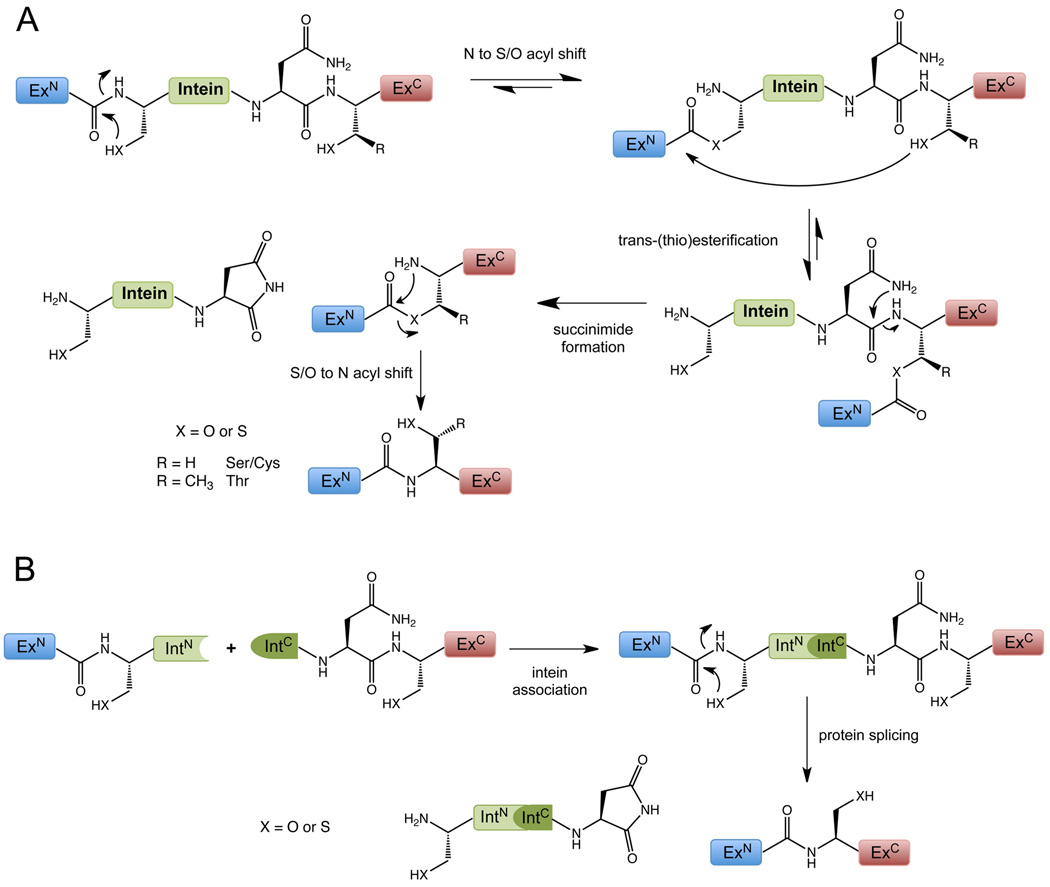
Protein splicing is an autocatalytic process in which an intervening protein domain (intein) excises itself from the polypeptide in which it is embedded, concomitantly creating a new peptide bond between its two flanking regions (exteins). In a sense, intein-mediated protein splicing is the protein equivalent of RNA splicing involving self-splicing introns. Several hundred inteins have been identified in unicellular organisms from all three phylogenetic domains, all share conserved sequence motifs and are derived from a common precursor (for a complete listing see; www.neb.com/neb/inteins.html). Thus, protein splicing is presumed to have an ancient evolutionary origin. Parenthetically, although intein-mediated protein splicing is not known to occur in multicellular organisms, protein automodification processes do occur that involve intein-like domains, most notably the hedgehog-like proteins that are essential for embryonic development (Paulus, 2000). A biological role for protein splicing in unicellular organisms has proven elusive; modern inteins seem to be parasitic genetic elements that are inserted into the open reading frames of (usually) essential genes. This frustration aside, the process has found a multitude of applications in biotechnology (Noren et al., 2000) and quickly attracted the interest of the peptide chemistry community, as α-thioesters were identified as crucial intermediates in the reaction mechanism (Figure 1). Several engineered inteins have been developed that allow access to recombinant protein α-thioester derivatives by thiolysis of the corresponding C-terminal intein fusions (Figure 2). Moreover, inteins have also been engineered to allow the introduction of an N-terminal cysteine (Cys) moiety into recombinant proteins. Simple access to reactive proteins without any size restriction, through molecular biology techniques suddenly enabled the application of NCL to the modification of a much larger fraction of the proteome. Indeed, the approach has been used to generate semi-synthetic derivatives of members of essentially every major class of protein including antibodies, integral membrane proteins, cytoplasmic signaling proteins, metabolic enzymes, and transcription factors (Muir, 2003; Muralidharan and Muir, 2006).
Figure 1. Mechanism of Protein Splicing.
Protein splicing (A) and its variant protein trans-splicing (B).
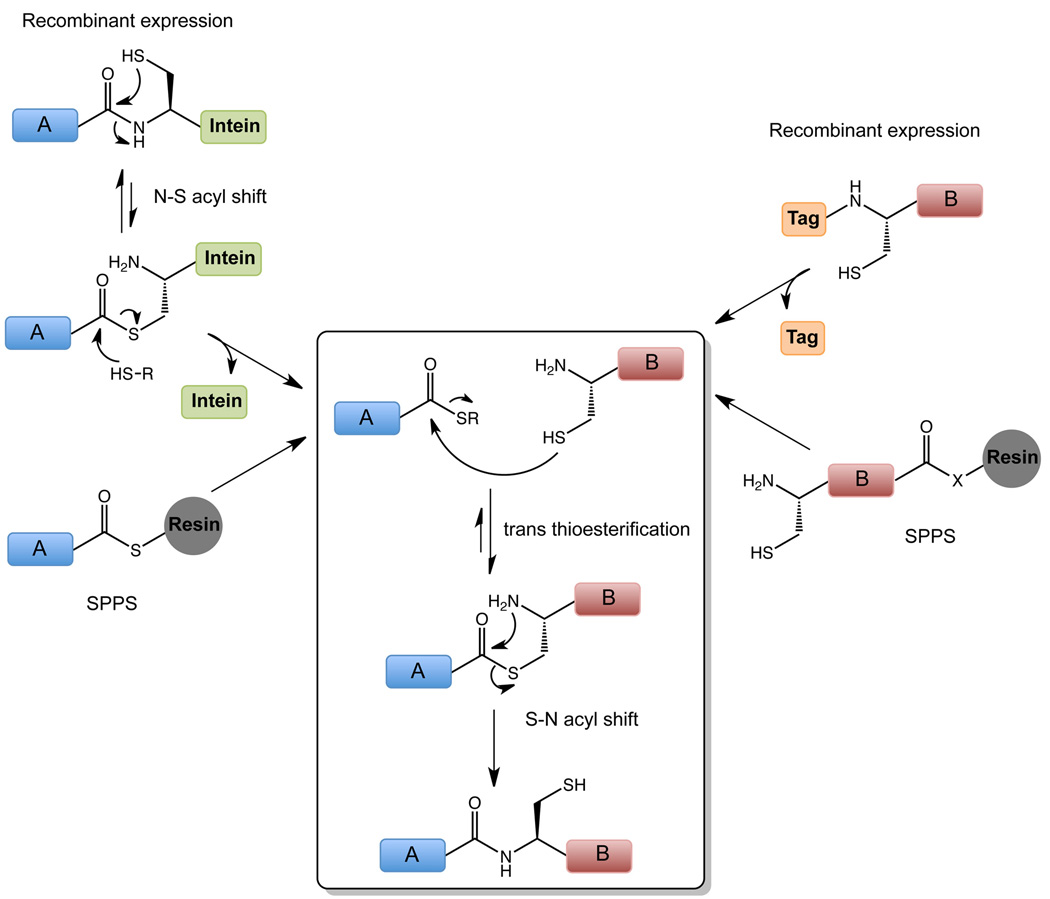
Figure 2. Expressed Protein Ligation.
The boxed region designated the native chemical ligation (NCL) reaction in which transthioesterification of the protein α-thioester by the N-terminal Cys polypeptide is followed by an S- to N-acyl shift to generate a new peptide bond linking the two polypeptides. α-thioesters can be obtained recombinantly, using engineered inteins, or by chemical synthesis. N-terminal Cys polypeptides can also be produced recombinantly or made using standard solid-phase peptide synthesis (SPPS) methods.
Protein Trans-Splicing
A technology related to EPL, also based on the use of inteins, is protein trans-splicing (PTS, Figure 1). In PTS, artificially or naturally split inteins are used to create a new peptide bond between their flanking exteins. Split inteins are characterized by the fact that their primary sequence is cut into two polypeptides giving an N-terminal fragment (IntN) and a C-terminal fragment (IntC). Fragment complementation leads to reconstitution of the canonical intein fold, recovery of protein splicing activity and ligation of the exteins. Importantly, several split-inteins have been described in which one of the two fragments is small enough to be obtained by peptide synthesis thus allowing splicing reactions to be performed between a recombinant fragment and a synthetic one (Table 1) (Mootz, 2009). This allows the generation of a semi-synthetic protein derivative upon PTS. Use of these auto-processing domains to carry out the ligation reaction precludes the need to isolate α-thioesters or N-terminal Cys peptides or proteins and, because the IntN and IntC fragments often have high affinity for one another, the reaction can be carried out at very low concentrations (low micromolar) under native conditions. This should be contrasted with EPL, which being a bimolecular process usually requires high concentrations of reactants (ideally high micromolar range) to be efficient.
Table 1.
Split-inteins commonly used for PTSa
| Sizeb | Half- life c (min) |
Comments | ||
|---|---|---|---|---|
| IntN | IntC | |||
| Naturally split inteins | ||||
| Ssp DnaE | 123 | 36 | 35–175 | One of the better studied and broadly used split inteins. Requires three native extein residues (Cys-Phe-Asn) at the C-terminal junction. IntC is accessible to SPPSd. |
| Npu DnaE e | 102 | 36 | ~1 | Most efficient split intein described so far. Active with a broad set of residues at the splicing junction. IntC is accessible to SPPS. |
| Artificially split inteins | ||||
| Mtu RecA | 105 | 38 | 60–120 | Reconstitution of the active intein requires co-refolding of previously denatured fragments. |
| Sce VMA | 184 | 55 | 6 | Requires induced fragment complementation by auxiliary dimerization domains. Has been used to control protein function in conditional protein splicing systems in vivo and in vitro. |
| Ssp DnaB-S0 f | 104 | 47 | 12 | Active under native conditions. Ser-Gly required at the N-terminal junction. |
| Ssp DnaB-S1 f | 11 | 143 | 280 | The short IntN is amenable to SPPS and has been used for the labeling of protein N-termini. Same sequence requirements than as DnaB-S0. |
| Ssp GyrB-S11 f | 150 | 6 | 170 | Active under native conditions. Ser-Ala-Asp used at the N-terminal junction. |
Ssp: Synechotcystis sp.; Npu: Nostoc punctiforme; Sce: Saccharomyces cerevisiae; Mtu: Mycobacterium tuberculosis. (Mootz, 2009) and references therein.
Number of residues.
Half-lives calculated from reported first order rate constants.
Solid-phase peptide synthesis.
Artificial variants of the Npu DnaE, with shorter IntC (15 and 6 residues), have been designed by shifting the split site closer to the C-terminus.
S0, S1 and S11 indicate the site at which the intein is split. S0 corresponds to the split site of naturally split inteins.
Applications of EPL and PTS
The simplest application of EPL or PTS is the modification of the N- or C-terminal regions of a protein since this can be achieved in a single ligation step involving a synthetic peptide fragment, containing the desired chemical probe(s), and a recombinant protein fragment. Central regions of the protein of interest can also be labeled, but a three-piece ligation strategy is then required (Muir, 2003), which is more technically challenging. It should be noted that EPL and PTS can be used to link a recombinant protein to a non-peptidic moiety, provided it has the necessary reactive handles for ligation. Examples of this include the attachment of proteins to surfaces, polymers, and nucleic acids (Cheriyan and Perler, 2009). Ligation of two fully recombinant protein domains is also possible and has been used to generate toxic proteins that cannot normally be expressed (Evans et al., 1998), as well as to label specific domains within large proteins with isotopes for structural studies using NMR (nuclear magnetic resonance) spectroscopy (Muralidharan and Muir, 2006).
A key decision when performing EPL and PTS is the selection of the ligation site. Obviously, this must be chosen such that the region of interest in the protein corresponds to the synthetic building block in the semi-synthesis scheme. The only sequence requirement for the standard EPL strategy is the Cys residue at the ligation site—this makes EPL virtually traceless compared with protein labeling methods involving the use of reactive tags (Lin and Wang, 2008). Furthermore, recent developments in the use of ligation auxiliaries as well as desulfurization methods have broadened the scope of EPL to include other residues such as glycine (Gly), alanine (Ala), valine (Val), and phenylalanine (Phe) at the ligation site; these more sophisticated methods employ a Cys surrogate for the ligation step which is later converted into the native residue (Hackenberger and Schwarzer, 2008). As an alternative to the use of traceless ligation methods, it is also possible to simply mutate in a Cys residue at a convenient site in the protein. Although a commonly used strategy, care must be taken to minimize the structural and functional impact of the mutation on the protein; a serine (Ser)/Ala→Cys mutation is often a good starting point (Valiyaveetil et al., 2006a). An additional criterion to be considered for EPL is the identity of the residue immediately upstream of the Cys at the ligation site (which will be the residue adjacent to the α-thioester in the N-terminal building block). Bulky, β-branched amino acids, such as threonine (Thr), isoleucine (Ile), and Val, slow-down the rate of the NCL reaction and should be avoided, if possible.
The sequence requirements associated with PTS are somewhat more nebulous than those for EPL and depend to a great extent on the exact split intein being used (Table 1) (Mootz, 2009). The mechanism of protein splicing dictates that, at a minimum, the reaction will result in a Ser/Thr/Cys residue being placed at the splice junction (Figure 1). However, in many cases there will be additional sequence requirements immediately adjacent to this site. In particular, the commonly used cyanobacterial DnaE split inteins prefer to have three native C-extein residues (Cys-Phe-Asn) for optimal splicing efficiency (Mootz, 2009). This restriction can be relaxed by using mutant split inteins evolved to splice at non-native splice junctions, although the Ser/Thr/Cys at the splice junction is still obligate (Lockless and Muir, 2009).
A final consideration when choosing a ligation site is its position within the secondary and tertiary structure of the protein. Where possible the protein should be dissected between modular domains as this will afford fragments that are well behaved in terms of solubility and, importantly, preclude the need for any folding step following the ligation reaction. The need for well-behaved fragments is especially important when using PTS because the process must be performed under native-like conditions. If the protein can be efficiently refolded then one naturally has more flexibility in choosing the ligation site. In this case, EPL may be the method of choice given that the actual ligation step can be can be performed in the presence of a variety of additives, including chemical denaturants and detergents (Muralidharan and Muir, 2006). Indeed, use of denaturants is often beneficial for EPL reactions as it allows high concentrations of reactants to be achieved, thereby improving the efficiency of the bimolecular reaction.
The principal bottleneck of any project involving EPL or PTS is the generation, by synthetic or recombinant means, of the reactive protein fragments. As is usually the case in protein chemistry, each protein target presents it own set of (often unique) challenges and so some investment in strategy optimization will be required for every system. Fortunately, after many years of methodology development, an extensive array of tools is now available for the generation of protein reactants for EPL and PTS. An overview of commonly used approaches is given in Tables 1 and Supplemental Table 2. These have allowed a large number of systems to be interrogated through semi-synthesis including proteins that might, at first pass, seem beyond the reach of organic chemistry such as integral membrane proteins.
EPL and PTS have been used to incorporate a variety of modifications into proteins (Figure 3) to answer biological questions that could not be addressed through more traditional approaches. In the following sections we discuss examples of these efforts and the biological insight they have revealed.
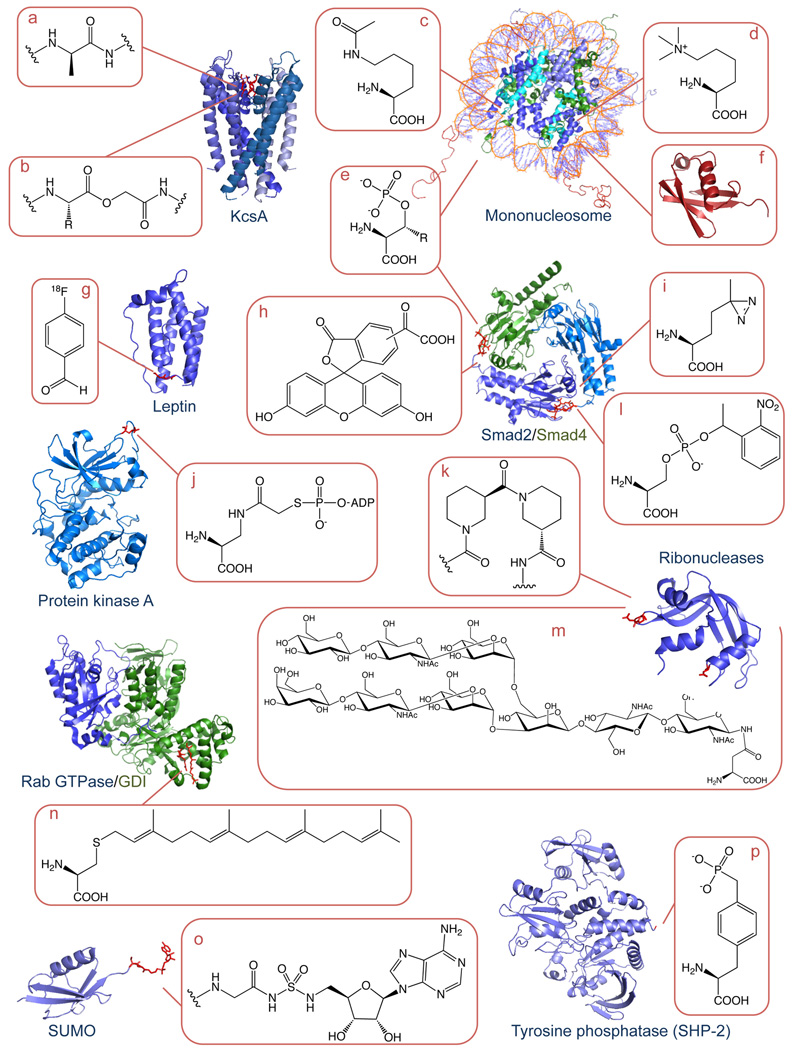
Figure 3. Examples of Proteins Modfiied by EPL and PTS.
Expressed protein ligation (EPL) and protein trans-splicing (PTS) can be used site-specifically modify a wide variety of structurally and functionally diverse proteins, as the examples given in the figure illustrate. Modifications range from naturally-occurring post-translational modifications (PTMs) to unnatural moieties and include: A) D-amino acids (D-Ala), B) ester bonds, C) acetylated (N-acetyl-Lys) and D) methylated amino acids (N-tri-methyl-Lys), E) phospho-Ser/Thr, F) ubiquitination, G) isotopes (PET emitting 18F), H) fluorophores (fluorescein), I) photo-crosslinkers (photo-Met), J) Ser-ATP bi-substrate transition state analogues, K), β-turn mimics (nipecotic acid), L) photo-caging groups (photo-caged phospho-Ser), M) glycosylated and N) prenylated amino acids O) non-hydrolysable analogues of AMP and P) non-hydrolysable phosphomimics (Tyr phosphonate).
Semi-synthesis of Proteins Containing Post-Translational Modifications
The most common application of EPL is in the semi-synthesis of post-translationally modified proteins. PTMs are used to regulate the activity of most proteins, and to fully understand how this is achieved inevitably requires access to these modified proteins for biochemical or structural studies. As noted earlier, standard site-directed mutagenesis provides limited possibilities in this regard. Thus, a clear opportunity exists for using more chemically-driven approaches. EPL, in particular, has helped fill this void, aided by the availability of robust methods for the chemical synthesis of peptides containing PTMs. Indeed, EPL has been used to generate proteins modified through phosphorylation, glycosylation, lipidation, ubiquitylation, acetylation, as well as several other classes of modification (Muir, 2003; Chatterjee and Muir, 2010). Below we focus on specific studies that highlight important themes.
Phosphorylation
Phosphorylation is one of the most common and extensively-studied PTMs. It should not be surprising then that EPL has been heavily utilized for the preparation of proteins containing this modification. Indeed, the first report of EPL described the semi-synthesis of a phospho-tyrosine (pTyr) containing analogue of the protein kinase Csk (Muir et al., 1998). Subsequently, EPL has been used to create several phosphorylated proteins for detailed functional and structural studies (Schwarzer and Cole, 2005; Muralidharan and Muir, 2006). This is exemplified by biochemical and crystallographic analyses of semi-synthetic versions of the transcription factors Smad2 and Smad3, which explain how bis-phosphorylation activates them through homo- and heterotrimerization (Wu et al., 2001; Chacko et al., 2004). This system has also served as a useful proving ground for several EPL-based technologies, including the incorporation into proteins of new amino acid crosslinkers (Vila-Perelló et al., 2007) and various photo-activation strategies, including caged phosphates (Hahn and Muir, 2004). One of the powers of applying chemistry for the study of proteins is the ability to tweak the covalent structure of the PTM. Cole and co-workers have exploited this freedom to introduce various non-hydrolyzable analogues of Ser/Thr/Tyr phosphorylation (termed phosphonates) into proteins (Schwarzer and Cole, 2005). This strategy is particularly powerful in systems where the native phospho-amino acid species is too short-lived to permit detailed mechanistic studies. For example, a semi-synthetic version of the protein tyrosine phosphatase, SHP-2, was prepared containing a stable tyrosine phosphonate in place of the native pTyr (Lu et al., 2001). Subsequent microinjection of this protein into cells helped define a role for this phosphorylation event in activation of the mitogen-activated kinase pathway.
Lipidation
In terms of ease of chemical synthesis, O-phosphorylation is among the lower hanging fruit of the PTM tree – this is equally true for N-acetylation and N-methylation, which have also been introduced into semi-synthetic proteins (Chatterjee and Muir, 2010). Certain modifications such as lipidation, glycosylation and ubiquitylation, however, present an altogether different level of synthetic challenge due to their complexity and/or physical attributes. Nonetheless, even these have yielded to the EPL and PTS approaches in recent years. Accordingly, a variety of lipid modifications have been introduced into proteins by EPL/PTS, including prenyl groups and glycophosphatidylinositol (GPI)-anchors (Brunsveld et al., 2006). This is nicely illustrated by the work of Goody and co-workers who have used semi-synthesis in conjunction with structural and functional approaches to study how lipidation regulates the function of members of the Ras superfamily, including, most recently, elucidation of the mechanism of membrane targeting of geranylgenanylated versions of a Rab GTPase (Wu et al., 2010).
Glycosylation
In terms of shear chemical complexity, glycosylation is arguably the winner among the PTMs. The attached sugars can be composed of several different mono-saccharide building blocks linked together in elaborate branched structures whose tailoring can differ from molecule to molecule (Bertozzi and Kiessling, 2001). Studying the structural and functional consequences of protein glycosylation is thus complicated by the inability to isolate well-defined glycosylated proteins from natural sources. Carbohydrate chemists have amassed an impressive arsenal for the synthesis of complex oligosaccharides (Lepenies et al., 2010). Recent years have seen this synthetic know-how integrated with EPL for the preparation of homogeneous glycoproteins (Buskas et al., 2006). An impressive recent example of this is the work of Unverzagt and co-workers who synthesized ribonuclease C, a 15 kDa enzyme with 4 disulfides and a biantennaric nonasaccharide, using a three-piece EPL strategy (Piontek et al., 2009). In the coming years, we expect that the semi-synthesis of homogeneous glycoproteins will become more routine, thereby allowing the role of this modification in the storage and transfer of biological information to be examined in greater detail than has hitherto been possible.
Ubiquitination
Ubiquitination is another example of a PTM difficult to study using proteins isolated from natural sources. Ubiquitin (Ub) is a 76 amino acid protein that is attached through its C-terminus to the ε-amino group of a lysine residue in a target protein. Proteins can be mono-ubiquitinated, multi-ubiquitinated or poly-ubiquitinated, with the precise nature of this conjugation dictating the functional consequences of the modification. The E1–E3 protein ligase family is responsible for the attachment of Ub to target proteins. Understanding the substrate specificity and enzymology of these enzymes is an area of very active study. Nonetheless, the details remain sufficiently obscure to make in vitro ubiquitination of a target protein impractical (at least on a preparative scale) in all but a few cases. Protein semi-synthesis provides an alternative source of ubiquitinated proteins. Indeed, recent years have seen a flurry of reports describing chemical methods to attach Ub to specific sites on a target protein (McGinty et al., 2008; Li et al., 2009; Ajish Kumar et al., 2009; Chatterjee et al., 2010; Chen et al., 2010). All of these strategies employ inteins at one stage or another and allow the conjugation of Ub to proteins through both native and non-native linkages. Armed with these approaches, investigators have studied the function of ubiquitination in several systems, including the role of the PTM in regulating the activity of PCNA (involved in translesion DNA synthesis) (Chen et al., 2010) and histones (McGinty et al., 2008; Chatterjee et al., 2010). These examples further highlight a unique power of semi-synthesis, namely the ability to manipulate the structure of the PTM in ways that would be impossible using an enzymatic approach. In particular, the chemical approach permits Ub to be substituted for related proteins (so-called ubiquitin-like proteins, Ubls), thereby allowing structure-activity relationships to be explored. In the histone example, the generation of a series of Ubl-modified mononucleosomes aided in defining the mechanism by which ubiquitination of histone H2B stimulates methylation of histone H3 by the methyltransferase hDot1L (Chatterjee et al., 2010). More generally, the biochemical analysis of histone modifications appears to be particularly fertile ground for the application of protein semi-synthesis. The majority of PTMs in histones are localized in the flanking regions, making them readily accessible to EPL and PTS. Indeed, several insights have already emerged from the study of semi-synthetic histones bearing chemically installed PTMs (Chatterjee and Muir, 2010). We anticipate that this area will continue to blossom in the years ahead.
Site-Specific Incorporation of Unnatural Building Blocks
EPL and PTS have been heavily utilized in the site-specific incorporation of unnatural amino acids into proteins. The ability to precisely tune the steric and electronic properties of amino acid side-chains is a powerful way to explore the details of protein function; nowhere is this truer than for the study of enzymes. Indeed, analogs of a number of enzymes (and their substrates) have been prepared by EPL. These studies have furnished mechanistic insights by manipulating various properties of key amino acid side-chains, including redox potential (as in the case ribonuclease reductase), nucleophilicity (such as the protein tyrosine kinase, Src) and steric bulk (for instance the GyrA intein) (Schwarzer and Cole, 2005; Frutos et al., 2010). Semi-synthesis has also been used to incorporate transition state analogues into enzymes and their substrates. This is exemplified by the development of bi-substrate inhibitors (i.e. simultaneously targeting two substrate binding sites) of protein kinases based on ATP-peptide conjugates that mimic the phosphoryl-transfer transition state (Schwarzer and Cole, 2005). This strategy was recently used to study the mechanism of autophosphorylation of full-length protein kinase A (PKA) (Pickin et al., 2008). PKA has two regulatory phosphorylation sites: one in its activation loop, installed by PDK1 (pyruvate dehydrogenase kinase 1), and the other one at Ser338, thought to be autocatalyzed. Semi-synthesis of PKA with a pSer338-ATP analogue was used to investigate if the autophosphorylation reaction was intra- or intermolecular. A combination of biochemical and computational experiments demonstrates that the pSer338-ATP moiety is docked into the PKA active site in an intramolecular fashion, arguing that Ser338 phosphorylation is an intramolecular event.
A related concept has recently been applied to the study of E1 ubiquitin ligases (Lu et al., 2010; Olsen et al., 2010). These enzymes activate Ub and Ubls through adenylation (AMP) of their C-terminus followed by thioesterification of a conserved Cys residue in the enzyme. To probe the first half-reaction, EPL was used to generate a reversible inhibitor by incorporating a non-hydrolyzable analog of AMP, 5‘-O-sulfamoyladenosine (AMSN), at the C-terminus of Ub and the Ubl, SUMO (Lu et al., 2010). A similar EPL approach was used to obtain a covalent inhibitor of the second half-reaction, in this case by incorporating a vinyl-sulfonamide electrophilic trap into the moiety (AVSN). These elegant chemical biology studies have been followed up by an equally impressive structural biology analysis (Olsen et al., 2010). Specifically, the crystal structures of both SUMO-AMSN and SUMO-AVNS in complex with the SUMO E1 were solved, revealing that a major reorganization of the enzyme active site accompanies the second half-reaction, that is, thioesterification of the E1. Examples like this highlight the utility of semi-synthesis in the study of enzymes. Nonetheless, we have barely scratched the surface in terms of what is possible in this area. There remain many exciting directions that have been largely or wholly unexplored, including the notion of creating new catalysts by integrating EPL/PTS with concepts and tools emanating from the fields of computational protein design and synthetic organic chemistry (e.g. novel organic catalysts).
Amino acid side-chains account for ~50% of the mass of a typical protein, the remainder is composed of the main-chain. Backbone hydrogen-bonding is, of course, critical to stabilizing the secondary and tertiary structure of proteins and frequently plays a direct role in enzyme catalysis and the recognition of ligands. Unfortunately, the protein main-chain constitutes a ‘blind-spot’ for standard mutagenesis and consequently the effects of backbone modifications on protein structure and function is relatively unexplored compared to side-chain alterations. Chemistry-driven protein engineering approaches such as EPL do allow changes to be made to the backbone of a protein and there are several excellent examples of this (Muralidharan and Muir, 2006; Kent, 2009). For instance, Raines and co-workers prepared analogs of the enzyme ribonuclease A in which an entire unit of secondary structure, a β-turn, was replaced with a reverse-turn mimetic called nipecotic acid (Arnold et al., 2002). The resulting “prosthetic” protein displayed wild-type enzymatic activity, but was thermodynamically more stable than the native protein. Another approach to stabilizing a protein involving a backbone change, is through head-to-tail cyclization. This can be achieved using either EPL or PTS and there are several examples of cyclic proteins exhibiting increased stability (Muir, 2003).
As noted above, backbone interactions can play a direct role in protein function, a point that is clearly illustrated by the selectivity filter of potassium ion channels. This is a narrow, 12 Å long pore lined with backbone carbonyl oxygen atoms that allows K+ ions to pass through, but not other mono-valent cations such as Na+. Access to this region in a semi-synthetic version of the bacterial channel, KcsA, allowed the electronegativity of these carbonyl groups to be attenuated through an amide-to-ester substitution (Valiyaveetil et al., 2006b). Crystallographic and electrophysiology studies on the resulting protein reveal alterations in ion occupancy and conductance consistent with a model of concerted ion conduction through the channel. The work on KcsA provides another nice example of how chemistry can be used to engineer the backbone of a protein, namely by engineering the chirality of the polypeptide. Specifically, substitution of a highly conserved Gly in the selectivity filter for a D-Ala shows that the ability of the amino acid at that position to adopt a left-handed helical conformation is absolutely required for activity and hence the native Gly residue acts as a D-amino acid surrogate (Valiyaveetil et al., 2006a). Electrophysiology and crystallographic studies demonstrate that the D-Ala containing channel is locked in an open conformation able to conduct Na+ in the absence of K+. The work shows that selectivity is due in part to the ability of the channel to structurally adapt in an ion-specific manner to K+.
Site-Specific Incorporation of Biophysical Probes
EPL and PTS have proven to be extremely powerful for the site-specific incorporation of spectroscopic probes into proteins (Muralidharan and Muir, 2006; Mootz, 2009). After PTMs, the incorporation of optical probes is the next most common application of semi-synthesis. In most cases, these semi-synthetic proteins are used to study ligand binding events or internal conformational changes in proteins, either by monitoring changes in the fluorescence of a single strategically placed probe in the protein, or by employing multiple probes and using fluorescence resonance energy transfer (FRET) between them. These spectroscopic approaches are nicely showcased by the work of Lorsch and co-workers who carried out a series of detailed thermodynamic and kinetic studies on the association of fluorescent derivatives of eukaryotic initiation factors with the ribosome (Maag et al., 2005). The generation of FRET-based reporter proteins via EPL has also been used for the screening of small molecule inhibitors of biomedically important proteins such as Abl kinase (Hofmann et al., 2001) and histone acetyltransferases (Xie et al., 2009). The former example highlights a key attribute of EPL/PTS, namely the ability to incorporate multiple non-coded elements, in this case two different fluorophores, into a protein. This capacity is taken a step further by a study in which five non-coded elements are incorporated into the protein Smad2, namely; two phosphoserines, a fluorophore, a fluorescent quencher, and a photo-cleavable trigger of activity (Hahn et al., 2007). This protein was designed to be inactive and non-fluorescent until irradiated with UV light whereupon the protein activates (through trimerization) and simultaneously becomes fluorescent. Microinjection of this caged protein into mammalian cells allowed the levels of the biologically active form of the protein to be precisely titrated (as quantified by fluorescence) by varying the amount of irradiation (Hahn et al., 2007).
EPL and PTS have also aided in the development of methods for the structural characterization of proteins in solution using NMR spectroscopy. NMR is a very powerful tool for the study of protein structure and dynamics; however, spectral overlap associated with large proteins limits its application. Both EPL and PTS have been used to isotopically-label specific regions, or even atoms, of a protein in order to obtain simplified spectra for detailed structural studies (Muralidharan and Muir, 2006). In one recent example, which evokes the symbology of the Uroboros (a mythical serpent that consumes its own tail), inteins were actually used (via EPL) to make inteins containing site-specific 15N and 13C isotopes (Frutos et al., 2010). NMR studies on these proteins reveal that formation of the branched intermediate in the splicing reaction drastically alters the dynamic properties of the scissile amide bond between the intein and the C-extein, rendering it more susceptible to nucleophilic attack. In the so-called segmental isotopic labeling strategy (Muralidharan and Muir, 2006), the target protein is divided up into appropriate fragments, which are then expressed individually, allowing uniform isotopic labeling of only the domain of interest. EPL or PTS are then used to put the protein back together again via one or more ligation steps. This strategy has been applied to study specific domains (flanking as well as internal) in the context of larger proteins as well as to identify intramolecular interactions or explore enzymatic mechanisms (Muralidharan and Muir, 2006). For example, Allain and co-workers prepared several segmental labeled versions of the polypyrimidine tract binding protein and used these in conjunction with transverse-relaxation optimized NMR spectroscopy to define domain-domain interfaces within the protein required for RNA binding (Vitali et al., 2006). The majority of segmental labeling studies have employed samples generated in vitro using individually expressed building blocks. This is often a technically demanding undertaking due to the large amounts of protein needed for NMR studies. To address this, Iwai and co-workers have demonstrated the feasibility of performing segmental labeling within Escherichia coli cells (Züger and Iwai, 2005). This employs a clever combination of PTS and orthogonal promoter systems to allow the in vivo reaction of a non-labeled and labeled fragment of the protein. This important advance not only promises easier access to segmental labeled proteins for traditional structural studies, but also could have application in the emerging field of cell-based protein NMR analysis.
In Vivo Applications of EPL and PTS
As we have already discussed, semi-synthetic proteins prepared in the test-tube can be injected into cells for the purposes of studying cell biological processes. This approach can be extended to animal studies. For instance, EPL was recently used to prepare a version of the protein hormone leptin containing an 18F-probe for PET (positron emission tomography) imaging (Ceccarini et al., 2009). This molecule was used to study the systemic biodistribution of the hormone in rodents and primates, revealing, among other things, high level uptake in tissues undergoing hematopoiesis. This strategy aside, there are many situations where it might be advantageous to perform the protein chemistry inside the living cell or animal. In this regard, PTS is especially powerful due to the availability of naturally split cyanobacterial inteins that have no cross-reactivity with any endogenous proteins in eukaryotic cells. Early work from our own group demonstrated the potential of PTS for the in vivo labeling of proteins by using the naturally split Ssp DnaE intein for the traceless ligation of synthetic probes to heterologously-expressed proteins in mammalian cells (Giriat and Muir, 2003). The efficiently of this cell-based semi-synthesis approach is sure to be improved by utilizing the recently described Nostoc punctiforme DnaE intein, which possesses a series of remarkably properties, including being the current record holder for splicing kinetics (t1/2 ~1 min.) (Mootz, 2009).
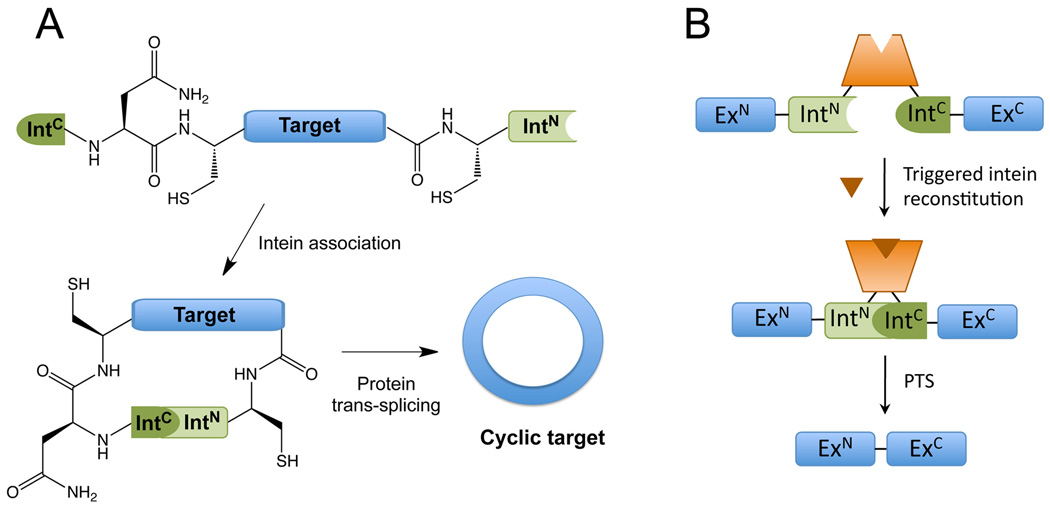
One of the most exciting uses of PTS is in the generation of cyclic peptides in cells. Peptide cyclization is commonly used in medicinal chemistry (and in nature) to improve peptide stability and bioactivity. The ability to biosynthesize cyclic peptides in vivo offers the possibility of generating large genetically-encoded libraries for high-throughput screening purposes. This can be accomplished by nesting the sequence (or library of sequences) to be cyclized between the IntC and IntN intein fragments. Flipping the order of the intein fragments in the precursor ensures that PTS spits out a cyclic peptide (Scott et al., 1999). This nifty technology, often referred to as SICLOPPS (split intein-mediated circular ligation of peptides and proteins, Figure 4A), has been used to screen for inhibitors of several processes (Cheriyan and Perler, 2009), including most recently the selection of cyclic peptide inhibitors of α-synuclein toxicity in a yeast model of Parkinson‘s Disease (Kritzer et al., 2009).
Figure 4. in vivo Applications of Protein Splicing.
A) Schematic representation of intein mediated peptide or protein cyclization. The target polypeptide is expressed flanked by IntC and IntN at the N- and C-termini, respectively. Protein trans-splicing (PTS) results in the formation of a new peptide bond between the N- and C-termini of the target and thus generates a circularized peptide or protein. B) Control of protein splicing through ligand-induced intein complementation. The splicing activity of artificially split inteins can be controlled by fusion to exogenous auxiliary domains (in the figure, a ligand binding domain). A triggering event (in the figure, ligand binding) causes a conformational change in the auxiliary domain, which induces intein reconstitution and subsequent protein splicing.
PTS results in a full-length active protein being generated from two inactive split fragments. This functional output can be harnessed for a variety of purposes. Umezawa and coworkers have developed several cell-based biosensors based on the activity of split inteins and used these for a variety of purposes, including the identification of mitochondrial proteins (Ozawa et al., 2003) and the monitoring of caspase activity (Kanno et al., 2007). PTS has also found application in the area of gene therapy. In a recent study, Li et al. expanded the scope of adeno-associated virus (AAV) as a delivery vehicle for therapeutic genes (Li et al., 2008). AAV has several advantageous properties as a vector, but is handicapped by its limited usable DNA capacity (~4 kb). To overcome this, the authors created two AAV vectors each carrying half of a therapeutic gene fused in-frame to a split intein coding sequence. Co-infection of target cells with the two AAV vectors leads to production of the therapeutic protein after PTS. As a proof of principle, the authors demonstrate the production of a therapeutic dystrophin protein upon co-delivery of appropriate AAV vectors in a mouse model of muscular dystrophy.
There are no known natural regulators of protein splicing. Rather, the process appears to occur spontaneously after translation of the precursor protein. The idea of controlling protein splicing is, nonetheless, attractive as this would provide a way to trigger the post-translational synthesis of a target protein. In principle, such a system would be fast (compared to inducible genes), tunable (allowing protein levels to be adjusted) and portable (many inteins are remarkably promiscuous). With this in mind, several conditional protein splicing systems have been reported that respond to changes in temperature, light, protease activity, and the presence of various small molecules (Cheriyan and Perler, 2009; Mootz, 2009). These have been used to control the activity of proteins both in cultured cells and in living animals. Examples include the control of Notch signaling in Drosophila melanogaster via a temperature-inducible intein mutant (Zeidler et al., 2004) and the control of hedgehog signaling in mammalian cells via a tamoxifen-inducible engineered intein (Figure 4B) (Yuen et al., 2006). It should be stressed, however, that conditional protein splicing does not work in every context due to the, as yet, poorly understood functional interplay between the intein and the surrounding extein sequences. Nevertheless, the big advantage of this approach over mainstream small molecule screening initiatives is that different conditional intein constructs can be easily surveyed using standard molecular biology techniques.
Outlook
EPL and PTS have proven remarkably useful for studying protein function in vitro and in vivo. The number of proteins studied by semi-synthesis is constantly growing, as is the complexity of the modifications that can be introduced. In this Primer, our aim is to provide a broad overview of the techniques and to introduce selected systems that have been instrumental in unlocking biological puzzles. As with any approaches, EPL and PTS have their strengths and weaknesses. The approaches are unparalleled in terms of the range and number of non-coded elements that can be introduced into large proteins. However, they are at their most practical when the regions to be modified are within ~50 amino acids of the N- or C-terminus of the protein of interest, given that this allows a single ligation step to be performed. The interior of proteins is far more difficult to access via semi-synthesis, requiring the use of technically demanding sequential ligation reactions. This should be contrasted with the nonsense suppression mutagenesis method. Although more restricted in the types of modification that can be introduced, it does provide general access to any part of the protein primary sequence (Wang et al., 2006). Thus, EPL/PTS and nonsense suppression are complementary protein engineering approaches and the decision to use one or the other will depend on the question at hand. Moreover, there is no reason why the two approaches cannot be used in combination, a tactic that we are now beginning to see (Li et al., 2009) and that will surely be more common in the future.
A defined biological role for protein splicing has so far eluded investigators – we currently know of no intein whose activity is naturally regulated, something that would point the way to a biological purpose. Inteins are, however, very ancient proteins and so such regulation may have been lost during evolution. What we can say about inteins is that they are an amazingly malleable platform for technology development. It is a fair bet that the chemical biology community will continue to find new uses for inteins in both the basic and applied biomedical sciences. Thus, these remarkable protein devices will continue to be a part of the thread that stitches together the fields of chemistry and biology.
Supplementary Material
Acknowledgements
We thank B. Fierz and N. Shah for valuable input. Some of the work discussed in this review was performed in the Muir laboratory and was supported by the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ajish Kumar KS, Haj-Yahya M, Olschewski D, Lashuel HA, Brik A. Highly Efficient and Chemoselective Peptide Ubiquitylation. Angew Chem Int Ed Engl. 2009;48:8090–8094. doi: 10.1002/anie.200902936. [DOI] [PubMed] [Google Scholar]
- Arnold U, Hinderaker MP, Nilsson BL, Huck BR, Gellman SH, Raines RT. Protein prosthesis: a semisynthetic enzyme with a beta-peptide reverse turn. J Am Chem Soc. 2002;124:8522–8523. doi: 10.1021/ja026114n. [DOI] [PubMed] [Google Scholar]
- Bertozzi CR, Kiessling LL. Chemical glycobiology. Science. 2001;291:2357–2364. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- Blanco-Canosa JB, Dawson PE. An Efficient Fmoc-SPPS Approach for the Generation of Thioester Peptide Precursors for Use in Native Chemical Ligation. Angew Chem Int Ed Engl. 2008 doi: 10.1002/anie.200705471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunsveld L, Kuhlmann J, Alexandrov K, Wittinghofer A, Goody R, Waldmann H. Lipidated ras and rab peptides and proteins--synthesis, structure, and function. Angew Chem Int Ed Engl. 2006;45:6622–6646. doi: 10.1002/anie.200600855. [DOI] [PubMed] [Google Scholar]
- Buskas T, Ingale S, Boons GJ. Glycopeptides as versatile tools for glycobiology. Glycobiology. 2006;16:113–136. doi: 10.1093/glycob/cwj125. [DOI] [PubMed] [Google Scholar]
- Ceccarini G, Flavell RR, Butelman ER, Synan M, Willnow TE, Bar-Dagan M, Goldsmith SJ, Kreek MJ, Kothari P, Vallabhajosula S, et al. PET imaging of leptin biodistribution and metabolism in rodents and primates. Cell Metab. 2009;10:148–159. doi: 10.1016/j.cmet.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacko B, Qin B, Tiwari A, Shi G, Lam S, Hayward L, de Caestecker M, Lin K. Structural Basis of Heteromeric Smad Protein Assembly in TGF-β Signaling. Molecular Cell. 2004;15:813–823. doi: 10.1016/j.molcel.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Chatterjee C, McGinty RK, Fierz B, Muir TW. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nature chemical biology. 2010;6:267–269. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- Chatterjee C, Muir TW. Chemical approaches for studying histone modifications. J Biol Chem. 2010;285:11045–11050. doi: 10.1074/jbc.R109.080291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ai Y, Wang J, Haracska L, Zhuang Z. Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nature chemical biology. 2010;6:270–272. doi: 10.1038/nchembio.316. [DOI] [PubMed] [Google Scholar]
- Cheriyan M, Perler FB. Protein splicing: A versatile tool for drug discovery. Adv Drug Deliv Rev. 2009 doi: 10.1016/j.addr.2009.04.021. [DOI] [PubMed] [Google Scholar]
- Erlanson DA, Chytil M, Verdine GL. The leucine zipper domain controls the orientation of AP-1 in the NFAT.AP-1.DNA complex. Chem Biol. 1996;3:981–991. doi: 10.1016/s1074-5521(96)90165-9. [DOI] [PubMed] [Google Scholar]
- Evans TC, Benner J, Xu MQ. Semisynthesis of cytotoxic proteins using a modified protein splicing element. Protein Sci. 1998;7:2256–2264. doi: 10.1002/pro.5560071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frutos S, Goger M, Giovani B, Cowburn D, Muir TW. Branched intermediate formation stimulates peptide bond cleavage in protein splicing. Nature chemical biology. 2010;6:527–533. doi: 10.1038/nchembio.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George EA, Novick RP, Muir TW. Cyclic peptide inhibitors of staphylococcal virulence prepared by Fmoc-based thiolactone peptide synthesis. J Am Chem Soc. 2008;130:4914–4924. doi: 10.1021/ja711126e. [DOI] [PubMed] [Google Scholar]
- Giriat I, Muir TW. Protein semi-synthesis in living cells. J Am Chem Soc. 2003;125:7180–7181. doi: 10.1021/ja034736i. [DOI] [PubMed] [Google Scholar]
- Hackenberger CP, Schwarzer D. Chemoselective ligation and modification strategies for peptides and proteins. Angew Chem Int Ed Engl. 2008;47:10030–10074. doi: 10.1002/anie.200801313. [DOI] [PubMed] [Google Scholar]
- Hahn ME, Muir TW. Photocontrol of Smad2, a multiphosphorylated cell-signaling protein, through caging of activating phosphoserines. Angew Chem Int Ed Engl. 2004;43:5800–5803. doi: 10.1002/anie.200461141. [DOI] [PubMed] [Google Scholar]
- Hahn ME, Pellois JP, Vila-Perelló M, Muir TW. Tunable photoactivation of a post-translationally modified signaling protein and its unmodified counterpart in live cells. Chembiochem. 2007;8:2100–2105. doi: 10.1002/cbic.200700404. [DOI] [PubMed] [Google Scholar]
- Hermanson GT. Bioconjugate techniques. 2nd edn. San Diego: Academic Press; 2008. [Google Scholar]
- Hofmann RM, Cotton GJ, Chang EJ, Vidal E, Veach D, Bornmann W, Muir TW. Fluorescent monitoring of kinase activity in real time: development of a robust fluorescence-based assay for Abl tyrosine kinase activity. Bioorg Med Chem Lett. 2001;11:3091–3094. doi: 10.1016/s0960-894x(01)00650-3. [DOI] [PubMed] [Google Scholar]
- Kanno A, Yamanaka Y, Hirano H, Umezawa Y, Ozawa T. Cyclic luciferase for real-time sensing of caspase-3 activities in living mammals. Angew Chem Int Ed Engl. 2007;46:7595–7599. doi: 10.1002/anie.200700538. [DOI] [PubMed] [Google Scholar]
- Kent SB. Total chemical synthesis of proteins. Chemical Society reviews. 2009;38:338–351. doi: 10.1039/b700141j. [DOI] [PubMed] [Google Scholar]
- Kritzer JA, Hamamichi S, McCaffery JM, Santagata S, Naumann TA, Caldwell KA, Caldwell GA, Lindquist S. Rapid selection of cyclic peptides that reduce alpha-synuclein toxicity in yeast and animal models. Nature chemical biology. 2009;5:655–663. doi: 10.1038/nchembio.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepenies B, Yin J, Seeberger PH. Applications of synthetic carbohydrates to chemical biology. Current opinion in chemical biology. 2010;14:404–411. doi: 10.1016/j.cbpa.2010.02.016. [DOI] [PubMed] [Google Scholar]
- Li J, Sun W, Wang B, Xiao X, Liu XQ. Protein trans-splicing as a means for viral vector-mediated in vivo gene therapy. Human gene therapy. 2008;19:958–964. doi: 10.1089/hum.2008.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Fekner T, Ottesen JJ, Chan MK. A pyrrolysine analogue for site-specific protein ubiquitination. Angewandte Chemie (International ed in English) 2009;48:9184–9187. doi: 10.1002/anie.200904472. [DOI] [PubMed] [Google Scholar]
- Lin MZ, Wang L. Physiology. Vol. 23. Bethesda, Md: 2008. Selective labeling of proteins with chemical probes in living cells; pp. 131–141. [DOI] [PubMed] [Google Scholar]
- Lockless SW, Muir TW. Traceless protein splicing utilizing evolved split inteins. Proc Natl Acad Sci U S A. 2009;106:10999–11004. doi: 10.1073/pnas.0902964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Gong D, Bar-Sagi D, Cole PA. Site-specific incorporation of a phosphotyrosine mimetic reveals a role for tyrosine phosphorylation of SHP-2 in cell signaling. Mol Cell. 2001;8:759–769. doi: 10.1016/s1097-2765(01)00369-0. [DOI] [PubMed] [Google Scholar]
- Lu X, Olsen SK, Capili AD, Cisar JS, Lima CD, Tan DS. Designed semisynthetic protein inhibitors of Ub/Ubl E1 activating enzymes. J Am Chem Soc. 2010;132:1748–1749. doi: 10.1021/ja9088549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Molecular Cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz HD. Split inteins as versatile tools for protein semisynthesis. Chembiochem. 2009;10:2579–2589. doi: 10.1002/cbic.200900370. [DOI] [PubMed] [Google Scholar]
- Muir TW. Semisynthesis of proteins by expressed protein ligation. Annu Rev Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- Muir TW, Sondhi D, Cole PA. Expressed protein ligation: a general method for protein engineering. Proc Natl Acad Sci USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan V, Muir TW. Protein ligation: an enabling technology for the biophysical analysis of proteins. Nat Methods. 2006;3:429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- Noren CJ, Wang J, Perler FB. Dissecting the Chemistry of Protein Splicing and Its Applications. Angewandte Chemie (International ed in English) 2000;39:450–466. [PubMed] [Google Scholar]
- Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010;463:906–912. doi: 10.1038/nature08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T, Sako Y, Sato M, Kitamura T, Umezawa Y. A genetic approach to identifying mitochondrial proteins. Nature biotechnology. 2003;21:287–293. doi: 10.1038/nbt791. [DOI] [PubMed] [Google Scholar]
- Paulus H. Protein splicing and related forms of protein autoprocessing. Annual Review of Biochemistry. 2000;69:447–496. doi: 10.1146/annurev.biochem.69.1.447. [DOI] [PubMed] [Google Scholar]
- Pickin KA, Chaudhury S, Dancy BC, Gray JJ, Cole PA. Analysis of protein kinase autophosphorylation using expressed protein ligation and computational modeling. J Am Chem Soc. 2008;130:5667–5669. doi: 10.1021/ja711244h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piontek C, Varón Silva D, Heinlein C, Pöhner C, Mezzato S, Ring P, Martin A, Schmid FX, Unverzagt C. Semisynthesis of a homogeneous glycoprotein enzyme: ribonuclease C: part 2. Angew Chem Int Ed Engl. 2009;48:1941–1945. doi: 10.1002/anie.200804735. [DOI] [PubMed] [Google Scholar]
- Schwarzer D, Cole PA. Protein semisynthesis and expressed protein ligation: chasing a protein's tail. Current opinion in chemical biology. 2005;9:561–569. doi: 10.1016/j.cbpa.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ. Production of cyclic peptides and proteins in vivo. Proc Natl Acad Sci U S A. 1999;96:13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiyaveetil FI, Leonetti M, Muir TW, Mackinnon R. Ion selectivity in a semisynthetic K+ channel locked in the conductive conformation. Science. 2006a;314:1004–1007. doi: 10.1126/science.1133415. [DOI] [PubMed] [Google Scholar]
- Valiyaveetil FI, Sekedat M, Mackinnon R, Muir TW. Structural and functional consequences of an amide-to-ester substitution in the selectivity filter of a potassium channel. J Am Chem Soc. 2006b;128:11591–11599. doi: 10.1021/ja0631955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila-Perelló M, Pratt MR, Tulin F, Muir TW. Covalent capture of phospho-dependent protein oligomerization by site-specific incorporation of a diazirine photo-cross-linker. J Am Chem Soc. 2007;129:8068–8069. doi: 10.1021/ja072013j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali F, Henning A, Oberstrass FC, Hargous Y, Auweter SD, Erat M, Allain FH. Structure of the two most C-terminal RNA recognition motifs of PTB using segmental isotope labeling. EMBO J. 2006;25:150–162. doi: 10.1038/sj.emboj.7600911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CT, Garneau-Tsodikova S, Gatto GJ. Protein posttranslational modifications: the chemistry of proteome diversifications. Angewandte Chemie (International ed in English) 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- Wang L, Xie J, Schultz PG. Expanding the genetic code. Annual review of biophysics and biomolecular structure. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- Wu JW, Hu M, Chai J, Seoane J, Huse M, Li C, Rigotti DJ, Kyin S, Muir TW, Fairman R, et al. Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling. Mol Cell. 2001;8:1277–1289. doi: 10.1016/s1097-2765(01)00421-x. [DOI] [PubMed] [Google Scholar]
- Wu YW, Oesterlin LK, Tan KT, Waldmann H, Alexandrov K, Goody R. Membrane targeting mechanism of Rab GTPases elucidated by semisynthetic protein probes. Nature chemical biology. 2010;6:534–540. doi: 10.1038/nchembio.386. [DOI] [PubMed] [Google Scholar]
- Xie N, Elangwe EN, Asher S, Zheng YG. A dual-mode fluorescence strategy for screening HAT modulators. Bioconjug Chem. 2009;20:360–366. doi: 10.1021/bc800467a. [DOI] [PubMed] [Google Scholar]
- Yuen CM, Rodda SJ, Vokes SA, McMahon AP, Liu DR. Control of transcription factor activity and osteoblast differentiation in mammalian cells using an evolved small-molecule-dependent intein. J Am Chem Soc. 2006;128:8939–8946. doi: 10.1021/ja062980e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler MP, Tan C, Bellaiche Y, Cherry S, H√§der S, Gayko U, Perrimon N. Temperature-sensitive control of protein activity by conditionally splicing inteins. Nature biotechnology. 2004;22:871–876. doi: 10.1038/nbt979. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nature reviews Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- Züger S, Iwai H. Intein-based biosynthetic incorporation of unlabeled protein tags into isotopically labeled proteins for NMR studies. Nat Biotechnol. 2005;23:736–740. doi: 10.1038/nbt1097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.