Abstract
Influenza A virus is a negative-strand segmented RNA virus in which antigenically distinct viral subtypes are defined by the hemagglutinin (HA) and neuraminidase (NA) major viral surface proteins. An ideal inactivated vaccine for influenza A virus would induce not only highly robust strain-specific humoral and T-cell immune responses but also cross-protective immunity in which an immune response to antigens from a particular viral subtype (e.g., H3N2) would protect against other viral subtypes (e.g., H1N1). Cross-protective immunity would help limit outbreaks from newly emerging antigenically novel strains. Here, we show in mice that the addition of cationic lipid/noncoding DNA complexes (CLDC) as adjuvant to whole inactivated influenza A virus vaccine induces significantly more robust adaptive immune responses both in quantity and quality than aluminum hydroxide (alum), which is currently the most widely used adjuvant in clinical human vaccination. CLDC-adjuvanted vaccine induced higher total influenza virus-specific IgG, particularly for the IgG2a/c subclass. Higher levels of multicytokine-producing influenza virus-specific CD4 and CD8 T cells were induced by CLDC-adjuvanted vaccine than with alum-adjuvanted vaccine. Importantly, CLDC-adjuvanted vaccine provided significant cross-protection from either a sublethal or lethal influenza A viral challenge with a different subtype than that used for vaccination. This superior cross-protection afforded by the CLDC adjuvant required CD8 T-cell recognition of viral peptides presented by classical major histocompatibility complex class I proteins. Together, these results suggest that CLDC has particular promise for vaccine strategies in which T cells play an important role and may offer new opportunities for more effective control of human influenza epidemics and pandemics by inactivated influenza virus vaccine.
Influenza A virus is an enveloped negative-sense single-stranded RNA virus with eight segments in its genome. The viral hemagglutinin (HA) and neuraminidase (NA) surface glycoproteins, which are encoded on separate viral genome segments, are the most important targets for antibody-mediated protection from infection (15). These HA and NA viral glycoproteins are classic T-cell-dependent antigens for which antibody responses depend on influenza virus-specific CD4 T-cell help in the form of surface expression of CD154 (11) and secretion of cytokines, such as interleukin 21 (IL-21) (13). The six remaining viral genome segments encode internal matrix, nucleoprotein, polymerase components, and nonstructural immunomodulatory proteins (44). In the event that influenza A virus eludes any preexisting neutralizing antibody and establishes a productive infection, T-cell immunity, particularly CD8 cytotoxic T lymphocytes (CTL) directed against major histocompatibility complex (MHC) class I-restricted viral peptides, are likely important for the reduction of viral load and for limiting spread within infected tissues (8, 35). CTL activity may also reduce influenza A virus shedding in nasal secretions and transmission to uninfected individuals (52).
The HA and NA surface proteins are used for dividing influenza A virus into 16 and 9 antigenically distinct subtypes, respectively, e.g., H1N1 and H3N2, that encompass genetically related proteins (67). Even within subtypes there can be a high degree of sequence diversity due to amino acid substitutions, sometimes referred to as major intrasubtypic diversity (37), which is a reflection of the relatively error-prone nature of the influenza virus RNA-dependent polymerase during viral replication (69). This sequence diversity combined with the immune selection pressure for HA and NA proteins that avoid neutralization by previously generated antibodies (38) results in antigenic drift in which strains with new antigenic determinants emerge during epidemics. Although internal viral proteins of circulating viruses are also subject to changes in amino acid sequence, this is less pronounced than for HA and NA (2, 7). This relative conservation is most likely because such amino acid substitutions in internal proteins mainly influence immune recognition by T cells (8), which may exert immune pressure on viral replication but does not appear by itself to prevent the establishment of infection (52). In contrast, neutralizing antibodies, particularly those that are directed against HA (22, 67), not only prevent infection, but in the event of infection, they also participate along with CTL in viral clearance (8, 63). Amino acid substitutions in internal viral proteins may also be constrained by less flexibility in avoiding adverse impacts on viral fitness than substitutions of the surface glycoproteins, although these constraints are not absolute (58).
The segmented nature of the influenza A virus genome permits reassortment when two or more subtypes or distinct clades of a subtype simultaneously infect a host cell (44, 53). Viral reassortment can result in the emergence of viruses that have acquired novel HA and NA subtypes or clade antigenic determinants from nonhuman viral sources, such as from birds (e.g., in the 1957 and 1968 influenza pandemics) or pigs (e.g., in the 2009 novel H1N1 swine influenza pandemic). Pandemics can arise from reassortment, since most of the human population worldwide may lack neutralizing antibody against these new strains. Nevertheless, cross-reactive T-cell immunity in this context might limit disease morbidity and mortality in cases of established infection. For example, healthy human individuals who have no prior exposure to H5N1 avian influenza virus have frequently been found to have memory CD4 and CD8 T cells generated from seasonal influenza A virus exposure that cross-react with internal proteins from multiple subtypes (6), including H5N1 (43) as well as other avian and pig-derived strains (33).
Inactivated influenza virus vaccine currently approved for clinical use in most of the world is unadjuvanted and is produced in eggs using recombinant viruses in which only the HA and NA proteins are derived from currently circulating strains (20). The virus used in vaccine production is typically “split” to partially purify the HA and NA components from other viral proteins and RNA (20) and thereby reduce local reactions to immunization. Thus, vaccine efficacy depends largely on the induction of neutralizing antibody directed against the HA and NA proteins and can be abrogated by mismatch between the amino acid sequences of the HA and NA proteins used in the vaccine and those encoded by antigenically shifted or reassortant strains that subsequently circulate. Current inactivated vaccines are also less immunogenic in select populations, such as young children (28), the elderly (20), and the immunocompromised (39).
Given the propensity of human influenza strains for unpredictable antigenic drift and viral genome reassortment and given the limitations in the immunogenicity of current inactivated vaccines, particularly in certain high-risk populations, there is considerable interest in developing new vaccines with improved efficacy despite antigen mismatch. One potential approach for improving cross-protection and overall inactivated vaccine efficacy is to include antigens that are more conserved among strains and subtypes for inducing either antibody, e.g., the membrane-proximal stem region of HA (16, 62), the external domain of the matrix 2 protein (18), or T-cell immunity, e.g., nucleoprotein (17). Alternatively, whole inactivated vaccines, in which all components of the mature virion are included along with viral RNA (24), can be used to provide a diverse set of antigens. Another approach is to add adjuvants (3) in order to induce or augment antibody responses that can provide cross-protection against antigenically drifted strains or even full heterosubtypic immunity, in which vaccination with antigens from one viral subtype (e.g., H3N2) protects from a different subtype (e.g., H1N1 or H5N1) (27). Such strategies might also allow dose sparing, i.e., whereby the amount of antigen and the number of administered doses could be reduced to achieve a protective immune responses (29).
The most commonly used adjuvants in humans for inactivated vaccines for T-cell-dependent antigens, e.g., diphtheria and tetanus toxoids, are aluminum salts (alum), including aluminum hydroxide. Humoral immune responses to influenza virus surface proteins as assessed by hemagglutination inhibition (HAI) or neutralizing antibody titers, are clearly augmented by the addition of alum, particularly in the setting of whole inactivated vaccines (66), resulting in improved cross-protection and dose sparing. Although the mechanisms for alum adjuvant activity remain poorly understood, Toll-like receptors (TLRs) are apparently not involved (23). Alum-adjuvanted protein vaccines induce human memory CD4 T cells with a nonpolarized cytokine profile (secretion of IL-2 but not gamma interferon [IFN-γ] [14]), whereas such immunization in mice tends to induce T helper 2 (Th2) type CD4 T cells that secrete IL-4 but not IFN-γ (48), accounting for the predominance of expression of the IgG1 and IgE isotypes (19). In contrast to humoral or CD4 T-cell immunity, alum-adjuvanted inactivated vaccines are weak inducers of CD8 T-cell immunity, particularly for primary responses (51), although repeated vaccination may be able to induce CD8 T-cell responses capable of protecting mice from lethal influenza virus challenge (50).
Cationic lipid/DNA complexes (CLDC), which were originally developed as a transient liposomal gene therapy delivery system (71), have more recently been evaluated as an adjuvant for inactivated vaccines in preclinical studies in mice and rhesus macaques (41). CLDC are also currently being evaluated in humans for seasonal inactivated influenza virus vaccination as part of phase I and phase II trials. The DNA component of CLDC consists of pMB75.6, a double-stranded plasmid that was originally used as an “empty vector” negative control for gene therapy studies, which contains a cytomegalovirus (CMV) immediate/early promoter segment but lacks any downstream cDNA sequence (26). In the CLDC adjuvant, this DNA component is combined with the cationic lipid DOTIM (octadecenolyoxy[ethyl-2-heptadecenyl-3-hydroxyethyl] chloride) and cholesterol at a 1:1 molar ratio (26). For vaccination, CLDC and the protein antigen of interest are mixed together just prior to administration. In initial studies in mice, CLDC mixed with model protein antigens, such as egg white ovalbumin, resulted in a dramatic adjuvant effect, with a robust expansion of antigen-specific CD4 and CD8 T cells as determined by MHC/antigenic peptide tetramer staining (68). More recently, we found that the addition of CLDC to an unadjuvanted trivalent inactivated split influenza vaccine (TIV) for two influenza A virus strains (H1N1 and H3N2) and an influenza B virus strain resulted in substantially higher levels of influenza virus-specific antibody and T-cell immunity in outbred mice and rhesus macaques compared to recipients of unadjuvanted vaccine (41).
Here, we directly compared the immunization of mice with whole inactivated influenza A virus vaccine adjuvanted with either CLDC or alum for the induction of humoral and T-cell immunity and for their ability to confer protection in vivo, including from sublethal and lethal heterosubtypic influenza A virus challenge. The importance of MHC class I-restricted CD8 T cells in providing cross-protective immunity was also assessed.
MATERIALS AND METHODS
Mice.
C57BL/6J mice (8 to 10 weeks old) were purchased from Jackson Laboratory (Bar Harbor, ME). KbDb−/− mice (64) maintained on a C57BL/6J background were kindly provided by R. Ahmed (Emory University, Atlanta, GA). Mice were housed in specific-pathogen-free animal facilities at Stanford University or the Molecular Medicine Research Institute (MMRI) (Sunnyvale, CA) (former affiliate Juvaris BioTherapeutics, Inc.). All experiments were approved by the Institution Animal Care and Use Committees of Stanford University and of the Molecular Medicine Research Institute.
Viruses.
Influenza A virus strains PR/8/34 (H1N1) and HKx31 (H3N2), a recombinant virus that has the H3 and N2 segments derived from A/Aichi/2/68 and all other proteins from PR/8/34 (36), were grown in embryonated chicken eggs and were used in allantoic fluid or purified on a sucrose density gradient (both from Charles River, North Franklin, CT). Purified viruses used for vaccination were heat inactivated at 56°C for 30 min, with inactivation confirmed by failure to grow on Madin-Darby canine kidney (MDCK) cells (American Type Culture Collection, Manassas, VA).
Preparation of vaccines and vaccination.
CLDC (41) was prepared by reconstituting a lyophilized mixture of DOTIM/cholesterol liposomes and plasmid DNA (pMB75.6 [26]) with sterile endotoxin-free water at a final concentration of 300 μg/ml of DNA. An aliquot of 2.5 μg of purified, heat-inactivated PR/8/34 or HKx31 virus was added to 15.0 μg of CLDC or 300 μg, 600 μg, or 1,000 μg of alum (Alhydrogel [aluminum hydroxide]; Brenntag Biosector, Frederikssund, Denmark) for each dose of CLDC-adjuvanted or alum-adjuvanted influenza virus vaccine. Mice were given a primary (1°) and secondary (2°) vaccination intramuscularly (i.m.) in the posterior thigh in a total volume of 50 μl separated by 14 days. As we did not observe significant differences among the three alum doses for antibody titers or CD4 or CD8 T-cell responses, we employed the lowest dose of 300 μg for comparison with CLDC unless indicated otherwise.
Serum and splenocyte isolation.
Blood samples were collected from the tail vein 14 days after 1° and 2° vaccinations, and serum samples were frozen at −20°C. Spleen tissue was removed from mice euthanized with an intraperitoneal (i.p.) injection of phenytoin/pentobarbital (Beuthanasia-D; Schering-Plough, Union, NJ) 14 days after the 2° vaccination. Splenocytes were dissociated from spleen tissue using a GentleMacs tissue dissociator (Miltenyi Biotec, Auburn, CA) followed by the lysis of red blood cells using ammonium chloride solution (Sigma, St. Louis, MO).
Influenza A viral challenge.
Mice were anesthetized with isoflurane and inoculated intranasally (i.n.) with 30 μl of either 240 hemagglutination units of influenza A virus strain HKx31 for sublethal challenge or with four times the 50% lethal dose (LD50) of HKx31 or PR/8/34 for lethal challenge. For sublethal challenge, mice were euthanized 4 days after infection, and lung tissue was isolated. For lethal challenge experiments, animals were weighed and inspected each day for 14 to 21 days. Moribund animals were euthanized per Institutional Animal Care and Use Committee guidelines.
ELISA for influenza A virus-specific antibody.
Maxisorp enzyme-linked immunosorbent assay (ELISA) plates (Nunc, Rochester, NY) were coated with purified heat-inactivated influenza virus (HKx31 or PR/8/34) at 1.0 μg/ml diluted in phosphate-buffered saline (PBS) at 4°C for 18 to 24 h. Microtiter wells were blocked by incubation with 10% fetal calf serum (FCS) in PBS for 1 h, followed by the addition of 10-fold serial dilutions of serum performed in duplicate. After incubation for 2 h at room temperature, the wells were washed and incubated with a 1:4,000 dilution of the appropriate isotype-specific antibody conjugated to horseradish peroxidase (Southern Biotech, Birmingham, AL). After the wells were washed, tetramethylbenzidine (TMB) substrate (Kierkegaard and Perry Laboratories [KPL], Gaithersburg, MD) was added for 30 min followed by TMB stop solution (KPL). Absorbance was measured at 450 nm and 570 nm using a plate reader spectrophotometer, and the data were plotted to obtain a curve of the inverse of the dilution versus the A450 − A570 measurement. The antibody titer for each serum sample was calculated (GraphPad Prism, San Diego, CA) as the midpoint of the dilution curve as defined by the effective concentration at 50% of maximum (EC50).
HAI titer determination.
Hemagglutination inhibition (HAI) titers were determined as previously described (65). Briefly, serum was incubated at 37°C for 18 to 24 h with receptor destroying enzyme II (Denka Seiken, Tokyo, Japan). After enzyme inactivation at 56°C for 30 to 60 min, serial 1:2 dilutions (vol/vol) of serum in PBS were mixed with 4.0 hemagglutination units of purified influenza virus in V-bottom 96-well plates (Corning, Lowell, MA). Serum/virus mixtures were incubated at room temperature for 15 min, followed by the addition of an equal volume of 0.5% chicken red blood cells (Colorado Serum Company, Denver, CO). The plates were read at 1 h, with the HAI titer determined as the reciprocal dilution of the last well that showed complete inhibition of hemagglutination.
ELISA of cell culture supernatants.
Splenocytes (10 × 106/ml) were incubated with 1.0 μg/ml of heat-inactivated and purified HKx31 or PR/8/34 influenza A virus in RPMI 1640 medium with 10% FCS. Supernatants were collected after 48 h of stimulation, and the IFN-γ concentration was measured by ELISA (BD Biosciences, San Jose, CA) following the manufacturer's instructions.
Flow cytometric analysis of influenza A virus-specific T-cell responses.
All monoclonal antibodies (MAbs) used for flow cytometry were purchased from BD Biosciences unless otherwise indicated. For analysis of CD4 T-cell responses, splenocytes were stimulated with 1.0 μg/ml of heat-inactivated influenza A virus strain PR/8/34 for 24 h, with 25 μg/ml of brefeldin A (Sigma) during the last 5 h of incubation. The cells were then stained with anti-CD4-PE-Cy7 (anti-CD4 antibody conjugated to phycoerythrin [PE] and fluorochrome Cy7) and anti-CD3-APC-Alexa Fluor 750 (anti-CD3 antibody conjugated to allophycocyanin [APC] and to Alexa Fluor 750) MAbs (eBioscience, San Diego, CA) and dead cell discriminator (Miltenyi Biotec). The cells were then fixed, permeabilized with CytoPerm/CytoFix solution (BD Biosciences), and stained with IFN-γ conjugated to PE (IFN-γ-PE), tumor necrosis factor alpha conjugated to fluorescein isothiocyanate (TNF-α-FITC), and IL-2 conjugated to APC (IL-2-APC) at 4°C for 30 min, followed by flow cytometric analysis using an LSRII instrument (BD Biosciences). For CD8 T-cell IFN-γ staining, splenocytes (10 × 106/ml) were restimulated with 20 hemagglutination units/ml of live purified influenza A virus strain HKx31 for 48 h with brefeldin A (25 μg/ml) present during the final 5 h of incubation. The splenocytes were then stained with anti-CD69-FITC, anti-CD3-peridinin chlorophyll protein, and anti-CD8-APC MAbs, fixed, and permeabilized with CytoPerm/CytoFix solution, intracellularly stained with IFN-γ-PE, and analyzed using a FACSCalibur instrument (BD Biosciences). For CD8 T-cell degranulation assays, anti-CD107a-FITC and anti-CD107b-FITC were added along with monensin at the manufacturer's recommended concentration (GolgiStop; BD Biosciences) for the last 5 h of 48 h of incubation of splenocytes with live influenza virus. Surface staining was then performed with anti-CD107a/b-FITC, anti-CD8-PE-Cy7, and anti-CD3-APC-Alexa Fluor 750 MAbs, followed by fixation/permeabilization, IFN-γ-PE MAb intracellular staining, and analysis using an LSRII instrument. FlowJo software (Treestar, Ashland, OR) was used for the analysis of flow cytometric data.
Lung tissue influenza A virus titer.
Whole lungs were collected in ice-cold PBS and homogenized using a GentleMACS dissociator. Samples were centrifuged at 600 × g to remove particulate matter, and supernatants were collected and stored at −80°C for a later assay. To determine the 50% tissue culture infective dose (TCID50), 10-fold dilutions of lung homogenate supernatants prepared in minimal essential medium (MEM) (GIBCO Invitrogen, Carlsbad, CA) with 0.0002% trypsin (Worthington Biochemical Corporation, Lakewood, NJ) were plated in 96-well U-bottom plates with MDCK cells as previously described (65). The plates were incubated overnight, and the medium was replaced with fresh MEM without trypsin. After 72 h, 50 μl of 0.5% chicken red blood cells was added to each well and incubated at room temperature for 1 h, and hemagglutination was recorded. TCID50 was calculated using the Reed-Muench formula (57).
RESULTS
Antibody responses to vaccination.
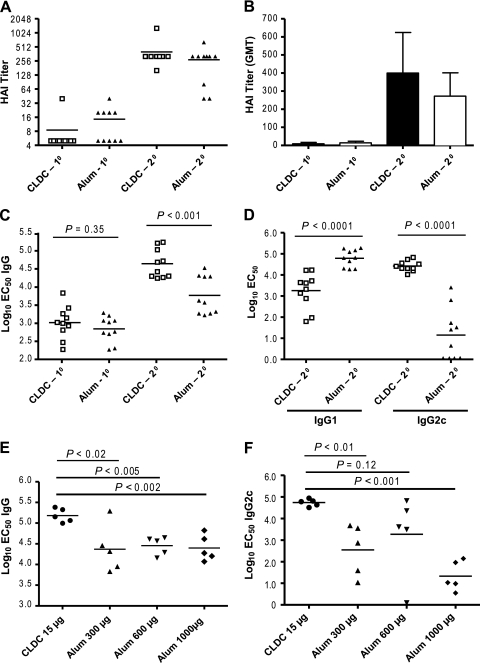
We compared the ability of primary (1°) and secondary (2°) intramuscular immunization with heat-inactivated whole influenza virus (PR/8/34) mixed with cationic lipid/noncoding DNA complexes (CLDC) or with alum (hereafter referred to as CLDC-influenza or alum-influenza vaccination, respectively) to induce influenza virus-specific antibody in C57BL/6J mice. Hemagglutination inhibition (HAI) titers after primary vaccination with either adjuvanted vaccine were well below a titer of 1:40, which based on human studies, is a serologic correlate of protection to homotypic infection (55). Protective titers were achieved following 2° vaccination with either adjuvant (Fig. 1A). The geometric mean titers in the CLDC-influenza-vaccinated group were higher than those in the alum-influenza-vaccinated group (Fig. 1B) but did not achieve statistical significance.
FIG. 1.
Vaccination with cationic lipid/noncoding DNA complexes (CLDC) as an adjuvant leads to a significantly higher IgG-specific influenza virus antibody titer highly skewed toward the IgG2c subclass compared to alum-adjuvanted (Alum) vaccine. C57BL/6J mice were immunized twice with CLDC-adjuvanted (15 μg of the DNA component) or alum-adjuvanted inactivated influenza A virus PR/8/34 vaccine. Alum was used at a dose of 300 μg per vaccination unless otherwise indicated. (A) Hemagglutination inhibition (HAI) titers from individual mice after primary (10) or secondary (20) immunization with CLDC-adjuvanted vaccine (open squares; n = 10) or alum-adjuvanted vaccine (closed triangles; n = 10). Each individual symbol represents the value for an individual mouse; the horizontal bar shows the mean value for the group of mice. (B) HAI geometric mean titer (GMT) after 10 or 20 immunization with CLDC-adjuvanted (closed bars) or alum-adjuvanted influenza vaccine (open bars). The geometric mean titers plus standard errors of the means (SEMs) (error bars) are shown. (C) Influenza virus-specific IgG levels after 10 or 20 immunization with CLDC-adjuvanted (open squares) or alum-adjuvanted vaccine (closed triangles). EC50, effective concentration at 50% of maximum. (D) IgG1 and IgG2c levels after 20 immunization with CLDC-adjuvanted vaccine (closed squares) or alum-adjuvanted vaccine (closed triangles). (E) Total influenza virus-specific IgG after 20 immunization with vaccine with CLDC adjuvant (15 μg) or escalating adjuvant doses of alum adjuvant (300 μg, 600 μg, and 1,000 μg). (F) Influenza virus-specific IgG2c after 20 immunization with CLDC-adjuvanted or escalating adjuvant doses of alum-adjuvanted vaccine. The P values were calculated by the two-tailed, unpaired Student t test. The results shown are representative of two independent experiments.
CLDC-influenza vaccination led to a significantly higher titer of total influenza virus-specific IgG antibody after two immunizations than alum-influenza vaccination at an alum dose of 300 μg did (Fig. 1C), with a dramatically (more than 1,000-fold) higher level of influenza virus-specific IgG2c antibody (Fig. 1D). Increasing the dose of alum to 600 μg or 1,000 μg did not consistently narrow the differences between CLDC and alum in the induction of influenza virus-specific IgG and IgG2c (Fig. 1E and F). This marked skewing of the antibody response toward influenza virus-specific IgG2c in C57BL/6J mice also applied to the BALB/c strain (data not shown), in which the IgG2a antibody rather than the IgG2c antibody is expressed (49). Although IgG2a and IgG2c differ by 16% at the amino acid sequence level (49), they can be considered functionally equivalent (e.g., both subclasses efficiently bind to Fc receptors and to fix complement compared to the IgG1 subclass [30]) and hereafter are collectively referred to as IgG2a/c. Alum-influenza vaccination resulted in a significantly higher level of influenza virus-specific IgG1 than CLDC-influenza vaccination did, although the fold difference in this case was less than for IgG2a/c (Fig. 1D).
CD4 T-cell responses to vaccination.
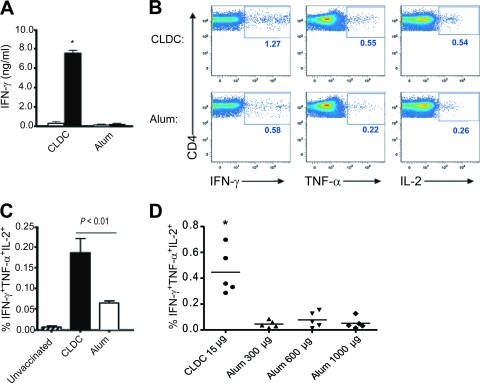
The CD4 T-cell response to immunization with CLDC-influenza vaccine versus alum-influenza vaccine was compared and revealed that CLDC-adjuvanted vaccine induced a stronger Th1 response based on IFN-γ detected in culture supernatants of splenocytes restimulated with homotypic inactivated influenza virus (Fig. 2A). CD4 T cells were the major source of IFN-γ in these assays, as magnetic bead depletion of CD4 T cells from splenocyte cultures abrogated IFN-γ production (data not shown). In contrast, IL-4 was not produced at appreciable levels (lower limit of detection of <10.0 pg/ml) under these conditions with either adjuvant, indicating that appreciable Th2 responses were not induced by vaccination.
FIG. 2.
Influenza A virus vaccination with CLDC as the adjuvant induces a quantitatively and qualitatively superior CD4 T-cell response compared to vaccination with alum as the adjuvant. Mice were immunized with two doses of CLDC-adjuvanted (CLDC) or alum-adjuvanted (Alum) influenza A virus strain PR/8/34 or were not immunized (Unvaccinated). Splenocytes were obtained 14 days following the second vaccine dose. (A) IFN-γ (IFN-g) content of cell culture supernatants of splenocytes stimulated with heat-inactivated PR/8/34 (filled bars) or with medium alone (open bars) using cells from recipients of CLDC-adjuvanted (n = 5) or alum-adjuvanted (n = 5) vaccine. The value for splenocytes stimulated with heat-inactivated PR/8/34 were significantly different (P < 0.05) from the value for stimulated splenocytes from mice vaccinated with alum and influenza virus (alum-influenza-vaccinated mice), as assessed by the two-tailed, unpaired Student t test and indicated by an asterisk. (B) Representative flow cytometric plots of CD4 T cells expressing IFN-γ (IFN-g), TNF-α (TNF-a), or IL-2 after heat-inactivated PR/8/34 stimulation of splenocytes. All events shown were positively gated for CD3 expression. The numbers in each panel indicate the frequency of cytokine-positive CD4 T cells. (C) Frequency of triple-cytokine-positive CD4 T cells simultaneously expressing IFN-γ, TNF-α, and IL-2 based on the gating parameters shown in panel B after PR/8/34 stimulation of splenocytes from unvaccinated mice (n = 3), CLDC-influenza-vaccinated mice (n = 5), and alum-influenza-vaccinated mice (n = 5). The means plus standard deviation (SDs) (error bars) are shown. The results shown are representative of two independent experiments. (D) Frequency of triple-cytokine-positive CD4 T cells from mice vaccinated with CLDC-influenza compared with mice vaccinated with PR/8/34 and 300 μg, 600 μg, or 1,000 μg of alum as the adjuvant. The value for splenocytes stimulated with CLDC-influenza-vaccinated mice was significantly different (P < 0.05) from the value for stimulated splenocytes from alum-influenza-vaccinated mice, as assessed by the two-tailed unpaired Student t test and indicated by an asterisk.
As polyfunctional CD4 T-cell responses to infection or vaccination have been associated with superior infection control (12, 21), it was of interest to determine whether CLDC and alum adjuvants differed in their ability to induce polyfunctional influenza virus-specific CD4 T cells. The frequency of splenic CD4 T cells that produced IFN-γ, tumor necrosis factor alpha (TNF-α), and/or IL-2 was determined following restimulation with inactivated influenza virus, using intracellular cytokine staining and the gating strategy and parameters shown in Fig. 2B. Flow cytometric analysis (Fig. 2C) revealed that splenic CD4 T cells from CLDC-influenza-vaccinated mice had a significantly higher frequency of triple-positive (IFN-γ+ TNF-α+ IL-2+) cytokine producers than these cells from mice vaccinated with 300 μg of alum adjuvant per dose (0.19% versus 0.06%, respectively). Increasing the alum dose to 600 μg or 1,000 μg failed to increase the polyfunctional CD4 T-cell response (Fig. 2D).
CD8 T-cell responses to vaccination.
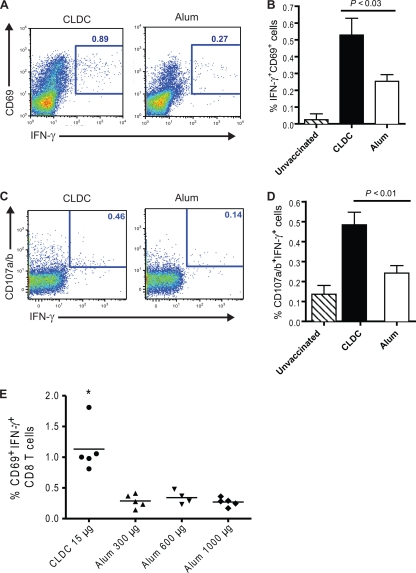
We next compared CLDC and alum adjuvant for their ability to induce influenza virus-specific CD8 T-cell responses to internal viral proteins. Splenocytes were collected from C57BL/6J mice following vaccination with inactivated PR/8/34 virus combined with either CLDC or alum. These cells were stimulated with live HKx31, a reassortant viral strain consisting of the internal proteins of H1N1 PR/8/34 and H3 and N2 surface proteins (36), and stained for CD3, CD8, IFN-γ, and CD69. As shown in Fig. 3A and B, CLDC-influenza vaccination induced a significantly higher frequency of IFN-γ+ CD69+ CD8 T cells than alum-influenza vaccination did (0.53% versus 0.25%, respectively) in response to live HKx31 virus. To determine whether this superior cross-reactive response also applied to the release of cytotoxic granules by influenza virus-specific CD8 T cells, splenocytes that were stimulated with viable HKx31 were subsequently surface stained for CD107a (lysosome-associated membrane glycoprotein 1 [LAMP1]) and C107b (LAMP2), CD3, and CD8 and internally stained for IFN-γ (Fig. 3C). This analysis (Fig. 3D) revealed that CLDC-influenza vaccine also induced a significantly greater percentage of influenza virus-specific IFN-γ-expressing CD8 T cells that had undergone cytotoxin granule secretion than alum-influenza vaccine did (0.48% versus 0.24%, respectively). The percentage of these cells was not increased by immunization with higher doses of alum (Fig. 3E).
FIG. 3.
CLDC-influenza vaccination induces greater ex vivo CD8 T-cell responses than alum-influenza vaccination. C57BL/6J mice were immunized twice with CLDC-adjuvanted (CLDC) or alum-adjuvanted (Alum) heat-inactivated PR/8/34 virus or were unimmunized (Unvaccinated). Splenic CD8 T-cell responses were evaluated after stimulation with viable HKx31 virus for 48 h. (A) Representative flow cytometric plots of IFN-γ and CD69 expression (gated on CD3+ and CD8+ events). The numbers in each panel indicate the frequency of IFN-γ+CD69+ CD8 T cells. (B) Percentage of splenic CD8 T cells coexpressing IFN-γ and CD69 (IFN-γ+ CD69+ cells) after no vaccination (n = 2), CLDC-adjuvanted vaccination (n = 4), and alum-adjuvanted vaccination (n = 5). The means plus SDs (error bars) are shown. The P value was calculated using the unpaired, two-tailed Student t test. (C) Representative flow cytometric plots of CD107a/b and IFN-γ expression. The numbers in each panel indicate the frequency of IFN-γ+CD107a/b+ CD8 T cells. (D) Percentage of IFN-γ+ CD107a/b+ splenic CD8 T cells (mean plus SD) after no vaccination (n = 3), CLDC-adjuvanted vaccination (n = 5), and alum-adjuvanted vaccination (n = 5). The P value was calculated using the unpaired, two-tailed Student t test. The results shown are representative of two independent experiments. (E) Percentage of IFN-γ+ CD69+ splenic CD8 T cells after vaccination with CLDC-influenza or influenza virus strain PR/8/34 with 300 μg, 600 μg, or 1,000 μg of alum as the adjuvant. The value for splenocytes from CLDC-influenza-vaccinated mice was significantly different (P < 0.05) from the value for stimulated splenocytes from alum-influenza-vaccinated mice, as assessed by the two-tailed, unpaired Student t test and indicated by the asterisk.
Cross-protection to sublethal heterosubtypic influenza A virus challenge after vaccination.
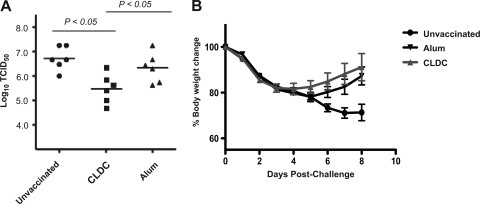
To determine whether CLDC-influenza vaccination provided superior cross-protection in vivo from infection with a viral strain of a completely different subtype, i.e., differing in both hemagglutinin (HA) and neuraminidase (NA) subtypes, C57BL/6J mice were vaccinated with influenza A virus strain PR/8/34 (H1N1) with either CLDC or alum adjuvant and challenged with a sublethal dose of influenza A virus strain HKx31 (H3N2). The amount of HKx31 virus in lung tissue on day 4 after challenge, the time at which viral load peaks in unvaccinated mice, was significantly and substantially reduced following CLDC-adjuvanted vaccine compared to either alum-adjuvanted vaccine recipients or unvaccinated mice (Fig. 4A). In addition, CLDC-adjuvanted vaccine recipients had a trend toward more rapid weight recovery than alum-adjuvanted vaccine recipients after sublethal heterosubtypic influenza virus challenge, although this difference was not statistically significant (Fig. 4B). Mice vaccinated twice with CLDC- or alum-adjuvanted PR/8/34 vaccine and challenged with PR/8/34 virus had no detectable virus in lung tissue on day 4 postinfection (data not shown), as expected given the effectiveness of either vaccine at inducing antibodies reactive to surface proteins (Fig. 1A and B).
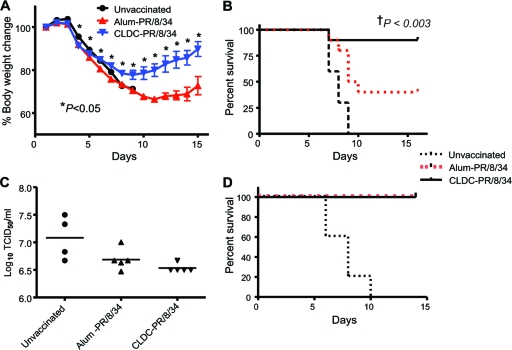
FIG. 4.
CLDC-adjuvanted vaccination confers greater protection than alum-adjuvanted vaccination from a sublethal heterosubtypic influenza virus challenge. Mice were vaccinated twice with influenza A virus strain PR/8/34 vaccine with alum or CLDC as the adjuvant and then challenged i.n. with a nonlethal dose of influenza A virus strain HKx31. (A) Lung virus titers were measured on day 4 after the infectious challenge for mice that previously had not been vaccinated (Unvaccinated) (closed circles; n = 6), CLDC-influenza-vaccinated mice (closed squares; n = 6), or alum-influenza-vaccinated mice (closed triangles; n = 6). Each symbol shows the TCID50 value for an individual mouse, with the mean value for each group indicated by a horizontal bar. The P values shown were calculated using the Tukey multiple-comparison test. (B) Percent body weight change assessed daily for 8 days following viral challenge.
Cross-protection to lethal heterosubtypic influenza A virus challenge.
The cross-protective effects of CLDC-influenza vaccination was further evaluated in a lethal challenge model. The LD50 of PR/8/34 and HKx31 viruses given by the i.n. route to unimmunized C57BL/6J mice were first determined. Influenza A virus strain HKx31 at four times this LD50 was administered i.n. to mice that had received either inactivated PR/8/34 with either CLDC or alum as an adjuvant. The mice were weighed daily for 2 weeks following the challenge and examined as to whether they were moribund, and if so, euthanized. CLDC-influenza-vaccinated mice had significant weight loss through day 9 postchallenge; however, they subsequently steadily regained weight through the end of the 15-day experiment (Fig. 5A). In contrast, alum-influenza-vaccinated mice had continued weight loss through day 11, with a delayed recovery compared to that of CLDC-influenza-vaccinated mice. Thus, the mean weight of the mice in the CLDC-influenza- and alum-influenza-vaccinated groups differed significantly starting on day 4, and this difference remained significant through the end of the experiment on day 15. There was almost complete protection in the CLDC-influenza-vaccinated animals, with 90% survival after lethal heterosubtypic challenge, compared with only 40% survival in the alum-influenza-vaccinated group (Fig. 5B). CLDC-influenza-vaccinated mice also exhibited modestly lower lung virus titers than alum-influenza-vaccinated mice at day 5 after lethal heterosubtypic challenge (Fig. 5C), although these differences were not statistically significant. Nevertheless, this reduced viral load was consistent with the protective effect of CLDC-adjuvanted immunization being due, at least in part, to more rapid control of viral lung replication. After a homotypic intranasal challenge with PR/8/34, both CLDC-influenza- and alum-influenza-vaccinated mice were completely protected with no appreciable weight loss (data not shown) and 100% survival (Fig. 5D), again consistent with the induction of robust HAI antibody using either adjuvant (Fig. 1A and B).
FIG. 5.
CLDC-adjuvanted influenza virus vaccine confers improved protection against a lethal heterotypic intranasal influenza virus challenge. C57BL/6J mice were vaccinated on days 1 and 7 with vaccine for influenza A virus strain PR/8/34 with CLDC as the adjuvant (CLDC-PR/8/34; n = 10) or with the same vaccine with alum as the adjuvant (Alum-PR/8/34; n = 10) or were not immunized (Unvaccinated; n = 10). Four times the LD50 of influenza A virus strain HKx31 or PR/8/34 was administered i.n. on day 10 following the second vaccination. Values for CLDC-PR/8/34-vaccinated mice were significantly different (P < 0.05) from the values for alum-PR/8/34-vaccinated mice by the two-tailed, unpaired Student t test as indicated by the asterisks. (A) Percent body weight change assessed daily from basal levels for 15 days after challenge. (B) Kaplan-Meier survival curve after a lethal HKx31 (heterosubtypic) viral challenge. †, P value comparing CLDC-PR/8/34 and Alum-PR/8/34 post-HKx31 challenge calculated using a log-rank (Mantel-Cox) test. (C) Lung influenza viral load on day 5 after a lethal HKx31 challenge. (D) Kaplan-Meier survival curve after a lethal PR/8/34 (homotypic) challenge. The results shown are representative of two independent experiments.
Role of CD8 T cells in CLDC-influenza vaccine-induced cross-protection.
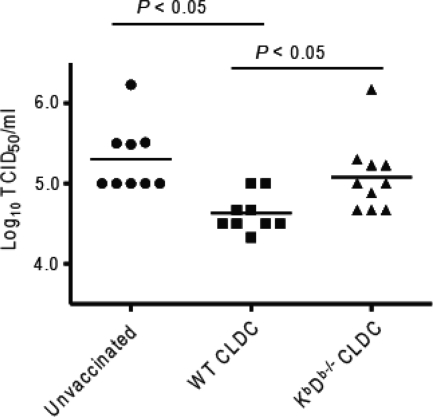
We next determined the importance of CD8 T-cell-mediated immunity for conferring cross-protection by CLDC-adjuvanted vaccination. C57BL/6 background wild-type or KbDb−/− mice, which lack MHC class I Kb or Db heavy chains for antigen recognition by CD8 T cells and have reduced numbers of CD8 T cells due to impaired MHC class I-mediated intrathymic positive selection (64), were immunized twice with CLDC-adjuvanted PR/8/34 vaccine. Four days after i.n. challenge with a sublethal dose of HKx31 virus, wild-type vaccinated mice showed a significant decrease in total lung virus titer compared with previously unvaccinated mice, whereas KbDb−/− animals showed lung virus titers similar to those of unvaccinated mice (Fig. 6). Thus, MHC class I-restricted CD8 T-cell responses are an important mechanism for the superior cross-protective immunity afforded by influenza virus vaccination using CLDC adjuvant.
FIG. 6.
MHC class I-restricted CD8 T cells significantly contribute to heterosubtypic protection after CLDC-adjuvanted vaccination. CLDC-PR/8/34 vaccine was administered twice i.m. to wild-type mice (WT CLDC; n = 9) or mice with targeted deletion for MHC class I (KbDb−/− CLDC; n = 10) of the C57BL6/J background. Mice were inoculated i.n. with 240 hemagglutination units of influenza A virus strain HKx31 on day 10 after the second vaccination. A separate control group of wild-type mice who had not been immunized (Unvaccinated; n = 9) were similarly inoculated. Each symbol shows the log10 of the TCID50/ml of lung homogenate from an individual mouse obtained on day 4 of infection, with the mean value for the group indicated by the horizontal bar. P values were calculated using the unpaired, two-tailed Student t test with Bonferroni's correction for multiple comparisons.
DISCUSSION
We directly compared the immunogenicity of CLDC-adjuvanted and alum-adjuvanted whole inactivated influenza A virus vaccines and their ability to confer in vivo protection after i.m. injection, the route most commonly used for inactivated influenza virus vaccination worldwide. This study revealed that CLDC induces significantly higher influenza virus-specific IgG2a/c antibody and CD4 and CD8 T-cell immune responses than alum-adjuvanted vaccine and, in contrast to alum, provides robust cross-protection from either a sublethal or lethal viral challenge. A significant proportion of this cross-protective immunity is dependent on classical MHC class I-restricted CD8 T cells. Together, these findings demonstrate potential application of CLDC in the design of more broadly protective influenza virus vaccines and other viral vaccine targets, in which robust CD8 T-cell immunity is likely to be desirable.
Both CLDC and alum adjuvants induce similar high levels of HAI titers after two vaccinations. These robust responses may contribute to the ability of vaccination with either adjuvant to provide complete protection of mice from infection following a homotypic sublethal challenge or death following a lethal viral challenge. The magnitude of the homotypic antibody response induced by vaccination with either adjuvant is likely similar to that induced by natural infection in mice, which also effectively prevents establishment of infection with subsequent homotypic viral challenges (9). The requirement for two doses of CLDC- or alum-adjuvanted vaccine to confer protective HAI levels is also similar to human vaccination of individuals with inactivated vaccines who are antigenically naïve for influenza virus antigens, i.e., antigens provided by natural infection or repeated vaccination, such as for young children (20).
In contrast to alum, CLDC adjuvant leads to a significantly higher titer of influenza virus-specific IgG, with more than a 100-fold mean level of IgG2c, whereas alum adjuvant results in a significantly higher level of influenza virus-specific IgG1. Similar to vaccination with CLDC as the adjuvant, the IgG response to natural viral infections is heavily skewed toward IgG2a/c (10), and this skewing after influenza virus infection is dependent on IFN-γ (60). There are several known mechanisms by which CLDC-adjuvanted vaccine, like natural influenza infection, might promote this dramatic skewing toward IgG2a/c isotype switching, none of which are mutually exclusive. First, CLDC induces relatively high levels of Th1 cell IFN-γ production, which is well established in promoting switching to IgG2a/c (19). Second, whole inactivated influenza virus vaccine includes viral RNA that could engage Toll-like receptor 7 (TLR7), and such engagement may indirectly promote IgG2a/c isotype switching by enhancing Th1 cell development from naïve CD4 T-cell precursors. This TLR7-dependent mechanism for Th1 differential bias appears to account for the ability of whole inactivated influenza virus vaccine to promote a higher level of IgG2a/c expression than subunit vaccines (25). Finally, CLDC includes double-stranded plasmid DNA, which could potentially engage the TLR9 in B cells, and directly promote switching to IgG2a/c (34).
Previous studies by others of BALB/c mice have shown that vaccination strategies that induce both influenza virus-specific IgG2a and IgG1 antibodies are superior to those in which IgG1 predominates, as these are associated with more rapid influenza virus clearance and protection from a lethal challenge (32). The IgG2a/c isotype has also been shown to be superior to IgG1, IgG2b, or IgG3 for passive antibody protection to virus-induced poliencephalomyelitis (47). The higher affinity of IgG2a/c antibodies for Fc receptors may be important in mediating antivirus protection, as gene knockout mice deficient in Fc receptors for IgG were highly susceptible to influenza virus infection after intranasal vaccination with a subunit vaccine compared to wild-type animals (31). Given these observations, it will be of interest in future experiments to determine the ability of influenza virus-specific antibody with a very high IgG2a/c-to-IgG1 ratio (e.g., antibody induced by CLDC adjuvant) to provide protection in vivo and ascertain whether this protection is mediated by antibody-dependent cell-mediated cytotoxicity versus opsonophagocytic mechanisms.
CLDC-adjuvanted vaccine induces a strong Th1 response compared to alum adjuvant, with the coexpression of IFN-γ, IL-2, and TNF-α in a significantly greater number of influenza virus-specific CD4 T cells. These antigen-specific multicytokine-producing CD4 T cells correlate with vaccine protection in models of Leishmania major (12) and Mycobacterium tuberculosis (21). Although the mechanisms that underlie their production are not clear, multicytokine-producing CD8 T cells following natural influenza virus infection have also have been identified, and in this context their accumulation appears to be dependent on strong activation of the T cell as a result of avidity of the αβ T cell receptor (TCR) for the antigen and optimal antigen availability (40). An earlier study (41) has shown that vaccine with CLDC also induces more influenza virus-specific multicytokine-producing CD4 and CD8 T cells in nonhuman primates than unadjuvanted vaccine does. This suggests that the induction of multicytokine T cells may apply to CLDC-adjuvanted influenza virus vaccination in humans as well; these clinical trials and immunogenicity studies are in progress.
The induction of antigen-specific CD8 T cells by influenza virus vaccination has been an area of intense research for several decades. A large body of experimental work indicates that after natural infection this response can potentially provide both homotypic and cross-protective immunity by recognition of conserved epitopes, most of which are derived from internal proteins that are less subject to variation in strains and subtypes (27). We found that CLDC adjuvant induces substantially greater numbers of influenza virus-specific CD8 T cells than alum based on their expression of IFN-γ and the activation-induced release of cytotoxic granules, as assessed by CD107a/b surface staining (5). Consistent with previous work examining the immunodominant hierarchy of the CD8 T-cell response to influenza virus infection (4), the viral peptide epitopes recognized by these CD8 T cells are clearly directed to internal epitopes, as stimulation was performed using a virus that differed in the HA and NA external glycoproteins but shared all internal proteins. In fact, HA and NA epitopes seem to have minimal contributions to CD8 T-cell immunity in the C57BL/6J (H-2b) mouse influenza model, as H-2b-restricted minor CD8 T-cell epitopes to HA and NA were only found recently using bioinformatics approaches (70). Importantly, this superior cross-protective CD8 T-cell response ex vivo is associated with CLDC adjuvant inducing significantly better protection in vivo after either sublethal or lethal heterosubtypic viral challenges. Interestingly, the time at which weight recovery begins (day 9 or 10) after lethal challenge mirrors the kinetics of the CD8 T-cell response in the spleen and mediastinal lymph node after secondary challenge with viral challenge (4), which is consistent with CD8 T cells mediating this protection. The importance of CD8 T cells in providing cross-protection in vivo is clear based on the results using gene knockout mice with impaired MHC class I-restricted CD8 T-cell immunity as a result of the lack of the classical MHC Kb and Db heavy-chain proteins. The importance of particular CD8 T-cell effector mechanisms in providing enhanced cross-protective immunity by CLDC-adjuvanted vaccination remains to be defined, but based on studies of experimental influenza virus infection (61), it is plausible that cytotoxicity by perforin and granzyme secretion contributes to immunity.
CD8 T-cell responses induced by vaccination do not prevent the establishment of infection, as indicated by the detectable levels of virus in the lung and associated morbidity after a heterosubtypic viral challenge. However, they clearly modify the severity of such infection by reducing viral load, which is consistent with studies of humans following natural influenza virus infection (52). Such immune control of viral replication would be particularly important in the event of an influenza pandemic where there was no preexisting humoral immunity to a novel influenza virus strain (8). Together, these findings suggest that CLDC has potent activity as an adjuvant for influenza virus vaccines with broad cross-reactivity by virtue of its ability to induce cytolytic CD8 T cells, and therefore, it is an attractive adjuvant for inclusion in vaccines directed against pandemic influenza virus, such as H5N1.
Our findings of a robust cross-protective T-cell immunity after CLDC-adjuvanted vaccination do not exclude a possible contribution from antibody or another effector cellular mechanism (27). The relative contributions of T cells, particularly of the CD8 cell subset, versus B cells and systemic or mucosa-associated antibody to such protection in mice following influenza virus infection is controversial (27, 45). This may in part reflect experimental differences in the tissues monitored for viral load, the viral strain, and the amount of virus used for primary infection and challenge, the sites used for primary infection and challenge, and the genetic background of the mice. Therefore, it is also not surprising that murine influenza virus vaccination, like natural infection, has also demonstrated cross-protection in which either T-cell immunity, particularly by CD8 T cells (27), or B-cell immunity (46, 54) is the predominant mechanism.
The mechanisms underlying the striking superiority of CLDC over alum adjuvant influenza vaccine immunogenicity, particularly for the robust induction of cytolytic CD8 T-cell responses, remain to be defined. As CLDC was originally designed as a means of intracellular delivery of plasmid DNA into eukaryotic cells (71), it is perhaps not surprising that our preliminary results suggest that both CLDC and protein antigen moieties rapidly enter conventional dendritic cells or mononuclear phagocytes and colocalize in an early endosomal compartment (C. M. Botham and D. B. Lewis, unpublished observations). Such a result is also not unexpected given other recent studies demonstrating the ability of liposomes using cationic lipids (56) to efficiently deliver nucleic acids, such as small interfering RNAs, into cells (1). Once proteins complexed with CLDC enter into the early endosome of conventional dendritic cells, these proteins may be cross-presented onto MHC class I molecules (59). Entry of the plasmid DNA moiety of CLDC also appears to be essential for its activity as an adjuvant (C. M. Botham, D. B. Lewis, and J. Fairman, unpublished results). It is plausible that this requirement may involve an interaction of the internalized DNA with intracellular innate immune receptors, of which there are both endosomal (e.g., TLR9) and cytoplasmic candidates. Internalization of CLDC is associated with activation of dendritic cells in vitro, including their secretion of IL-12 and type I IFNs (D. K. Hong, C. M. Botham, M. Draghi, and D. B. Lewis, unpublished observations), which may account for the robust Th1 type CD4 T-cell response and cytolytic CD8 T-cell response observed in vivo after immunization (42).
In summary, the inclusion of a novel vaccine adjuvant, CLDC, in inactivated influenza A virus vaccination augments cross-protection after either a sublethal or lethal viral challenge. CLDC-adjuvanted whole inactivated influenza virus vaccine induces robust IgG2a/c antibody responses, the accumulation of Th1 type CD4 T cells capable of producing multiple cytokines, and cytotoxin-secreting MHC class I-restricted CD8 T cells that recognize internal viral proteins. These CD8 T cells are major contributors to cross-protection against a mismatched influenza virus challenge. This combination of robust humoral and CD4 and CD8 T-cell immunity makes CLDC a promising adjuvant for influenza virus vaccines by improving cross-protection against antigenic drift or novel emerging subtypes or clades.
Acknowledgments
This work was supported by National Institutes of Health grants K08 AI-079269 (to D.K.H.), F32 AI-082936 (to C.M.B.), and U01 AI-074512 (to J.F. and D.B.L.), a MedImmune/Pediatric Infectious Diseases Society Research Development Award (to D.K.H.), and a Solvay Pharmaceuticals Influenza Research Grant (to D.B.L.).
We thank Lan Liu, Marla Lay, and Tim Carroll for technical assistance and the Stanford Shared FACS (fluorescence-activated cell sorter) facility for assistance with flow cytometry.
Footnotes
Published ahead of print on 13 October 2010.
REFERENCES
- 1.Aigner, A. 2008. Cellular delivery in vivo of siRNA-based therapeutics. Curr. Pharm. Des. 14:3603-3619. [DOI] [PubMed] [Google Scholar]
- 2.Assarsson, E., H. H. Bui, J. Sidney, Q. Zhang, J. Glenn, C. Oseroff, I. N. Mbawuike, J. Alexander, M. J. Newman, H. Grey, and A. Sette. 2008. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J. Virol. 82:12241-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atmar, R. L., and W. A. Keitel. 2009. Adjuvants for pandemic influenza vaccines. Curr. Top. Microbiol. Immunol. 333:323-344. [DOI] [PubMed] [Google Scholar]
- 4.Belz, G. T., W. Xie, and P. C. Doherty. 2001. Diversity of epitope and cytokine profiles for primary and secondary influenza A virus-specific CD8+ T cell responses. J. Immunol. 166:4627-4633. [DOI] [PubMed] [Google Scholar]
- 5.Betts, M. R., and R. A. Koup. 2004. Detection of T-cell degranulation: CD107a and b. Methods Cell Biol. 75:497-512. [DOI] [PubMed] [Google Scholar]
- 6.Boon, A. C., G. de Mutsert, D. van Baarle, D. J. Smith, A. S. Lapedes, R. A. Fouchier, K. Sintnicolaas, A. D. Osterhaus, and G. F. Rimmelzwaan. 2004. Recognition of homo- and heterosubtypic variants of influenza A viruses by human CD8+ T lymphocytes. J. Immunol. 172:2453-2460. [DOI] [PubMed] [Google Scholar]
- 7.Bragstad, K., L. P. Nielsen, and A. Fomsgaard. 2008. The evolution of human influenza A viruses from 1999 to 2006: a complete genome study. Virol. J. 5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, L. E., and A. Kelso. 2009. Prospects for an influenza vaccine that induces cross-protective cytotoxic T lymphocytes. Immunol. Cell Biol. 87:300-308. [DOI] [PubMed] [Google Scholar]
- 9.Christensen, J. P., P. C. Doherty, K. C. Branum, and J. M. Riberdy. 2000. Profound protection against respiratory challenge with a lethal H7N7 influenza A virus by increasing the magnitude of CD8(+) T-cell memory. J. Virol. 74:11690-11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coutelier, J. P., J. T. van der Logt, F. W. Heessen, G. Warnier, and J. Van Snick. 1987. IgG2a restriction of murine antibodies elicited by viral infections. J. Exp. Med. 165:64-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowe, J. E., Jr., E. C. Sannella, S. Pfeiffer, G. L. Zorn, A. Azimzadeh, R. Newman, G. G. Miller, and R. N. Pierson. 2003. CD154 regulates primate humoral immunity to influenza. Am. J. Transplant. 3:680-688. [DOI] [PubMed] [Google Scholar]
- 12.Darrah, P. A., D. T. Patel, P. M. De Luca, R. W. Lindsay, D. F. Davey, B. J. Flynn, S. T. Hoff, P. Andersen, S. G. Reed, S. L. Morris, M. Roederer, and R. A. Seder. 2007. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 13:843-850. [DOI] [PubMed] [Google Scholar]
- 13.Dienz, O., S. M. Eaton, J. P. Bond, W. Neveu, D. Moquin, R. Noubade, E. M. Briso, C. Charland, W. J. Leonard, G. Ciliberto, C. Teuscher, L. Haynes, and M. Rincon. 2009. The induction of antibody production by IL-6 is indirectly mediated by IL-21 produced by CD4+ T cells. J. Exp. Med. 206:69-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Divekar, A. A., D. M. Zaiss, F. E. Lee, D. Liu, D. J. Topham, A. J. Sijts, and T. R. Mosmann. 2006. Protein vaccines induce uncommitted IL-2-secreting human and mouse CD4 T cells, whereas infections induce more IFN-gamma-secreting cells. J. Immunol. 176:1465-1473. [DOI] [PubMed] [Google Scholar]
- 15.Doherty, P. C., S. J. Turner, R. G. Webby, and P. G. Thomas. 2006. Influenza and the challenge for immunology. Nat. Immunol. 7:449-455. [DOI] [PubMed] [Google Scholar]
- 16.Ekiert, D. C., G. Bhabha, M. A. Elsliger, R. H. Friesen, M. Jongeneelen, M. Throsby, J. Goudsmit, and I. A. Wilson. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epstein, S. L., W. P. Kong, J. A. Misplon, C. Y. Lo, T. M. Tumpey, L. Xu, and G. J. Nabel. 2005. Protection against multiple influenza A subtypes by vaccination with highly conserved nucleoprotein. Vaccine 23:5404-5410. [DOI] [PubMed] [Google Scholar]
- 18.Fiers, W., M. De Filette, K. El Bakkouri, B. Schepens, K. Roose, M. Schotsaert, A. Birkett, and X. Saelens. 2009. M2e-based universal influenza A vaccine. Vaccine 27:6280-6283. [DOI] [PubMed] [Google Scholar]
- 19.Finkelman, F. D., J. Holmes, I. M. Katona, J. F. Urban, Jr., M. P. Beckmann, L. S. Park, K. A. Schooley, R. L. Coffman, T. R. Mosmann, and W. E. Paul. 1990. Lymphokine control of in vivo immunoglobulin isotype selection. Annu. Rev. Immunol. 8:303-333. [DOI] [PubMed] [Google Scholar]
- 20.Fiore, A. E., C. B. Bridges, and N. J. Cox. 2009. Seasonal influenza vaccines. Curr. Top. Microbiol. Immunol. 333:43-82. [DOI] [PubMed] [Google Scholar]
- 21.Forbes, E. K., C. Sander, E. O. Ronan, H. McShane, A. V. Hill, P. C. Beverley, and E. Z. Tchilian. 2008. Multifunctional, high-level cytokine-producing Th1 cells in the lung, but not spleen, correlate with protection against Mycobacterium tuberculosis aerosol challenge in mice. J. Immunol. 181:4955-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia, J. M., S. Pepin, N. Lagarde, E. S. Ma, F. R. Vogel, K. H. Chan, S. S. Chiu, and J. S. Peiris. 2009. Heterosubtype neutralizing responses to influenza A (H5N1) viruses are mediated by antibodies to virus haemagglutinin. PLoS One 4:e7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gavin, A. L., K. Hoebe, B. Duong, T. Ota, C. Martin, B. Beutler, and D. Nemazee. 2006. Adjuvant-enhanced antibody responses in the absence of Toll-like receptor signaling. Science 314:1936-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geeraedts, F., L. Bungener, J. Pool, W. ter Veer, J. Wilschut, and A. Huckriede. 2008. Whole inactivated virus influenza vaccine is superior to subunit vaccine in inducing immune responses and secretion of proinflammatory cytokines by DCs. Influenza Other Respir. Viruses 2:41-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geeraedts, F., N. Goutagny, V. Hornung, M. Severa, A. de Haan, J. Pool, J. Wilschut, K. A. Fitzgerald, and A. Huckriede. 2008. Superior immunogenicity of inactivated whole virus H5N1 influenza vaccine is primarily controlled by Toll-like receptor signaling. PLoS Pathog. 4:e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gowen, B. B., J. Fairman, D. F. Smee, M. H. Wong, K. H. Jung, A. M. Pace, M. L. Heiner, K. W. Bailey, S. W. Dow, and R. W. Sidwell. 2006. Protective immunity against acute phleboviral infection elicited through immunostimulatory cationic liposome-DNA complexes. Antiviral Res. 69:165-172. [DOI] [PubMed] [Google Scholar]
- 27.Grebe, K. M., J. W. Yewdell, and J. R. Bennink. 2008. Heterosubtypic immunity to influenza A virus: where do we stand? Microbes Infect. 10:1024-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He, X. S., T. H. Holmes, C. Zhang, K. Mahmood, G. W. Kemble, D. B. Lewis, C. L. Dekker, H. B. Greenberg, and A. M. Arvin. 2006. Cellular immune responses in children and adults receiving inactivated or live attenuated influenza vaccines. J. Virol. 80:11756-11766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hehme, N., H. Engelmann, W. Kuenzel, E. Neumeier, and R. Saenger. 2004. Immunogenicity of a monovalent, aluminum-adjuvanted influenza whole virus vaccine for pandemic use. Virus Res. 103:163-171. [DOI] [PubMed] [Google Scholar]
- 30.Heusser, C. H., C. L. Anderson, and H. M. Grey. 1977. Receptors for IgG: subclass specificity of receptors on different mouse cell types and the definition of two distinct receptors on a macrophage cell line. J. Exp. Med. 145:1316-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huber, V. C., J. M. Lynch, D. J. Bucher, J. Le, and D. W. Metzger. 2001. Fc receptor-mediated phagocytosis makes a significant contribution to clearance of influenza virus infections. J. Immunol. 166:7381-7388. [DOI] [PubMed] [Google Scholar]
- 32.Huber, V. C., R. M. McKeon, M. N. Brackin, L. A. Miller, R. Keating, S. A. Brown, N. Makarova, D. R. Perez, G. H. Macdonald, and J. A. McCullers. 2006. Distinct contributions of vaccine-induced immunoglobulin G1 (IgG1) and IgG2a antibodies to protective immunity against influenza. Clin. Vaccine Immunol. 13:981-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jameson, J., J. Cruz, M. Terajima, and F. A. Ennis. 1999. Human CD8+ and CD4+ T lymphocyte memory to influenza A viruses of swine and avian species. J. Immunol. 162:7578-7583. [PubMed] [Google Scholar]
- 34.Jegerlehner, A., P. Maurer, J. Bessa, H. J. Hinton, M. Kopf, and M. F. Bachmann. 2007. TLR9 signaling in B cells determines class switch recombination to IgG2a. J. Immunol. 178:2415-2420. [DOI] [PubMed] [Google Scholar]
- 35.Kedzierska, K., N. L. La Gruta, S. J. Turner, and P. C. Doherty. 2006. Establishment and recall of CD8+ T-cell memory in a model of localized transient infection. Immunol. Rev. 211:133-145. [DOI] [PubMed] [Google Scholar]
- 36.Kilbourne, E. D. 1969. Future influenza vaccines and the use of genetic recombinants. Bull. World Health Organ. 41:643-645. [PMC free article] [PubMed] [Google Scholar]
- 37.Kilbourne, E. D., C. Smith, I. Brett, B. A. Pokorny, B. Johansson, and N. Cox. 2002. The total influenza vaccine failure of 1947 revisited: major intrasubtypic antigenic change can explain failure of vaccine in a post-World War II epidemic. Proc. Natl. Acad. Sci. U. S. A. 99:10748-10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knossow, M., and J. J. Skehel. 2006. Variation and infectivity neutralization in influenza. Immunology 119:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunisaki, K. M., and E. N. Janoff. 2009. Influenza in immunosuppressed populations: a review of infection frequency, morbidity, mortality, and vaccine responses. Lancet Infect. Dis. 9:493-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.La Gruta, N. L., S. J. Turner, and P. C. Doherty. 2004. Hierarchies in cytokine expression profiles for acute and resolving influenza virus-specific CD8+ T cell responses: correlation of cytokine profile and TCR avidity. J. Immunol. 172:5553-5560. [DOI] [PubMed] [Google Scholar]
- 41.Lay, M., B. Callejo, S. Chang, D. K. Hong, D. B. Lewis, T. D. Carroll, S. Matzinger, L. Fritts, C. J. Miller, J. F. Warner, L. Liang, and J. Fairman. 2009. Cationic lipid/DNA complexes (JVRS-100) combined with influenza vaccine (Fluzone) increases antibody response, cellular immunity, and antigenically drifted protection. Vaccine 27:3811-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Bon, A., N. Etchart, C. Rossmann, M. Ashton, S. Hou, D. Gewert, P. Borrow, and D. F. Tough. 2003. Cross-priming of CD8+ T cells stimulated by virus-induced type I interferon. Nat. Immunol. 4:1009-1015. [DOI] [PubMed] [Google Scholar]
- 43.Lee, L. Y., D. L. A. Ha, C. Simmons, M. D. de Jong, N. V. Chau, R. Schumacher, Y. C. Peng, A. J. McMichael, J. J. Farrar, G. L. Smith, A. R. Townsend, B. A. Askonas, S. Rowland-Jones, and T. Dong. 2008. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Invest. 118:3478-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis, D. B. 2006. Avian flu to human influenza. Annu. Rev. Med. 57:139-154. [DOI] [PubMed] [Google Scholar]
- 45.Liang, S., K. Mozdzanowska, G. Palladino, and W. Gerhard. 1994. Heterosubtypic immunity to influenza type A virus in mice. Effector mechanisms and their longevity. J. Immunol. 152:1653-1661. [PubMed] [Google Scholar]
- 46.Lu, X., L. E. Edwards, J. A. Desheva, D. C. Nguyen, A. Rekstin, I. Stephenson, K. Szretter, N. J. Cox, L. G. Rudenko, A. Klimov, and J. M. Katz. 2006. Cross-protective immunity in mice induced by live-attenuated or inactivated vaccines against highly pathogenic influenza A (H5N1) viruses. Vaccine 24:6588-6593. [DOI] [PubMed] [Google Scholar]
- 47.Markine-Goriaynoff, D., and J. P. Coutelier. 2002. Increased efficacy of the immunoglobulin G2a subclass in antibody-mediated protection against lactate dehydrogenase-elevating virus-induced polioencephalomyelitis revealed with switch mutants. J. Virol. 76:432-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marrack, P., A. S. McKee, and M. W. Munks. 2009. Towards an understanding of the adjuvant action of aluminum. Nat. Rev. Immunol. 9:287-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin, R. M., J. L. Brady, and A. M. Lew. 1998. The need for IgG2c specific antiserum when isotyping antibodies from C57BL/6 and NOD mice. J. Immunol. Methods 212:187-192. [DOI] [PubMed] [Google Scholar]
- 50.Mbawuike, I. N., S. B. Dillion, S. G. Demuth, C. S. Jones, T. R. Cate, and R. B. Couch. 1994. Influenza A subtype cross-protection after immunization of outbred mice with a purified chimeric NS1/HA2 influenza virus protein. Vaccine 12:1340-1348. [DOI] [PubMed] [Google Scholar]
- 51.McCormack, S., A. Tilzey, A. Carmichael, F. Gotch, J. Kepple, A. Newberry, G. Jones, S. Lister, S. Beddows, R. Cheingsong, A. Rees, A. Babiker, J. Banatvala, C. Bruck, J. Darbyshire, D. Tyrrell, C. Van Hoecke, and J. Weber. 2000. A phase I trial in HIV negative healthy volunteers evaluating the effect of potent adjuvants on immunogenicity of a recombinant gp120W61D derived from dual tropic R5X4 HIV-1ACH320. Vaccine 18:1166-1177. [DOI] [PubMed] [Google Scholar]
- 52.McMichael, A. J., F. M. Gotch, G. R. Noble, and P. A. Beare. 1983. Cytotoxic T-cell immunity to influenza. N. Engl. J. Med. 309:13-17. [DOI] [PubMed] [Google Scholar]
- 53.Nelson, M. I., L. Edelman, D. J. Spiro, A. R. Boyne, J. Bera, R. Halpin, N. Sengamalay, E. Ghedin, M. A. Miller, L. Simonsen, C. Viboud, and E. C. Holmes. 2008. Molecular epidemiology of A/H3N2 and A/H1N1 influenza virus during a single epidemic season in the United States. PLoS Pathog. 4:e1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen, H. H., F. W. van Ginkel, H. L. Vu, J. R. McGhee, and J. Mestecky. 2001. Heterosubtypic immunity to influenza A virus infection requires B cells but not CD8+ cytotoxic T lymphocytes. J. Infect. Dis. 183:368-376. [DOI] [PubMed] [Google Scholar]
- 55.Potter, C. W., and J. S. Oxford. 1979. Determinants of immunity to influenza infection in man. Br. Med. Bull. 35:69-75. [DOI] [PubMed] [Google Scholar]
- 56.Rao, N. M., and V. Gopal. 2006. Cell biological and biophysical aspects of lipid-mediated gene delivery. Biosci. Rep. 26:301-324. [DOI] [PubMed] [Google Scholar]
- 57.Reed, L. J., and H. Muench. 1938. A simple method of estimating 50% endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 58.Rimmelzwaan, G. F., R. A. Fouchier, and A. D. Osterhaus. 2007. Influenza virus-specific cytotoxic T lymphocytes: a correlate of protection and a basis for vaccine development. Curr. Opin. Biotechnol. 18:529-536. [DOI] [PubMed] [Google Scholar]
- 59.Rock, K. L., D. J. Farfan-Arribas, and L. Shen. 2010. Proteases in MHC class I presentation and cross-presentation. J. Immunol. 184:9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sarawar, S. R., M. Sangster, R. L. Coffman, and P. C. Doherty. 1994. Administration of anti-IFN-gamma antibody to beta 2-microglobulin-deficient mice delays influenza virus clearance but does not switch the response to a T helper cell 2 phenotype. J. Immunol. 153:1246-1253. [PubMed] [Google Scholar]
- 61.Stambas, J., C. Guillonneau, K. Kedzierska, J. D. Mintern, P. C. Doherty, and N. L. La Gruta. 2008. Killer T cells in influenza. Pharmacol. Ther. 120:186-196. [DOI] [PubMed] [Google Scholar]
- 62.Sui, J., W. C. Hwang, S. Perez, G. Wei, D. Aird, L. M. Chen, E. Santelli, B. Stec, G. Cadwell, M. Ali, H. Wan, A. Murakami, A. Yammanuru, T. Han, N. J. Cox, L. A. Bankston, R. O. Donis, R. C. Liddington, and W. A. Marasco. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Topham, D. J., and P. C. Doherty. 1998. Clearance of an influenza A virus by CD4+ T cells is inefficient in the absence of B cells. J. Virol. 72:882-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vugmeyster, Y., R. Glas, B. Perarnau, F. A. Lemonnier, H. Eisen, and H. Ploegh. 1998. Major histocompatibility complex (MHC) class I KbDb−/− deficient mice possess functional CD8+ T cells and natural killer cells. Proc. Natl. Acad. Sci. U. S. A. 95:12492-12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Webster, R., N. Cox, and K. Stohr. 2002. WHO manual on animal influenza diagnosis and surveillance. WHO publication no. WHO/CDS/CSR/NCS/2002.5. Department of Communicable Disease Surveillance and Response, WHO Global Influenza Programme, World Health Organization, Geneva, Switzerland.
- 66.Wu, J., H. H. Fang, J. T. Chen, J. C. Zhou, Z. J. Feng, C. G. Li, Y. Z. Qiu, Y. Liu, M. Lu, L. Y. Liu, S. S. Dong, Q. Gao, X. M. Zhang, N. Wang, W. D. Yin, and X. P. Dong. 2009. Immunogenicity, safety, and cross-reactivity of an inactivated, adjuvanted, prototype pandemic influenza (H5N1) vaccine: a phase II, double-blind, randomized trial. Clin. Infect. Dis. 48:1087-1095. [DOI] [PubMed] [Google Scholar]
- 67.Yen, H. L., and J. S. Peiris. 2009. Mapping antibody epitopes of the avian H5N1 influenza virus. PLoS Med. 6:e1000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zaks, K., M. Jordan, A. Guth, K. Sellins, R. Kedl, A. Izzo, C. Bosio, and S. Dow. 2006. Efficient immunization and cross-priming by vaccine adjuvants containing TLR3 or TLR9 agonists complexed to cationic liposomes. J. Immunol. 176:7335-7345. [DOI] [PubMed] [Google Scholar]
- 69.Zambon, M. C. 2001. The pathogenesis of influenza in humans. Rev. Med. Virol. 11:227-241. [DOI] [PubMed] [Google Scholar]
- 70.Zhong, W., P. A. Reche, C. C. Lai, B. Reinhold, and E. L. Reinherz. 2003. Genome-wide characterization of a viral cytotoxic T lymphocyte epitope repertoire. J. Biol. Chem. 278:45135-45144. [DOI] [PubMed] [Google Scholar]
- 71.Zhu, N., D. Liggitt, Y. Liu, and R. Debs. 1993. Systemic gene expression after intravenous DNA delivery into adult mice. Science 261:209-211. [DOI] [PubMed] [Google Scholar]